Abstract
Genome-wide expression profiling is a powerful tool for implicating novel gene ensembles in cellular mechanisms of health and disease. The most popular platform for genome-wide expression profiling is the Affymetrix GeneChip. However, its selection of probes relied on earlier genome and transcriptome annotation which is significantly different from current knowledge. The resultant informatics problems have a profound impact on analysis and interpretation the data. Here, we address these critical issues and offer a solution. We identified several classes of problems at the individual probe level in the existing annotation, under the assumption that current genome and transcriptome databases are more accurate than those used for GeneChip design. We then reorganized probes on more than a dozen popular GeneChips into gene-, transcript- and exon-specific probe sets in light of up-to-date genome, cDNA/EST clustering and single nucleotide polymorphism information. Comparing analysis results between the original and the redefined probe sets reveals ∼30–50% discrepancy in the genes previously identified as differentially expressed, regardless of analysis method. Our results demonstrate that the original Affymetrix probe set definitions are inaccurate, and many conclusions derived from past GeneChip analyses may be significantly flawed. It will be beneficial to re-analyze existing GeneChip data with updated probe set definitions.
INTRODUCTION
While extensive attention has been devoted to improving the accuracy and sensitivity of the statistical algorithms used to estimate gene expression levels and to detect differential expression in GeneChip-based expression analyses (1–4), problems related to probe and probe set identity likely lead to significant errors, especially under conditions where expression changes are not dramatic. GeneChips for expression analysis use probe sets containing 11–20 pairs of 25mer oligonucleotides to represent a target gene or transcript. Each oligonucleotide pair consists of an oligo with perfect match to a target sequence region (PM probe) and another oligo with a single base mismatch in the center of the oligo (MM probe) to the same target region. Although Affymetrix utilized the most complete information available at the time of GeneChip design, tremendous progress in genome sequencing and annotation in recent years renders existing GeneChip probe set designs suboptimal. For example, when the HG-U133 chip set was designed, the human UniGene Build 133 contained ∼2.8 million cDNA/EST sequences and the human genome sequence was only ∼25% complete (5). Currently, the human UniGene builds contain over 5 million sequences and the human genome build 35 has 99% of the euchromatic portion of the genome sequenced (6). Our analysis indicates that many of the old probe sets do not faithfully reflect the expression levels of a significant number of genes in a given tissue due to several informatics-related issues which impact probe identity. It should be pointed out that three recent papers also investigated some of the problems for the HG-U133A, HG-U95A and HG-U133 Plus 2.0 GeneChips but no systematic solution was provided (7–9). For example, Harbig et al. re-annotated 37% of the probes on the HG-U133 Plus 2.0 array based on BLAST sequence match. They also envisioned several ways to automate the GeneChip probe annotation process and called help from the bioinformatics community. Here, we present a user-friendly solution compatible with all existing GeneChip analysis software. The impact of the updated probe set definition on the interpretation of GeneChip data is also evaluated using a public domain data set.
METHODS
Generation of reorganized probe sets based on the UniGene database
We want to describe the procedure for generating the UniGene-based probe set definitions first since the UniGene database is the most widely used gene classification system and most researchers will first map the GeneChip results to the UniGene database in order to understand the biological significance of the GeneChip data. Generating UniGene-based probe sets is complicated because UniGene clusters often contain multiple sequences of unknown reliability and strand direction.
The following are the steps in our UniGene probe set re-organization process. These steps are applied in the order presented.
(i) Perform sequence alignments. There are two components in this step: (a) map all GeneChip probe sequences to individual sequences in UniGene, dbSNP and the genome sequence of the corresponding species. Only perfect matches are retained. (b) Align all sequences in the UniGene database to the most current genome assembly of the corresponding species. Since UniGene clusters can frequently contain sequences from other genes, we will use the genome alignment results in later steps to provide some easily automatable cleanups.
(ii) We require each probe in a probe set should only have one perfect match with the corresponding genome sequence. This may exclude probes that also match a non-transcribed region in the genome, but this filter is not dependent on the completeness of genome annotation or cDNA/EST sequence collection and, therefore, should be more stable in the long term. The ubiquitous presence of non-coding transcripts also supports the use of this somewhat more aggressive strategy (10). Given the fact that only 10% or less probes are eliminated by this criterion, we believe the slight drop in statistical power for each probe set is a worthwhile price for the gain in the confidence in the final results.
(iii) Because EST sequences are subject to a relatively high error rate, we require a probe to perfectly match a genomic region that can be aligned with mRNA/EST sequences collected in the UniGene database. Probes with perfect match only to EST sequences but not to the corresponding genomic sequence will not be included in the final probe sets. An exception to this rule is exon–exon junction probes with perfect match to the mRNA reference sequences in the same UniGene cluster. We add such exon–exon junction probes back to the corresponding probe set and assign the lowest probe pair number(s).
(iv) In order to ensure that a probe is specific for one UniGene cluster, we eliminate probes with multiple matching cDNA/EST sequences that can be assigned to more than one UniGene cluster. Our previous requirement for genomic sequence alignment can reduce false non-specific probes caused by erroneous EST sequencing or contaminating EST sequences. Although this filter may remove good probes due to errors in UniGene clustering, it will guarantee that every probe set is consistent with current UniGene clustering.
In theory, a probe with the potential to hybridize with transcripts from more than one gene is still useful if the unintended transcripts are not expressed at a level that leads to significant probe signal interference in a given tissue or sample. However, while tissue- or sample-dependent probe set definitions can increase GeneChip probe utilization, the simplicity and the consistency of a tissue/sample-independent probe set definition should be more advantageous in most situations.
In the ideal situation, a gene-specific probe set should only contain probes whose sequence will be present on the shared sequences of all splicing products from the same gene, as the signal level of such a probe set will not be influenced by the alternative splicing in different tissues or individuals. For most genes, current knowledge of potential alternative splicing products is far from complete. Thus, we choose to pool all probes targeting the same gene for the definition of a gene-specific probe set. We believe the gene-based probe set definition is useful for evaluating the overall transcription activity of a gene, which is in fact claimed in most microarray research papers. Potential alternative splicing events can conceivably be explored by our transcript- or exon-specific probe sets. Since some researchers prefer to examine probes targeting at the 3′ end of transcripts, we also created probe sets that contain no more than 11 probe pairs at the most 3′ end.
(v) Except for genes with known mRNA/reference sequences, we require all probes in each probe set be aligned in the same direction on the genome, as old probe sets representing the same gene can sometimes target different strands of the same transcript. This constraint ensures the directional homogeneity of newly defined probe sets during the merging of probes from multiple old probe sets. Probes with perfect match to the same genomic region but in different directions are separated into two probe sets if there is no mRNA/reference sequence in the UniGene cluster for determining the transcription orientation on the genome.
(vi) We also require that probes targeted to the same UniGene cluster be aligned continuously on the genomic sequence in the same direction. For example, if probes representing a UniGene cluster are distributed across different genomic regions or chromosomes, the largest continuous set of probes will be used to represent this UniGene cluster. All other probes intermingling with probes targeting different UniGene clusters will be omitted from the final probe set.
An exemption to this rule is when an mRNA reference sequence in a UniGene cluster can be aligned to different genomic locations, as mRNA reference sequences are probably more reliable than the current version of genome assembly.
(vii) Each probe set should contain at least three probe pairs. Targets that cannot be represented by at least three probe pairs are eliminated in the final probe set definition. This threshold is largely arbitrary, but a probe set with three probe pairs should satisfy the minimum requirements of most probe-level analysis algorithms. In our new UniGene probe set definitions, probe sets containing three or four probe pairs account for <10% of the all probe sets. The size of most probe sets are ∼1× or 2× of the original probe set size on a given GeneChip (e.g. ∼11 or 22), although some probe sets can have several dozen probe pairs due to the redundancy of original GeneChip probe sets described before.
Generation of Refseq, DoTS, Entrez gene, ENSEMBL gene, Transcript and Exon probe sets
Generation of custom probe sets for these target types is much easier since each target sequence and direction are well-defined in the corresponding databases. After identifying all perfect match probes on a GeneChip to the corresponding target sequences, we remove probes with more than one perfect hit on the corresponding genomic sequence and we also require each final probe set that contains more than three probes. The 3′-focused probe sets only contain the most 3′ eleven probes in the corresponding gene or transcript definitions.
Generation of allele-independent probe sets
In order to reduce the noise caused by single nucleotide polymorphisms (SNPs) in different samples, we also generated probe sets by removing all probe pairs known to have allele-specific base in the central 15 bp region of either the Perfect Match or the Mismatch probes. Of course, unknown high heterogeneity SNP sites may still cause high noise for some probe sets.
Naming of the final probe sets
If a probe passes our selection criteria, it is added to a preliminary pool of probes for the same target (gene, transcript or exon) based on the target definition in the corresponding databases. As described above, there can be additional criteria before generating the final probe set, such as only retaining the most 3′ 11 probes or the removal of allele-specific probes. An initial probe set does not lead to a final probe set if it only contains one or two probes. The final probe set will have the corresponding target name in the related database. Following Affymetrix's nomenclature, we add ‘_at’ at the end of the sequence ID name. Consequently, Hs.10000_at, Mm.1111_at, NM_12235_at, ENSG00003456_at, etc., can be probe set names in the corresponding custom CDF files.
Assignment of the best match accession number to probe sets
Since many of the non-GenBank accession number-based probe set Ids, such as UniGene ID and ENSEMBL transcript ID, are not very stable, we also assign a GenBank accession number to all gene- and transcript-specific probe sets, including the original GeneChip probe sets, by identifying the accession number for the most reliable short sequence that has the highest percentage of probe matches for the corresponding probe set. Among the sequences with the same top probe hit rate for a given probe set, the order of sequence selection is Refseq > cDNA > EST. If there is still a tie, the shortest sequence is selected. There are a number of situations where the above procedure still leads to multiple sequences and we simply pick the accession number with the lowest alphabetic order as the designated best accession number for a probe set.
All procedures described above are implemented on a 4× dual opteron/8 GB memory cluster and a dual Itanium Oracle server with 16 GB of memory. We usually generate a new CDF build every 3–4 months and each build takes ∼10 days to finish on our current setup. A total of six custom CDF builds have been generated since early last year.
Use of custom CDF
Custom CDF files can be easily selected based on species, Affymetrix GeneChip type, CDF file type and CDF file format on our CDF download grid. The following are three examples that cover all common GeneChip probe-level analysis situations.
Example 1. Use custom CDF in Affymetrix MAS5 or standalone dCHIP: the ASCII format CDF files are for Affymetrix MAS5 and standalone dCHIP programs. After unzipping the ASCII CDF package, an ASCII format custom CDF file can be used exactly the same way as an Affymetrix CDF file. Please note that the dCHIP program only accepts Affymetrix CDF names thus one has to change the name of a custom CDF file to the corresponding Affymetrix CDF file name.
Example 2. Use custom CDF in R environment by calling custom CDF packages directly on a computer with Internet link. The following is an example session:
library(affy)
data<-ReadAffy()
UMRepos<-getOption(“repositories2”)
UMRepos[“UMRepository”] = ‘http://arrayanalysis.mbni.med.umich.edu/repository’
options(‘repositories2’ = UMRepos)
data@cdfName<-“HS133A_HS_UG_5”
result<-rma(data)
write.exprs(result, file=‘output.txt’)
Strings in bold italic are the extra commands that a user need to add in an R session. The custom CDF file name ‘HS133A_HS_UG_5’ can be replaced with any custom CDF file name on the CDF download page (‘CDF file name’, the fourth column from left on the CDF download grid).
Example 3. Use of a custom CDF in R environment after downloading the corresponding custom CDF R package onto user's local computer.
Please notice there is an R package for LINUX/UNIX/MAC OS X and another R package for the Windows platform. After the correct package is downloaded, one needs to perform the following actions:
Under Linux/Unix/MAC OS X, use command ‘R CMD INSTALL ?.tar.gz’.
Under Windows, select menu ‘Packages->Install package(s) from local zip files’.
In order to use the custom CDF files in data analysis after installation, a single line of R command should be added to replace the default Affymetrix CDF file. The following are two examples for different chip and custom probe set combinations:
data←ReadAffy()
data@cdfName<-“HS133A_HS_UG_5”
data<-read.affybatch(‘1.cel’, ‘2.cel’);
data@cdfName<-‘HS133B_HS_ENSG_5’.
Again, the CDF name in the bold italic part can be replaced with the name of any custom CDF you download. The standard name for each custom CDF is in the fourth column of the CDF download grid for a given CDF version.
RESULTS
Problems in the original GeneChip probe set definition and annotation
Unreliable representative accession numbers
The prevailing method for associating the latest gene identity and function annotations to probe sets on GeneChips is to map the Affymetrix ‘Representative Public ID’ for each probe set to the current version of gene and annotation databases such as UniGene (11,12), LocusLink/Entrez Gene (11,12) and Gene Ontology (http://www.geneontology.org). While the use of one nucleic accession number to represent all probes in a set significantly simplifies the handling of GeneChip data, this approach implicitly assumes that all probes in a probe set are derived from the same gene as their ‘Representative Public IDs’. This assumption can be problematic because a significant percentage of probe sets were created based on the so-called ‘consensus sequence’ derived from merging several sequences in an old UniGene cluster. Probes excluded from the ‘Representative Public ID’ sequence can possibly be assigned to a different UniGene cluster because old clusters have been split in the more recent build. In addition, many of the representative accession numbers are no longer in the current version of UniGene/Refseq/EST databases. Our analysis indicates that between 10 and 40% of the original accession numbers assigned to probe sets on popular GeneChips either match less than half of the probes in the corresponding set or are retired from current databases. These probe sets are more likely to contain probes for non-associated genes or probes derived from untrustworthy sequences (Table 1).
Table 1.
Percent of potentially problematic GeneChip probe sets
| Chiptype | Unreliable representative public ID | UniGene redundancy | Containing probe(s) with multiple UniGene hit | Having probe(s) with multiple genome hit | With genomic location or strand issues | Including probe(s) with no known target | Containing allele-specific probe(s) |
|---|---|---|---|---|---|---|---|
| HG-U95Av2 | 27.9 | 21.1 | 36.6 | 16.2 | 8.8 | 4.6 | 40.5 |
| HG-U133A | 14.4 | 34.2 | 36.0 | 16.3 | 10.1 | 3.6 | 42.7 |
| HG-U133B | 22.2 | 31.4 | 22.3 | 9.3 | 10.4 | 5.0 | 35.2 |
| HG-U133 Plus | 18.2 | 47.2 | 26.1 | 11.6 | 12.0 | 4.8 | 37.6 |
| Human X3P | 21.0 | 50.8 | 22.8 | 10.6 | 10.3 | 4.8 | 32.7 |
| MG-U74Av2 | 42.7 | 18.8 | 28.8 | 16.1 | 8.8 | 10.0 | 11.7 |
| MOE430A | 13.3 | 38.6 | 30.9 | 15.0 | 10.4 | 4.1 | 11.0 |
| MOE430B | 28.5 | 31.2 | 16.5 | 5.5 | 9.9 | 11.6 | 4.6 |
| Mouse430 | 20.8 | 44.7 | 23.6 | 10.2 | 11.2 | 7.8 | 7.8 |
| Rn34A | 21.3 | 28.0 | 17.4 | 15.8 | 7.0 | 8.2 | 18.1 |
| RAE230A | 10.7 | 17.5 | 16.5 | 13.2 | 8.7 | 3.6 | 19.5 |
| RAE230B | 32.8 | 15.1 | 6.8 | 7.0 | 5.0 | 15.8 | 7.8 |
| Rat230 | 21.5 | 24.8 | 11.7 | 10.1 | 8.3 | 9.6 | 13.7 |
Probe set redundancy
The infusion of new cDNA/EST sequences results in the merger of some old UniGene clusters, the effect of which is obvious since 15–50% of UniGene IDs are represented by more than one probe set based on the ‘Representative Public ID’ assigned to each probe set (Table 1). Since understanding the real biological implication of each probe set (e.g. target transcripts or exons) is not straightforward, most researchers just use the latest UniGene ID associated with the probe set accession number as the identity of a given probe set, leading to high level of probe set redundancy. There is no standard way to deal with data from redundant probe sets. Some reports use the average signal of all probe sets representing the same gene while others focus on the probe set showing differential expression, regardless of the behavior of other probe sets representing the same gene. Redundant probe sets will also create bias in function category-based analysis, such as Fisher's Exact Test and Gene Set Enrichment Analysis utilizing Gene Ontology. For most analyses, a one probe set-to-one target relationship would be highly desirable.
Non-specific probes
A significant increase of cDNA/EST/genome sequence information leads to the possibility a probe thought to be specific for one gene can actually hybridize to transcripts from additional genes or non-coding transcripts. As shown in Table 1, according to the current version of the UniGene database, for most GeneChips 10–30% of probe sets contain at least one non-specific probe. Probe alignment to genomic sequences also reveals that 5–16% of probe sets contain a probe(s) with more than one genomic sequence hit(s). The difference between the UniGene- and genome-based criteria may largely be due to UniGene clustering or EST sequencing errors.
Deleted target sequence
Some probes no longer match any sequences in the current UniGene database or the genomic sequence of the corresponding species in either strand direction. The major cause is probably the removal of sequences used for probe design from the new UniGene database.
Genomic location issues
The alignment of single-hit probes to genomic sequence reveals additional issues at the probe set level. Some probe sets contain at least one probe with a perfect match to a unique sequence in another chromosome or to a different strand on the same chromosome. Other probes supposedly representative of different UniGene clusters are intermingled with each other on the same strand of a given chromosome. Sequence clustering problems and/or earlier genome assembly errors are probably the cause of these complications.
Furthermore, thousands of probes targeted to the opposite strand of the same genomic region can be aligned with cDNA/EST sequences assigned to particular UniGene IDs. This is likely due to the use of pure EST clusters in probe set design, since it is often hard to determine the transcription direction of a pure EST cluster without a known cDNA sequence. Probe sets affected by these issues are listed in Table 1 as ‘with genomic location or strand issues’. Although the current genome assemblies are by no means perfect, a large portion of such location problems is likely caused by shortcomings in earlier version of the UniGene databases and genome assemblies.
Allele-specific probes
The remarkable increase in known SNP sites in the human genome in the last few years creates another type of probe identity issue: some GeneChip probes are allele-specific, and therefore may behave differently across samples from different individuals. Our analysis indicates that between 30 and 40% of probe sets on popular human GeneChips contain at least one probe that overlaps with known SNP sites in the central 15 bp region of the probe. We focused solely on probes with allele-specific bases in the central 15 bp region because a mismatch in the central region is more likely to cause a significant change in binding energy than a mismatch near the end of a probe sequence (13,14).
Generation of updated probe set definitions and related utility functions
Given the extent of the probe identity problems in the existing GeneChip probe set definitions, we applied a series of probe selection and grouping criteria utilizing the latest sequence and annotation information. We generated new GeneChip library files (CDF files) for popular human, mouse and rat GeneChips according to different target definitions, such as UniGene (11,12), Refseq (11,12), DoTS (http://www.cbil.upenn.edu/downloads/DoTS/), ENSEMBL Gene, Transcript and Exon (15).
All the custom CDFs we generated as well as the statistics related to the most recently three versions of custom CDFs can be freely accessed at our custom CDF webpage at http://brainarray.mbni.med.umich.edu/CustomCDF. These CDF files are compatible with all popular R analysis packages (e.g. RMA, GCRMA, fitPLM, MAS5, dCHIP, three-step) as well as independent probe-level analysis programs, such as Affymetrix's MAS5, and Li and Wong's dCHIP. Since different programs and operating systems require different custom CDF data format, we provide custom CDF R packages for LINUX/MAC OS X, Windows R package and ASCII CDFs for use in non-R programs such as Affymetrix's MAS5 and the independent dCHIP program.
In addition to the updated probe set definitions, we provide four useful files related to each CDF. (i) The probe set package. This package is required for analysis methods such as GCRMA that utilize probe sequence in low-level signal modeling. (ii) Best accession number list for each gene and transcript in the corresponding CDF files. (iii) Probe-genome map file: list the genomic location of each probe in a probe set on the corresponding genomic sequences. (iv) Probe set group file: list the probe content of each probe set. Users can easily find redundancies in related probe set definitions, such as shared probes among different transcripts from the same gene. These files are all freely downloadable through our custom CDF page.
Furthermore, we developed a series of auxiliary functions that will help researchers to compare and explore the details of each probe set definition, such as mapping custom probe sets to entries in the corresponding target definition database, finding the genomic location for each probe in a list of probe sets, finding whether probes in a probe set overlap with known SNPs, examining the probe set content, match probe sets across different GeneChips, probe set definitions and species. These web functions are listed under the ‘download custom CDF’ link on our main custom CDF page.
Most importantly, users can test the effect of these custom CDF files on GeneChip analysis results through the ‘GeneChip Analysis Using Custom CDF Files’ link located on the custom CDF page. All popular GeneChip analysis functions in the BioConductor package (16) and GeneChip cel files deposited in the NCBI Gene Expression Omnibus database (17) are accessible through this function. As indicated in this function, ‘public’ is both the username and password for login.
Comparison of the new and old GeneChip probe sets based on the UniGene database
Since the UniGene database is the most widely used gene definition system and is also the foundation for existing GeneChip designs, we summarized the comparison of our updated UniGene-derived probe set definitions with the original GeneChip probe sets in Table 2. We present the SNP-containing version of UniGene probe set definitions here, as we are investigating the pure effect of gene definition change rather than including the additional effect of removing allele-specific probes. Table 2 shows that the new annotation impacts over 30% of the UniGene IDs for all GeneChips examined: this percentage includes the sum of all the completely reassigned UniGene IDs in the new probe set definitions, as well as the probe sets that retain their old UniGene IDs but with over 50% difference in probe content. Since this comparison is performed under the same UniGene build and does not involve the assignment of the same probe set to different UniGene in different UniGene builds, it is likely that at least 30% of genes will have very different absolute expression values under the new probe set definitions.
Table 2.
Probe set content comparison between Affymetrix probe sets and updated UniGene probe setsa
| Chiptype | Total Affymetrix probe sets | UGID shared by both definitions | 100% Identical probe sets | Probe set content difference ≥ 50% | Unique UGID in Affymetrix probe set definition | Unique UGID in updated UniGene probe sets |
|---|---|---|---|---|---|---|
| HG-U95Av2 | 12 558 | 6847 | 3275 | 1153 | 956 | 1355 |
| HG-U133A | 22 212 | 11 182 | 4800 | 1920 | 1612 | 657 |
| HG-U133B | 22 577 | 7924 | 2799 | 2155 | 4912 | 1052 |
| HG-U133 Plus | 54 613 | 18 555 | 5624 | 5450 | 5496 | 1483 |
| Human X3P | 61 297 | 18 250 | 6339 | 5673 | 5714 | 1507 |
| MG-U74Av2 | 12 422 | 6531 | 3056 | 1217 | 1455 | 1253 |
| MOE430A | 22 626 | 11 488 | 5732 | 1694 | 1461 | 753 |
| MOE430B | 22 511 | 7866 | 2834 | 1904 | 3751 | 1147 |
| Mouse430 | 45 037 | 17 215 | 6487 | 4074 | 3356 | 1507 |
| Rn34A | 8740 | 3934 | 1538 | 886 | 990 | 595 |
| RAE230A | 15 866 | 9296 | 4586 | 1354 | 2614 | 722 |
| RAE230B | 15 276 | 6379 | 2453 | 1141 | 3034 | 890 |
| Rat230 | 31 042 | 14 598 | 5899 | 2992 | 4303 | 1384 |
aThe UniGene build used in Table 2 is HsUG 183, MmUG 146 and RnUG 142. If several old probe sets are mapped to the same new UniGene ID, probes in these old probe sets are merged before comparing probe content with the corresponding new UniGene-based probe set.
Detailed statistics for differences between Refseq, ENSENBL gene, Entrez Gene and the original Affymetrix probe set definitions can be found on our website at http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/cdfreadme.htm#Statistics_of_Affymetrix_and_custom_CDF_files_. Information for exon-based probe set can also be found by following this link. Researchers interested in investigating the details of each probe set and comparing probe sets derived from different definitions can use the corresponding web functions on the main custom CDF page for querying probe set content, genomic location of probes, probe match to any cDNA sequences as well as cross-chip, cross-target definition and cross-species probe set match.
However, the key question is will the new custom probe set definitions lead to significant variation in the actual genes identified as differentially expressed in a GeneChip experiment? The ultimate purpose of most GeneChip-based expression profiling experiments is not to quantify absolute expression levels but to establish a reliable list of differentially expressed genes.
Impact of updated probe set definitions on GeneChip analysis results
Since the HG-U133A chip is one of the most widely used GeneChips, we use our internal as well as public domain HG-U133A data sets to examine the impact of updated probe set definitions on the differentially expressed gene lists under various analysis methods and cut-off thresholds. Our analyses suggest that updated CDF files under most situations can cause between 30 and 40% difference in the final lists of differentially expressed genes for various data sets derived from HG-U133A chips. Table 3 is a comparison of results derived from the old Affymetrix probe sets and various new probe set definitions using a heart tissue expression profiling data set deposited in the Gene Expression Omnibus database (GSE974) (18). We selected this data set since its use of paired samples from each individual significantly reduced the impact of allele-specific probes in paired t-tests or false discovery rate analysis, as our main goal here is to assess the pure effect of gene/transcript definition changes on the interpretation of GeneChip data. The R implementation of RMA was used to generate probe set-level data, which were analyzed by the SAMR package (19) for deriving differentially expressed gene lists under various false discovery rate thresholds for genes showing at least 20% expression changes. Differentially expressed gene/transcript lists derived from non-UniGene probe sets are mapped to UniGene IDs in the same version of the UniGene database using the best accession number we generated for each probe set. The average percentage of shared distinct UniGene IDs for a given probe set definition pair (e.g. Affymetrix and ENSG) is presented in Table 3, as the same Q-value thresholds usually leads to different number of unique genes under different probe set definitions. It can be seen that the consistency between the old and new probe set definition is usually ∼60–70%, regardless of the cut-off thresholds or custom probe set definitions used. Consequently, 30–40% of genes thought to be differentially expressed under the old probe set definitions can be problematic based on current gene and transcript definitions.
Table 3.
Percent of shared UniGene ID between Affymetrix and other probe set definitions under different FDR thresholds
| FDR (SAM Q-value) cut-off (%) | <1 | <2 | <5 | <10 | <20 |
|---|---|---|---|---|---|
| UG | 73.0 | 73.0 | 65.1 | 63.9 | 71.6 |
| 3UGa | 75.4 | 75.4 | 67.8 | 63.8 | 68.2 |
| ENTREZG | 64.4 | 54.8 | 64.1 | 62.3 | 71.8 |
| ENSG | 70.9 | 62.3 | 61.7 | 62.0 | 70.5 |
| REFSEQ | 66.0 | 58.1 | 70.4 | 62.2 | 72.2 |
| 3REFSEQa | 67.5 | 67.5 | 67.7 | 64.5 | 70.6 |
| ENST | 69.7 | 52.3 | 67.1 | 64.8 | 71.7 |
| 3ENSTa | 72.9 | 65.6 | 65.8 | 62.9 | 69.2 |
| DOTS | 56.7 | 61.1 | 65.6 | 65.2 | 67.7 |
| 3DOTSa | 60.6 | 61.4 | 65.0 | 65.5 | 69.3 |
aProbe set definition started with ‘3’ are those only containing the most 3′ 11 probes if there are more than 11 probes in a probe set.
In order to make sure that the observed probe set effect is not unique to our routine analysis approach, we tested other analysis methods, such as MAS5, dCHIP, affyPLM and GCRMA as well as t-test P-value based gene ranking. Table 4 is a summary of top-ranked gene list similarity between the Affymetrix and other probe set definitions. Each similarity value in Table 4 is the average of 50 similarity values for the corresponding probe set definition pair (e.g. Affymetrix versus UniGene) under five different analysis methods (RMA, MAS5, dCHIP, affyPLM and GCRMA), two gene ranking methods (P-value from t-test and Q-value from SAMR) and five different thresholds (top 10, 20, 50, 100, 200 genes based on P-value; SAMR Q-value cut-off at 1, 2, 5, 10 and 20%). In addition, we require all genes/transcripts in the differentially expressed list to show at least 20% expression change. Regardless of the analysis methods and cut-off thresholds, using an updated probe set definition always leads to 30–50% difference in the differentially expressed gene lists for HG-U133A data (data column 1 in Table 4), suggesting the difference in probe set content indeed caused the 30–50% difference in differentially expressed gene lists.
Table 4.
Average similarity between different probe set definitions based on differentially expressed gene lists derived under various cut-off thresholds and analysis methodsa
| Probe set definition | AFFY | UG | 3UG | ENTREZG | ENSG | REFSEQ | 3REFSEQ | ENST | 3ENST | DOTS | 3DOTS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AFFY | 100.0 | ||||||||||
| UG | 66.0 | 100.0 | |||||||||
| 3UG | 71.5 | 77.7 | 100.0 | ||||||||
| ENTREZG | 65.8 | 80.1 | 73.2 | 100.0 | |||||||
| ENSG | 66.4 | 78.4 | 72.6 | 87.8 | 100.0 | ||||||
| REFSEQ | 67.2 | 78.5 | 73.7 | 89.1 | 86.5 | 100.0 | |||||
| 3REFSEQ | 68.6 | 72.8 | 82.3 | 80.1 | 78.1 | 83.4 | 100.0 | ||||
| ENST | 66.0 | 74.9 | 71.8 | 83.7 | 87.8 | 87.4 | 78.4 | 100.0 | |||
| 3ENST | 68.7 | 68.9 | 79.6 | 76.3 | 79.8 | 78.2 | 84.4 | 82.5 | 100.0 | ||
| DOTS | 60.0 | 59.0 | 58.6 | 62.2 | 63.1 | 62.9 | 60.8 | 63.6 | 62.3 | 100.0 | |
| 3DOTS | 61.3 | 57.0 | 61.0 | 60.4 | 61.5 | 61.7 | 62.7 | 62.4 | 64.2 | 89.0 | 100.0 |
aSimilarity values <70% are in bold.
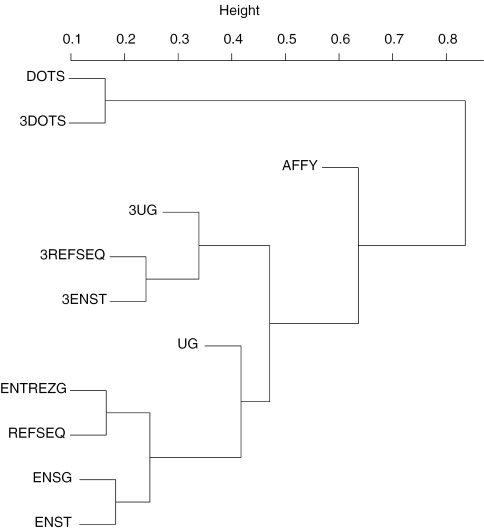
A closer scrutiny of Table 4 reveals that with the exception of DoTS-based transcript definitions, results from widely adopted gene and transcript definitions such as UniGene, Entrez Gene (originally LocusLink), ENSEMBL gene, transcript and mRNA reference sequences are often more similar to each other than those from the original Affymetrix probe set definition. Figure 1 is the dendrogram derived from the similarity data in Table 4 using the R hclust function at its default setting. It confirms the fact that the original Affymetrix probe set definition is very different from all widely used gene/transcript definitions at this time. While there must be problems in the current gene/transcript databases, that fact that the original Affymetrix probe set definition does not like any of them while most of the current gene/transcript definitions are more similar to each other suggest that the original Affymetrix probe set definition is no longer accurate in the presence of new genome and transcriptome information.
Figure 1.
Hierarchical clustering of probe set definition similarity based on differentially expressed gene lists derived from the GSE974 data set using different probe set definitions and analysis methods.
DISCUSSION
Our analyses suggest that the original GeneChip probe set definition is problematic in many aspects based on the current genome and transcriptome knowledge. We believe the reorganized probe set definitions should provide more accurate interpretation of GeneChip data.
Since HG-U133A is actually one of the GeneChips with relatively high probe set content consistency between the new and old probe set definitions (Table 2), it is conceivable that the use of updated probe set definitions for data from other chips, particularly those from HG-U133B, mouse_U74Av2 and rat Rn34A will lead to significantly higher gene-level differences. A significant alteration in the final differentially expressed gene list or ranking not only influences the selection of genes for follow-up studies but also changes the results for function category-based analysis, such as Gene Set Enrichment Analysis and the Fisher's Exact Test using functional categories (20,21).
We believe combining all probes for a gene provides the possibility of detecting the overall transcription activity of a gene. Given our limited understanding of the alternative splicing events related to various genes, the gene-based probe sets should be very useful in expression profiling. In addition, for all popular probe level analysis algorithms, such as MAS5, RMA and dChip, more probes in a probe set usually provide higher statistical power for detecting subtle changes.
Researchers interested in examining individual transcripts may want use our probe set definitions based on Refseq, ENSEMBL transcript and DoTS. These transcript-based definitions provide the possibility of detecting splicing variants as well as the corroboration of findings on different transcripts from the same gene. However, we have to point out that these transcript-targeted probe sets are not transcript-specific, as probe sets targeting transcripts from the same gene may share many or even all probes. Under many circumstances, it is not possible to generate transcript-specific probe sets containing at least three probes for genes with multiple transcripts based on probes available on the current generation of GeneChips.
If a researcher is more interested in alternative splicing, the most sensitive approach is to use our exon-based probe set definitions. Each exon-based probe set only contains probes in a particular exon without the ‘averaging’ effect caused by probes on shared exons between different transcripts. We believe the exon-based probe sets is superior to redundant Affymetrix probe sets for detecting alternative splicing, as Affymetrix redundant probe sets representing the same gene have very complex relationship with each other and they usually span a couple of exons as well as overlap with each other in different ways.
Whether the 3′-focused version of CDFs is better than their corresponding full probe set version is still debatable. Our experience suggests that 3′-focused probe sets usually lead to higher noise. It is also interesting to note that results from the 3′-focused CDFs showed lower consistency among themselves than results from the corresponding full probe set versions in Table 4.
We think gene-, transcript- and exon-targeted probe sets as well as the 3′-focused version of gene and transcript probe sets provide different views of the complex transcription activities related to individual genes. In the solution we developed, a researcher has the freedom of choosing any CDF or utilizing all CDFs for a more comprehensive analysis. Comparing results from Affymetrix probe sets and the custom probe sets may also lead to interesting findings and as mentioned previously, we provided various web functions to facilitate such exploration processes.
It is conceivable that mapping GeneChip probes to the latest sequence and annotation can facilitate the development of novel analysis methods for detecting alternative splicing and sequence polymorphism related to hundreds or thousands of genes in large GeneChip data sets. Without doubt, such re-analysis and re-interpretation of existing GeneChip data would not be possible if Affymetrix did not publish GeneChip probe sequences. The importance of making the actual probe sequences public was addressed in an open letter from the Microarray Gene Expression Data (MGED) Society recently and our results strongly support this vital request (http://www.mged.org/Workgroups/MIAME/MIAME_reporters.pdf).
The possibility of applying different probe set definitions for the same data set provides a very good way for confirming analysis results under different gene/transcript models. Although consistency does not equal to truth, the fact that a set of genes or transcripts can always pass a cut-off threshold regardless of the probe set definitions used will strongly suggest the reliability of the detected expression changes.
It would also be interesting to estimate how much ‘real’ improvement that these custom CDFs may bring to GeneChip analysis. Although we believe all current gene/transcript definitions are more accurate than the information Affymetrix used in existing GeneChip designs, gene/transcript models from different databases are not 100% identical, thus some of the differences between the new and old CDF may due to problems in current databases. Comparing the consistency of results from different probe set definitions will give us a rough idea about the ‘real’ improvements from these CDF. However, a reliable estimation should be based on the comparison of results from Affymetrix CDF and custom CDFs that are based on more stringent gene/transcript definitions, as definitions using aggressive rules, such as UniGene and DoTS, may contain significant noises. It can be derived from Table 4 that the average consistency of results from the Affymetrix CDF with results from the full probe set version (i.e. not the 3′-focused version) of Entrez Gene, ENSG, ENST and Refseq CDFs is 66.4%, while the average consistency of results from the later four CDFs is 87.1%, suggesting ∼20% ‘real’ improvement in using CDFs based on more stringent gene and transcript definitions. We would also like to point out that the impact of custom CDFs on other GeneChips is likely to be bigger since HG-U133A mainly represents known genes and transcripts. In the long run, we expect different gene/transcript definitions will converge but their differences with the genome and transcriptome information used by Affymetrix several years ago is likely to increase. Consequently, updating probe set definitions based on the latest genome and transcriptome information will bring more real improvements for GeneChip analysis in the future.
In summary, our analyses show that a significant percentage of existing GeneChip probe set definitions on popular human, mouse and rat GeneChips are no longer consistent with gene and transcript models in major public databases. The probe identity issue is of critical importance, as it can dramatically influence the interpretation and the understanding of expression data derived from GeneChips. We therefore recommend re-analysis of previous GeneChips data using the more accurate annotation we have made publicly available, and which will need to be continuously updated with additional improvement in genome and transcriptome informatics.
Acknowledgments
We want to thank Dr Manuel Lopez-Figueroa and Mr Ross Bersot at the Pritzker Neuropsychiatric Disorders Research Consortium for insightful suggestions and comments. M.D., P.W., E.G.J., W.E.B., R.M.M., T.P.S., H.A., S.J.W. and F.M. are members of the Pritzker Neuropsychiatric Disorders Research Consortium, which is supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. This work was partly supported by the Office of Naval Research grant N00014-02-1-0879 to H.A., A.D.B., G.K. and B.A. gratefully acknowledge the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000819). Funding to pay the Open Access publication charges for this article was provided by the Pritzker Neuropsychiatric Disorders Research Consortium.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 3.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 5.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J., Finney R.P., Clifford R.J., Derr L.K., Buetow K.H. Detecting false expression signals in high-density oligonucleotide arrays by an in silico approach. Genomics. 2005;85:297–308. doi: 10.1016/j.ygeno.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Gautier L., Moller M., Friis-Hansen L., Knudsen S. Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinformatics. 2004;5:111. doi: 10.1186/1471-2105-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbig J., Sprinkle R., Enkemann S.A. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler D.L., Church D.M., Federhen S., Lash A.E., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., Tatusova T.A., et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler D.L., Church D.M., Edgar R., Federhen S., Helmberg W., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mei R., Hubbell E., Bekiranov S., Mittmann M., Christians F.C., Shen M.M., Lu G., Fang J., Liu W.M., Ryder T., et al. Probe selection for high-density oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 2003;100:11237–11242. doi: 10.1073/pnas.1534744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee I., Dombkowski A.A., Athey B.D. Guidelines for incorporating non-perfectly matched oligonucleotides into target-specific hybridization probes for a DNA microarray. Nucleic Acids Res. 2004;32:681–690. doi: 10.1093/nar/gkh196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbard T., Andrews D., Caccamo M., Cameron G., Chen Y., Clamp M., Clarke L., Coates G., Cox T., Cunningham F., et al. Ensembl 2005. Nucleic Acids Res. 2005;33:D447–D453. doi: 10.1093/nar/gki138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T., Suzek T.O., Troup D.B., Wilhite S.E., Ngau W.C., Ledoux P., Rudnev D., Lash A.E., Fujibuchi W., Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall J.L., Grindle S., Han X., Fermin D., Park S., Chen Y., Bache R.J., Mariash A., Guan Z., Ormaza S., et al. Genomic profiling of the human heart before and after mechanical support with a ventricular assist device reveals alterations in vascular signaling networks. Physiol. Genomics. 2004;17:283–291. doi: 10.1152/physiolgenomics.00004.2004. [DOI] [PubMed] [Google Scholar]
- 19.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Shahrour F., Diaz-Uriarte R., Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 21.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]