Abstract
Abscisic acid (ABA) is a hormone that modulates a variety of agronomically important growth and developmental processes and various stresses responses, but its signal transduction pathways remain poorly understood. ROP10, a member of ROP small GTPases in Arabidopsis (Arabidopsis thaliana), is a plasma membrane-associated protein specifically involved in negative regulation of ABA responses. To dissect the ROP10-mediated ABA signaling, we carried out transcriptome analysis using the Arabidopsis full-genome chip. Our analysis revealed a total of 262 and 125 genes that were, respectively, up- and down-regulated (≥2-fold cutoff) by 1 μm ABA in wild type (Wassilewskija [Ws]); 42 up-regulated and 38 down-regulated genes have not been identified in other studies. Consistent with the nonpleiotropic phenotypes of rop10-1, only three genes were altered in rop10-1 in the absence of ABA treatment. In response to 1 μm ABA, 341 and 127 genes were, respectively, activated and repressed in rop10-1. Interestingly, a particular subset of 21 genes that were not altered by 1 μm ABA in Ws but only activated in rop10-1 was identified. Reverse transcription-polymerase chain reaction analysis revealed the existence of three distinct categories of ABA dose-response patterns. One novel category is characterized by their ABA unresponsiveness in Ws and activation in rop10-1 at 1 μm but not 10 and 100 μm of ABA. This indicates that ROP10 gates the expression of genes that are specific to low concentrations of ABA. Furthermore, almost all of these 21 genes are known to be highly induced by various biotic and abiotic stresses. Consequently, we found that rop10-1 enhanced the sensitivity of seed germination inhibition to mannitol and sodium chloride. Our results suggest that ROP10 negatively regulates ABA responses by specifically and differentially modulating the ABA sensitivity of a subset of genes including protein kinases and zinc-finger family proteins.
Abscisic acid (ABA) is a hormone that modulates a variety of agronomically important growth and developmental processes (such as the synthesis of seed storage proteins and lipids, and the control of seed maturation, dormancy, and germination) and responses to various stresses, including drought, salt, and cold (for a recent comprehensive review, see Finkelstein et al., 2002). Despite significant progress in the understanding of ABA biosynthesis (Xiong and Zhu, 2003), ABA perception and signal transduction pathways remain poorly defined (Finkelstein et al., 2002; Himmelbach et al., 2003). Since it was compiled three years ago that nearly 50 genes in Arabidopsis (Arabidopsis thaliana) alone can directly or indirectly affect ABA responses (Finkelstein et al., 2002), novel candidate ABA signaling genes have been identified through either forward or reverse genetic approaches. Together, these include genes encoding intracellular signaling proteins, such as heterotrimeric and monomeric GTP-binding proteins; G-protein-coupled receptors; (receptor-like) protein kinases and protein phosphatases; transcription factors; proteins involved in RNA metabolism, protein modification/degradation, and lipid metabolism; and proteins of unknown biochemical functions (for review, see Finkelstein et al., 2002; Himmelbach et al., 2003; for some of the most recent representative reports, see Abe et al., 2003; Coursol et al., 2003; Kim et al., 2003, 2004; Lopez-Molina et al., 2003; Smalle et al., 2003; Brocard-Gifford et al., 2004; Fujita et al., 2004; He and Gan, 2004; Lee et al., 2004; Leonhardt et al., 2004; Pandey and Assmann, 2004; Pandey et al., 2004; Zhang et al., 2004; Osakabe et al., 2005). However, how these genes are connected in ABA signaling remains unclear because of the lack of complete epistatic relationships for most of the ABA signaling mutants. The difficulty could be accounted for by the complex cross-talks with other pathways, the functional redundancy of some ABA signaling components possibly acting in a linear pathway, the existence of a signaling network composed of interacting modules for ABA signaling instead of a linear signaling pathway, and/or that signaling components sufficiently involved in a specific linear pathway remain to be identified.
Among the signaling proteins known to regulate ABA responses, the plasma membrane (PM)-associated receptor-like kinases, heterotrimeric G-proteins and G-protein-coupled receptors, and monomeric or small GTP-binding proteins (Lemichez et al., 2001; Li et al., 2001; Wang et al., 2001; Zheng et al., 2002; Pandey and Assmann, 2004; Osakabe et al., 2005) are particularly interesting. These proteins might interact with an unknown PM-localized ABA receptor(s), and some of the receptor proteins (such as RPK1) could be the ABA receptors. By cycling between GDP- and GTP-bound forms, ROP GTPases seem to relay the signal from the PM to the cytoplasm (Zheng and Yang, 2000; Yang, 2002). Three ROPs have been implicated in ABA responses (Lemichez et al., 2001; Li et al., 2001; Zheng et al., 2002). Transgenic plants expressing constitutively active ROP2 (Li et al., 2001) or AtRac1/ROP6 (Lemichez et al., 2001) reduced ABA sensitivity in seed dormancy/germination and stomatal closure, respectively. This is consistent with the enhanced effect of dominant negative mutations of ROP2 and AtRac1/ROP6. However, as demonstrated in the case of Drosophila Rac small GTPases (Hakeda-Suzuki et al., 2002), such dominant mutations might interfere with the signaling pathways unrelated to ROP GTPases. Therefore, the functions of ROP2 and AtRac1/ROP6 in ABA remain to be demonstrated by the loss-of-function mutations.
ROP10 is the first ROP member whose function has been demonstrated by the loss-of-function mutation. The T-DNA knockout mutant rop10-1 has been shown through genetic and physiological studies to specifically and negatively regulate a variety of ABA responses (Zheng et al., 2002). These include the control of seed dormancy, inhibition of seed germination, suppression of root growth, and promotion of stomatal closure and reduction of water loss in detached leaves (Zheng et al., 2002). Importantly, the negative regulation of ABA signaling by ROP10 is dependent upon its PM localization. ABA perception is believed to occur in the cytoplasm and the PM (Finkelstein et al., 2002), and very recently the receptor-like kinase RPK1 has been shown to regulate ABA signaling in Arabidopsis (Osakabe et al., 2005). Together with an earlier report that one or more members of ROP GTPases are probably associated with the CLAVATA1 (CLV1) receptor complex (Trotochaud et al., 1999), these observations raise an interesting possibility that ROP10 GTPase might situate in close proximity to an unknown ABA receptor. However, signaling components that link ROP10 from the PM to the cytoplasm and to the nucleus remain to be identified.
How does ROP10 regulate ABA signaling in a complex network? It has been proposed that ROP10 might act either at an early step or in a common pathway of ABA signaling (Zheng et al., 2002). This is mostly supported by the observations that multiple aspects of ABA responses are affected in rop10-1 yet the rop10-1 mutant has an ABA-specific phenotype, the ROP10 promoter activity in the root tips is specifically suppressed by ABA, and transgenic plants expressing constitutively active or dominant negative mutants of rop10 lack any pleiotropic phenotypes (Zheng et al., 2002). However, the constitutively active rop10 transgenic plants only partially reduced ABA sensitivity, and dominant negative rop10 partially suppressed the strong ABA insensitivity phenotypes of abi2-1 (Zheng et al., 2002). Although we could not rule out the possibility of the functional redundancy of ROP10 with other closely related ROPs, this could indicate that ROP10 more likely acts at an early step of an ABA signaling branch that modulates the ABA sensitivity, instead of participating in a common pathway leading to various ABA signaling branches.
To test this hypothesis and help identify the signaling components that link ROP10 to gene expression, we decided to investigate the global transcriptional profile of ABA responses in rop10-1. DNA microarray has been used to profile ABA- or drought- and cold-regulated gene expression in several reports (Seki et al., 2001, 2002a, 2002b; Chen et al., 2002; Fowler and Thomashow, 2002; Hoth et al., 2002; Kreps et al., 2002; Abe et al., 2003; Suzuki et al., 2003; Bray, 2004; Kawaguchi et al., 2004; Leonhardt et al., 2004; Takahashi et al., 2004; Sanchez et al., 2004; Osakabe et al., 2005). However, these studies were mostly performed using a partial genome array, and more importantly very high concentrations (10–100 μm) of ABA were used in their corresponding studies. We chose a low ABA concentration, 1 μm, to identify genes that are modulated by ROP10 and very sensitive to ABA. Analysis of our full-genome gene chip data and comparative analysis using the large datasets available to the public have led us to identify novel genes in ABA responses that have not been revealed in other studies. More importantly, our results support that ROP10 is a specific negative regulator of ABA signaling, likely by differentially modulating the expression of a particular subset of genes in response to different levels of ABA. These ROP10-regulated genes are also highly induced by various abiotic and biotic stresses, and we show that ROP10 negatively regulates osmotic and salt stress response. Several of these ROP10-regulated genes encode regulatory proteins, such as GCN5-related N-acetyltransferase (GNAT) N-acetyltransferase, protein kinases, and zinc-finger proteins. Most interestingly, they are only activated in rop10-1 by 1 μm but not higher concentrations of ABA. Our results suggest the existence of a low concentration of ABA-specific pathway that is gated by the ROP10 small GTPase.
RESULTS
System Validation and Data Assessment
To identify novel genes that are regulated by ABA and modulated by ROP10, we profile the transcripts from 7-d-old whole seedlings treated by the low concentration of ABA (1 μm) using the ATH1 Arabidopsis GeneChip. This chip contains more than 22,500 probe sets representing approximately 24,000 unique genes. An earlier study showed that 4-h treatments of 1 and 50 μm ABA activated MYB2 transcription more dramatically in rop10-1 seedlings than in its wild-type ecotype Wassilewskija (Ws; Zheng et al., 2002). We therefore chose 1 μm ABA, which already enhanced ABA responses in rop10-1 (Zheng et al., 2002) and is either closer to its physiological range or an indicator of the mild stresses, to treat both Ws and rop10-1 for 4 h in this DNA microarray study. Three biological replicates were designed to reduce the experimental variations.
To determine genes that are significantly regulated by ABA and/or affected by the ROP10 mutation, pair-wise comparisons of the linearly scaled/normalized data of Ws and rop10-1 that were treated without and with ABA were performed using a modified t test (also called the S-test), which is the basis of the software Significant Analysis of Microarrays (SAM; Tusher et al., 2001). SAM was run on the log2 normalized data with a 2-fold cutoff and at a median false discovery rate of ≤6%. These analyses resulted in sets of significantly increased or decreased probe sets/genes for each pair-wise comparison. These sets of genes were then filtered by the Affymetrix GCOS Absence (“A”) call, which means the level is below the detection limit, resulting in sets with only those genes that could be detected in all of the 12 chips and whose expression differed by at least 2-fold.
We then examined the ROP10 and MYB2 expression levels in the chip for the indication of the chip data reliability and resolution. In all of three replicates, ROP10/Arac8 was called “A” by the Affymetrix GCOS software in rop10-1, regardless of ABA treatment, but was called Present (“P”) in Ws. The fact that ROP10 gene expression is below the detection limit in rop10-1 but above in Ws is consistent with the likely null mutation of ROP10. Furthermore, ROP10 transcripts were nearly identical between ABA-treated and untreated Ws, consistent with the earlier report that ROP10 is not transcriptionally regulated by ABA at the whole-seedling level (Zheng et al., 2002). However, MYB2 expression was called “A” in all of 12 samples, although MYB2 expression was determined by reverse transcription (RT)-PCR analysis as significantly higher in rop10-1 than Ws (Zheng et al., 2002). This indicates that genes with low abundance could not be readily detected by the chip. In our chip analysis, there were a total of 14,991 probe sets determined as “P” or “M” (marginal), suggesting that about 70% of genes could be analyzed in our experimental conditions.
Significantly Induced and Suppressed Genes by ABA in Wild Type
We first examined the ABA up-regulated genes in Ws. A total of 248 probe sets likely representing 262 unique genes (because some probe sets represent two or more highly homologous genes) were determined significantly increased (2-fold cutoff) by 1 μm ABA. Functional categorical analysis of this set of genes reveals that the largest group of genes is related to metabolism, followed by cellular transport, cell rescue, defense and virulence, transcription, signaling, and biogenesis of cellular components (Table I). These genes are listed in Supplemental Table I.
Table I.
Functional categories of ABA-activated and -repressed genes in Ws
The numbers of genes (and their respective percentages) that belong to different functional categories are listed.
| Functional Category | Activated Genes | Repressed Genes |
|---|---|---|
| Metabolism | 48 (18.6%) | 14 (11.3%) |
| Amino acid metabolism | 8 (3.1%) | 1 (0.8%) |
| Nitrogen and sulfur metabolism | 3 (1.2%) | |
| C-compound and carbohydrate metabolism | 21 (8.2%) | 7 (5.7%) |
| Lipid, fatty acid, and isoprenoid metabolism | 12 (4.7%) | 1 (0.8%) |
| Secondary metabolism | 9 (3.5%) | 4 (3.3%) |
| Energy | 7 (2.7%) | 1 (0.8%) |
| Storage protein | 3 (1.2%) | |
| Transcription | 9 (3.5%) | 6 (4.9%) |
| Protein fate (folding, modification, destination) | 6 (2.3%) | 1 (0.8%) |
| Protein degradation | 4 (1.6%) | 1 (0.8%) |
| Protein with binding function | 1 (0.4%) | 2 (1.6%) |
| Cellular transport | 14 (5.4%) | 2 (1.6%) |
| Ion transport | 5 (1.9%) | 1 (0.8%) |
| Amino acid transport | 2 (0.8%) | |
| ABC transporters | 3 (1.2%) | |
| Signal transduction mechanism | 8 (3.1%) | 3 (2.4%) |
| Cell rescue, defense, and virulence | 12 (4.7%) | 7 (5.7%) |
| Stress response | 5 (1.9%) | 3 (2.4%) |
| Interaction with the cellular environment | 2 (0.8%) | |
| Cellular sensing and response | 2 (0.8%) | |
| Interaction with the environment (systemic) | 2 (0.8%) | 1 (0.8%) |
| Development (systemic) | 3 (1.2%) | |
| Biogenesis of cellular components | 8 (3.1%) | 2 (1.6%) |
| Cell wall | 7 (2.7%) | 2 (1.6%) |
| Subcellular localization | 8 (3.1%) | 4 (3.3%) |
| Cell wall | 4 (1.6%) | 2 (1.6%) |
| Nucleus | 3 (1.2%) | |
| Classification not yet clear-cut | 21 (8.2%) | 20 (16.2%) |
| Unclassified proteins | 139 (54.0%) | 77 (62.6%) |
| Genes that match MAtDB (with 26,642 annotated genes) | 257 (out of 262) | 123 (out of 125) |
Compared to the ABA up-regulated genes, a smaller number of genes were down-regulated by ABA in Ws. By the 2-fold cutoff, a total of 118 probe sets likely representing 125 unique genes were determined significantly decreased by 1 μm ABA (Supplemental Table II). Functional categorical analysis of this set of genes shows that similar groups of genes were down-regulated as for the up-regulated genes, with the largest group of genes being associated with metabolism, followed by cellular transport, cell rescue, defense and virulence, transcription, and signaling, with more than 60% being unclassified at present (Table I).
To help determine whether the genes that were identified in this study were specifically involved in response to low concentrations of ABA, these genes were compared with those data analyzed on the GENEVESTIGATOR Web site (https://www.genevestigator.ethz.ch/), which collects publicly available chip data from at least 750 ATH1 and 121 AG arrays and also some unpublished work (Zimmermann et al., 2004). ABA responses in these datasets include seed imbibitions and seedlings treated with high ABA concentrations. Because the GENEVESTIGATOR tool takes into account all the experiments, some genes that are altered in only one or two experiments might be considered as no change or a change smaller than 2-fold. Therefore, we further filtered our data with the three published datasets that cover the full genome (Hoth et al., 2002; Suzuki et al., 2003; Sanchez et al., 2004). In the first study (Hoth et al., 2002), 660 ABA up-regulated and 694 down-regulated genes were identified in 4-week-old seedlings of the wild-type Landsberg erecta treated with 50 μm ABA for 3 to 6 h by Massively Parallel Signature Sequencing analysis (≥3-fold cutoff). In the second study, 1,490 up-regulated and 1,470 down-regulated genes were identified by the ATH1 GeneChip analysis with a 2-fold cutoff in 50 μm ABA-treated 2- to 3-week-old Landsberg erecta seedlings for 2 or 6 h (Sanchez et al., 2004). In the third report, a lower concentration (5 μm) of ABA was applied to both abi3 and 35S:VP1/abi3 for 12 h, and a total of 158 and 87 genes were, respectively, activated and repressed by ABA that acts alone or together with VP1 (Suzuki et al., 2003). In a recent DNA microarray study (Osakabe et al., 2005), a different full-genome oligoarray was used to profile gene expression changes in the RPK1 knockout and antisense plants, but the ABA regulation data in wild type is not available. Therefore, this dataset was not used in this comparative analysis.
As a result, our analysis revealed a total of 42 ABA up-regulated (Table II) and 38 down-regulated (Table III) genes that do not seem to be regulated by high concentrations of ABA. It is possible that this set of genes affected by 1 μm ABA in our study is functional in particular in responding to low concentrations of ABA or mild stresses. While about 60% of these genes belong to the unclassified functional category, about 25% of them are involved in metabolism in both up- or down-regulated genes. Compared to 11% or 18% of metabolism genes among all of ABA-activated or -repressed genes in Ws, this is a slight overrepresentation of metabolism genes uniquely identified in our study that used a low concentration of ABA.
Table II.
ABA-activated genes in wild type (Ws) identified in this study but not in other studies
The fold changes listed are the averages of three biological replicates. For comparison, the fold changes for rop10-1 are also listed and most of the ABA-activated genes show similar induction.
| Probe Set | Locus | Ws | rop10-1 | Annotation |
|---|---|---|---|---|
| Metabolism | ||||
| 262780_at | AT1G13090 | 2.2 | 2.2 | Cytochrome P450 71B28 |
| Amino acid metabolism | ||||
| 258322_at | AT3G22740 | 2.4 | 1.9 | Homocysteine S-methyltransferase 3 |
| Carbohydrate metabolism | ||||
| 264553_s_at | AT1G09480 | 2.1 | 2.1 | Cinnamyl-alcohol dehydrogenase family |
| 264553_s_at | AT1G09490 | 2.1 | 2.1 | Cinnamyl-alcohol dehydrogenase family |
| 264078_at | AT2G28470 | 2.5 | 2.7 | Putative β-galactosidase |
| Lipid metabolism | ||||
| 256860_at | AT3G23840 | 2.1 | 1.8 | Transferase family protein |
| 245612_at | AT4G14440 | 2.1 | 1.8 | Enoyl-CoA hydratase/isomerase family protein |
| Secondary metabolism | ||||
| 252123_at | AT3G51240 | 2.3 | 1.7 | Naringenin 3-dioxygenase |
| 252943_at | AT4G39330 | 2.0 | 1.9 | Mannitol dehydrogenase |
| Cellular transport | ||||
| 259081_at | AT3G05030 | 2.2 | 2.3 | Sodium proton exchanger |
| 255674_at | AT4G00430 | 2.3 | 2.5 | PM intrinsic protein |
| 245399_at | AT4G17340 | 2.8 | 2.4 | Major intrinsic family protein |
| 250261_at | AT5G13400 | 2.5 | 2.0 | Proton-dependent oligopeptide transport family |
| Cellular communication | ||||
| 250428_at | AT5G10480 | 2.1 | 2.0 | Protein tyrosine phosphatase-like protein |
| Plant hormonal regulation | ||||
| 259596_at | AT1G28130 | 2.0 | 2.4 | Auxin-responsive GH3 family protein |
| Biogenesis/localization of cell wall | ||||
| 267590_at | AT2G39700 | 2.0 | 2.2 | α-Expansin gene family |
| Classification not clear-cut | ||||
| 253048_at | AT4G37560 | 2.0 | 1.9 | Putative formamidase |
| Unclassified proteins | ||||
| 259430_at | AT1G01610 | 2.0 | 2.0 | Phospholipid/glycerol acyltransferase family protein |
| 255786_at | AT1G19670 | 2.2 | 1.1 | Coronatine-induced protein 1 |
| 260943_at | AT1G45145 | 2.1 | 2.0 | Thioredoxin H-type 5 (TRX-H-5) |
| 245629_at | AT1G56580 | 2.3 | 1.8 | Expressed protein |
| 260234_at | AT1G74460 | 3.0 | 3.0 | GDSL-motif lipase/hydrolase family protein |
| 265511_at | AT2G05540 | 2.4 | 2.1 | Gly-rich protein |
| 263318_at | AT2G24762 | 2.1 | 1.7 | Expressed protein |
| 266418_at | AT2G38750 | 2.0 | 1.6 | Annexin 4 |
| 266989_at | AT2G39330 | 3.7 | 1.5 | Jacalin lectin family protein |
| 260547_at | AT2G43550 | 2.1 | 1.4 | Putative trypsin inhibitor |
| 262349_at | AT2G48130 | 3.7 | 3.9 | Protease inhibitor/seed storage/LTP family protein |
| 258751_at | AT3G05890 | 4.9 | 3.9 | Hydrophobic protein (RCI2B) |
| 259040_at | AT3G09270 | 2.2 | 2.1 | Putative glutathione S-transferase |
| 256994_s_at | AT3G25820 | 2.5 | 2.4 | Putative myrcene/ocimene synthase |
| 256994_s_at | AT3G25830 | 2.5 | 2.4 | Putative myrcene/ocimene synthase |
| 255243_at | AT4G05590 | 2.0 | 1.9 | Expressed protein |
| 254697_at | AT4G17970 | 2.1 | 2.1 | Expressed protein |
| 254283_s_at | AT4G22870 | 5.0 | 1.9 | Putative leucoanthocyanidin dioxygenase |
| 250794_at | AT5G05270 | 2.1 | 1.3 | Chalcone-flavanone isomerase family protein |
| 249545_at | AT5G38030 | 2.4 | 2.7 | MATE efflux family protein |
| 248790_at | AT5G47450 | 2.2 | 2.3 | Major intrinsic family protein |
| 248466_at | AT5G50720 | 2.1 | 1.8 | ABA-responsive protein (HVA22e) |
| 247965_at | AT5G56540 | 2.4 | 2.3 | Arabinogalactan protein (AGP14) |
| 247882_at | AT5G57785 | 3.1 | 2.4 | Expressed protein |
| 247884_at | AT5G57800 | 2.3 | 2.2 | CER1 protein, putative (WAX2) |
Table III.
ABA-repressed genes in wild type (Ws) identified in this study but not in other studies
The fold changes listed are the averages of three biological replicates. For comparison, the fold changes for rop10-1 are also listed and most of the ABA-repressed genes show similar suppression.
| Probe Set | Locus | Ws | rop10-1 | Annotation |
|---|---|---|---|---|
| Metabolism | ||||
| 262793_at | AT1G13110 | −2.3 | −1.9 | Cytochrome P450 71B7 |
| Amino acid metabolism | ||||
| 258941_at | AT3G09940 | −2.3 | −2.3 | Putative monodehydroascorbate reductase |
| Carbohydrate metabolism | ||||
| 260914_at | AT1G02640 | −2.9 | −2.4 | Glycosyl hydrolase family 3 protein |
| 263183_at | AT1G05570 | −2.2 | −2.2 | Callose synthase 1 (CALS1) |
| 249477_s_at | AT5G38930 | −2.6 | −3.1 | Putative germin-like protein |
| 249477_s_at | AT5G38940 | −2.6 | −3.1 | Putative germin-like protein |
| Secondary metabolism | ||||
| 251124_s_at | AT5G01040 | −2.4 | −2.3 | Laccase family protein |
| 251124_s_at | AT5G01050 | −2.4 | −2.3 | Laccase family protein |
| 251144_at | AT5G01210 | −2.1 | −1.2 | Transferase family protein |
| Transcription | ||||
| 252081_at | AT3G51910 | −2.3 | −2.1 | Heat shock transcription factor family protein |
| Metal binding | ||||
| 257197_at | AT3G23800 | −2.6 | −2.7 | Selenium-binding family protein |
| Classification not clear-cut | ||||
| 266693_at | AT2G19800 | −2.1 | −2.2 | Expressed protein |
| 254785_at | AT4G12730 | −2.3 | −2.1 | Fasciclin-like arabinogalactan-protein (FLA2) |
| 245392_at | AT4G15680 | −2.6 | −3.5 | Glutaredoxin family protein |
| 246149_at | AT5G19890 | −6.7 | −6.6 | Putative peroxidase |
| Unclassified proteins | ||||
| 263182_at | AT1G05575 | −2.5 | −1.9 | Expressed protein |
| 262603_at | AT1G15380 | −2.1 | −1.7 | Lactoylglutathione lyase family protein |
| 260668_at | AT1G19530 | −3.3 | −3.0 | Expressed protein |
| 261937_at | AT1G22570 | −2.0 | −2.7 | Proton-dependent oligopeptide transport (POT) family protein |
| 256525_at | AT1G66180 | −2.2 | −1.5 | Aspartyl protease family protein |
| 260425_at | AT1G72440 | −2.1 | −1.5 | CCAAT-box-binding transcription factor-related |
| 263126_at | AT1G78460 | −2.1 | −1.9 | SOUL heme-binding family protein |
| 260297_at | AT1G80280 | −2.6 | −2.2 | Hydrolase, α/β fold family protein |
| 267523_at | AT2G30600 | −2.0 | −1.9 | BTB/POZ domain-containing protein |
| 260496_at | AT2G41700 | −2.0 | −2.0 | ABC transporter family protein |
| 266711_at | AT2G46740 | −2.2 | −2.3 | FAD-binding domain-containing protein |
| 259000_at | AT3G01860 | −2.0 | −1.9 | Expressed protein |
| 258965_at | AT3G10530 | −2.0 | −1.7 | WD-40 repeat family protein |
| 256661_at | AT3G11964 | −2.1 | −1.5 | S1 RNA-binding domain-containing protein |
| 256788_at | AT3G13730 | −2.2 | −2.0 | Cytochrome P450 |
| 258338_at | AT3G16150 | −4.0 | −4.7 | Putative l-asparaginase |
| 251538_at | AT3G58660 | −2.0 | −1.4 | 60S ribosomal protein-related |
| 255652_at | AT4G00950 | −2.4 | −2.0 | Expressed protein |
| 254043_at | AT4G25990 | −3.1 | −2.9 | Expressed protein |
| 246562_at | AT5G15580 | −2.6 | −1.9 | Expressed protein |
| 246919_at | AT5G25460 | −2.0 | −1.9 | Expressed protein |
| 249037_at | AT5G44130 | −2.4 | −2.7 | Fasciclin-like arabinogalactan protein |
The ROP10 Mutation Does Not Greatly Alter Gene Expression without ABA Treatment
We did not observe any visible phenotype in rop10-1 in the absence of exogenous ABA (Zheng et al., 2002). We therefore were interested whether the ROP10 mutation alters gene expression in the absence of ABA treatment. Consistent with the phenotype observed, only three genes were altered in rop10-1 compared to Ws by a 2-fold cutoff. Two of them were higher in rop10-1: the expansin-related protein 3 precursor (EXPR3/At2g18660, 2.2-fold of that in Ws) and an expressed protein (At5g02150, 2.3-fold of that in Ws) with an unknown function. While At5g02150 was unresponsive to ABA in both Ws and rop10-1, EXPR3 in rop10-1 was slightly induced by ABA in Ws (1.9-fold), but the induction was much higher in rop10-1 (3.4-fold). Expansins are involved in cell wall relaxation, but it was reported they were frequently repressed by ABA in several stress microarray studies (Bray, 2004). In our chip analysis, we also found two expansin genes, EXP16 and EXP1, were, respectively, activated and repressed in Ws and rop10-1 (Supplemental Tables I–IV).
The down-regulated (2.0-fold) gene in rop10-1 is At5g02100 that encodes an oxysterol-binding family protein similar to yeast SWH1. However, its expression was not responsive to ABA in both Ws and rop10-1. The exact function of yeast SWH1/OSH1 is unknown, although its homologs in humans are involved in cholesterol homeostasis. At5g02100 does not contain the PH domain in SWH1 that specifies the late Golgi targeting (Levine and Munro, 2002), so it remains elusive regarding its function in ROP10 GTPase signaling.
The ROP10 Mutation Activates a Subset of Genes in Response to ABA
In contrast to the observation that only three genes were altered in rop10-1 without ABA treatment, 1 μm ABA resulted in larger number of genes up-regulated (323 probe sets representing 341 genes; Supplemental Table III) in rop10-1 than in Ws. However, a similar number of genes were down-regulated (119 probe sets representing 127 genes; Supplemental Table IV) by the 2-fold cutoff in rop10-1. When the ABA-induced or -suppressed datasets of rop10-1 and Ws were compared, 40 genes were induced in Ws but not in rop10-1. However, 2-fold more genes (119 in total) were induced in rop10-1 but not in Ws. A careful examination of these 40 (Supplemental Table V) and 119 (Supplemental Table VI) genes, respectively, showed that the majority of them were simply less dramatically induced in rop10-1 or Ws, but they were filtered out by the 2-fold cutoff and/or failed to pass the statistical test in case the average fold changes were ≥2-fold. On the average, the 40 genes induced in Ws were also increased by 1.7-fold in rop10-1, and similarly 119 genes induced by ABA in rop10-1 were also ABA up-regulated by about 1.7-fold in Ws (Supplemental Fig. 1). Tables II and III also reflect this trend. This result indicates that while there are some genes that are ABA activated in Ws but not induced in rop10-1 (Supplemental Table V), the majority of these sets of genes were only slightly less responsive to ABA in rop10-1 and Ws, respectively, and hence not analyzed further.
The most interesting subset of genes was obtained from the comparison between ABA-treated Ws and ABA-treated rop10-1. A total of 42 genes showed at least a 2-fold increase in ABA-treated rop10-1 compared to ABA-treated Ws (Supplemental Table II). Interestingly, a particular subset of 21 genes in this set of 42 genes did not exhibit obvious ABA regulation in Ws (Table IV). The remaining 21 genes did not exhibit obvious ABA activation in rop10-1 (Supplemental Table VII) and thus were not chosen for further analysis. Among the subset of 21 genes that were ABA activated in rop10-1, 10 of them were significantly activated by 1 μm ABA by 2.0-fold in rop10-1, while 11 of them showed a slightly weaker activation (1.5- to 1.9-fold). All of these genes did not have much difference between untreated Ws and untreated rop10-1. Functional categorical analysis indicated that a large number of these genes encode regulatory proteins, including six protein kinases, two zinc-finger proteins, and two transcription factors. This result suggests that ROP10 small GTPase likely modulates the expression of this particular set of genes in modulating ABA signaling, and we focus our analysis on these genes.
Table IV.
A list of 21 genes with higher expression in ABA-treated rop10-1 than in ABA-treated Ws
These genes are activated by 1 μm ABA in rop10-1 compared to the no-ABA control, but they are not induced in Ws. Results from the comparative analysis of their expression with the GENEVESTIGATOR database are also shown. Bold text indicates that there are no statistical differences. AGI, Arabidopsis Genome Initiative.
| AGI Locus
|
Annotation
|
ABAarop10/Ws
|
rop10 ABA/CK
|
Ws ABA/CK
|
CK rop10/Ws
|
1b
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
11
|
12
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.2c | 3.3 | 3 | 2.3 | 9.9 | 5.3 | 9.7 | 42.1 | 12.3 | 3.7 | 4.9 | 5.7 | ||||||
| AT3G28580 | AAA-type ATPase | 6.2 | 2.5 | 0.6 | 1.4 | 0.6 | 0.8 | 5.3 | 1.7 | 16.6 | 5.9 | 8.2 | 95.0 | 35.8 | 13.0 | 7.2 | 9.0 |
| AT3G26830 | Cytochrome P450 71B15 | 3.3 | 2.1 | 0.9 | 1.4 | 0.9 | 3.2 | 1.1 | 4.2 | 8.5 | 9.7 | 23.5 | 1.5 | 15.7 | 22.8 | 8.8 | 21.3 |
| AT1G01560 | MAPK (MPK11)d | 3.1 | 2.9 | 1.5 | 1.6 | 1.1 | 3.7 | 6.5 | 1.9 | 3.6 | 3.3 | 8.2 | 27.4 | 5.9 | 1.8 | 2.4 | 0.9 |
| AT4G01370 | MAPK (MPK4)d | 3.1 | 2.9 | 1.5 | 1.6 | 1.1 | 3.7 | 6.5 | 1.9 | 3.6 | 3.3 | 8.2 | 27.4 | 5.9 | 1.8 | 2.4 | 0.9 |
| AT5G01540 | Lectin receptor kinase | 2.9 | 2.3 | 1.0 | 1.3 | 0.8 | 1.7 | 2.2 | 3.7 | 5.4 | 6.9 | 4.2 | 62.6 | 2.6 | 0.5 | 2.3 | 2.0 |
| AT3G28210 | AN1-like zinc-finger protein | 2.7 | 2.0 | 1.1 | 1.6 | 1.7 | 7.8 | 8.4 | 3.2 | 8.8 | 3.1 | 30.4 | 12.3 | 85.5 | 0.8 | 11.6 | 3.4 |
| AT2G17040 | NAM family protein | 2.5 | 3.1 | 1.3 | 1.0 | 1.0 | 1.4 | 1.4 | 2.9 | 8.4 | 9.8 | 3.6 | 68.8 | 0.7 | 0.2 | 0.7 | 1.2 |
| AT1G66880 | Ser/Thr protein kinase | 2.4 | 2.3 | 1.1 | 1.2 | 1.3 | 6.7 | 2.1 | 1.7 | 3.1 | 3.2 | 13.5 | 40.5 | 2.5 | 8.6 | 1.6 | 1.9 |
| AT2G41380 | Embryo-abundant protein | 2.4 | 2.0 | 1.2 | 1.5 | 1.3 | 3.7 | 1.4 | 1.5 | 1.4 | 1.4 | 3.8 | 0.3 | 10.9 | 0.2 | 29.7 | 6.5 |
| AT5G18470 | Lectin family protein | 2.1 | 2.4 | 1.3 | 1.1 | 2.1 | 3.3 | 1.7 | 4.3 | 2.9 | 4.5 | 17.8 | 30.6 | 0.9 | 2.8 | 2.5 | 5.2 |
| AT3G50930 | AAA-type ATPase | 3.9 | 1.7 | 0.8 | 1.8 | 1.3 | 2.5 | 7.1 | 2.4 | 52.6 | 7.2 | 18.0 | 58.5 | 22.4 | 1.5 | 3.2 | 2.0 |
| AT2G32020 | GNAT family protein | 3.9 | 1.6 | 1.0 | 2.3e | 1.5 | 2.2 | 5.9 | 1.4 | 27.9 | 3.6 | 2.3 | 74.7 | 9.5 | 2.8 | 2.6 | 2.0 |
| AT5G52750 | Heavy-metal-associated | 3.3 | 1.9 | 0.9 | 1.6 | 1.9 | 12.7 | 6.7 | 3.8 | 15.6 | 9.4 | 12.5 | 121.3 | 4.1 | 1.2 | 1.6 | 2.7 |
| AT5G27420 | C3HC4-type zinc-finger protein | 3.2 | 1.7 | 0.8 | 1.5 | 1.5 | 2.3 | 2.8 | 3.5 | 6.3 | 3.3 | 6.3 | 137.3 | 5.1 | 2.8 | 3.0 | 4.1 |
| AT4G23260 | Protein kinase | 2.9 | 1.8 | 0.8 | 1.2 | 1.0 | 1.2 | 0.5 | 0.5 | 0.6 | 1.1 | 2.5 | 3.4 | 0.5 | 0.3 | 0.9 | 1.2 |
| AT2G36790 | UDP-glucosyl transferase | 2.9 | 1.5 | 0.8 | 1.6 | 0.9 | 2.0 | 0.4 | 2.0 | 3.9 | 6.6 | 6.4 | 37.1 | 4.2 | 0.2 | 3.9 | 1.3 |
| AT3G22060 | Receptor protein kinase | 2.6 | 1.9 | 1.1 | 1.5 | 1.1 | 1.3 | 1.1 | 1.1 | 1.0 | 3.7 | 13.3 | 9.3 | 0.8 | 4.3 | 1.1 | 4.1 |
| AT3G26210 | Cytochrome P450 71B23 | 2.2 | 1.9 | 0.9 | 1.0 | 0.8 | 4.1 | 0.6 | 1.8 | 0.8 | 2.0 | 3.2 | 2.5 | 3.3 | 1.0 | 1.5 | 7.0 |
| AT2G04070 | MATE efflux family protein | 2.1 | 1.6 | 0.9 | 1.3 | 0.9 | 2.5 | 1.0 | 0.9 | 24.7 | 17.6 | 3.7 | 22.4 | 13.4 | 2.3 | 3.2 | 1.5 |
| AT4G18880 | HSF21 | 2.0 | 1.6 | 0.8 | 1.0 | 1.1 | 2.1 | 2.5 | 2.1 | 3.4 | 2.1 | 4.0 | 34.9 | 6.0 | 0.9 | 1.4 | 1.4 |
| AT5G38900 | DSBA-like oxidoreductase | 2.0 | 1.6 | 1.1 | 1.4 | 1.3 | 1.1 | 1.0 | 1.5 | 1.9 | 2.0 | 8.7 | 1.9 | 15.6 | 6.9 | 9.1 | 35.3 |
ABA, rop10/Ws: rop10-1 versus Ws in ABA; rop10, ABA/CK: ABA versus control (no ABA) in rop10-1; Ws, ABA/CK: ABA versus control in Ws; CK, rop10/Ws: rop10-1 versus Ws in control.
Different treatments: 1, ABA; 2, SA; 3, cold; 4, osmotic; 5, salt; 6, UV-B; 7, ozone; 8, cycloheximide; 9, syringolin; and 10 to 12, infection of A. tumefaciens, B. cinerea, and P. infestans.
The average fold changes of 21 genes in each treatment. Fold changes for each gene in each treatment are listed in the table.
MPK11 and MPK4 are two highly homologous genes represented by the same probe set, and, thus, they have the same gene expression fold changes.
The 2.3-fold change was determined statistically insignificant by the SAM analysis of the whole-chip data. However, we found there was a real difference in three out of four treatments. A representative RT-PCR analysis is shown in Figure 1D.
Identification of Significantly ABA-Regulated Genes That Are Common to All Studies
Prior to this study, several DNA microarray studies on Arabidopsis ABA responses had been reported, but most of these experiments had different ABA treatments, such as different ABA doses and durations, or different organ/cellular types and developmental stages (Hoth et al., 2002; Seki et al., 2002a; Abe et al., 2003; Suzuki et al., 2003; Bray, 2004; Leonhardt et al., 2004; Takahashi et al., 2004; Sanchez et al., 2004; Osakabe et al., 2005). However, we reason that if some genes are commonly regulated in all or most of these experiments, these genes are probably very sensitive to ABA and thus are critical to ABA response or likely to function in the major ABA response pathway(s). Therefore, we compared the genes that are, respectively, up- or down-regulated by ABA in both Ws and rop10-1 with GENEVESTIGATOR and the three data sources (Hoth et al., 2002; Suzuki et al., 2003; Sanchez et al., 2004). Surprisingly, only one gene was commonly down-regulated by ABA, and a total of 16 genes were up-regulated by ABA (Supplemental Table VIII, with those common to all of the experiments marked with asterisks). In comparison to a total of 222 and 79 genes that were, respectively, activated and suppressed by 1 μm ABA in this study (Supplemental Table VIII), this indicates that the biological variations between different experiments are quite dramatic. The small number of genes overlapped in all of these studies is similar to a recent comparative analysis (Bray, 2004). In that study, only 1.4% and 0.2% of the genes were commonly induced and suppressed in three microarray studies on ABA and water deficit stress (Bray, 2004).
Verification of Chip Data with RT-PCR Analysis
To verify the chip data, we first carried out regular RT-PCR analysis (Fig. 1). A total of 16 genes were selected to determine their expression at 0 and 1 μm ABA. Five genes represent those that are similarly and dramatically activated by ABA in Ws and rop10-1 (PP2C/At5g59220, RD20, ABF3, auxin-responsive family protein/At4g17280, and MYB74; see Fig. 1A), and one gene (protein kinase/At4g21410) is suppressed in both Ws and rop10-1 (Fig. 1B). Another gene is At1g63840 that encodes a C3HC4-type zinc-finger family protein (Fig. 1C). According to the chip data (Supplemental Table VI), it was activated by 1 μm ABA in rop10-1 (2.0-fold) but less than 2-fold in Ws (actually 1.5-fold on the average), and thus was filtered out by the 2-fold cutoff in ABA activation in Ws. In our RT-PCR analysis, it also seemed to have a small increase in Ws by 1 μm ABA treatment, consistent with the overall pattern shown in Supplemental Figure 1A. Finally, a total of nine genes from the particular subset of 21 genes that were induced (by the 2-fold cutoff) only in rop10-1 but not in Ws (Table IV) were also selected (Fig. 1, D–F). All of these 16 genes seemed to exhibit an expression pattern consistent with the chip data. Indeed, when five genes were selected for real-time quantitative PCR analysis, they all showed consistent patterns (Fig. 2). The real-time PCR analysis revealed that these genes exhibited slightly higher induction in 1 μm ABA-treated rop10-1 versus ABA-treated Ws, compared to the chip hybridization assays (Table IV). This is consistent with an extensive report showing that real-time RT-PCR is more sensitive to the GeneChip hybridization (Czechowski et al., 2004). Taken together, these results confirm that that our chip data involving three biological replicates are reliable.
Figure 1.
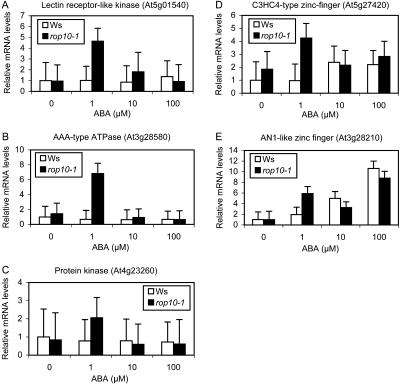
RT-PCR analysis of gene expression in response to ABA doses. A total of 19 genes were investigated using 7-d-old wild-type (Ws) and rop10-1 (r10) seedlings treated with 0, 1, 10, and 100 μm of ABA for 4 h. A, Five genes that show similar ABA activation in Ws and rop10-1. B, One gene that exhibits similar ABA repression in Ws and rop10-1. C, One gene that shows ≥2-fold activation by 1 μm ABA in rop10-1 but weakly in Ws. D to F, Nine genes from the particular subset of 21 genes that show ABA activation in rop10-1. D, Four genes in the first category that is not responsive to all concentrations of ABA in Ws but induced in rop10-1 only by 1 μm ABA. E, Three genes in the second category that is not responsive to 1 μm ABA but only very weakly induced by higher doses of ABA in Ws. These genes are induced in rop10-1 only by 1 μm ABA. F, Two genes in the third category that is significantly activated by 1 μm ABA in rop10-1 but not greatly induced by 1 μm ABA in Ws. However, it is induced by higher ABA concentrations in both Ws and rop10-1. G, MYB2 and two MYB2 closely related MYB genes that exhibit differential expression patterns. H, ACT2, an internal control. Three biological replicates showed the similar patterns, and one of the representative RT-PCR gel pictures was shown for each gene.
Figure 2.
Real-time PCR analysis of transcript changes in response to ABA doses. Samples were essentially the same as in Figure 1. Three genes in the first category (A–C) and one gene each for the second (D) and third (E) categories were selected for quantitative analysis. Relative mRNA levels were determined by first normalizing their PCR threshold cycle numbers with those of the reference (ACT2) and setting the relative mRNA levels for each gene in nontreated Ws at 1. The bar indicates the sd (n = 3).
Distinct ABA Dose-Response Patterns of Gene Expression
To further understand whether the most interesting subset of 21 genes induced by ABA only in rop10-1 is responsive to higher ABA concentrations, dose response was analyzed with RT-PCR and five of the genes were further selected for the analysis using real-time PCR assay. Those five genes that were similarly activated by 1 μm ABA in Ws and rop10-1 (see above) continued to increase with ABA concentrations (Fig. 1A). However, those nine genes tested here among 21 that had higher expression levels in ABA-treated rop10-1 than in ABA-treated Ws showed different response types. They could be further classified into three categories of ABA responses. The first category represents those genes that were neither induced by 1 μm ABA in Ws nor responsive at all to 10 and 100 μm ABA in Ws (Figs. 1D and 2, A–C). Surprisingly, higher concentrations of ABA reduced their expression in rop10-1 to their wild-type level. These include genes encoding a lectin receptor kinase (At5g01540), AAA-type ATPase (At3g28580), protein kinase (At4g23260), and GNAT family protein (At2g32020). The lectin receptor kinase family has a total of 42 members in Arabidopsis (Herve et al., 1996). This family of genes contains an extracellular lectin domain, and some of them have been shown to be autophosphorylated and possess Ser/Thr kinase activity (Herve et al., 1996; He et al., 2004). One member of this family, LecRK2, has been revealed to be induced by salt stress (He et al., 2004). Some AAA-type ATPases have been shown to be subunits of 26S proteasome (Fu et al., 1999), and one such ATPase has also been found to be highly induced upon genotoxic stress (Chen et al., 2003). Increasing evidence shows that protein degradation is involved in ABA responses (such as Lopez-Molina et al., 2003; Smalle et al., 2003), so it will be interesting to test whether these two ATPases identified in this research have a role in protein degradation and whether this regulatory process is controlled by ROP10 GTPase in ABA signaling. GCN5, a member of GNAT family protein, is a histone acetyltransferase that modulates chromatin activity. It has been shown to regulate the expression of some genes, including the cold-responsive CBF1 transcription factor and many cold-regulated (COR) genes (Vlachonasios et al., 2003). The function of this GNAT family protein in ABA signaling remains to be determined.
The second category includes those genes that were not induced by 1 μm ABA in Ws but became slightly responsive to higher concentrations of ABA (Figs. 1E and 2D). This category includes at least a C3HC4-type zinc-finger protein (At5g27420), a putative receptor-like kinase (At3g22060), and a cytochrome P450 (71B15/At3g26830). Similar to the first category, expression of the genes in this category at 10 and 100 μm ABA did not increase further in rop10-1, and instead was reduced to lower levels. There are a large number of zinc-finger family proteins that play regulatory roles, such as transcription, protein degradation, or protein-protein interactions. One zinc-finger protein has been suggested to mediate ABA-regulated seed dormancy (He and Gan, 2004), so it will be interesting to test whether this zinc-finger protein has any function in ROP10-controlled ABA signaling. The function of the putative receptor-like kinase remains to be determined. Very recently, the Leu-rich repeat receptor-like kinase RPK1 has been demonstrated to function in ABA signaling and to regulate many but not all of the genes that are responsive to ABA (Osakabe et al., 2005). Because the PM localization is required for ROP10 function, it has been speculated that ROP10 might interact with receptor-like kinases (Zheng et al., 2002), such as RPK1 or the one identified here. The identification of cytochrome P450 genes is also interesting given that it is one of the largest gene families in Arabidopsis (Werck-Reichhart et al., 2002). CYP71 is the largest P450 family, and the CYP71B subfamily forms the largest cluster of 13 P450s on chromosome III. Two genes in the specific subset of 21 genes encode the CYP71B subfamily, CYP71B15 and CYP71B23. In addition, two other isoforms of CYP71B were also induced (CYP71B28) or repressed (CYP71B7) by ABA in both Ws and rop10-1 (Tables II and III). Thus, it seems that ABA and ROP10 differentially regulate the expression of various CYP71B isoforms. CYP71B15 is identical to PAD3, which is involved in the biosynthesis of camalexin (Zhou et al., 1999). CYP71B15/PAD3 has been shown to be induced by salicylic acid (SA) and involved in the fungal pathogen resistance. In our experiment, it is slightly induced by 100 μm ABA in Ws and shows dramatic activation in rop10-1 by 1 μm ABA (Fig. 1E). The functional implication of SA induction in ROP10-mediated ABA signaling remains to be investigated.
The third category of genes, including an AN1-like zinc-finger protein (At3g28210) and a mitogen-activated protein kinase (MAPK; MPK11/At1g01560), was slightly induced by 1 μm ABA in Ws and further responsive to higher concentrations of ABA (Figs. 1F and 2E). Unlike the first two categories, expression level in rop10-1 was mostly not reduced back to the original level. Instead, they were similarly induced by higher ABA concentrations as in Ws, indicating an enhanced ABA sensitivity of these genes in rop10-1. AN1-like zinc-finger protein in Arabidopsis has not been studied, but the AN1 domain in the mammalian zinc-finger protein ZNF216 is responsible for interaction with components that regulate the transcription factor NFkappaB, such as in the control of apoptosis (Huang et al., 2004). The other interesting gene is MAPK. In this microarray study, two highly homologous MAPKs, MPK11 (At1g01560) and MPK4 (At4g01370), were identified to be induced by ABA and suppressed by ROP10. These two MAPKs belong to the B1 subgroup of MAPK (MAPK Group, 2002). While MPK11 has not been characterized, MPK4 is involved in both biotic (systemic acquired resistance) and abiotic (osmotic, cold, and salt) stresses (Ichimura et al., 2000; Petersen et al., 2000; Droillard et al., 2004). Furthermore, the MPK4 activation is regulated by MKK2, the upstream MAPK kinase (Teige et al., 2004). However, MPK4 is not activated by ABA at both transcriptional and kinase activity levels (Ichimura et al., 2000; Lu et al., 2002). The MAPK that has been shown to be activated by ABA is MPK3, a member of the A1 subgroup of MAPK (Lu et al., 2002). Interestingly, a small increase (about 2-fold) in MPK3 transcript and kinase activity in the 35S:MPK3 transgenic plants is sufficient to significantly enhance ABA responses in postgermination growth arrest, possibly by activating the ABI5 transcription factor (Lu et al., 2002). Therefore, it will be interesting to test whether the receptor-like kinase and MPK11 have any function in ABA signaling and whether their functions involve ROP10 GTPase, like the mammalian Ras-MAPK cascade.
In addition, two MYB genes (MYB78 and MYB112) that are closely related to MYB2 (Stracke et al., 2001) and are below the chip detection limit were also chosen to reveal their ABA response patterns (Fig. 1G). The MYB2 expression pattern is similar to the quantitative analysis result reported in rop10-1 (Zheng et al., 2002). While MYB112 exhibited a similar pattern as MYB2, MYB78 did not appear to be greatly regulated by ABA in Ws but was significantly up-regulated by ABA in rop10-1. Thus, among four MYB genes tested (including MYB74, see above), they showed various ABA response patterns. MYB2 has been implicated as a positive regulator, based on the expression analysis and overexpression phenotypes (Abe et al., 1997, 2003). The functions of these MYB genes that are responsive to ABA and/or modulated by ROP10 small GTPase remain to be dissected.
ROP10-Specific ABA-Responsive Genes Are Activated by Stresses
To gain further insights into the biological functions of the specific subset of 21 genes that are modulated by ROP10, we investigated whether their expression is mediated by other stimuli. Interestingly, the search using the GENEVESTIGATOR tool revealed that almost all of these genes (except the putative protein kinase, At4g23260) are regulated by a variety of abiotic and biotic stresses, but they are not affected by other hormones except SA. Specifically, they are highly induced by stresses of salt and UV-B, by treatments of ozone, cycloheximide, and syringolin, and by infection with Agrobacterium tumefaciens, Botrytis cinerea, and Phytophthora infestans (Table IV). They are also slightly induced by cold and osmotic stresses. As mentioned above, one of these genes, PAD3/CYP71B15, has been shown to be activated by SA and important for the fungal pathogen resistance (Zhou et al., 1999). In this GENEVESTIGATOR comparative analysis, PAD3 exhibited a slight SA induction and strong activation by other biotic and abiotic stresses.
rop10-1 Increases Sensitivity of Seed Germination Inhibition to Mannitol and NaCl
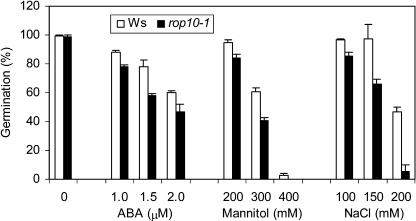
Because ABA also mediates several abiotic stresses, we decided to assess whether this type of comparative data analysis has any functional implications. Both Ws and rop10-1 seeds were sown on plates containing ABA, mannitol, or NaCl. As shown in Figure 3, while rop10-1 slightly enhanced ABA responses in inhibiting seed germination similar to an earlier study (Zheng et al., 2002), rop10-1 also increased sensitivity to mannitol. Interestingly, rop10-1 showed a slightly more dramatic effect in increasing the NaCl sensitivity of seed germination inhibition. At 200 mm NaCl, while Ws exhibited an about 50% inhibition, rop10-1 showed a 95% reduction, an almost 2-fold increase of sensitivity. This is more dramatic than those in 2 μm ABA and 300 mm mannitol. Under these conditions where Ws showed 40% to 50% inhibition, rop10-1 exhibited only 1.3- or 1.5-fold higher sensitivity to ABA or mannitol than Ws. The enhanced sensitivity to NaCl in rop10-1 could be complemented by transgenic expression of the ROP10 cDNA (data not shown). In summary, our results indicate that the specific subset of genes induced by ABA in rop10-1 is likely functional in the ROP10 GTPase-controlled ABA and stress signaling.
Figure 3.
rop10-1 enhances the sensitivity of seed germination inhibition to mannitol and NaCl. Both wild-type (Ws) and rop10-1 seeds were sown on the half-strength MS medium supplemented with ABA, mannitol, or NaCl. Germination was scored 2 d after the cold treatment. Each data point represents the average and sd of germination percentage for three replicates, each with 60 to 90 seeds.
DISCUSSION
Genetic and physiological studies have shown that the PM-associated Arabidopsis ROP10 small GTPase is a specific negative regulator in ABA responses (Zheng et al., 2002). However, how ROP10 regulates ABA signaling remains unclear despite the establishment of the mammalian Ras small GTPase signaling paradigm and the identification of at least 50 Arabidopsis genes involved in ABA response. Several factors have contributed to the difficulty in dissecting the ROP10-mediated pathway in ABA signaling. First and most critically, our understanding on how plants distinguish low versus high magnitudes and/or transient versus sustained changes in ABA and stress status is still lacking (Verslues and Zhu, 2005). To our knowledge, ABA dose-specific responsive genes or signaling pathways have not been reported. Using the low concentration of ABA transcriptome analysis, we have identified three subsets of genes that are likely specific for low concentrations of ABA (Tables II–IV). In particular, most of the subset of 21 regulatory genes (Table IV) is specifically gated by ROP10 in response to low dose of ABA (Figs. 1, D and E, and 2). However, this subset of genes would not have been revealed without using the rop10-1 mutant background. Additionally, two other factors increase the difficulty of illustrating the ROP10-mediated ABA signal transduction. Homologs of yeast or mammalian Ras signaling components exist in large number in Arabidopsis, for example, about 90 genes involved in the MAPK cascade (MAPK Group, 2002). Most of these homologs have not been extensively characterized for their possible functions in ABA response. ROP GTPase signaling in plants might also have evolved with some novel components such as ROP-interactive CRIB motif-containing proteins (Yang, 2002; Fu et al., 2005). Furthermore, the currently fragmented ABA signaling remains to be networked, although many ABA signaling proteins have been identified (Finkelstein et al., 2002; Himmelbach et al., 2003). Our work that ROP10 modulates a particular subset of genes that are ABA dose-specific will provide new insights into the complex ABA signaling network.
ROP10 Does Not Directly Regulate Major ABA Response Pathways
ROP10 is suggested to act at an early step or in a common pathway of ABA signaling because of its PM localization and its involvement in various ABA responses (Zheng et al., 2002). However, analysis of our full-genome chip data and comparison with the published and the publicly available large-scale gene chip datasets indicate that ROP10 does not directly regulate a major ABA response pathway(s). We have found that only 17 genes regulated by 1 μm ABA are common to many other experiments involving higher concentrations of or longer exposures to ABA. Because these genes are identified in many different biological experiments, their roles are likely critical to a major or default ABA response pathway(s), regardless of ABA levels and developmental stages. Many of these genes have been characterized at the expression or the functional levels (such as Himmelbach et al., 2002; Bray, 2004; Fujita et al., 2004; Olsson et al., 2004; Takahashi et al., 2004; for earlier studies, see Finkelstein et al., 2002, and refs. therein). Interestingly, none of these 17 genes was altered in the RPK1 knockout or antisense plants (Osakabe et al., 2005). However, this is in sharp contrast to ABI1 that affects all of 16 ABA-activated genes, and indeed the expression of about 75% of ABA-regulated genes in wild type is abolished by the ABI1 mutation in their studies (Hoth et al., 2002). Although it has not been reported how many ABA-regulated genes in wild type are affected by the RPK1 mutation, 44% of genes repressed in rpk1-2 are ABA inducible (Osakabe et al., 2005). In comparison, 70% (301 out of 420) of ABA-regulated genes in either Ws or rop10-1 are not altered in rop10-1. Therefore, both ABI1 and RPK1 seem to affect more ABA-regulated genes than ROP10. These results strongly suggest that the phenotypic severity is associated with the extent of ABA-regulated transcriptomes being affected by the mutations. These results also explain why the dominant negative rop10 mutant only partially suppressed the abi2-1 mutant phenotypes (Zheng et al., 2002). Taken together, it is less likely that ROP10 directly acts in or directly modulates a major and common signaling pathway(s), although ROP10 can also possibly act in a redundant fashion with other closely related ROPs, such as ROP9 (Zheng and Yang, 2000; Yang, 2002), as the PM-associated components of the major ABA signaling pathway(s).
ROP10 Specifically Modulates a Subset of Genes in Response to ABA Doses
Importantly, we have identified a subset of at least 21 genes that are modulated by ROP10. Moreover, they are not induced by other hormones (except SA), as revealed by comparative analysis using the publicly available large datasets of microarray studies. Furthermore, only three genes are altered (by a 2-fold cutoff) in rop10-1 without ABA treatment. Taken together, our results are in strong support that ROP10 specifically regulates ABA response (Zheng et al., 2002). The particular subset of 21 genes is not, or at most very weakly, altered in rop10-1 in the absence of 1 μm ABA, but is significantly up-regulated by ABA in rop10-1. Two novel categories of genes do not respond or respond only very slightly to high concentrations (10 and 100 μm) of ABA, and the third category of genes is responsive to high concentrations of ABA in wild type. However, the strongest expression of these genes is achieved only in rop10-1 and by 1 μm ABA treatment (Figs. 1 and 2, A–D). This result suggests that the restricted transcriptional regulation of these genes by low levels of ABA is gated by ROP10.
During plant growth and development or as the external environments (such as water, salt, and temperature status) fluctuate, the concentrations of endogenous and physiologically active ABA also vary from a small to a large magnitude of changes. This dynamic distribution of active ABA pools has been observed through in vivo ABA imaging that is aided by the RD29B and HB6 promoter-reporter systems (Christmann et al., 2004). Moreover, plants can differentially respond to different concentrations of ABA. For example, root growth is slightly promoted by low ABA concentrations (≤1 μm) but then becomes inhibitory when ABA concentrations increase up to 100 μm (Ghassemian et al., 2000; Zheng et al., 2002). Therefore, plants need to interpret these dynamic changes in order to make a most appropriate decision to respond and adapt to the fluctuating internal and external environments. Hence, the function of this ROP10-exerted gating control could be to faithfully relay the signals to downstream events from a to-be-identified PM-associated ABA receptor(s) that precisely perceives low ABA levels. Because the majority of these genes encode regulatory proteins such as a GNAT family N-acetyltransferase, various protein kinases, and zinc-finger proteins, this gating would enhance or tighten the control of the regulatory process that is specific to low concentrations of ABA signaling. This mechanism can be important for plants not to overreact to low levels of ABA or mild stresses.
The identification of receptor kinases, MAPKs, and transcription factors or regulators that are modulated by ROP10 is exciting because it is well established that mammalian Ras GTPases relay the extracellular signals from the PM-bound receptors via the MAPK cascade to activate transcription in the nucleus. Certain receptor-like kinases, MAPKs, and MYB transcription factors are known to function in ABA responses (Lu et al., 2002; Abe et al., 2003; Osakabe et al., 2005). A certain member(s) of ROP GTPases probably interacts with some receptor-like kinases because ROP(s) has been found in the active but not the inactive CLV1 receptor-like kinase complex (Trotochaud et al., 1999). Therefore, it will be interesting to test whether this conserved signaling mechanism in mammals also acts in plant ABA signaling.
Role of ROP10-Modulated ABA Signaling in Abiotic and Biotic Stress Responses
Interestingly, almost all of this particular subset of 21 genes is highly induced by a variety of biotic and abiotic stresses. We show here that ROP10 does play a negative role in osmotic and salt stresses. It is well established that some pathways of osmotic and salt stresses are mediated by or interact with ABA (Chinnusamy et al., 2004), and it was recently shown that SOS2, a key protein kinase in the salt stress signaling pathway, can interact directly with ABI2 (Ohta et al., 2003). Hence, it is possible that ROP10 gates the ABA sensitivity threshold for these genes under various stress conditions. As discussed above, it is important for plants to distinguish mild from severe stresses. In the case of salt stress, fewer rop10-1 seeds germinated in the presence of medium to high concentrations of NaCl, which, if not removed, subsequently arrested seedling growth. Therefore, ROP10 plays an important role as a negative regulator in the balance of promoting versus preventing germination. A similar scenario has been illustrated by the MAPK or ABI5-mediated ABA inhibition in postgermination growth (Lopez-Molina et al., 2001; Lu et al., 2002).
Regarding the biotic stress response, we initially were surprised that these ROP10-modulated genes are so highly induced by several biotic stresses. However, this is possible given that ROP/Rac GTPases in rice (Oryza sativa) and barley (Hordeum vulgare) have been shown to be important for disease responses (Ono et al., 2001; Schultheiss et al., 2003). The rice Rac GTPase acts through the control of reactive oxygen species (ROS) production, and Arabidopsis ROP can also regulate ROS production in hypoxia responses or cell death (Baxter-Burrell et al., 2002; Park et al., 2004). Ample evidence has shown that ROS mediates ABA responses (Mustilli et al., 2002; Desikan et al., 2004; Verslues and Zhu, 2005). In addition, accumulating evidence suggests that ABA is also involved in biotic stresses/plant-pathogen interactions that have not been well recognized before (for a recent review, see Mauch-Mani and Mauch, 2005). ABA signaling can interact with response pathways of some hormones that are important for biotic stress response, such as SA (Mauch-Mani and Mauch, 2005). One of these 21 genes tested in RT-PCR analysis is cytochrome P450 71B15/PAD3, which is SA inducible and important for resistance to certain pathogen attack (Zhou et al., 1999). Another gene, MPK4, has been demonstrated to function in a variety of biotic and abiotic stresses (Ichimura et al., 2000; Petersen et al., 2000; Droillard et al., 2004). Therefore, it is perceivable that the activation of these genes under various stresses is at least partly controlled by the ROP10-mediated ABA signaling pathway. It will be interesting to determine whether ROP10 also modulates the induction of these genes by biotic stresses independently of ABA.
In summary, we not only provide genomic evidence to support that ROP10 is a specific regulator of ABA response, but more importantly show that ROP10 likely modulates the ABA sensitivity of a particular subset of regulatory genes, such as protein kinases, zinc-finger family proteins, and transcription factors. The existence of three categories of genes modulated by ROP10 enables its differential regulation of their expression in response to various levels of ABA and, thus, severity of stresses. Apparently, ROP10 can also modulate the expression of some genes that are activated by increasing ABA concentrations, such as certain C3HC4 zinc-finger proteins and MYB2/MYB112. It remains to be investigated whether ROP10 regulates genes specific for high concentrations of ABA, but our finding that ROP10 can gate the sensitivity of several regulatory genes specific to a low ABA concentration provides novel insights into the complex ABA signaling. We plan to investigate in the future whether these genes are functional in particular in response to low ABA level or mild stress, and, if so, how they are subjected to the control of ROP10. The dissection of this potential ABA dose-specific ROP10 small GTPase pathway(s) will advance our understanding on how plants fine tune the ABA and stress-responsive network. By this mechanism, plants, as immotile organisms, can make a most appropriate decision to respond and adapt to the dynamic internal and external environments during growth and development.
MATERIALS AND METHODS
Plant Growth and ABA Treatment
Seeds of the Arabidopsis (Arabidopsis thaliana) T-DNA knockout mutant rop10-1 (Zheng et al., 2002) and its wild type (ecotype Ws) were incubated in 50 mL of liquid medium of Murashige and Skoog (MS) basal salts, pH 5.7, supplemented with 3% (w/v) Suc at 4°C for 4 d and then allowed to germinate and grow at 22°C for 7 d under 16-h-day and 8-h-night regime with continuous shaking at the speed of 150 rpm. After three times of rinse with freshly prepared MS liquid medium, young seedlings were treated with 0 and 1 μm ABA (mixed isomers; Sigma) that was dissolved with dimethyl sulfoxide (with the final concentration in the medium of 0.1%, v/v) prepared in the MS medium. After 4 h of treatment, the seedlings were immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation. Three biological replicates were performed at different times for all genotype/treatment combinations.
RNA Extraction, Labeling, and Hybridization to Arabidopsis GeneChip
These procedures followed the recommendations of the University of California, Irvine DNA Microarray Facility, which were also described by Price et al. (2004). Briefly, total RNA was extracted with TRIzol (Invitrogen) and purified by RNeasy kit (Qiagen) following the manufacturers' instructions. A total of 12 ATH1 Arabidopsis GeneChips (Affymetrix) were used for hybridization with fragmented and biotinylated cRNA that was synthesized from total RNA. After washing and staining with streptavidin phycoerythrin, the chips were scanned by measuring light emitted at 570 nm (with an excitation wavelength at 488 nm). The GeneChip data were then compiled using the Affymetrix GCOS software.
Statistical and Bioinformatic Analysis of GeneChip Data
The raw data were linearly scaled/normalized using the RMAExpress (http://stat-www.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html) after they were determined to be linearly distributed. After log2 transformation, pair-wise comparisons between Ws/rop10-1 and ABA treated/untreated were performed using a modified t test (also called the S-test), which is the basis of the software SAM (Tusher et al., 2001). Significantly regulated genes were identified by running SAM on the log2 normalized data with a 2-fold cutoff and at a median false discovery rate of ≤6% by manually adjusting the Delta factor. The resulting significantly increased and decreased genes were further filtered out by the Affymetrix detection calls, eliminating those genes called “A” in any one of the 12 chips. Table merge and intersection were performed using the SAS 8.2 software package. Annotation of gene function was done through the Gene Ontology Web site (http://www.geneontology.org/). Functional categorical (FunCat) analysis was conducted by MIPS (http://mips.gsf.de/proj/funcatDB/search_main_frame.html). Expression comparison of the selected genes with the publicly available chip datasets was performed using the GENEVESTIGATOR bioinformatics tool (https://www.genevestigator.ethz.ch/; Zimmermann et al., 2004).
Confirmation of Transcript Levels and Determination of ABA Dose-Response Patterns with RT-PCR Analysis
Seedlings grown for 7 d after cold treatment were similarly prepared as above but they were subjected to treatments of 0, 1, 10, and 100 μm ABA for 4 h. Total RNA was reverse transcribed by Superscript III reverse transcriptase (Invitrogen). PCR analysis was conducted using the Taq DNA polymerase (GenScript), with ACT2 used as the internal control as described (Li et al., 2001). Gene-specific primers were synthesized and their oligonucleotide sequences listed in Supplemental Table IX. Real-time quantitative PCR analysis was also performed for selected genes using the QuantiTect SYBR Green PCR kit (Qiagen). Real-time PCR was carried out in the MasterCycler II (Cypheid) according to the manufacturer's protocol. The primers were designed using the software provided online by GenScript (http://www.genscript.com/bioinformatics.html) while the ACT2 primers were designed previously (Zheng et al., 2002), and the primer sequences were listed in Supplemental Table IX.
Phenotypic Analysis of Abiotic Stresses
Seed germination assay was performed as described previously (Li et al., 2001). Briefly, seeds were sown on the agar medium of half-strength MS basal salts supplemented with various concentrations of ABA, mannitol, or sodium chloride. ABA was dissolved in 3 m KOH as the 100 mm stock. The plates were cold treated for 4 d and then incubated at 22°C in the light. Seed germination was scored after 2 d, with the criterion being set as the complete protrusion of the radicle.
All microarray data from this work are available from NCBI GEO (www.ncbi.nlm.nih.gov/geo) under the series entry GSE3454.
Supplementary Material
Acknowledgments
We are grateful to Zhenbiao Yang (University of California, Riverside) for encouragement and insightful discussions of this work and constructive comments on the manuscript. We greatly appreciate John Price and Jyan-Chyun Jang (Ohio State University) for stimulating discussion and technical help in microarray design and data analysis. We thank Nam-Hai Chua (Rockefeller University) for providing the supplemental data of their published DNA microarray studies (Sanchez et al., 2004). We also thank William Tramontano (Lehman College, CUNY) for critical reading of the manuscript.
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant no. 2004–35304–14911) and in part by the City University of New York (start-up fund to Z.-L.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhi-Liang Zheng (zhiliang.zheng@lehman.cuny.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068064.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J (2002) RopGAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science 296: 2026–2028 [DOI] [PubMed] [Google Scholar]
- Bray EA (2004) Genes commonly regulated by water deficit stress in Arabidopsis thaliana. J Exp Bot 55: 2331–2341 [DOI] [PubMed] [Google Scholar]
- Brocard-Gifford I, Lynch TJ, Garcia ME, Malhotra B, Finkelstein RR (2004) The Arabidopsis thaliana ABSCISIC ACID-INSENSITIVE8 encodes a novel protein mediating abscisic acid and sugar responses essential for growth. Plant Cell 16: 406–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I-P, Haehnel U, Altschmied L, Schubert I, Puchta H (2003) The transcriptional response of Arabidopsis to genotoxic stress—a high-density colony array study (HDCA). Plant J 35: 771–786 [DOI] [PubMed] [Google Scholar]
- Chen W, Provart NJ, Glazebrook J, Katagiri F, Chang HS, Eulgem T, Mauch F, Luan S, Zou G, Whitham SA, et al (2002) Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Christmann A, Hoffmann T, Teplova I, Grill E, Muller A (2004) Generation of active pools of abscisic acid revealed by in vivo imaging of water-stressed Arabidopsis. Plant Physiol 137: 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Droillard MJ, Boudsocq M, Barbier-Brygoo H, Lauriere C (2004) Involvement of MPK4 in osmotic stress response pathways in cell suspensions and plantlets of Arabidopsis thaliana: activation by hypoosmolarity and negative role in hyperosmolarity tolerance. FEBS Lett 574: 42–48 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CR (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Doelling JH, Rubin DM, Vierstra RD (1999) Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteosome. Plant J 18: 529–539 [DOI] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z-L, Wasteneys G, Yang Z (2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeda-Suzuki S, Ng J, Tzu J, Dietzl G, Sun Y, Harms M, Nardine T, Luo L, Dickson BJ (2002) Rac function and regulation during Drosophila development. Nature 416: 438–442 [DOI] [PubMed] [Google Scholar]
- He XJ, Zhang ZG, Yan DQ, Zhang JS, Chen SY (2004) A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor Appl Genet 109: 377–383 [DOI] [PubMed] [Google Scholar]
- He Y, Gan S (2004) A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol Biol 54: 1–9 [DOI] [PubMed] [Google Scholar]
- Herve C, Dabos P, Galaud JP, Rouge P, Lescure B (1996) Characterization of an Arabidopsis thaliana gene that defines a new class of putative plant receptor kinases with an extracellular lectin-like domain. J Mol Biol 258: 778–788 [DOI] [PubMed] [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Yang Y, Grill E (2003) Relay and control of abscisic acid signaling. Curr Opin Plant Biol 6: 470–479 [DOI] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Huang J, Teng L, Li L, Liu T, Li L, Chen D, Xu LG, Zhai Z, Shu HB (2004) ZNF216 is an A20-like and IkappaB kinase gamma-interacting inhibitor of NFkappaB activation. J Biol Chem 279: 16847–16853 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S (2003) CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15: 411–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Choi HI, Ryu HJ, Park JH, Kim MD, Kim SY (2004) ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol 136: 3639–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16: 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemichez E, Wu Y, Sanchez JP, Mettouchi A, Mathur J, Chua NH (2001) Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev 15: 1808–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S (2002) Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol 12: 695–704 [DOI] [PubMed] [Google Scholar]
- Li H, Shen JJ, Zheng ZL, Lin Y, Yang Z (2001) The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol 126: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV (2002) Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 99: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson AS, Engstrom P, Soderman E (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55: 663–677 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17: 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Assmann SM (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D'Angelo C, Weinl S, Kudla J, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Gu Y, Lee Y, Yang Z, Lee Y (2004) Phosphatidic acid induces leaf cell death in Arabidopsis by activating the Rho-related small G protein GTPase-mediated pathway of reactive oxygen species generation. Plant Physiol 134: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, et al (2000) Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JP, Duque P, Chua NH (2004) ABA activates ADPR cyclase and cADPR induces a subset of ABA-responsive genes in Arabidopsis. Plant J 38: 381–395 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Huckelhoven R (2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36: 589–601 [DOI] [PubMed] [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. a) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002. b) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD (2003) The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li QB, McCarty DR (2003) Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol 132: 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Seki M, Ishida J, Satou M, Sakurai T, Narusaka M, Kamiya A, Nakajima M, Enju A, Akiyama K, et al (2004) Monitoring the expression profiles of genes induced by hyperosmotic, high salinity, and oxidative stress and abscisic acid treatment in Arabidopsis cell culture using a full-length cDNA microarray. Plant Mol Biol 56: 29–55 [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11: 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ (2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochrome P450. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/10.1199/tab.0028, http://www.aspb.org/publications/arabidopsis
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell (Suppl) 14: S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X (2004) Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-L, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14: 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-L, Yang Z (2000) The Rop GTPase: an emerging signaling switch in plants. Plant Mol Biol 44: 1–9 [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.