Abstract
Native major surface protein 1 (MSP1) of Anaplasma marginale, composed of covalently associated MSP1a and MSP1b proteins, stimulates protective immunity in cattle against homologous and heterologous strain challenge. Protective immunity against pathogens in the family Anaplasmataceae involves both CD4+ T cells and neutralizing immunoglobulin G. Thus, an effective vaccine should contain both CD4+ T- and B-lymphocyte epitopes that will elicit strong memory responses upon infection with homologous and heterologous strains. Previous studies demonstrated that the predominant CD4+ T-cell response in MSP1 vaccinates is directed against the MSP1a subunit. The present study was designed to identify conserved CD4+ T-cell epitopes in MSP1a presented by a broadly represented subset of major histocompatibility complex (MHC) class II molecules that would be suitable for inclusion in a recombinant vaccine. Transmembrane protein prediction analysis of MSP1a from the Virginia strain revealed a large hydrophilic domain (HD), extending from amino acids (aa) 1 to 366, and a hydrophobic region extending from aa 367 to 593. The N terminus (aa 1 to 67) includes one 28-aa form A repeat and one 29-aa form B repeat, which each contain an antibody neutralization-sensitive epitope [Q(E)ASTSS]. In MSP1 vaccinates, recombinant MSP1a HD (aa 1 to 366) stimulated recall proliferative responses that were comparable to those against whole MSP1a excluding the repeat region (aa 68 to 593). Peptide mapping determined a minimum of five conserved epitopes in aa 151 to 359 that stimulated CD4+ T cells from cattle expressing DR-DQ haplotypes common in Holstein-Friesian breeds. Peptides representing three epitopes (aa 231 to 266, aa 270 to 279, and aa 290 to 319) were stimulatory for CD4+ T-cell clones and restricted by DR. A DQ-restricted CD4+ T-cell epitope, present in the N-terminal form B repeat (VSSQSDQASTSSQLG), was also mapped using T-cell clones from one vaccinate. Although form B repeat-specific T cells did not recognize the form A repeat peptide (VSSQS_EASTSSQLG), induction of T-cell anergy by this peptide was ruled out. The presence of multiple CD4+ T-cell epitopes in the MSP1a HD, in addition to the neutralization-sensitive epitope, supports the testing of this immunogen for induction of protective immunity against A. marginale challenge.
Anaplasma marginale is a tick-transmitted obligate intraerythrocytic pathogen of cattle that causes high levels of rickettsemia, hemolytic anemia, and often death. Although effective non-blood-derived commercial vaccines are not yet available, the feasibility of developing efficacious vaccines is evidenced by the ability to achieve protection against disease or infection by immunization with purified A. marginale outer membranes or outer membrane proteins (18, 42, 43, 45, 48, 54). Protection in cattle immunized with outer membranes or purified protein is associated with the development of T-lymphocyte-dependent gamma interferon (IFN-γ) production and high-titer immunoglobulin G (IgG) responses (18, 43, 54).
The importance of CD4+ T cells and antibody in protective immunity against pathogens in the family Anaplasmataceae has also been demonstrated in mouse models of infection with Anaplasma phagocytophila and Ehrlichia chaffeensis. Through production of IFN-γ, CD4+ T cells promote Ig class switching to protective IgG isotypes and activate phagocytic cells to produce nitric oxide, which is toxic for these organisms (3, 6, 11, 31, 34, 36, 53, 59). Direct evidence for the importance of CD4+ T cells in protective immunity against E. chaffeensis infection was recently demonstrated (31). In this study, major histocompatibility complex (MHC) class II gene knockout mice, which do not develop functional CD4+ T cells, were unable to clear E. chaffeensis and remained persistently infected during the 3-month study. To date, the E. chaffeensis and A. phagocytophila targets of this protective response have not been identified. We have characterized the CD4+ T-cell response to A. marginale surface proteins with a goal of identifying protective CD4+ T-cell epitopes that are conserved among otherwise antigenically distinct strains (16-19).
The protective outer membrane fraction of A. marginale includes six well-characterized major surface proteins (MSPs), designated MSP1a, MSP1b, and MSP2 to MSP5 (47, 54, 58). In protected cattle immunized with outer membranes, predominant T-lymphocyte and antibody responses are directed against MSP1, MSP2, and MSP3 (18, 19, 54). MSP2 and MSP3 are encoded by multiple genes and contain large central hypervariable regions (4, 8, 10, 13, 14, 46; P. F. M. Meeus and A. F. Barbet, Am. Soc. Rickettsiol.-Bartonella Emerg. Pathog. Group 2001 Joint Conf., abstr. 82, 2001). Continual emergence of novel antigenically variant MSP2s, and potentially MSP3s, that elicit new primary immune responses during infection is believed to be responsible for the organism persistence that characterizes anaplasmosis (28-30, 45). In contrast, MSP1a is encoded by a single msp1α gene in all A. marginale strains, and through covalent association with MSP1b, composes the high-molecular-weight heteromeric MSP1 complex (7, 9, 20, 44, 56, 57). Importantly, native MSP1 has been shown previously to confer significant protection against homologous and heterologous strain challenge (20, 42, 43), indicating that epitopes that stimulate protective immune responses are conserved among heterologous strains of A. marginale. We recently demonstrated that the predominant CD4+ T-lymphocyte response in MSP1-immunized cattle is directed against MSP1a, with transient or undetectable responses to MSP1b (17).
MSP1a is composed of an N-terminal region consisting of 28- or 29-amino-acid (aa) serine-rich repeats that vary in number and sequence among strains and a highly conserved C-terminal region (5, 12, 41, 49). Despite sequence variation in the N-terminal repeat region, all strains contain a conserved neutralization-sensitive epitope defined by monoclonal antibodies (MAbs) and represented by the linear sequence Q(E)ASTSS (5, 39, 49). Conserved serine-rich motifs have also been identified in the repeat units of the high-molecular-weight proteins of A. phagocytophila (130-kDa protein) and the related organisms E. chaffeensis and Ehrlichia canis (120- and 140-kDa proteins, respectively) (52, 60, 61). In MSP1-immunized cattle, the predominant CD4+ T-cell proliferative and IFN-γ responses are directed against the MSP1a region lacking the N-terminal repeats, although one of the two N-terminal repeat forms present in the immunizing Florida (FL) strain (5) also stimulated memory T-cell responses in one vaccinate (17).
Because of the importance of MSP1a-specific CD4+ T-cell responses in generating both cell-mediated and antibody effector mechanisms against A. marginale, the present study was undertaken to more precisely define the CD4+ T-cell epitopes in MSP1a that stimulate memory responses in MSP1-immunized cattle. It was also important to identify epitopes presented by MHC class II haplotypes represented broadly in the population. Knowing that MSP1a-specific responses were predominantly directed to the highly conserved MSP1a region lacking the N-terminal repeats, we focused on mapping T-lymphocyte epitopes in the large hydrophilic domain (HD) extending from aa 1 to 366 in the Virginia (VA) strain. The HD contains a neutralization-sensitive epitope(s) in the repeat region and, because it is highly hydrophilic, is amenable for vaccine development using recombinant DNA expression vectors. We report that the sequence of aa 151 to 366 within the HD of MSP1a in the VA strain, which is completely conserved in the FL strain, contains multiple T-cell epitopes. Seven overlapping peptides within this region defined a minimum of five T-cell epitopes that stimulated strong responses of all vaccinates in the study, regardless of MHC class II haplotype. Three of these epitopes were restricted by DR alleles commonly expressed in Holstein and Friesian cattle. The identification of these epitopes supports proceeding to test recombinant expressed MSP1a HD for its ability to induce protection against challenge.
MATERIALS AND METHODS
Anaplasma strains and preparation of homogenates and MSP1 antigen.
The A. marginale strains used in this study are designated by their original location of isolation and include FL, VA, and St. Maries, Idaho (St. M). The Dubois strain of Anaplasma ovis, isolated in Idaho, was also used. These have been described or referenced previously (16, 17, 50). Anaplasma strains were maintained as liquid nitrogen-cryopreserved stabilates of infected bovine erythrocytes in dimethyl sulfoxide-phosphate-buffered saline (PBS). Antigen was prepared for in vitro assays by resuspending organisms in PBS in the presence of protease inhibitors and homogenization either by sonication or by two passages through a French pressure cell (SLM Instruments, Urbana, Ill.) as described elsewhere (18). Native MSP1 protein was isolated from the FL strain of A. marginale by MAb affinity chromatography with MAb ANA15D2 or ANA22B1 (42, 43).
Recombinant MSP1 proteins and peptides.
Recombinant MSP1a (FL strain) was expressed in vaccinia virus (40), and recombinant MSP1b was expressed in Escherichia coli (9). Proteins were isolated by affinity chromatography (43) with MAb ANA15D2 (MSP1a) and MAb AMR38A6 (MSP1b). The C-terminal region of MSP1a (aa 242 to 767 in the FL strain or aa 68 to 593 in the VA strain), which lacks the N-terminal repeat region, was expressed from the FL strain and prepared as a recombinant maltose-binding protein (MBP) fusion protein (17). MBP purchased from New England Biolabs (Beverly, Mass.) was used as a control antigen. Protein concentrations were determined by the Bradford assay (Bio-Rad, Hercules, Calif.).
The hydropathic profile and membrane orientation of MSP1a were predicted with TMPred software (www.ch.embnet.org/software/Tmpred-form.html). A large HD extending from aa 1 to 366 in the VA strain was predicted by this analysis (Fig. 1). MSP1a HD was expressed as a fusion with the human CD5 secretory signal to direct extracellular secretion. The details of protein expression are reported elsewhere (40a). Briefly, the open reading frame encoding aa 1 to 366 of A. marginale MSP 1a was amplified by PCR from a recombinant plasmid (pVAr1) containing a genomic DNA fragment of the VA strain msp1a gene (5). The forward primer (5′-ATACTGCAGATGTCAGCAGAGTATGTGTCTACC-3′) introduced a PstI restriction site (in boldface) at the 5′ end, and the reverse primer (5′-TGGATCCTACTGTGTAGTAGTGTCCGAAGG-3′) introduced a BamHI restriction site (in italics) at the 3′ end. To direct secretion of the MSP1a HD protein, the human CD5 secretory signal sequence was added (27). The CD5 signal sequence was amplified by PCR, and the product was subcloned into PCR-Blunt vector (Invitrogen, Carlsbad, Calif.) to generate pCD5ss. The MSP1a HD product was subcloned in frame as a PstI-BamHI fragment into pCD5ss to generate pCD5MSP1aHD. The CD5MSP1aHD open reading frame was released as an EcoRV-BamHI fragment and then subcloned into VR-1055 eukaryotic expression vector (Vical, San Diego, Calif.). The resultant construct, pVR-CD5MSP1aHD, was sequenced from both directions with an ABI PRISM automated fluorescence DNA sequencer (Applied Biosystems, Foster City, Calif.).
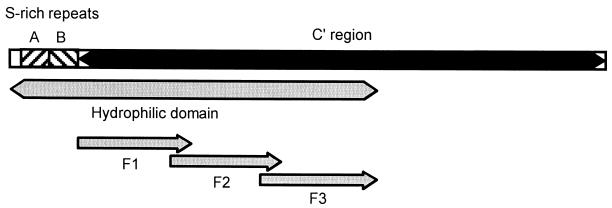
FIG. 1.
Schematic diagram of full-length A. marginale MSP1a (VA strain). Depicted are the two N-terminal serine (S)-rich form A and B repeats (hatched bars) which each contain one MAb neutralization-sensitive epitope, the C-terminal region extending from aa 68 to 593 (black bar); the large HD extending from aa 1 to 366 (gray bar); and proteins F1 (aa 68 to 180), F2 (aa 160 to 270), and F3 (aa 250 to 366) (arrows).
To map T-cell epitopes present in the MSP1a HD excluding the repeats (aa 68 to 366 in the VA strain), three recombinant proteins spanning this region (designated MSP1a HD F1 to F3) that overlap by 30 aa were generated from PCR-cloned fragments (Table 1). Forward primers used to generate these fragments introduced a PstI restriction site (in boldface) at the 5′ end, whereas reverse primers introduced a BamHI restriction site (in italics) at the 3′ end. In addition, the reverse primers introduced a His tag at the C terminus (in boldface italics). The primers were as follows: F1 primers, forward, 5′-ATACTGCAGATGACTGATTGGCGGCAAGAGATGCGC-3′; reverse, 5′-TGGATCCTAGTGGTGGTGGTGGTGGTGCTGAACATCAGCCGCAACCACCAC-3′; F2 primers, forward, 5′-ATACTGCAGATGGGGTACGCCACCTATCTCGCG-3′; reverse, (5′-TGGATCCTAGTGGTGGTGGTGGTGGTGGTCAGCATCAAGCTTATCCAGCTG-3′; and F3 primers, forward, 5′-ATACTGCAGATGCTTGCCGGGCACGTCGATGC-3′; reverse, 5′-TGGATCCTAGTGGTGGTGGTGGTGGTGCTGTGTAGTAGTGTCCGAAGGC-3′. The three PCR products were subcloned into pCD5ss as PstI-BamHI fragments to generate pCD5MSP1aHDF1, pCD5MSP1aHDF2, and pCD5MSP1aHDF3, respectively. The CD5MSP1aHDF1, CD5MSP1aHDF2, and CD5MSP1aHDF3 open reading frames were released as EcoRV-BamHI fragments and then subcloned into the VR-1055 eukaryotic expression vector. The resultant constructs F1, F2, and F3 were sequenced from both directions with VR-1055-specific primers. Large-scale plasmid DNA was prepared with an Endo-Free Plasmid Maxiprep kit (Qiagen, Valencia, Calif.) and sterilized with 100% ethanol.
TABLE 1.
Proteins and peptides spanning the predicted HD of MSP1a
| Protein or peptidea | Amino acid positionb | Sequencec |
|---|---|---|
| Protein | ||
| MSP1a C-terminal region | End of repeats (68)-593 | Reference 5 |
| HD | 1-366 | Reference 5 |
| F1 | 68-180 | |
| F2 | 160-270 | |
| F3 | 250-366 | |
| Peptide | ||
| F2-1 | 151-180 | GLQGLKIGEGYATYLAQAFADSVVVAADVQ |
| F2-2 | 171-200 | DSVVVAADVQSSGACSASLDSAIANVETSW |
| F2-3 | 191-220 | SAIANVETSWSLHGGLVSKGFDRDTKVERG |
| F2-4 | 211-240 | FDRDTKVERGDLEAFVDFMFGGVSYNDGNA |
| F2-5 | 231-260 | GGVSYNDGNASAARSVLETLAGHVDALGIS |
| F3-1 | 250-279 | LAGHVDALGISYNQLDKLDADTLYSVVSFS |
| F3-2 | 270-299 | DTLYSVVSFSAGSAIDRGAVSDAADKFRVM |
| F3-3 | 290-319 | SDAADKFRVMMFGGAPAGQEKTAEPEHEAA |
| F3-4 | 310-339 | KTAEPEHEAATPSASSVPSTVHGKVVDAVD |
| F3-5 | 330-359 | VHGKVVDAVDRAKEAAKQAYAGVRKRYVAK |
| F3-6 | 350-366 | AGVRKRYVAKPSDTTTQ |
MSP1a was expressed as a fusion protein with MBP in E. coli (17). The HD and F1 to F3 proteins were expressed as His-tagged proteins in COS-7 cells.
Amino acid designation is based on the position in the VA strain, which contains two N-terminal repeats.
The peptides overlap by 10 or 11 aa, indicated by underlined sequences.
MSP1a HD and F1 to F3 proteins were expressed in COS-7 cells. A total of 20 μg of DNA was transfected into one 100-mm-diameter plate of COS-7L cells (Life Technologies, Gaithersburg, Md.) with 30 μl of Lipofectamine reagent and 20 μl of PLUS reagent (Life Technologies) according to the manufacturer's instructions. Mock- and VR-1055-transfected COS-7L cells were included as negative controls. Protein expression was verified by in situ immunocytochemistry. To produce protein for T-cell proliferation assays, COS-7 cell monolayers were transfected as described above, and 1 day posttransfection, the medium was replaced with serum-free VP-SFM medium (Life Technologies) supplemented with 4 mM l-glutamine. Supernatants were harvested 96 h posttransfection and concentrated 10-fold with a Centriprep centrifugal filter device with a 10-kDa (HD) or 5-kDa (F1 to F3) molecular mass cutoff (Millipore Corporation, Bedford, Mass.). To estimate the amount of HD or F1 to F3 proteins in the concentrated supernatants, serial dilutions of supernatants were used to generate dot blots that were probed respectively with MAb ANA22B1 or MAb specific for poly-His (His probe H3; Santa Cruz Biotechnology, Santa Cruz, Calif.).
Peptides spanning the F2 and F3 regions of the MSP1a HD (Table 1) and peptides that compose the MSP1a N-terminal form A and B repeats (see Table 5) were synthesized by Gerhardt Munske (Laboratory for Biotechnology and Bioanalysis I, Washington State University, Pullman). The form A repeat (DDSSSASGQQQESSVSSQS_EASTSSQLG) is present one time in the FL and VA strains, and the form B repeat (ADSSSAGGQQQESSVSSQSDQASTSSQLG) is tandemly repeated seven times in the FL strain and is present once in the VA strain (5). Peptides were resuspended in PBS and stored at −20°C.
TABLE 5.
Identification of the epitope in the N-terminal form B repeat of MSP1a stimulatory for CD4+ T cells from MSP1-immunized calf 87
| Expt no. and antigen or peptide | Proliferative response (SI) of T cellsa:
|
|||
|---|---|---|---|---|
| 87 CL | 87.2C5 | 87.2D11 | 87.4G10 | |
| 1 | ||||
| A. marginale | 11.1 | 9.8 | 1.3 | 11.7 |
| MSP1a | 11.7 | 80.3 | 7.0 | NDb |
| Peptide B | 5.4 | 1,790.2 | 277.0 | 242.8 |
| 2 | ||||
| Peptides | ||||
| ADDSSSASGQQQESSVSSQSEASTSSQLGc | ND | ND | ND | ND |
| BADSSSAGGQQQESSVSSQSDQASTSSQLG | 11.3 | 1,450.5 | 65.8 | 241.1 |
| B1ADSSSAGGQQQESSV | 1.0 | 1.3 | 1.5 | 0.8 |
| B2AGGQQQESSVSSQSD | 1.8 | 1.0 | 1.4 | 0.9 |
| B3QESSVSSQSDQASTS | 0.9 | 0.8 | 1.1 | 1.3 |
| B4VSSQSDQASTSSQLG | 11.0 | 1,532.9 | 61.0 | 267.8 |
| B5VSSQSDEASTSSQLG | 0.9 | 1.3 | ND | ND |
| B6VSSQSDQASTSSQ | 1.1 | 1.3 | ND | ND |
| B7VSSQSGQASTSSQLGG | 6.8 | 710.3 | ND | ND |
| A4VSSQSEASTSSQLG | 1.4 | 1.1 | 1.1 | 1.6 |
| B8SSQSDQASTSSQLGADSSSAGGQQQESS | ND | 381.4 | 14.7 | 41.4 |
A T-cell line (CL) was obtained following stimulation of calf 87 PBMC for 1 week with recombinant MSP1a and 1 week with A. marginale homogenate (experiment 1) or for 1 week with A. marginale homogenate (experiment 2), and clones were obtained by limiting dilution. T cells were cultured for 3 days with APC and antigen. Results are presented as the SI comparing the mean counts per minute of duplicate cultures of T cells cultured with medium with those cultured with antigen, using 25 μg of A. marginale per ml, 5 μg of MSP1a per ml or 25 μg of peptide B per ml (experiment 1), 1 μg of peptides per ml (experiment 2, T-cell line), or 10 μg of peptides per ml (experiment 2, T-cell clones). Background counts per minute for the T-cell line 87 were 6,683 ± 1,015 cpm (experiment 1) and 4,047 ± 709 cpm (experiment 2) and for the T-cell clones ranged from 63 ± 7 to 251 ± 3 cpm. Numbers in bold have an SI of >3.0 and are significantly greater (P < 0.05) than medium control values.
ND, not determined in this experiment.
Underlined amino acids or spaces indicate different amino acids or deletions when compared with peptide B.
Immunization of calves with MSP1.
Three 6-month-old Holstein calves, animals 87, 93, and 96, were immunized intramuscularly four times at 2-week intervals with 20 μg of native MSP1 per injection emulsified in Freund's complete adjuvant for the first injection and in Freund's incomplete adjuvant for subsequent injections (17). The bovine lymphocyte antigen A (BoLA-A) class I alleles of the calves were defined by serological typing (22), and DRB3 alleles were defined by PCR-restriction fragment length polymorphism analysis of exon 2 (55). The BoLA-DQ haplotypes were inferred from BoLA-A and DRB3 typing on the basis of haplotypes defined in the Seventh International BoLA Workshop (33) (BoLA nomenclature website: http://www2.ri.bbsrc.ac.uk/bola/). The BoLA-A and DR-DQ haplotypes for all cattle used in this study are shown in Table 2. For steer 2216, used as a negative control, the DQ alleles were not inferred because of insufficient information on this animal (Hereford crossbred).
TABLE 2.
MHC class I (BoLA-A) and class II (BoLA-DR, DQ) haplotypes of the cattle used in this study
| Animal no. | BoLA-A type | DRB3 allele | DQA allele(s) | DQB allele(s) |
|---|---|---|---|---|
| 87a | 12 | ∗16 | ∗11A | ∗11C |
| 15 | ∗22 | ∗9B | ∗9B | |
| 93a | 12 | ∗16 | ∗11A | ∗11C |
| 10 | ∗3 | ∗10 | ∗10 | |
| 96a | 15 | ∗22 | ∗9B | ∗9B |
| 10 | ∗3 | ∗10 | ∗10 | |
| 62 | 12 | ∗16 | ∗11A | ∗11C |
| 15 | ∗22 | ∗9B | ∗9B | |
| 77 | 12 | ∗16 | ∗11A | ∗11C |
| 14 | ∗10 | ∗14 or ∗11Bb | ∗14 or ∗11Ab | |
| 75 | 10 | ∗7 | ∗2 | ∗2 |
| 15 | ∗22 | ∗9B | ∗9B | |
| 2216 | 2 | ∗7 | NDc | ND |
| 6 | ∗28 | ND | ND |
Calves 87, 93, and 96 were immunized with native MSP1.
The DQ alleles could not be precisely inferred from the BoLA-A and DRB3 restriction fragment length polymorphism analyses.
ND, not determined.
A. marginale-specific T-lymphocyte lines and clones.
Short-term T-lymphocyte lines were repeatedly established from peripheral blood mononuclear cells (PBMC) of A. marginale MSP1-immunized calves 87, 93, and 96 from shortly after immunization to more than 1 year later. In all experiments, cell lines were propagated by stimulation with homogenate prepared from the FL strain of A. marginale, with native MSP1, or with peptide B as described elsewhere (17). Briefly, 4 × 106 PBMC were cultured per well in 24-well plates (Costar, Cambridge, Mass.) in a volume of 1.5 ml of complete RPMI 1640 medium (15) with 5 to 25 μg of antigen per ml. After 7 days and weekly thereafter, cells were subcultured to a density of 7 × 105 cells per well (cpw) and cultured with 2 × 106 irradiated (3,000 rads) autologous PBMC as a source of antigen-presenting cells (APC) with or without antigen, which was often given on alternate weeks to lower background proliferation. T-lymphocyte lines were maintained for up to 3 weeks, and cells were assayed for antigen-dependent proliferation 7 days following the last stimulation. In many experiments, γδ T lymphocytes and CD8+ T lymphocytes were depleted by incubating either PBMC or cell lines with 15 μg of γδ TCR1-specific MAb CACT61A and CD8-specific MAb CACT80C per ml followed by washing and incubation with rabbit complement (Sigma, St. Louis, Mo.) diluted 1:8 (16).
T-lymphocyte clones were obtained from MSP1-specific cell lines by limiting dilution and were propagated with A. marginale homogenate and 10% bovine T-cell growth factor (TCGF) as described elsewhere (17). T-lymphocyte clones specific for the form B repeat (peptide B) were also obtained by limiting dilution cloning from calf 87. In one experiment, PBMC from calf 87 were cultured for 1 week with 5 μg of peptide B per ml and for 2 weeks with A. marginale (FL) homogenate and then cloned by plating 0.3 or 1 cpw in 96-well round-bottomed plates (Costar) with 5 × 104 APC per well, 5 μg of A. marginale per ml, and 10% TCGF. In a second experiment, PBMC depleted of γδ T cells were cultured with 5 μg of peptide B per ml for 7 days, and lymphoblasts were plated at 0.3 or 1 cpw with APC, TCGF, and 1 μg of peptide B per ml. Frequencies of positive wells plated at 1 cpw were 6% (experiment 1) and 22% (experiment 2) and at 0.3 cpw were 3%.
Cell surface phenotypic analysis.
Differentiation markers on T-lymphocyte lines and clones were analyzed by flow cytometry (19). The MAbs used were specific for bovine CD2 (MAb MUC2A), CD3 (MAb MM1A), CD4 (MAb CACT138A), CD8 (MAb CACT80C and BAT82B), and the δ chain of the γδ T-cell receptor (MAb CACT61A), purchased from the Washington State University Monoclonal Antibody Center, Pullman.
Lymphocyte proliferation assays.
Proliferation assays were carried out in replicate wells of round-bottomed 96-well plates for 3 to 4 days with short-term T-lymphocyte lines or T-lymphocyte clones, as described previously (17). T cells (2 × 104 cells) were cultured in duplicate wells in a total volume of 100 μl of complete medium containing antigen and 2 × 105 APC. APC consisted of irradiated PBMC from the autologous donor or from calves fully matched for both DR-DQ haplotypes, matched for one DR-DQ haplotype, or mismatched for both haplotypes. Antigens consisted of 0.2 to 25.0 μg of homogenate prepared from FL or St. M strains of A. marginale or A. ovis per ml, native MSP1 protein, recombinant MSP1a and MSP1b proteins, and 0.1 to 10 μg of peptide per ml. Negative control antigens included membranes prepared from uninfected red blood cells, recombinant MBP, and a 30-mer peptide (MSP2-1) derived from MSP2 (16). COS-7 cell supernatants from untransfected cells or cells expressing MSP1a HD or F1 to F3 proteins and diluted from 1:4 to 1:16 (one experiment) or 1:50 to 1:6,250 (second experiment) were also used. Different dilutions were used because the amount of expressed antigen varied between transfection experiments. Cells were radiolabeled for the last 18 h of culture with 0.25 μCi of [3H]thymidine, harvested with an automated cell harvester (TomTec, Orange, Conn.), and counted with a liquid scintillation counter. Results are presented as the mean counts per minute of replicate cultures ± 1 standard deviation (SD) or, for ease of presentation, as the stimulation index (SI), which represents the mean counts per minute of replicate cultures of cells plus antigen divided by the mean counts per minute of replicate cultures of cells plus medium.
To determine whether T-cell clones were DR restricted or DQ restricted, 2 × 105 autologous APC were preincubated in 96-well plates for 1 to 2 h with 20 μg of MAb per ml against bovine MHC class II molecules DRα (MAb TH14B) or DQα (MAb TH22A) (1, 2, 26). These IgG2a MAbs and an isotype control MAb (AV213A) were obtained from the Washington State University Monoclonal Antibody Center and purified by affinity to protein G with the Equilibrate Hi Trap protein G column (Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's protocol. This amount of MAb was determined to provide optimal blocking of proliferation without nonspecific effects.
The Student one-tailed t test was used to determine statistically significant differences in proliferation induced by using different antigens or APC. An SI of ≥3.0 was considered statistically significant.
Peptide competition-antagonist assays.
Experiments were designed to determine whether nonstimulatory N-terminal serine-rich form A repeat peptide A4 (VSSQS_EASTSSQLG) had any competitive or antagonistic effect on the response of peptide B-specific T-cell clones to agonistic form B repeat peptide B4 (VSSQSDQASTSSQLG). First, proliferation assays were performed with peptide B-specific T-cell clones, APC, and a suboptimal amount of agonist peptide B4 (1 μg/ml) without or with doubling amounts (0.125 to 8 μg/ml) of nonstimulatory peptide A4. Second, APC were prepulsed for 1.5 to 2 h with a suboptimal concentration (1 or 5 μg/ml) of agonist peptide B4, washed, and plated with CD4+ T-cell clones and no peptide or 1 to 100 μg of peptide A4 per ml by a protocol similar to that described by De Magistris et al. (25). Finally, T-cell clones were cultured for 7 days in 24-well plates with 10% TCGF, APC, and 5 μg of peptide A4 or peptide B4 per ml or no peptide. Cells were then washed and tested in a standard proliferation assay with 0.1 to 10 μg of agonist peptide B4 per ml.
Detection of IFN-γ in supernatants of T-lymphocyte clones.
T-cell clones were stimulated for 48 to 72 h in 24-well plates with APC, 10% TCGF, and antigen consisting of 5 μg of A. marginale (FL strain) per ml or 10 μg of peptide A or peptide B per ml, and supernatants were tested for IFN-γ production by enzyme-linked immunosorbent assay as described elsewhere (17). Control supernatants were obtained from APC cultured with antigen and TCGF.
RESULTS
Identification of CD4+ T-cell epitopes in the HD of MSP1a.
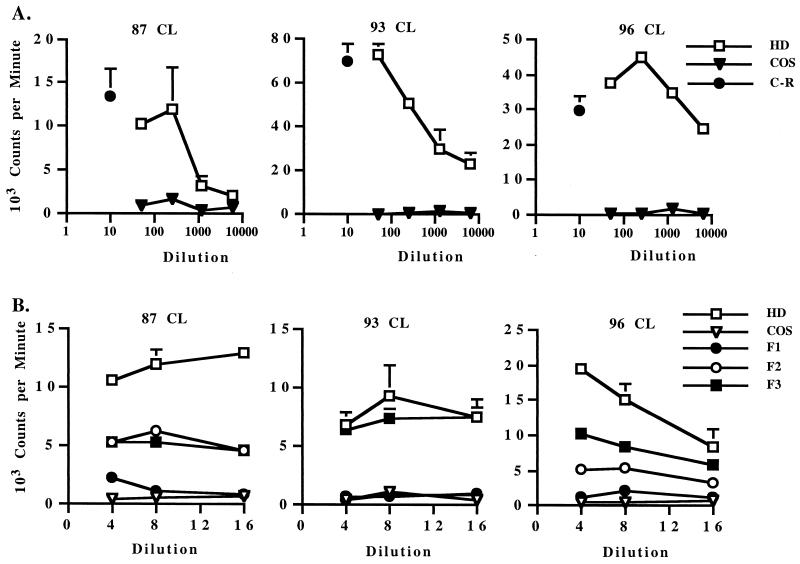
Transmembrane protein prediction analysis of MSP1a revealed an HD extending from aa 1 to 366 in the VA strain (aa 1 to 540 in the FL strain) and a hydrophobic domain extending from aa 367 to 593 in the VA strain (aa 541 to 767 in the FL strain) (Fig. 1). Included in the HD are the N-terminal repeats (two in the VA strain and eight in the FL strain) that contain defined antibody neutralization-sensitive epitopes QASTSS and EASTSS (5, 43). Furthermore, the HD region excluding the repeats (aa 68 to 366 in the VA strain) is completely conserved (100% identical) between FL and VA strains (5); 98% identical between FL and St. M strains (49); and 89% identical among FL, Washington Okanogan (WA-O), and South Idaho (SI) strains (5). To determine whether the highly conserved HD contained T-cell epitopes, this region from the VA strain was expressed in COS-7 cells and tested for elicitation of memory CD4+ T-cell responses in FL strain MSP1-immunized calves. The VA strain HD sequence was used since the C-terminal region nonrepeat sequence is identical to that in FL strain MSP1a and the N-terminal repeat region contains the same types of repeats found in the FL strain MSP1a (form A and B) but is shorter. A 1:50 or 1:250 dilution of COS-7 cell supernatant expressing MSP1a HD stimulated levels of proliferation of short-term T-cell lines comparable to that for 10 μg of MSP1a C-terminal region protein (aa 68 to 593) per ml, whereas control COS-7 supernatant was not stimulatory (Fig. 2A). To further define the location of the dominant conserved T-helper (Th) cell epitopes in this immunostimulatory region, three fusion proteins (F1 to F3) spanning the HD region lacking the two N-terminal repeats were constructed and expressed in COS-7 cells (Fig. 1 and Table 1). Repeated testing of these proteins on short-term CD4+ T-cell lines demonstrated significant levels of stimulation by the F2 and F3 region proteins, with no or weak stimulation by the F1 region (Fig. 2B). Dot blot analysis of His-tagged F1, F2, and F3 proteins with a MAb specific for C-terminal poly-His showed similar levels of expression of all three proteins in COS-7 cell supernatants, indicating that the lack of response to the F1 protein was not due to insufficient levels of antigen (data not shown).
FIG. 2.
Proliferative responses of short-term T-lymphocyte lines from MSP1-immunized calves against the C-terminal region of MSP1a (aa 68 to 593) or the predicted large HD (aa 1 to 366) and proteins F1 to F3 spanning this region. Short-term T-cell lines were propagated from PBMC for 1 to 2 weeks with A. marginale homogenate. The calf number is indicated for each panel. (A) T cells were stimulated with 10 μg of MSP1a C-terminal region protein (C-R) per ml or 1:50 to 1:6,250 dilutions of supernatants from untreated COS-7 cells (COS) or cells transfected with the HD of MSP1a, and results are representative of four experiments with different cell lines from each calf. (B) T cells were stimulated with 1:4 to 1:16 dilutions of COS-7 cell supernatants from untreated cells (COS) or cells transfected with the HD or F1, F2, or F3 protein spanning this region. Results are representative of two experiments performed with different cell lines from each calf. Data are presented as the mean counts per minute of duplicate cultures in response to antigen. Error bars, SD.
Identification of peptides in MSP1a HD that elicit recall proliferative responses by MSP1a-specific T-lymphocyte lines.
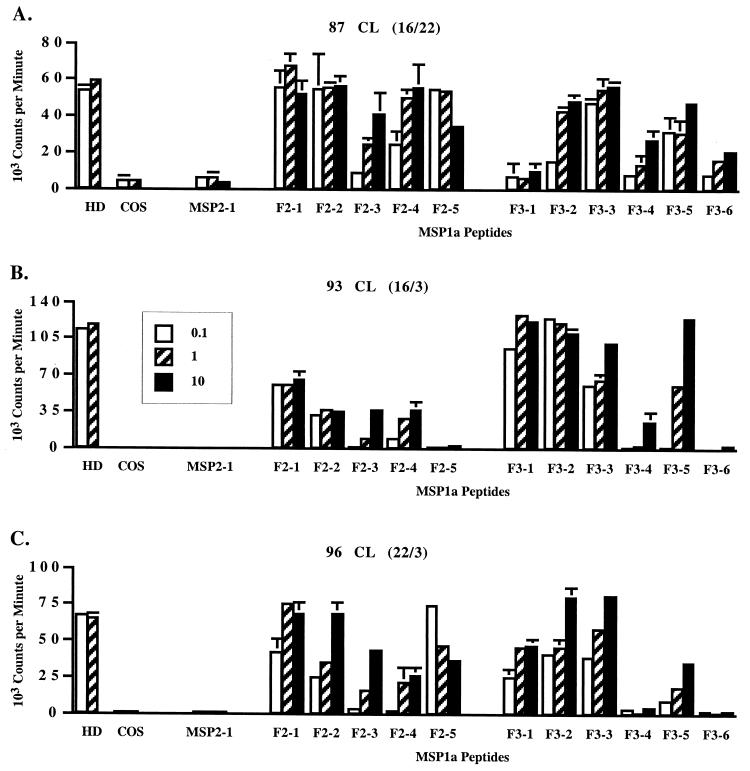
Short-term CD4+ T-cell lines were then used to identify stimulatory peptides spanning the HD F2-F3 region of MSP1a. The cell lines were depleted of CD8+ T cells and γδ T cells, cultured with A. marginale homogenate, and tested after 1 or 2 weeks of culture. As shown in Fig. 3, the cell lines from all three immunized calves responded to multiple peptides spanning the F2-F3 region comprising aa 151 to 366 in the VA strain, although different response patterns were observed for the calves that expressed different class II haplotypes. It was consistently observed in three or more experiments that all five overlapping peptides spanning the F2 region stimulated T cells from calves 87 and 96, whereas all but one (F2-5) stimulated T cells from calf 93. Calves 93 and 96 responded in a similar manner to F3 region peptides, with strong responses to peptides F3-1 to F3-3 and F3-5, relatively weak responses to peptide F3-4, and no response to peptide F3-6. In contrast, calf 87 responded strongly to peptides F3-2 to F3-5 and weakly to peptide F3-6 but did not proliferate against peptide F3-1. Because the response to a peptide could be to the 10- or 11-aa overlap with a contiguous peptide, a minimum of eight T-cell epitopes were recognized by each calf.
FIG. 3.
Proliferative responses of short-term T-lymphocyte lines from MSP1-immunized calves against peptides spanning the F2-F3 regions of the predicted HD of MSP1a. Cell lines were propagated from γδ- and CD8+ T-lymphocyte-depleted PBMC by stimulation with A. marginale homogenate for 1 (A) or 2 (B and C) weeks. The calf number and DR haplotype (parentheses) are indicated for each panel. Antigens consisted of a 1:10 (hatched bars) or 1:100 (white bars) dilution of supernatants from untreated COS-7 cells (COS) or COS-7 cells transfected with the HD of MSP1a (HD) or 0.1 (white bars), 1 (hatched bars), or 10 (black bars) μg of peptides spanning F2 and F3 regions of the HD per ml. Peptide MSP2-1 was used as a negative control peptide. Results are representative of two or three assays using different cell lines from each calf. Error bars, SD.
Identification of peptides in the MSP1a HD that contain epitopes stimulatory for CD4+ T-cell clones.
Previous work reported deriving a panel of MSP1a-specific CD4+ T-cell clones from cell lines of MSP1 vaccinates 87, 93, and 96 (17). Table 3 shows that representative clones respond specifically to MSP1a and to A. marginale derived from FL and St. M strains but not to A. ovis. When stimulated with A. marginale homogenate and APC, the clones produced from 23 to 125 U of IFN-γ per ml, whereas APC cultured without T cells produced ≤2 U of IFN-γ per ml (data not shown). The response of six clones to the HD and peptides spanning the HD F2 and F3 regions was then examined. Interestingly, one clone from calf 87 and two clones from calf 96 responded to peptide F2-5, and a third clone from calf 96 responded to peptide F3-3, whereas two clones from calf 93 responded to peptides F3-1 and F3-2 (Table 4). The response pattern of the clones was consistent with that of uncloned short-term T-cell lines from these calves. For example, a low concentration (0.1 μg/ml) of peptide F2-5 stimulated strong proliferative responses from calves 87 and 96, whereas peptides F3-1 and F3-2 stimulated strong proliferative responses from calf 93 (Fig. 3). Clones 87.2A1, 96.4D8, and 96.2.4G4 responded to peptide F2-5, but not F2-4 or F3-1, each of which overlaps peptide F2-5 by 10 or 11 aa, indicating that the core epitope for these clones was not represented by the 10- to 11-aa sequences on the N and C termini of F2-5 (Table 1). Similarly, the core epitope in peptide F3-3 recognized by clone 96.1.4G4 did not likely consist of the 10 aa on the N- or C-terminal ends. However, the response of clones 93.3A8 and 93.4E4 to peptides F3-1 and F3-2 indicates that the epitope DTLYSVVSFS shared by these peptides constitutes the core epitope for these clones. In agreement with the response of clones to antigen prepared from both FL and St. M strains (Table 3), the sequence of peptide F2-5 and the overlapping sequence of peptides F3-1 and F3-2 (DTLYSVSFS) are each completely conserved in these A. marginale strains.
TABLE 3.
Proliferative responses of A. marginale MSP1a-specific CD4+ T-cell clones are conserved in two A. marginale strains but not in A. ovis
| Expt no. and antigen | Proliferation (mean cpm ± 1 SD) of T-cell clonea:
|
|||
|---|---|---|---|---|
| 87.2A1 | 93.4E4 | 96.4D8 | 96.2.4G4 | |
| 1 | ||||
| None | 242 ± 10 | 2,861 ± 34 | 951 ± 426 | 5,070 ± 731 |
| A. marginale (FL) | 205,660 ± 6,771 | 49,866 ± 2,221 | 19,775 ± 1,848 | 43,243 ± 787 |
| MSP1a | 81,439 ± 13,143 | 35,600 ± 335 | 28,116 ± 3,200 | 22,581 ± 890 |
| MSP1b | NDb | 3,963 ± 335 | 1,993 ± 119 | 5,630 ± 129 |
| 2 | ||||
| None | 259 ± 43 | 948 ± 147 | 892 ± 152 | 239 ± 33 |
| URBCc | 511 ± 208 | 437 ± 5 | 614 ± 101 | 293 ± 45 |
| MSP1 (FL) | 16,434 ± 79 | 56,563 ± 5,762 | 39,890 ± 3,345 | 79,744 ± 1,733 |
| A. marginale (St. M) | 5,802 ± 1,036 | 25,089 ± 2,863 | 20,298 ± 918 | 84,181 ± 2,425 |
| A. ovis | 160 ± 23 | 747 ± 346 | 644 ± 151 | 288 ± 39 |
CD4+ T-cell clones were cultured in duplicate wells with APC and the indicated antigens. Results are presented for 25 μg (clone 87.2A1) or 10 μg (other clones) of A. marginale (FL) homogenate per ml and 5 μg of MSP1a or MSP1b per ml (experiment 1) or 10 μg of all antigens per ml (experiment 2). Numbers in bold have an SI of >3.0 and are significantly greater (P < 0.01) than medium control values. Results are representative of at least two independent assays.
ND, not determined in this assay.
URBC, uninfected red blood cells.
TABLE 4.
Proliferation of MSP1a-specific CD4+ T-cell clones to peptides spanning the F2 and F3 regions of the predicted HD of MSP1a
| Antigen | Proliferative response (SI) of T-cell clonea:
|
|||||
|---|---|---|---|---|---|---|
| 87.2A1 | 93.3A8 | 93.4E4 | 96.1.4G4 | 96.4D8 | 96.2.4G4 | |
| MSP1a C-terminal region | 46.7 | 4.6 | 19.7 | 4.7 | 18.8 | 68.8 |
| MBP | 0.6 | 0.9 | 1.0 | 0.8 | 0.8 | 1.0 |
| HD | 9.6 | 3.3 | 23.1 | 3.6 | 9.7 | 34.6 |
| COS sup | 1.0 | 0.8 | 0.6 | 0.4 | 1.3 | 1.2 |
| F2-4 | 1.2 | 0.9 | NDb | ND | 0.9 | 1.5 |
| F2-5 | 120.1 | 0.7 | ND | ND | 50.5 | 97.7 |
| F3-1 | 1.0 | 4.4 | 6.7 | 0.9 | 1.0 | 0.6 |
| F3-2 | 1.2 | 5.1 | 8.0 | 0.7 | 1.1 | 1.1 |
| F3-3 | 1.1 | 1.8 | 2.6 | 67.7 | 1.3 | 0.8 |
| F3-4 | 1.6 | 1.4 | 1.2 | 1.2 | 0.9 | 1.3 |
Results are presented as the SI comparing the mean counts per minute of duplicate cultures of T cells cultured with APC and antigen with those cultured with medium. Antigen concentrations were a 1:10 dilution of COS-7 cell supernatants (COS sup) or 10 μg of recombinant protein or peptide per ml, except for clone 87.2A1, where peptide F2-5 was used at 0.1 μg/ml; for clone 93.4E4, where peptide F3-2 was used at 1 μg/ml; and for clones 96.4D8 and 96.2.4G4, where peptide F2-5 was used at 1 μg/ml. Background proliferation ranged from 142 ± 107 to 559 ± 254 cpm. The data are representative of three to five separate assays for each clone except 93.4E4, where one assay was performed. Numbers in bold have an SI of >3.0 and are significantly greater (P < 0.05) than medium control values.
ND, not determined in this experiment.
Identification of the epitope in the N-terminal form B repeat recognized by CD4+ T cells from calf 87.
We previously reported that PBMC and CD4+ T-cell lines from MSP1-immunized calf 87, but not calf 93 or 96, responded specifically to the form B repeat, but not the form A repeat of the FL strain MSP1a (17). To more precisely define the epitope(s) within the form B repeat sequence, five CD4+ T-cell clones specific for this sequence (peptide B) were generated. All clones produced IFN-γ, ranging from 50 to 210 U/ml, in response to culture with APC and peptide B, compared with APC cultured with peptide B, which produced 1 U of IFN-γ per ml (data not shown). T-cell clones and short-term cell lines were then tested for proliferation to truncated peptides spanning the 29-aa peptide B (Table 5). All T-cell lines and clones tested gave the same response pattern and recognized peptide B4 consisting of amino acids VSSQSDQASTSSQLG. This peptide contains the neutralization-sensitive epitope Q(E)ASTSS defined by two MAbs (5, 39, 43). Peptide B7 (VSSQSGQASTSSQLGG), which represents the sequence present in the form C repeat found in the WA-O strain (5), was immunostimulatory, although the D-to-G alteration at position 6 of the peptide (underlined) resulted in suboptimal responses. To determine the importance of a Q-to-E alteration at position 7, which is present in the form A repeat (Table 5), peptide B5 (VSSQSDEASTSSQLG) representing this sequence was also tested but did not stimulate any of the T-cell lines or clones. Similarly, the sequence representing the exact epitope in the form A repeat, VSSQS_EASTSSQLG (peptide A4), which has a deletion of residue D at position 6 and a Q-to-E alteration at position 7 (underlined), was also unable to elicit any T-cell proliferation of the cell lines or clones. To verify the lack of response by peptide B-specific T cells to peptide A4, T-cell clone 87.2C5 cells were incubated with APC and 10 μg of agonist peptide B4 or peptide A4 per ml for 48 h and secreted IFN-γ was measured. Peptide B4 stimulated 47 U of IFN-γ per ml, whereas peptide A4 was unable to stimulate production of detectable levels of IFN-γ.
T-cell clones specific for peptide B responded weakly to MSP1a and A. marginale homogenate (Table 5). To determine whether this weak response was due to the nonstimulatory form A repeat in the whole protein, the antagonistic potential of the form A repeat (peptide A4) on the response to the form B repeat agonist epitope (peptide B4) was examined. First, peptide A4 was mixed with a stimulatory but suboptimal amount of peptide B4 at ratios of A4 to B4 ranging from 1:8 to 8:1 during the proliferation assay. Second, APC were prepulsed with a suboptimal amount of agonist peptide B4, washed, and then assayed with peptide B-specific T-cell clones with either no additional antigen or increasing amounts of peptide A4. Finally, T-cell clones and APC were cultured with peptide A4 or B4 or medium for 2 h or 7 days, washed, and stimulated with agonistic peptide B4 in the proliferation assay. None of these treatments resulted in a diminution of the response to any concentration of agonist peptide B4 or to baseline levels of B4 on prepulsed APC (experiment 2), indicating the lack of an antagonistic effect of the form A repeat on the agonist form B epitope (data not shown).
MHC class II DR- and DQ-restricted responses to MSP1a epitopes.
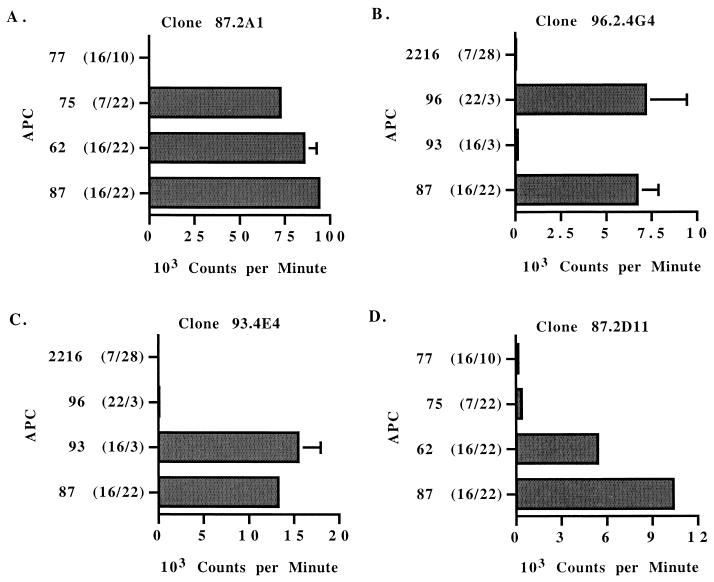
In cattle, DR and DQ genes are closely associated on chromosome 23. Each haplotype expresses one nonpolymorphic DRα molecule, one polymorphic DRβ molecule (DRB3), and one or more polymorphic DQα and DQβ molecules when DQ is duplicated (reviewed in reference 33). Furthermore, in cattle DQα and DQβ chains can form both inter- and intrahaplotype pairing (32, 33), resulting in increased class II polymorphism. To determine whether responses of individual Th-cell clones were restricted by DR or DQ molecules, T cells were cultured with specific antigen and autologous APC, APC fully matched for both DR-DQ haplotypes, APC matched for one DR-DQ haplotype, or APC mismatched for both haplotypes (Table 2). Clones specific for T-cell epitopes in the F2-F3 HD region of MSP1a responded to antigen presented by APC from a calf matched for one of the two DR-DQ haplotypes (Fig. 4A to C). Clones 87.2A1 and 96.2.4G4 specific for peptide F2-5 responded to antigen only in the presence of APC expressing haplotype DRB3∗22-DQA∗9B-DQB∗9B (Fig. 4A and B and Table 2). The inability of APC from donor calf 93 to present antigen to peptide F2-5-specific T-cell clones (Fig. 4B) is consistent with the lack of response to this peptide by T cells from this calf (Fig. 3). Clone 93.4E4 specific for peptides F3-1-F3-2 responded to antigen only in the presence of APC expressing haplotype DRB3∗16-DQA∗11A-DQB∗11C (Fig. 4C).
FIG. 4.
MHC restriction of T-lymphocyte clones specific for epitopes present in the predicted HD of MSP1a. T-lymphocyte clones were stimulated with 25 μg of A. marginale homogenate (A to C) per ml or 10 μg of peptide B (D) per ml in the presence of autologous APC, APC fully matched for both DR-DQ haplotypes, APC matched for one DR-DQ haplotype, or APC mismatched for both haplotypes (animal 2216). APC and corresponding DR alleles are indicated on the y axis. Results are presented for individual clones as the mean counts per minute of duplicate cultures. Error bars, SD. The results are representative of at least two experiments performed with each T-cell clone.
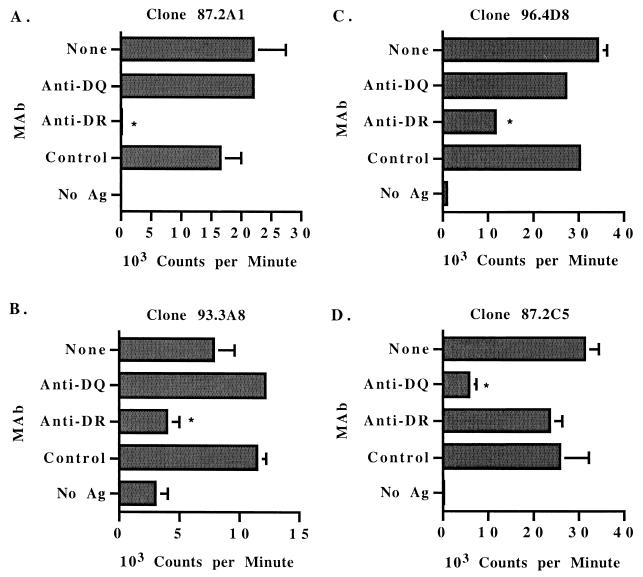
Because DR and DQ genes are closely linked, these results with different APC cannot determine which class II molecule is presenting antigen. Therefore, MAbs specific for DRα or DQα were used to block stimulation by antigen. Clones specific for epitopes in the F2-F3 HD region of MSP1a were stimulated by peptide presented by DR molecules, because MAbs to DRα but not DQα significantly inhibited the response to antigen (Fig. 5A to C). Based on the results with APC half-matched for one haplotype (Fig. 4), the epitope in peptide F2-5 is presented by the DRB3∗22 gene product to T-cell clones 87.2A1 and 96.4G4 (Fig. 5A and B), whereas the epitope in peptides F3-1 and F3-2 is presented by the DRB3∗16 gene product to clone 93.3A8 (Fig. 5C).
FIG. 5.
DR or DQ restriction of MSP1a-specific CD4+ T-lymphocyte clones. T-lymphocyte clones were cultured with autologous APC and medium (no antigen) or either 25 μg of A. marginale homogenate per ml (A), 2 μg of native MSP1 per ml (B and C), or 10 μg of peptide B per ml (D) and either no MAb or 20 μg of MAb specific for DR-α or DQ-α per ml. An IgG2a isotype control MAb was also included. Results are presented as the mean counts per minute of duplicate cultures. Error bars, SD. Asterisks indicate that the response is significantly lower than the response in the presence of isotype control MAb (P < 0.05). Results are representative of at least two experiments for each clone. Ag, antigen.
In contrast to the results with clones specific for epitopes in the F2-F3 HD region of MSP1a, clones from calf 87 specific for the form B repeat responded to peptide B only in the presence of APC expressing both donor haplotypes, DRB3∗16-DQA∗11A-DQB∗11C/DRB3∗22-DQA∗9B-DQB∗9B (Fig. 4D). APC from donors 75 and 77 expressing only one of these DR-DQ haplotypes were ineffective at presenting antigen (Fig. 4D). Similarly, APC from donor calves 93 and 96 were unable to present antigen to peptide B-specific T-cell clone 87.2C5 (data not shown). These results suggested that the response to peptide B was not restricted by DR. Antibody blocking studies confirmed that DQ molecules presented the form B repeat peptide to T-cell clones from calf 87 (Fig. 5D).
The finding that APC derived from cattle 87 and 62 completely matched at DQ can present peptide B, whereas APC from cattle that each express only one set of the DQ alleles found on cattle 87 and 62 APC cannot, strongly suggests that peptide B is presented by DQ molecules formed by intrahaplotype pairing of DQα and DQβ chains. Thus, for calves 87 and 62, the product of the DQα 11A allele could pair with that of the DQβ 9B allele, or the product of the DQα 9B allele could pair with that of the DQβ 11C allele (Table 2). Neither of these potential intrahaplotype pairs is present in cattle 75, 77, 93, and 96, and we were unable to identify additional cattle with the potential to express these DQα-DQβ chain dimers to further test this possibility.
DISCUSSION
MSP1 is a candidate for inclusion in an A. marginale vaccine because of its ability to induce protection in vaccinates, defined as a significant reduction in rickettsemia and anemia upon homologous and heterologous strain challenge (20, 42, 43). Protection against ehrlichial pathogens is associated with both neutralizing and opsonizing IgG antibody and IFN-γ-mediated activation of phagocytes (3, 6, 11, 21, 34, 48, 53, 59). CD4+ T cells, which are required for clearance of ehrlichial pathogens (31), orchestrate both of these effector mechanisms through IFN-γ production. Identification of MSP1 Th-cell epitopes conserved among A. marginale strains that are presented by MHC class II molecules represented broadly in the population is important for designing epitope-based vaccines that incorporate both Th-cell and attachment-invasion-blocking antibody epitopes. Research described in the present study focused on characterizing Th-cell epitopes in the highly conserved HD of the MSP1a protein that elicited the dominant CD4+ T-cell response in MSP1-immunized cattle (17).
MSP1a and MSP1b appear to be erythrocyte adhesins, because binding of recombinant E. coli expressing MSP1a or MSP1b as well as hemagglutination by recombinant E. coli or A. marginale was blocked with immune sera specific for each protein (37, 38). Furthermore, it was recently reported that recombinant E. coli expressing MSP1a binds to tick cells, suggesting that MSP1a facilitates invasion of tick midgut epithelial cells (23, 24). Interestingly, we observed that, even though IgG titers specific for MSP1a and MSP1b were comparable in MSP1-immunized cattle, CD4+ T-cell responses were reproducibly detected only against MSP1a (17). This suggested that MSP1a-specific T cells provide cognate help to both MSP1b-specific B cells and MSP1a-specific B cells, which could occur since MSP1a and MSP1b are associated in the native protein through disulfide bonds (56). The apparent importance of MSP1a-specific Th-cell responses in eliciting IgG antibody production to both of these proteins, and the presence of erythrocyte- and tick cell-binding domains on the MSP1a protein blocked by immune sera, led us to target the large, predicted surface-exposed HD for more detailed analysis of Th-cell recognition.
CD4+ T-cell-enriched oligoclonal lines in culture with whole A. marginale for only 1 to 3 weeks were used as responder cells to obviate a potential selection of cells specific for immunodominant epitopes in longer-term culture. A combination of recombinant proteins F1 to F3 spanning the HD lacking the repeat region (aa 68 to 366) and peptides spanning the most immunostimulatory F2-F3 region (aa 151 to 366) was used to map T-cell epitopes that would elicit recall responses from cattle with different MHC class II haplotypes. A minimum of five T-cell epitopes contained within overlapping peptides F2-1 to F2-4 (aa 151 to 240), F3-2 to F3-3 (aa 270 to 319), and F3-5 (aa 330 to 359) stimulated significant T-lymphocyte recall responses from all three calves, which express, in total, three different DR-DQ haplotypes (Table 2). These DR-DQ haplotypes are common in Holstein and Friesian cattle, such that a predicted 50% of cattle in a given Holstein or Friesian herd would express at least one of the DR-DQ haplotypes evaluated in this study (51; H. A. Lewin, unpublished observations). Thus, inclusion in a recombinant vaccine construct of the sequences represented by peptides that stimulate memory T-cell responses in the three MSP1-immunized cattle described here would predictably stimulate effective MSP1a-specific CD4+ T-cell responses in a large proportion of Holstein-Friesian cattle.
An additional T-cell epitope was defined on the N-terminal form B repeat, present in FL, VA, St. M, and WA-O strains (5, 49). This epitope contained a MAb neutralization-sensitive B-cell epitope defined as either EASTSS (form A repeat) or QASTSS (form B repeat). Whereas the alteration of a Q to an E did not affect antibody neutralization (5), this single-amino-acid substitution completely prevented T-cell recognition (Table 5). Unlike the Th cells specific for epitopes in the HD F2-F3 region which responded strongly to native A. marginale antigen, CD4+ T cells specific for the form B repeat epitope recognized native protein very weakly. Altered peptide ligand antagonism by the form A repeat peptide did not account for the poor response of these T cells to native protein, suggesting that, even though there is no recognition of the form A repeat, its presence in the native protein is not inhibitory. A second potential explanation for the weak response of form B repeat-specific Th-cell clones to MSP1a is that the T-cell epitope is inefficiently processed from the native protein or poorly presented. Alternatively, the form B repeat-specific T-cell clones may represent the “type B” T cells described by Lindner and Unanue that are derived from peptide-immunized mice and recognize peptide but not processed protein antigen (reviewed in reference 35). Type B T cells apparently bind a peptide-MHC complex with a unique conformation, formed when exogenous peptide interacts with class II molecules in an early endosome, that differs conformationally from the peptide-MHC class II complex formed by processing whole polypeptide antigen in deep endosomal compartments (35). Such type B T cells could develop if the MSP1 immunogen was partially degraded so that form B repeat peptides were present.
It is also interesting that the form B repeat epitope could be presented only by autologous APC or APC from a donor expressing the identical set of DR-DQ alleles but not by APC from cattle with one of the two sets of parental DR-DQ alleles. Although not directly tested, this result could be explained by intrahaplotype pairing of DQα and DQβ chains, which has been previously observed for cattle (32).
IFN-γ is required for protective immunity against ehrlichial pathogens. The production of IFN-γ by memory-effector CD4+ T-cell lines from MSP1-immunized cattle stimulated with MSP1a C-terminal region protein (17), as well as by HD F2-F3 region-specific Th-cell clones stimulated with A. marginale, indicates that native MSP1 is naturally processed for T-cell epitope presentation by APC that initiate IFN-γ-producing CD4+ T-cell responses. Therefore, immunization with a construct containing these immunostimulatory T-cell epitopes should similarly prime Th cells to recognize these epitopes processed and presented from whole organisms. The Th-cell epitope-rich region of MSP1a defined here can be incorporated into a vaccine construct designed to link CD4+ Th-cell epitopes broadly recognized by class II at the population level with B-cell epitopes recognized by antibody blocking tick or erythrocyte attachment or invasion, such as the epitope(s) present in the N-terminal repeat region.
Acknowledgments
We are grateful to Emma Karel, Bev Hunter, Shelley Whidbee, and Colleen Olmstead for excellent technical assistance and to Kelly Brayton for critical review of the manuscript.
This research was supported by National Institutes of Health grant R01-AI44005, by U.S.-Israel Binational Agricultural Research and Development Fund grant US-2799-96C, by U.S. Department of Agriculture (USDA) NRICGP grant 99-35204-8274, and by USDA cooperative agreement 58-5348-044.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Ababou, A., W. C. Davis, and D. Levy. 1993. The DA6-147 monoclonal antibody raised against the HLA-DR alpha chain identifies a cryptic epitope on the BoLA-DR alpha chain. Vet. Res. 24:402-407. [PubMed] [Google Scholar]
- 2.Ababou, A., J. Goyeneche, W. C. Davis, and D. Levy. 1994. Evidence for the expression of three different BoLA-class II molecules on the bovine BL-3 cell line: determination of a non-DR non-DQ gene product. J. Leukoc. Biol. 56:182-186. [DOI] [PubMed] [Google Scholar]
- 3.Akkoyunlu, M., and E. Fikrig. 2000. Gamma interferon dominates the murine cytokine response to the agent of human granulocytic ehrlichiosis and helps to control the degree of early rickettsemia. Infect. Immun. 68:1827-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alleman, A. R., G. H. Palmer, T. C. McGuire, T. F. McElwain, L. E. Perryman, and A. F. Barbet. 1997. Anaplasma marginale major surface protein 3 is encoded by a polymorphic, multigene family. Infect. Immun. 65:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 87:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee, R., J. Anguita, and E. Fikrig. 2000. Granulocytic ehrlichiosis in mice deficient in phagocyte oxidase or inducible nitric oxide synthase. Infect. Immun. 68:4361-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbet, A. F., and D. R. Allred. 1991. The msp1β multigene family of Anaplasma marginale: nucleotide sequence analysis of an expressed copy. Infect. Immun. 59:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of the ehrlichia Anaplasma marginale by expression of MSP2 sequence mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbet, A. F., G. H. Palmer, P. J. Myler, and T. C. McGuire. 1987. Characterization of an immunoprotective protein complex of Anaplasma marginale by cloning and expression of the gene coding for polypeptide Am 105L. Infect. Immun. 55:2428-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbet, A. F., J. Yi, A. Lundgren, B. R. McEwen, E. F. Blouin, and K. M. Kocan. 2001. Antigenic variation of Anaplasma marginale: major surface protein 2 diversity during cyclic transmission between ticks and cattle. Infect. Immun. 69:3057-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnewall, R. E., and Y. Rikihisa. 1994. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect. Immun. 62:4804-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowie, M. V., J. de la Fuente, K. M. Kocan, E. Blouin, and A. F. Barbet. 2002. Conservation of major surface protein 1 genes of Anaplasma marginale during cyclic transmission between ticks and cattle. Gene 282:95-102. [DOI] [PubMed] [Google Scholar]
- 13.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 15.Brown, W. C., K. S. Logan, G. G. Wagner, and C. L. Tetzlaff. 1991. Cell-mediated immune responses to Babesia bovis antigens in cattle following infection with tick-derived or cultured parasites. Infect. Immun. 59:2418-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown, W. C., T. C. McGuire, D. Zhu, H. A. Lewin, J. Sosnow, and G. H. Palmer. 2001. Highly conserved regions of the immunodominant major surface protein 2 (MSP2) of the ehrlichial pathogen Anaplasma marginale are rich in naturally derived CD4+ T lymphocyte epitopes that elicit strong recall responses. J. Immunol. 166:1114-1124. [DOI] [PubMed] [Google Scholar]
- 17.Brown, W. C., G. H. Palmer, H. A. Lewin, and T. C. McGuire. 2001. CD4+ T lymphocytes from calves immunized with Anaplasma marginale major surface protein 1 (MSP1), a heteromeric complex of MSP1a and MSP1b, preferentially recognize the MSP1a carboxyl terminus that is conserved among strains. Infect. Immun. 69:6853-6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, W. C., V. Shkap, D. Zhu, T. C. McGuire, W. Tuo, T. F. McElwain, and G. H. Palmer. 1998. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown, W. C., D. Zhu, V. Shkap, T. C. McGuire, E. F. Blouin, K. M. Kocan, and G. H. Palmer. 1998. The repertoire of Anaplasma marginale antigens recognized by CD4+ T-lymphocyte clones from protectively immunized cattle is diverse and includes major surface protein 2 (MSP-2) and MSP-3. Infect. Immun. 66:5414-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camacho-Nuez, M., M. de Lourdes Muñoz, C. E. Suarez, T. C. McGuire, W. C. Brown, W. C., and G. H. Palmer. 2000. Expression of polymorphic msp1β genes during acute Anaplasma marginale rickettsemia. Infect. Immun. 68:1946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantor, G. H., C. H. Pontzer, and G. H. Palmer. 1993. Opsonization of Anaplasma marginale mediated by bovine antibody against surface protein MSP-1. Vet. Immunol. Immunopathol. 37:343-350. [DOI] [PubMed] [Google Scholar]
- 22.Davies, C. J., I. Joosten, D. Bernoco, M. A. Arriens, J. Bester, G. Ceriotti, S. Ellis, E. J. Hensen, H. C. Hines, P. Horin, B. Kristensen, H. A. Lewin, D. Meggiolaro, A. L. G. Morgan, M. Morita, P. H. R. Nilsson, R. A. Oliver, A. Orlova, H. Østergard, C. A. Park., H.-J. Schuberth, M. Simon, R. L. Spooner, and J. A. Stewart. 1994. Polymorphism of bovine MHC class I genes. Joint report of the Fifth International Bovine Lymphocyte Antigen (BoLA) Workshop, Interlaken, Switzerland, 1 August 1992. Eur. J. Immunogenet. 21:259-289. [DOI] [PubMed] [Google Scholar]
- 23.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2001. Differential adhesion of major surface proteins 1a and 1b of the ehrlichial cattle pathogen Anaplasma marginale to bovine erythrocytes and tick cells. Int. J. Parasitol. 31:145-153. [DOI] [PubMed] [Google Scholar]
- 24.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, B. R. McEwen, D. Clawson, and K. M. Kocan. 2001. Major surface protein 1a effects tick transmission of Anaplasma marginale. Int. J. Parasitol. 31:1705-1714. [DOI] [PubMed] [Google Scholar]
- 25.De Magistris, M. T., J. Alexander, M. Coggeshall, A. Altman, F. C. A. Gaeta, H. M. Grey, and A. Sette. 1992. Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell 68:625-634. [DOI] [PubMed] [Google Scholar]
- 26.Dutia, B. M., L. MacCarthy-Morrough, E. J. Glass, R. L. Spooner, and J. Hopkins. 1995. Discrimination between major histocompatibility complex class II DQ and DR locus products in cattle. Anim. Genet. 26:111-114. [DOI] [PubMed] [Google Scholar]
- 27.Edwards, C. P., and A. Aruffo. 1993. Current applications of COS cell based transient expression systems. Curr. Opin. Biotechnol. 4:558-563. [DOI] [PubMed] [Google Scholar]
- 28.Eid, G., D. M. French, A. M. Lundgren, A. F. Barbet, T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1996. Expression of major surface protein 2 variants during acute Anaplasma marginale rickettsemia. Infect. Immun. 64:836-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.French, D., W. C. Brown, and G. H. Palmer. 1999. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect. Immun. 67:5834-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.French, D. M., T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1998. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect. Immun. 66:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganta, R. R., M. J. Wilkerson, C. Cheng, A. M. Rokey, and S. K. Chapes. 2002. Persistent Ehrlichia chaffeensis infection occurs in the absence of functional major histocompatibility complex class II genes. Infect. Immun. 70:380-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glass, E. J., R. A. Oliver, and G. C. Russell. 2000. Duplicated DQ haplotypes increase the complexity of restriction element usage in cattle. J. Immunol. 165:134-138. [DOI] [PubMed] [Google Scholar]
- 33.Lewin, H. A., G. C. Russell, and E. J. Glass. 1999. Comparative organization and function of the major histocompatibility complex of domesticated cattle. Immunol. Rev. 167:145-158. [DOI] [PubMed] [Google Scholar]
- 34.Li, J. S.-Y., E. Yager, M. Reilly, C. Freeman, G. R. Reddy, A. A. Reilly, F. K. Chu, and G. A. Winslow. 2001. Outer membrane protein-specific monoclonal antibodies protect SCID mice from fatal infection by the obligate intracellular bacterial pathogen Ehrlichia chaffeensis. J. Immunol. 166:1855-1862. [DOI] [PubMed] [Google Scholar]
- 35.Lindner, R., and E. R. Unanue. 1996. Distinct antigen MHC class II complexes generated by separate processing pathways. EMBO J. 15:6910-6920. [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, M. E., K. Caspersen, and J. S. Dumler. 2001. Immunopathology and ehrlichial propagation are regulated by IFN-γ and interleukin-10 in a murine model of human granulocytic ehrlichiosis. Am. J. Pathol. 158:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGarey, D. J., and D. R. Allred. 1994. Characterization of hemagglutinating components of the Anaplasma marginale initial body surface and identification of possible adhesins. Infect. Immun. 62:4587-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGarey, D. J., A. F. Barbet, G. H. Palmer, T. C. McGuire, and D. R. Allred. 1994. Putative adhesins of Anaplasma marginale: major surface polypeptides 1a and 1b. Infect. Immun. 62:4594-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuire, T. C., G. H. Palmer, W. L. Goff, M. I. Johnson, and W. C. Davis. 1984. Common and isolate restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect. Immun. 45:697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuire, T. C., E. B. Stephens, G. H. Palmer, T. F. McElwain, C. A. Lichtensteiger, S. R. Leib, and A. F. Barbet. 1994. Recombinant vaccinia virus expression of Anaplasma marginale surface protein MSP-1a: effect of promoters, leader sequences and GPI anchor sequence on enhancement of antibody response. Vaccine 12:465-471. [DOI] [PubMed] [Google Scholar]
- 40a.Mwangi, W., W. C. Brown, H. A. Lewin, C. Howard, T. V. Baszler, P. Caplazi, J. A. Abbott, and G. H. Palmer. Flt3 ligand and GM-CSF increase dendritic cell recruitment to the inoculation site and enhance antigen-specific CD4+ T cell responses induced by DNA vaccination of outbred animals. J. Immunol., in press. [DOI] [PubMed]
- 41.Oberle, S. M., G. H. Palmer, A. F. Barbet, and T. C. McGuire. 1988. Molecular size variation in an immunoprotective protein complex among isolates of Anaplasma marginale. Infect. Immun. 56:1567-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer, G. H., A. F. Barbet, G. H. Cantor, and T. C. McGuire. 1989. Immunization of cattle with a 105-kDa surface protein complex induces protection against a structurally variant Anaplasma marginale isolate. Infect. Immun. 57:3666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299-1302. [DOI] [PubMed] [Google Scholar]
- 44.Palmer, G. H., A. F. Barbet, K. L. Kuttler, and T. C. McGuire. 1986. Detection of an Anaplasma marginale common surface protein present in all stages of infection. J. Clin. Microbiol. 23:1078-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer, G. H., W. C. Brown, and F. R. Rurangirwa. 2000. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Palmer, G. H., G. Eid, A. F. Barbet, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62:3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer, G. H., and T. C. McGuire. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 133:1010-1015. [PubMed] [Google Scholar]
- 48.Palmer, G. H., F. R. Rurangirwa, K. M. Kocan, and W. C. Brown. 1999. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol. Today 15:281-286. [DOI] [PubMed] [Google Scholar]
- 49.Palmer, G. H., F. R. Rurangirwa, and T. F. McElwain. 2001. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J. Clin. Microbiol. 39:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rurangirwa, F. R., D. Stiller, and G. H. Palmer. 2000. Strain diversity in major surface protein 2 expression during tick transmission of Anaplasma marginale. Infect. Immun. 68:3023-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharif, S., B. A. Mallard, B. N. Wilkie, J. M. Sargeant, H. M. Scott, J. C. M. Denkers, and K. E. Leslie. 1998. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim. Genet. 29:185-193. [DOI] [PubMed] [Google Scholar]
- 52.Storey, J. R., L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, T. N. Mather, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 1998. Molecular cloning and sequencing of three granulocytic Ehrlichia genes encoding high-molecular-weight immunoreactive proteins. Infect. Immun. 66:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun, W., J. W. Ijdo, S. R. Telford III, E. Hodzic, Y. Zhang, S. W. Barthold, and E. Fikrig. 1997. Immunization against the agent of human granulocytic ehrlichiosis in a murine model. J. Clin. Investig. 100:3014-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tebele, N., T. C. McGuire, and G. H. Palmer. 1991. Induction of protective immunity using Anaplasma marginale initial body membranes. Infect. Immun. 59:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Eijk, M. J. T., J. A. Stewart-Haynes, and H. A. Lewin. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23:483-496. [DOI] [PubMed] [Google Scholar]
- 56.Vidotto, M., T. C. McGuire, T. F. McElwain, G. H. Palmer, and D. P. Knowles. 1994. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect. Immun. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viseshakul, N., S. Kamper, M. V. Bowie, and A. F. Barbet. 2000. Sequence and expression analysis of a surface antigen gene family of the rickettsia Anaplasma marginale. Gene 253:45-53. [DOI] [PubMed] [Google Scholar]
- 58.Visser, E. S., T. C. McGuire, G. H. Palmer, W. C. Davis, V. Shkap, E. Pipano, and D. P. Knowles. 1992. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect. Immun. 60:5139-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winslow, G. A., E. Yager, K. Shilo, E. Volk, A. Reilly, and F. K. Chu. 2000. Antibody-mediated elimination of the obligate intracellular bacterial pathogen Ehrlichia chaffeensis during active infection. Infect. Immun. 68:2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, X.-J., P. Croquet-Valdes, and D. H. Walker. 1997. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene 184:149-154. [DOI] [PubMed] [Google Scholar]
- 61.Yu, X.-J., J. W. McBride, M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]