Abstract
During the symbiotic interaction between Medicago truncatula and the arbuscular mycorrhizal (AM) fungus Glomus intraradices, an endogenous increase in jasmonic acid (JA) occurs. Two full-length cDNAs coding for the JA-biosynthetic enzyme allene oxide cyclase (AOC) from M. truncatula, designated as MtAOC1 and MtAOC2, were cloned and characterized. The AOC protein was localized in plastids and found to occur constitutively in all vascular tissues of M. truncatula. In leaves and roots, MtAOCs are expressed upon JA application. Enhanced expression was also observed during mycorrhization with G. intraradices. A partial suppression of MtAOC expression was achieved in roots following transformation with Agrobacterium rhizogenes harboring the MtAOC1 cDNA in the antisense direction under control of the cauliflower mosaic virus 35S promoter. In comparison to samples transformed with 35S∷uidA, roots with suppressed MtAOC1 expression exhibited lower JA levels and a remarkable delay in the process of colonization with G. intraradices. Both the mycorrhization rate, quantified by fungal rRNA, and the arbuscule formation, analyzed by the expression level of the AM-specific gene MtPT4, were affected. Staining of fungal material in roots with suppressed MtAOC1 revealed a decreased number of arbuscules, but these did not exhibit an altered structure. Our results indicate a crucial role for JA in the establishment of AM symbiosis.
Arbuscular mycorrhizal (AM) fungi have been found to exist for more than 400 million years, suggesting that plants became evolutionarily associated with these fungi during land colonization (Harrison, 1999). More than 80% of terrestrial plant species form symbiotic associations with AM fungi, which belong exclusively to the phylum Glomeromycota (Schüssler et al., 2001). Today, AM is the most widespread type of mycorrhizal association worldwide. The main feature of this mutual symbiosis is the exchange of nutrients between both partners: the fungus supplies the plant with mineral nutrients from the soil and receives carbohydrates in return (Smith and Read, 1997). Other features of mycorrhizal-associated plants are an increased resistance to root pathogens (Gianinazzi-Pearson et al., 1996; Cordier et al., 1998) and abiotic stresses like drought and heavy metals (Augé, 2001; Schützendübel and Polle, 2002).
Two classes of AM were described on the basis of structural differences in forming intracellular hyphal branches, the so-called Arum and the Paris type (Smith and Smith, 1997). In the Arum type, the AM fungus invades the root cortex and forms intraradical hyphae. Subsequently, these hyphae enter cortex cells and form highly branched tree-like structures, called arbuscules. Arbuscules are the main symbiotic organs, which serve for the exchange of nutrients between the fungus and the plant (Harrison, 1999).
AM have a significant ecological importance. However, our understanding of the development and maintenance of functional symbiosis is still limited. Although screening of several nonmycorrhizal mutants has led to the identification of putative plant receptors of fungal signals (Kistner and Parniske, 2002), nothing is known about such factors and the molecular dialogue between the two symbiotic partners that leads to the establishment of AM (Hause and Fester, 2005).
Phytohormones such as cytokinins, auxin, gibberellins, and abscisic acid are assumed to participate in this communication (Ludwig-Müller, 2000), but their precise role in the interaction is still unknown. Evidence for phytohormone involvement in the establishment of AM has been drawn mainly from application experiments or from increases in their endogenous levels observed during mycorrhization (Bothe et al., 1994; Regvar et al., 1996; Barker and Tagu, 2000). For jasmonic acid (JA), a rise in endogenous levels correlating with the mycorrhization of barley (Hordeum vulgare) and Medicago truncatula has been shown (Hause et al., 2002; Stumpe et al., 2005). In the case of barley, the content of JA and its amino acid conjugates increased concomitantly with the cell-specific expression of genes coding for JA-biosynthetic enzymes and of jasmonate-induced genes within arbuscule-containing cells (Hause et al., 2002). Since JA levels increased after the initial step of the plant-fungal interaction, the development of mycorrhiza rather than the recognition of the interacting partners may be linked to the expression of JA-biosynthetic genes and to elevated JA levels.
Free JA, its methyl ester (JAME), and amino acid conjugates (commonly named jasmonates) are signals in various plant responses to biotic and abiotic stresses as well as of distinct stages of plant development (for review, see Wasternack and Hause, 2002). In particular, jasmonates may activate genes that are involved in plant defense in response to wounding and pathogen attack (Creelman and Rao, 2002; Ryan and Moura, 2002). However, the regulation of the rise in JA upon wounding or pathogen attack is still only partially understood. In tomato (Lycopersicon esculentum) and Arabidopsis (Arabidopsis thaliana), most genes encoding the enzymes of JA biosynthesis are also JA inducible (Strassner et al., 2002), suggesting feed-forward regulation. The rise in JA, however, precedes the accumulation of corresponding mRNAs, and enzymes of JA biosynthesis occur constitutively in tomato and Arabidopsis leaves (Hause et al., 2000; Stenzel et al., 2003b). Furthermore, JA formation seems to be dependent on substrate availability, since in plants that overexpress one of the biosynthetic enzymes, elevation of JA in leaves only occurs upon additional stimulation (Laudert et al., 2000; Stenzel et al., 2003a).
The biosynthesis of JA originates from α-linolenic acid, which is oxygenated by 13-lipoxygenase. The activities of allene oxide synthase and allene oxide cyclase (AOC) lead to cis-(+)-12-oxo-phytodienoic acid (OPDA). Among these biosynthetic enzymes, AOC is responsible for the formation of the cis-(+)-enantiomer (9S,13S) of OPDA. This enantiomer is thought to be the unique precursor for the naturally occurring (+)-7-iso-JA, which is synthesized via reduction by an OPDA reductase (OPR3) and three subsequent steps of β-oxidation (Vick and Zimmerman, 1983). The AOC is therefore regarded to be of prime importance in JA biosynthesis.
The AOC has been cloned from a variety of plant species including tomato (Ziegler et al., 2000) and Arabidopsis (Stenzel et al., 2003b). In tomato, AOC is specifically expressed in all vascular bundles and the surrounding parenchymatic cells (Hause et al., 2000), leading to preferential formation of JA in at least the main veins and to amplification of the wound response (Stenzel et al., 2003a). In contrast, in Arabidopsis AOC protein occurs constitutively in all leaf tissues (Stenzel et al., 2003b).
To study the role of JA in mycorrhization by a reverse genetic approach, we cloned two cDNAs coding for AOC from M. truncatula. In M. truncatula, the AOC protein occurs constitutively in all vascular bundles. We show here that mycorrhization of this plant causes an accumulation of MtAOC1 transcripts and protein, which is located in arbuscule-containing cells. The cDNA of MtAOC1 was used in antisense direction for the transformation of roots to partially suppress MtAOC1 expression. Our data clearly show that this suppression markedly decreases the rate of colonization and arbuscule formation by the AM fungus Glomus intraradices. The results are discussed in terms of the possible role of jasmonates in the establishment of AM symbiosis.
RESULTS
Two MtAOC-Encoding cDNAs: Cloning and Characterization
A cDNA library from mycorrhizal M. truncatula roots was screened for a full-length cDNA coding for AOC and resulted in isolation of one cDNA coding for AOC, designated as MtAOC1 (deposited in GenBank accession no. AJ308489). A second cDNA coding for MtAOC2 (deposited in GenBank accession no. AJ866733) was isolated by RACE using RNA isolated from M. truncatula roots and specific primers deduced from the tentative consensus sequence 90433 (The Institute for Genomic Research Gene Index). A search of expressed sequence tag (EST) databases did not produce any sequences that might correspond to additional AOC genes. The coding regions of both cDNAs encompass 956 bp and 1,084 bp corresponding to proteins containing 252 and 250 amino acid residues, respectively. The calculated molecular masses were 27.98 kD and 27.49 kD. Alignment of the deduced protein sequences indicated an identity of 75.3% between both proteins and an identity of 64.6% and 64.7% to the tomato AOC (Ziegler et al., 2000). Using computer analyses by ChloroP and TargetP programs, a location in plastids was predicted for both proteins. The putative cleavage site for MtAOC1 lies between amino acids 56 and 57 and for MtAOC2 lies between amino acids 79 and 80. The predicted pI of the cloned MtAOCs without the putative chloroplast signal peptide was calculated as 7.09 and 6.74, respectively.
To experimentally observe the subcellular location of MtAOC, we used an immunocytological approach using an antibody directed against recombinant LeAOC. In M. truncatula preparations and in those containing purified recombinant MtAOC1, this antibody recognizes the MtAOC1 (Supplemental Fig. 1). Due to the high identity of both MtAOCs, the anti-LeAOC antibody was able to recognize both MtAOCs (data not shown). No cross-reactivity with other plant proteins was observed in separated proteins of M. truncatula. Cross sections of petioles of M. truncatula probed with this antibody showed significant fluorescence label within the chloroplasts (Fig. 1A), whereas cross sections treated with preimmune serum did not (Fig. 1B). This confirms our assumption, based on the sequence analysis, that MtAOCs are plastid-located proteins.
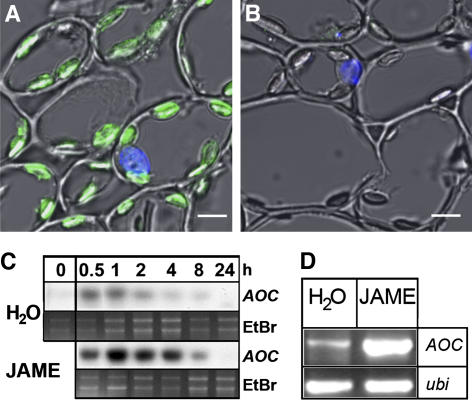
Figure 1.
Subcellular localization of MtAOC protein in M. truncatula and jasmonate-induced accumulation of MtAOC1 mRNA in leaves and roots of M. truncatula. A and B, Cross sections of petioles were probed with anti-AOC antibody raised against purified recombinant LeAOC (A) or with preimmune serum (B), followed by a fluorescence-labeled secondary antibody. Micrographs were taken by confocal laser scanning microscopy showing immunodecorated AOC (green), DAPI-stained nuclei (blue), and the bright-field image (gray). The strong green fluorescence label within chloroplasts in A is indicative of the AOC protein and is not visible upon treatment with preimmune serum. The bars represent 5 μm. C, Water- or JAME-treated leaves (50 μm) were subjected to northern analysis. Twenty micrograms of total RNA were loaded per lane and hybridized with the full-length cDNA coding for MtAOC1. Loading control is given by the ethidium bromide (EtBr) staining of gels. D, Accumulation of MtAOC1 transcripts in roots analyzed by RT-PCR. Intact plants were treated by application of 50 μm JAME to the roots, which were harvested 2 h later. Total RNA was isolated from root material and used for RT-PCR. Ubiquitin transcripts were used as a control to confirm constant levels of amplified fragments for all samples.
Heterologous expression of the MtAOC1 cDNA without the predicted chloroplast-targeting signal in Escherichia coli resulted in an additional band of about 26 kD, which was absent in control bacteria transformed with the empty vector (Supplemental Fig. 1). Enzymatic assays for AOC activity were performed with extracts of bacteria carrying the MtAOC1-containing vector, and cis-(+)-OPDA was formed exclusively, which is indicative of AOC activity (Ziegler et al., 1999; Ziegler et al., 2000).
JA, OPDA, and their derivatives are known to induce AOC expression in tomato and Arabidopsis (Stenzel et al., 2003a, 2003b). To test JA- and OPDA-dependent MtAOC expression, leaves of M. truncatula were floated on water or 50 μm solutions of JA, JAME, and OPDA or on 1 m sorbitol, known to induce endogenous JA biosynthesis. There was a slight accumulation of MtAOC mRNA in the water-treated control leaves at early time points, presumably due to wounding of the leaves during sample preparation (Fig. 1C). As shown for JAME, all treatments with the compounds mentioned produced up-regulation of MtAOC expression, which reached a maximal level after 1 to 2 h of treatment. MtAOC1 expression was analyzed by reverse transcription (RT)-PCR using MtAOC1-specific primers in JAME-treated roots (Fig. 1D). MtAOC1 was found to be expressed at a low level in nontreated roots. After 2 h of treatment, a clear increase in transcript accumulation was observed (Fig. 1D).
Organ- and Tissue-Specific Occurrence of AOC in M. truncatula
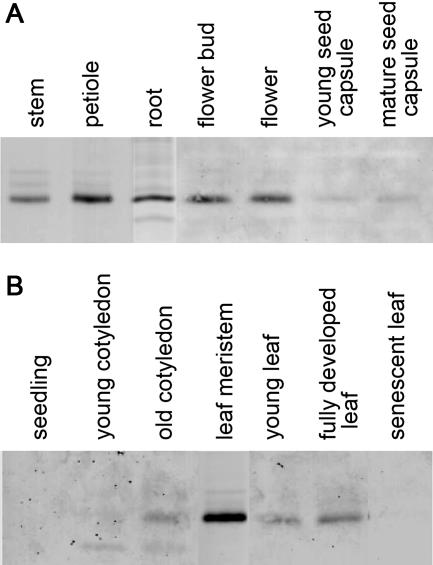
The occurrence of MtAOC protein was analyzed by immunoblot analysis in different organs of the adult plant and in different stages of leaf development (Fig. 2). The MtAOC protein was detectable in all organs of the flowering plant (Fig. 2A). The highest level of MtAOC protein was observed in organs containing a relatively high portion of vascular tissues, such as stems, petioles, and roots, and in flowers. Only minor amounts of MtAOC could be detected in seed capsules. In contrast, not all developmental stages of the leaf showed accumulation of the MtAOC protein (Fig. 2B). Here, the highest level of AOC protein was detected in leaf meristems, but AOC was not found in seedlings, young cotyledons, or senescent (yellow) leaves. Young leaves contained only minor amounts of MtAOC (Fig. 2B).
Figure 2.
Accumulation of MtAOC protein in different organs and developmental stages of M. truncatula. Immunoblot analyses in different organs of the flowering plant (A) and in different developmental stages of leaves (B) were performed using material pooled from five plants. Ten micrograms of total protein were loaded per lane, and blots were probed with anti-LeAOC antibody in a dilution of 1:5,000.
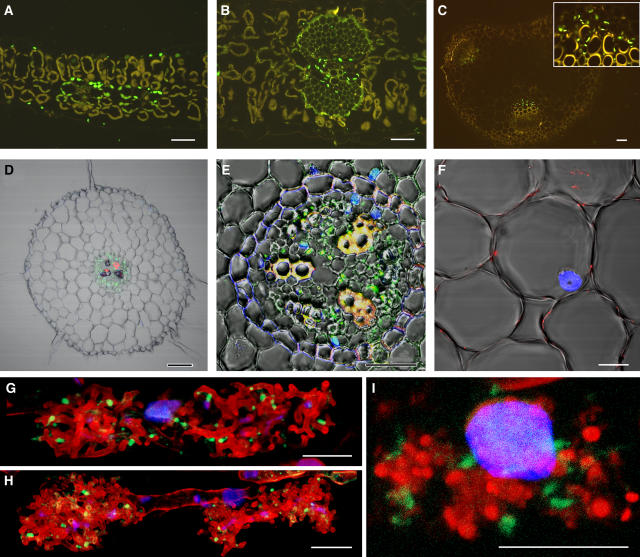
To investigate the cell-specific occurrence of MtAOC, we analyzed 2-μm-thick cross sections of different organs of M. truncatula using immunohistological techniques. In all vegetative tissues, MtAOC was clearly detectable by the anti-LeAOC antibody in parenchymatic cells of the vascular bundles as shown for the intercostal region of a leaflet (Fig. 3A), a minor vein enclosed by sclerenchyma (Fig. 3B), and the stem (Fig. 3C). When the tissues were probed with a preimmune serum, no signal was detectable in any of the tissues (data not shown). Cross sections of roots exhibited a strong label within the phloem cells of the central cylinder (Fig. 3, D and E). The cortex cells of nonmycorrhizal roots were free of label (Fig. 3F). Upon mycorrhization, however, MtAOC was detectable in cortex cells harboring an arbuscule (Fig. 3, G–I). Here, the signal was clearly located in plastids located near the arbuscule. Moreover, the AOC protein seemed to be present in arbuscule-containing cells independent of the developmental stage of arbuscules.
Figure 3.
Tissue-specific expression of MtAOC in leaves (A and B), stems (C), and nonmycorrhizal (D–F) as well as mycorrhizal roots (G–I) of 2-month-old M. truncatula plants. For immunocytochemical analysis, semithin cross sections of the respective organs were probed with rabbit anti-AOC antibody raised against recombinant LeAOC followed by a goat anti-rabbit IgG antibody conjugated with Alexa Fluor 488 resulting in a green fluorescence. In all vegetative tissues, MtAOC protein was defined in chloroplasts of the parenchymatic cells of the vascular bundles, as shown for the intercostal region of the leaf showing two minor veins (A), a sclerenchymatic vein of the leaf (B), and the stem (C, inset). Nonstained chloroplasts appear in brown due to autofluorescence. The central cylinder of the root exhibited strong MtAOC accumulation (D and E), whereas cortex cells did not show any label (F). In cortex cells harboring an arbuscule, however, AOC protein was detectable (G–I). Independent of the developmental stage of arbuscule shown by staining with wheat germ agglutinin conjugated with tetramethylrhodamine isothiocyanate (red) as developing (G), fully developed (H), and collapsing (I) arbuscules, AOC protein was defined to plastids surrounding the arbuscule (green). The cortex cell nucleus and fungal nuclei were stained with DAPI (blue). The superposition of 15 single optical sections obtained by confocal laser scanning microscopy is shown in G to I. Bars represent 25 μm (A, B, and E), 50 μm (C and D), and 10 μm (F–I).
Partial Suppression of MtAOC in Transformed Roots of M. truncatula
A binary vector containing the MtAOC1 cDNA in antisense direction under the control of the cauliflower mosaic virus (CaMV) 35S promoter was used to transform roots of M. truncatula by Agrobacterium rhizogenes. Transformation of roots by A. rhizogenes can lead to 75% of hairy roots expressing the transgene (Vieweg et al., 2004). Roots transformed with uidA under the control of CaMV 35S promoter were used as a control to exclude effects due to the formation of hairy roots. As tested by β-glucuronidase activity staining in an independent approach, the frequency of hairy root formation at the inoculation sites was about 65%, with about 75% of roots expressing the transgene (Supplemental Fig. 2). The A. rhizogenes-transformed roots exhibited a similar morphology to untransformed roots.
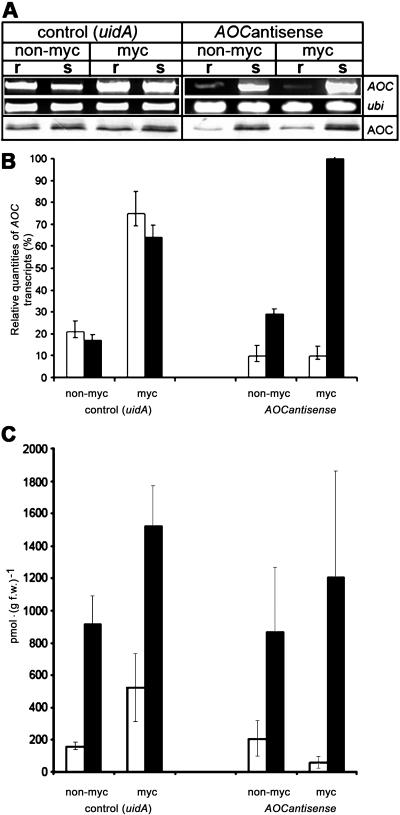
To test the efficiency of the antisense suppression, the MtAOC1 expression levels from nonmycorrhizal plants were analyzed by RT-PCR, real-time RT-PCR, and immunoblot analyses using plants 21 d after hairy root formation and transfer to new pots (Fig. 4, non-myc). In MtAOC1antisense-transformed roots, a 2-fold decrease of MtAOC1 transcript level compared to uidA root-transformed plants was detected. The transcript levels of MtAOC2 were also reduced by this antisense approach (data not shown). The amount of AOC protein was also reduced in MtAOC1antisense-transformed roots. Nearly constant levels of MtAOC1 transcript and protein were observed for roots and shoots of uidA root-transformed plants as well as shoots of the MtAOC1antisense root-transformed plants. This indicates the successful suppression of MtAOC expression occurring specifically within roots.
Figure 4.
Accumulation of MtAOC1 mRNA and protein as well as content of JA in roots and shoots after transformation of roots with 35S∷uidA and 35S∷MtAOC1antisense, respectively. Plants were harvested 21 d after inoculation, and the same pool of 20 plants was used for all RNA extractions. A, Total RNA isolated from roots (r) and shoots (s) was subjected to RT-PCR as described in “Materials and Methods.” Equal volumes of the RT-PCR products were separated in a 1.5% agarose gel and stained with ethidium bromide. Ubiquitin transcripts were used as a control to confirm constant levels of amplified fragments for all samples. For western-blot analysis, 10 μg of total protein were loaded per lane and blots were probed with anti-LeAOC antibody in a dilution of 1:5,000. B, Quantification of MtAOC1 expression examined by real-time RT-PCR. Two extractions of total RNA from roots (□) and shoots (▪) were analyzed in triplicates with a TaqMan probe specific for MtAOC1. The level of transcripts coding for elongation factor α served as control. n = 6. C, JA content. Roots (□) and shoots (▪) of four different pools of root-transformed plants were subjected for extraction of JA. n = 4.
Effects of Mycorrhization in Control (uidA Root-Transformed) and MtAOC1antisense Root-Transformed Plants
The MtAOC1 expression was 3.5-fold higher in mycorrhizal roots when compared with nonmycorrhizal uidA-transformed roots (Fig. 4, A and B), indicating an induction of MtAOC1 expression by mycorrhization with G. intraradices. In contrast, roots transformed with 35S∷MtAOC1antisense exhibited nearly constant levels of MtAOC1 transcript in nonmycorrhizal as well as of mycorrhizal plants (Fig. 4, A and B). Here, in mycorrhizal MtAOC1antisense roots, MtAOC1 mRNA accumulation was suppressed down to 15% of the level measured in mycorrhizal control roots. This is clearly reflected in endogenous JA levels (Fig. 4C). The JA level increased in the mycorrhizal uidA-transformed roots up to 3-fold 21 d after inoculation. In shoots of those plants, the JA levels were constitutively higher than in roots and increased only slightly in mycorrhizal plants in comparison to nonmycorrhizal plants (Fig. 4C). In plants containing MtAOC1antisense-transformed roots, however, JA level decreased in roots upon mycorrhization, but was also slightly increased in shoots.
Effect of Partial AOC Suppression on Mycorrhization
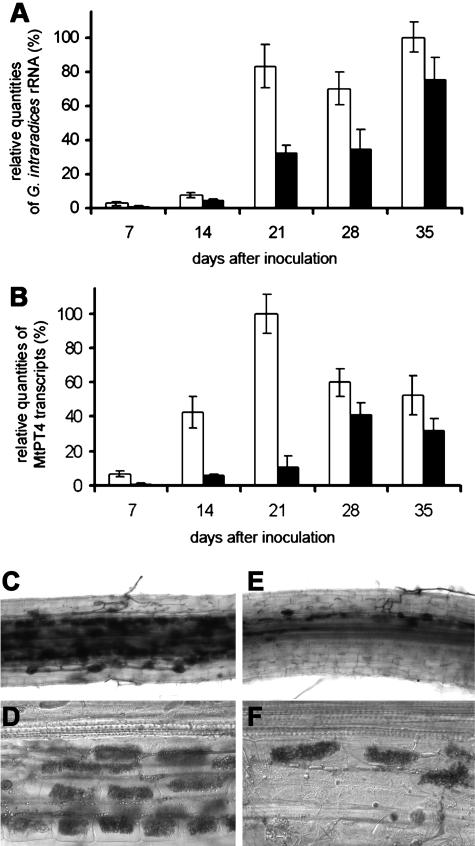
To study the effect of partial MtAOC1 suppression on mycorrhization, the fungus was quantified via its rRNA in uidA-transformed and MtAOC1antisense-transformed roots by real-time RT-PCR. Even small differences in earlier stages of colonization could be detected, when the mycorrhization level determined by staining was about 2%. The quantitative RT-PCR data indicated that the levels of G. intraradices rRNA were positively correlated with the time of growth and progressive inoculation of plants. The hairy root plants derived from the transformation with 35S∷uidA and 35S∷MtAOC1antisense, respectively, exhibited clear differences in the G. intraradices rRNA accumulation upon mycorrhization. In 35S∷MtAOC1antisense roots about 2- to 1.5-fold less fungal rRNA amplicons compared to uidA-transformed roots were detectable (Fig. 5A).
Figure 5.
Effects of partial suppression of MtAOC1 in roots of M. truncatula on mycorrhization. Plants after transformation of roots with 35S∷uidA (□) and 35S∷MtAOC1antisense (▪), respectively, were inoculated with G. intraradices, harvested at the indicated time points, and total RNA was isolated two times independently and subjected to real-time RT-PCR analysis in triplicates each. A, Relative quantities of G. intraradices rRNA. B, Relative quantities of transcripts coding for MtPT4. In both cases, the level of transcripts coding for elongation factor α served as control. n = 6. C to F, Representative micrographs of ink-stained fungal structures in roots transformed with empty pRedRoot (C and D) and pRedRoot containing MtAOC1antisense (E and F), respectively. Note the less dense appearance of arbuscules in MtAOC1antisense-tranformed roots in comparison to the empty vector control, whereas the structure of arbuscules did not change.
The arbuscular development was examined by detection of transcripts for the arbuscule-specific M. truncatula phosphate transporter (MtPT4; Harrison et al., 2002). MtPT4 transcripts showed highest levels at 21 d after inoculation in uidA-transformed roots and at 28 d after inoculation in MtAOC1antisense roots (Fig. 5B). In particular, at early time points after inoculation the MtAOC1antisense-transformed roots exhibited 1.5- to 5-fold lower levels of MtPT4 amplicons in comparison to uidA-transformed roots.
To check the mycorrhizal phenotype in MtAOC1antisense-transformed roots versus control-transformed roots, we used a vector containing a nondestructive marker (DsRed) for transient transformation of roots according to Limpens et al. (2004). Using DsRed fluorescence, transgenic roots could be selected 2 weeks after formation of hairy roots (Supplemental Fig. 2) and were then inoculated with G. intraradices. Ink staining of mycorrhizal roots transformed with the empty vector revealed a strong mycorrhization 3 weeks after inoculation (Fig. 5C), whereas roots transformed with pRedRoot-MtAOC1antisense exhibited the same degree of mycorrhization over the root length but exhibited less dense mycorrhizal structures (Fig. 5E). The structure of arbuscules did not show any alterations on a light microscope level (Fig. 5, D and F). Although these plants were inoculated later than all the plants described before, there was no difference in the delay of mycorrhization due to the decreased JA content.
DISCUSSION
Among the lipid-derived compounds, octadecanoids and jasmonates have a crucial role in plant responses to biotic and abiotic stresses (León et al., 2001; Wasternack and Hause, 2002; Weber, 2002). JA is suggested to be one of several signals that lead to the expression of plant defense genes, thereby acting as an intra- and intercellular as well as interorganismic signal (Farmer, 2001). Here, we analyzed the role of JA in the establishment of the AM symbiosis in M. truncatula because a rise in JA has been detected in mycorrhizal M. truncatula roots (Stumpe et al., 2005). We first characterized the AOC from M. truncatula, which is believed to catalyze a crucial step in JA biosynthesis. We then showed that a partial suppression of MtAOC1 expression in roots and the subsequent decrease in JA biosynthesis delays root colonization and arbuscule formation.
During mycorrhization of M. truncatula with G. intraradices, MtAOC1 transcripts accumulated at higher levels in mycorrhizal roots in comparison to nonmycorrhizal roots. Interestingly, increased MtAOC protein accumulation was not detectable in total protein extracts of roots, but the AOC protein was clearly observed by immunocytology in arbuscule-containing cells. This discrepancy in detection of the AOC protein could be due to the high amount of AOC protein in the central cylinder overlaying the AOC protein located in arbuscule-containing cells after extraction of total roots. The occurrence of AOC in arbuscule-containing cells is similar to mycorrhizal barley roots, where allene oxide synthase and a jasmonate-induced protein could be detected cell specifically in arbuscule-containing cells (Hause et al., 2002). Therefore, it is tempting to speculate that JA biosynthesis occurs in the arbuscule-containing cells.
To decrease the endogenous JA level in roots, we performed a partial suppression of MtAOC1 expression using an antisense approach. A potential complication for this experiment was that we isolated a second cDNA coding for AOC (MtAOC2). MtAOC2 shows high identity to MtAOC1, and therefore its expression is also suppressed by the 35S∷MtAOC1antisense construct. According to all EST databases available, there are no hints of other additional genes coding for AOC in M. truncatula.
This antisense approach was promising because a previous study showed that changes in the transcript level of a plant regulatory gene involved in the control of the mycorrhizal symbiosis can lead to alterations in mycorrhizal colonization (Staehelin et al., 2001). Furthermore, induction of hairy roots with A. rhizogenes can be achieved with a transformation efficiency of about 70% (this study; Vieweg et al., 2004). By using the root transformation system, MtAOC1 expression could be suppressed in roots without affecting the shoots of the plant. The amounts of MtAOC1 transcript and protein in the MtAOC1antisense lines were clearly reduced in roots, as shown in Figure 4.
The reduction in the amount of MtAOC protein resulted in a decrease in endogenous JA level in mycorrhizal roots. Moreover, this decrease in endogenous JA level was accompanied by a delay in colonization of M. truncatula roots. The most obvious effect was visible at 21-d postinoculation: the amount of fungal rRNA dropped to about 40% in MtAOC1antisense-transformed roots, and the amount of MtPT4 transcript decreased to 10%, in comparison with uidA-transformed roots. This suggests that a lower JA level in mycorrhizal roots reduces the amount of fungal material within the roots and affects arbuscule formation.
Evaluation of the fungal structures in transgenic roots requires a selection system like pRedRoot, and this was used to determine the fungal structures by staining of transgenic roots with ink. The suppression of MtAOC1 within M. truncatula roots resulted in a reduced but regular appearance of fungal structures. Therefore, reduced JA levels in roots led to an overall reduction of arbuscule frequency rather than to an abnormal or even aborted infection process.
The data point to a fundamental function of jasmonates in the interaction of roots with mycorrhizal fungi, and the following mechanisms might be involved.
Jasmonates might act to induce flavonoid biosynthesis during mycorrhization. Flavonoids have been shown to stimulate the growth of AM fungi, thereby possibly acting as signals to stimulate fungal growth (Harrison, 1999). Moreover, in M. truncatula, transcripts encoding enzymes of the flavonoid biosynthetic pathway, such as Phe ammonia lyase (PAL) and chalcone synthase, are induced specifically in cells containing arbuscules (Harrison and Dixon, 1994). It is well known that application of jasmonates leads to increases in PAL mRNA accumulation and in PAL enzyme activity (Gundlach et al., 1992; Thoma et al., 2003).
Jasmonates might contribute to the alterations of the microtubular pattern during mycorrhization. An extensive remodeling of the microtubular cytoskeleton was observed in the early stages of arbuscule development, and this continues until the arbuscule senesces and collapses (Genre and Bonfante, 1997, 1998). As recently shown for M. truncatula, changes in the microtubular pattern occurred not just in arbuscule-containing cells but also in adjacent noncolonized cortical cells (Blancaflor et al., 2001). Furthermore, the gene coding for β-tubulin (MtTubb1) is transcriptionally up-regulated in mycorrhizal roots (Manthey et al., 2004) and in nonmycorrhizal roots upon JA treatment (S. Isayenkov, unpublished data). Furthermore, microtubules are known to change their organization in response to application of jasmonates (Matsuki et al., 1992; Koda, 1997).
Jasmonates could enhance the sink strength of mycorrhizal roots and thereby stimulate carbohydrate biosynthesis in the shoots and their transport into the roots. The maintenance of mycorrhizal symbiosis requires a carbohydrate supply for the fungus. Jasmonates are known to contribute to a redistribution of nutrients (Creelman and Mullet, 1997). This is supported by data that demonstrated that genes coding for enzymes with function in sink/source relationships are JA responsive. LIN6, a gene coding for an extracellular invertase from tomato, is inducible by JA (Thoma et al., 2003) and is expressed in tissues that require a high carbohydrate supply (Godt and Roitsch, 1997). Interestingly, invertases and Suc synthase, both known to supply a sink tissue with Glc, are specifically expressed in mycorrhizal roots (Blee and Anderson, 2002).
Jasmonates could play an indirect role in mycorrhization via the action of cytokinins. Interaction of plant roots with AM fungi elevates the levels of cytokinins (Barker and Tagu, 2000), which are well known factors in cell division and growth (Haberer and Kieber, 2002). Mycorrhizal plants exhibited an improved growth that was independent of their nutrient status, probably mediated by cytokinins (Drüge and Schönbeck, 1992). Here, an increased flux of cytokinins into the shoots may enhance shoot growth (Allen et al., 1980; Baas and Kuiper, 1989). Interestingly, levels of active cytokinins increased in potato (Solanum tuberosum) plants upon treatment with JA (Dermastia et al., 1994).
Jasmonates might contribute to an increase in plant fitness. One aspect of plant fitness is the higher defense status of mycorrhizal plants against pathogens and drought stress (Cordier et al., 1998; Augé, 2001). The altered defense status might be mediated by a JA-induced expression of genes coding, among others, for defense-related proteins (Wasternack and Hause, 2002). In particular, JA produced in mycorrhizal roots could serve as a systemic signal. JA itself or compounds derived from it could be transported into the shoots to induce the genes mentioned above, thereby improving the general fitness of the plant. Long-range signaling effects of JA, e.g. in the systemic wound response of tomato, are clearly indicated (Li et al., 2002). In M. truncatula, such a long-range signaling effect might contribute to shoot responses to root mycorrhization, as reflected in the systemic accumulation of MtAOC1 transcripts and the slightly enhanced JA levels in shoots of mycorrhizal plants.
In summary, it appears that jasmonates affect mycorrhization, possibly in multiple ways. Analyses of transcript and metabolite patterns by cDNA microarrays and metabolite profiling, both in wild type and transgenic roots, can help determine exactly which processes during mycorrhization are mediated by jasmonates. Our results provide strong indication for a crucial role of jasmonates in mycorrhizal roots.
MATERIALS AND METHODS
Plant Material
Medicago truncatula L. Gaertn. var. Jemalong (obtained from Austra Hort Pty) was grown in expanded clay (Lecaton, 2 to 5 mm particle size; Fibo Exclay) in 250-mL plastic pots under a 16/8-h photoperiod at 210 μE m−2 s−1, 25°C, and 50% relative humidity in controlled chambers (Percival Scientific). Fungal inoculum of Glomus intraradices Schenk and Smith (isolate 49 [Maier et al., 1995], enriched by previous cocultivation with leek [Allium porrum]), was used to achieve mycorrhization by cocultivation of plants with the inoculum mixed with sterile expanded clay (1.5:8.5, v/v).
Isolation of AOC cDNA and Expression in Escherichia coli
The sequence of the EST AW225613 (National Center for Biotechnology Information database) was used to deduce primers for amplifying an MtAOC-specific fragment from JAME-treated M. truncatula leaves. With that, a cDNA expression library from roots of M. truncatula (genotype A17) infected with Glomus versiforme (kindly provided by M. Harrison) was screened for a full-length cDNA coding for MtAOC resulting in cDNA coding for MtAOC1. cDNA of MtAOC2 was isolated by RACE. For this, RNA was isolated from roots of M. truncatula using the NucleoSpin RNA plant kit (Macherey-Nagel) according to the manufacturer's instructions. First-strand cDNA synthesis was performed using BD SMART RACE cDNA amplification kit (BD Biosciences Clontech) starting with 1 μg of total RNA in a total volume of 10 μL with 5′-CDS primer, BD SMART II A oligo for 5′ RACE-Ready cDNA, and with 3′-CDS primer A for 3′ RACE-Ready cDNA. RACE was performed with 2.5 μL of experimental cDNA (5′ and 3′ RACE-Ready cDNA) in a total volume of 50 μL according to the manufacturer's instructions. The PCR was performed as follows: 5 cycles (94°C for 30 s, 72°C for 3 min), 5 cycles (94°C for 30 s, 72°C for 30 s, and 72°C for 3 min), and 25 cycles (94°C for 30 s, 68°C for 30 s, 72°C for 3 min). To amplify 3′ and 5′ RACE fragments, primers were designed according to the partial sequence of MtAOC2 (TC90433, The Institute for Genomic Research Gene Index, http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=medicago) and used for 3′ RACE (3AOC2RACE_for: 5′-CATTATGGCGGGGACACAAGCTAGTAAG-3′) and 5′ RACE (5AOC2RACE_rev: 5′-CCCAATTTGATGCAACTTCACTTGACC-3′), respectively. Amplification was performed using BD SMART RACE cDNA amplification kit (BD Biosciences Clontech). 3′ RACE amplification resulted in a fragment of 1,192 bp and 5′ RACE in a fragment 740 bp, respectively. Both cDNA fragments were cloned into pGEM-T Easy vector (Promega) and subsequently sequenced by MWG Biotech.
Computer analysis of the first 100 amino acids of both cDNAs was performed with the ChloroP version 1.1 program (http://www.dtu.dk/services/ChloroP; Emanuelsson et al., 1999) and the TargetP program version 1.0 (http://www.cbs.dtu.dk/services/TargetP; Emanuelsson et al., 2000). For heterologous expression of MtAOC1, a sequence corresponding to amino acid 60 to amino acid 238, thus lacking the putative chloroplast target sequence, was subcloned into pQE30. pQE30 with or without insert was transformed into the host strain Escherichia coli M15 (Qiagen). Total protein of isopropyl-β-thiogalactopyranoside-induced or noninduced cultures was isolated and purified. The overexpressed MtAOC1 protein was purified using the Ni-NTA spin kit (Qiagen) as described (Maucher et al., 2000). The resulting supernatants were used for AOC activity tests (Maucher et al., 2004) and immunoblot analyses (Supplemental Fig. 1).
Plasmid Construction and Root Transformation
The cDNA coding for MtAOC1 (SmaI/XhoI fragment) was inserted into the CaMV 35S promoter cassette of the pGreen series using SmaI restriction sites (Hellens et al., 2000; www.pgreen.ac.uk). A fragment containing the 35S∷AOCantisense fusion was integrated into the T-DNA of the pGreen0029 binary vector (Hellens et al., 2000). In parallel, the uidA gene (SmaI/EcoRI fragment) was fused with the CaMV 35S promoter and also transferred into the T-DNA of pGreen0029. Agrobacterium rhizogenes Arqua1, pretransformed with the helper plasmid pSoup (Hellens et al., 2000), was transformed by electroporation.
To create a pRedRoot vector (Limpens et al., 2004) containing the MtAOC1antisense construct, the MtAOC1 cDNA was PCR amplified (primers used: 5′-GCACTAGTTCGCACGAGTTCATCATC-3′ and 5′-GCGCTAGCTTTTTTTTTTTTTTTTTTCC-3′) and cloned into pGEM-T Easy (Promega). The 998-bp SpeI-NcoI fragment of this vector was cloned into pRNAi (Limpens et al., 2004). For plant transformation the resulting 35S∷MtAOC1antisense construct was inserted as KpnI-PacI fragment into the binary vector pRedRoot.
The induction of transgenic hairy roots was performed using A. rhizogenes Arqua1 (Quandt et al., 1993) according to standard procedures (Vieweg et al., 2004). After development of hairy roots, all roots that did not emerge from the infection site were removed. Subsequently, in the case of transformation with pGreen vector, the 23-d-old seedlings were transferred into new pots for fungal inoculation. In case of pRedRoot vectors, seedlings were allowed to grow 2 weeks more and were then screened by using a fluorescence stereomicroscope (Leica). Five-week-old plantlets with fluorescent roots were then transferred into new pots for fungal inoculation. Twenty-one days after inoculation, red fluorescent roots were selected and stained with ink according to Vierheilig et al. (1998). The complete mycorrhization kinetics (see Fig. 5) were done twice.
Northern-Blot Analysis and Immunoblot Analysis
Total RNA was extracted from frozen tissues and subjected to northern-blot analysis as described (Stenzel et al., 2003a). Twenty micrograms of total RNA were loaded per lane and gel loading was checked by comparing ethidium bromide-stained rRNAs. Proteins were isolated from the phenolic phase of RNA extraction or were extracted from frozen plant material according to Meyer et al. (1988). Protein separation and immunoblot analysis were performed as described (Hause et al., 2000). The antibody raised against recombinant LeAOC (Ziegler et al., 2000) was used in a dilution of 1:5,000.
Extraction and Quantification of JA
Fresh roots and shoots from at least 20 plants (21 d after inoculation) were pooled to minimize biological variation and were immediately frozen in liquid nitrogen. Roots and shoots (0.5 g fresh weight) were homogenized in a mortar and extracted with 5 mL 80% (v/v) methanol. For quantification of JA, (2H6)-JA was added in an appropriate amount before extraction. Ion-exchange chromatography on DEAE Sephadex A-25 cartridges, reversed-phase HPLC, and gas chromatography-mass spectrometry/selected ion monitoring analyses were performed as described (Hause et al., 2002). Extractions were done from four biological replicates.
Immunocytochemistry
Immunocytochemical analysis of aboveground tissues was performed as described (Hause et al., 2000) using a cross section (2 μm thickness) of polyethylene glycol-embedded material of 2-month-old plants. For roots (nonmycorrhizal and mycorrhizal), material was embedded in PEG1000 (Xu et al., 1998), sectioned transversely (nonmycorrhizal roots) or longitudinally (mycorrhizal roots) into 30-μm-thick sections, and immunostained according to Hans et al. (2004). The antibody raised against recombinant LeAOC (Ziegler et al., 2000) was used in a dilution of 1:1,000 for aboveground tissues and 1:500 for roots, respectively. The use of preimmune serum at the same dilutions served as control. As secondary antibody, goat anti-rabbit IgG conjugated with Alexa Fluor 488 (Molecular Probes) was used according to the manufacturer's instructions. Counterstaining was performed with 4,6-diamidino-2-phenylindol (Sigma) and for arbuscule-containing roots with wheat germ agglutinin conjugated with tetramethylrhodamine isothiocyanate (Molecular Probes). Sections were analyzed by epifluorescence microscopy using a Zeiss Axioskop equipped with a CCD camera (Sony) or by confocal laser scanning microscopy using a LSM510 META (Zeiss). All micrographs were processed through the Photoshop 8.0.1 program (Adobe).
RT-PCR and Real-Time RT-PCR
RNA was isolated separately from roots and shoots. All extractions (three independent measurements each) were done twice. For all individual measurements at least 20 plants were pooled. Plant material (100 mg f.w.) was homogenized in liquid nitrogen. Total RNA from roots and shoots was extracted using the NucleoSpin RNA plant kit (Macherey-Nagel) according to the manufacturer's instructions.
cDNA synthesis was performed using Superscript II First-Strand Synthesis system for RT-PCR (Invitrogen) starting with 1 μg of total RNA in a total volume of 10 μL with oligo(dT)15 primer and 5′ RACE Anchor primer (Invitrogen) at 42°C for 60 min.
Initially, the expression level of AOC was measured by semiquantitative RT-PCR. Primers were designed from MtAOC1 and ubiquitin (Salzer et al., 2000; accession no. AJ245511) and used in the following combinations: MtAOC1, 5′-ATGGCATCCATGAGTTCTTTG-3′ and 5′-TGATCAGTTGGTGAAGTTTGG-3′ resulting in a fragment of 758 bp; and ubiquitin, 5′-GTAGAATCATCCGACACAATCG-3′ and 5′-GGAGACGGAGAACAAGGTG-3′ resulting in a fragment of 172 bp.
The PCR reaction contained one-fifth of the reverse transcription reaction, 0.2 mm 2′-deoxynucleoside 5′-triphosphates, 40 pmol primers, 1× PCR reaction buffer (10 mm Tris-HCl, 1.5 mm MgCl2, 50 mm KCl, pH 8.3), and 1 unit of Taq polymerase (Invitrogen) in a total volume of 50 μL. The PCR was performed as follows: 95°C for 2 min, 28 cycles (denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and elongation at 72°C for 2 min), and termination at 72°C for 10 min.
TaqMan probes and primers for real-time PCR were designed using the Primer Express software (Applied Biosystems). Purified PCR primers with TaqMan probe were purchased from Applied Biosystems (Assays-by-Design service) and contained a 6-FAM reporter dye connected to the 5′ end and the nonfluorescent quencher to the 3′ end. To increase the melting temperature (Tm) without increasing probe length, the nonfluorescent quencher was connected with minor groove binder. Primers for MtPT4, for elongation factor α-1, and for the G. intraradices rRNA were used as described by Isayenkov et al. (2004). A fragment from the MtAOC1 transcript was amplified using the primers MTAOC1-ASEF (5′-TTGAAGGGTGTTGCTGATTTGC-3′), MTAOC1-ASER (5′-GCAGCAGTAGATGGTTCAACATG-3′), and MTAOC1-ASEM2 FAM (5′-AAAGCCTGTTGATCCTTC-3′). Real-time PCR was carried out using the ABI Prism 7,000 sequence detection system, optical caps, and optical tubes (Applied Biosystems). PCR amplification mixtures (20 μL) contained 20 ng template cDNA or 50 ng sample DNA, 2× TaqMan Master Mix buffer (10 μL, Applied Biosystems), and 20× TaqMan probe with primers (1 μL). The cycling conditions and method of calculation were used according to Isayenkov et al. (2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AJ308489 (MtAOC1) and AJ866733 (MtAOC2).
Supplementary Material
Acknowledgments
The authors thank Ulrike Huth and Carola Tretner for dependable technical assistance, Conrad Dorer for determination of AOC enzyme activity, and Maria Harrison for providing the M. truncatula cDNA library. Claus Wasternack and Margaret Rice are acknowledged for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP 1084 MolMyk: Molecular Basics of Mycorrhizal Symbioses, project Ha–2655/4–2).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Bettina Hause (bhause@ipb-halle.de).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.069054.
References
- Allen M, Moore TM Jr, Christensen M (1980) Phytohormone changes in Bouteloua gracilis infected by vesicular-arbuscular mycorrhizae. I. Cytokinin increases in the host plant. Can J Bot 58: 371–374 [Google Scholar]
- Augé R (2001) Water relations, drought and vesicular-arbuscular mycorrhizal symbiosis. Mycorrhiza 11: 3–42 [Google Scholar]
- Baas R, Kuiper D (1989) Effects of vesicular-arbuscular mycorrhizal infection and phosphate on Plantago major ssp. pleiosperma in relation to internal cytokinin concentrations. Physiol Plant 76: 211–215 [Google Scholar]
- Barker S, Tagu D (2000) The roles of auxins and cytokinins in mycorrhizal symbioses. J Plant Growth Regul 19: 144–154 [DOI] [PubMed] [Google Scholar]
- Blancaflor E, Zhao L, Harrison M (2001) Microtubule organization in root cells of Medicago truncatula during development of an arbuscular mycorrhizal symbiosis with Glomus versiforme. Protoplasma 217: 154–165 [DOI] [PubMed] [Google Scholar]
- Blee K, Anderson A (2002) Transcripts for genes encoding soluble acid invertase and sucrose synthase accumulate in root tip and cortical cells containing mycorrhizal arbuscules. Plant Mol Biol 50: 197–211 [DOI] [PubMed] [Google Scholar]
- Bothe H, Klingner A, Kaldorf M, Schmitz O, Esch H, Hundeshagen B, Kernebeck H (1994) Biochemical approaches to the study of plant-fungal interactions in arbuscular mycorrhiza. Experientia 50: 919–925 [Google Scholar]
- Cordier C, Pozo M, Barea J, Gianinazzi S, Gianinazzi-Pearson V (1998) Cell defense responses associated with localized and systemic resistance to Phytophthora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol Plant Microbe Interact 11: 1017–1028 [Google Scholar]
- Creelman R, Mullet J (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- Creelman R, Rao M (2002) The oxylipin pathway in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Dermastia M, Ravnikar M, Vilhar B, Kovac M (1994) Increased levels of cytokinin riboside in jasmonic acid-treated potato (Solanum tuberosum) stem node cultures. Physiol Plant 92: 241–246 [Google Scholar]
- Drüge U, Schönbeck F (1992) Effects of vesicular-arbuscular mycorrhizal infection on transpiration, photosynthesis and growth of flax (Linum usitatissimum L.) in relation to cytokinin levels. J Plant Physiol 141: 40–48 [Google Scholar]
- Emanuelsson O, Nielsen H, Bonnak S, von Heijne G (2000) Predicting subcellular localization of proteins based on N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E (2001) Surface-to-air signals. Nature 411: 854–856 [DOI] [PubMed] [Google Scholar]
- Genre A, Bonfante P (1997) A mycorrhizal fungus changes microtubule orientation in tobacco root cells. Protoplasma 199: 30–38 [Google Scholar]
- Genre A, Bonfante P (1998) Actin versus tubulin configuration in arbuscule-containing cells from mycorrhizal tobacco roots. New Phytol 140: 745–752 [DOI] [PubMed] [Google Scholar]
- Gianinazzi-Pearson V, Dumas-Gaudot E, Gollotte A, Tahiri-Alaoui A, Gianinazzi S (1996) Cellular and molecular defence-related root responses to invasion by arbuscular mycorrhizal fungi. New Phytol 133: 45–57 [Google Scholar]
- Godt D, Roitsch T (1997) Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol 115: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach H, Müller M, Kutchan T, Zenk M (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci USA 89: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Kieber J (2002) Cytokinins: new insights into a classic phytohormone. Plant Physiol 128: 345–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans J, Hause B, Strack D, Walter M (2004) Cloning, characterization, and immunolocalization of a mycorrhiza-inducible 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) in arbuscule-containing cells of maize. Plant Physiol 134: 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M (1999) Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu Rev. Plant Physiol. Plant Mol. Biol. 50: 361–389 [DOI] [PubMed] [Google Scholar]
- Harrison M, Dixon R (1994) Spation patterns of expression of flavonoid/isoflavonoid pathway genes during interactions between roots of Medicago truncatula and the mycorrhizal fungus Glomus versiforme. Plant J 6: 9–20 [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Fester T (2005) Molecular and cell biology of arbuscular mycorrhizal symbiosis. Planta 221: 184–196 [DOI] [PubMed] [Google Scholar]
- Hause B, Maier W, Miersch O, Kramell R, Strack D (2002) Induction of jasmonate biosynthesis in arbuscular mycorrhizal barley roots. Plant Physiol 130: 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C (2000) Tissue-specific oxylipin signature of tomato flowers—Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J 24: 113–126 [DOI] [PubMed] [Google Scholar]
- Hellens R, Edwards E, Leyland N, Bean S, Mullineaux P (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Isayenkov S, Fester T, Hause B (2004) Rapid determination of fungal colonisation and arbuscule formation in roots of Medicago truncatula using real-time (RT) PCR. J Plant Physiol 161: 1379–1383 [DOI] [PubMed] [Google Scholar]
- Kistner C, Parniske M (2002) Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Koda Y (1997) Possible involvement of jasmonates in various morphogenic events. Physiol Plant 100: 639–646 [Google Scholar]
- Laudert D, Schaller F, Weiler E (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211: 163–165 [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Lee G, Howe G (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J (2000) Hormonal balance in plants during colonization by mycorrhizal fungi. In Y Kapulnik, D Douds, eds, Arbuscular Mycorrhizas: Physiology and Function. Kluwer Academic Publishers, Amsterdam, pp 263–283
- Maier W, Peipp H, Schmidt J, Wray V, Strack D (1995) Levels of a terpenoid glycoside (blumenin) and cell wall-bound phenolics in cereal mycorrhizas. Plant Physiol 109: 465–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick A, Küster H (2004) Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant Microbe Interact 17: 1063–1077 [DOI] [PubMed] [Google Scholar]
- Matsuki T, Tazaki H, Fujimori T, Hogetsu T (1992) The influences of jasmonic acid methyl ester on microtubules in potato cells and formation of potato tubers. Biosci Biotechnol Biochem 56: 1329–1330 [Google Scholar]
- Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C (2000) Allene oxide synthases of barley (Hordeum vulgare cv. Salome): tissue specific regulation in seedling development. Plant J 21: 199–213 [DOI] [PubMed] [Google Scholar]
- Maucher H, Stenzel I, Miersch O, Stein N, Prasa M, Zierold U, Schweizer P, Dorer C, Hause B, Wasternack C (2004) The allene oxide cyclase of barley (Hordeum vulgare L.)—cloning and organ-specific expression. Phytochemistry 65: 801–811 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Grosset J, Chartier Y, Cleyet-Marel J-C (1988) Preparation by two-dimensional electrophoresis of proteins for antibody production: antibodies against proteins whose synthesis is reduced by auxin in tobacco mesophyll. Electrophoresis 9: 704–712 [DOI] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact 6: 699–706 [Google Scholar]
- Regvar M, Gogala N, Zalar P (1996) Effects of jasmonic acid on mycorrhizal Allium sativum. New Phytol 134: 703–707 [DOI] [PubMed] [Google Scholar]
- Ryan C, Moura D (2002) Systemic wound signaling in plants: a new perception. Proc Natl Acad Sci USA 99: 6519–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer P, Bonanomi A, Beyer K, Vögeli-Lange R, Aeschbacher R, Lange J, Wiemken A, Kim D, Cook D, Boller T (2000) Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection. Mol Plant Microbe Interact 13: 763–777 [DOI] [PubMed] [Google Scholar]
- Schüssler A, Schwarzott D, Walker C (2001) A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res 105: 1413–1421 [Google Scholar]
- Schützendübel A, Polle A (2002) Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J Exp Bot 53: 1351–1365 [PubMed] [Google Scholar]
- Smith F, Smith S (1997) Structural diversity in (vesicular)-arbuscular mycorrhizal symbiosis. New Phytol 137: 373–388 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ (1997) Mycorrhizal Symbiosis, Ed 2. Academic Press, San Diego
- Staehelin C, Charon C, Boller T, Crespi M, Kondorosi A (2001) Medicago truncatula plants overexpressing the early nodulin gene enod40 exhibit accelerated mycorrhizal colonization and enhanced formation of arbuscules. Proc Natl Acad Sci USA 98: 15366–15371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Maucher H, Pitzschke A, Miersch O, Ziegler J, Ryan C, Wasternack C (2003. a) Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato—amplification in wound signaling. Plant J 33: 577–589 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C (2003. b) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Strassner J, Schaller F, Frick U, Howe G, Weiler E, Amrhein N, Macheroux P, Schaller A (2002) Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductase reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 32: 585–601 [DOI] [PubMed] [Google Scholar]
- Stumpe M, Carsjens J-G, Stenzel I, Gobel C, Lang I, Pawlowski K, Hause B, Feussner I (2005) Lipid metabolism in arbuscular mycorrhizal roots of Medicago truncatula. Phytochemistry 66: 781–791 [DOI] [PubMed] [Google Scholar]
- Thoma I, Loeffler C, Sinha A, Gupta M, Krischke M, Steffan B, Roitsch T, Mueller M (2003) Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J 34: 363–375 [DOI] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC (1983) The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem Biophys Res Commun 111: 470–477 [DOI] [PubMed] [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U, Piché Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieweg M, Frühling M, Quandt H-J, Heim U, Bäumlein H, Pühler A, Küster H, Perlick A (2004) The promoter of the Vicia faba L. leghemoglobin gene VfLb29 is specifically activated in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots from different legume and nonlegume plants. Mol Plant Microbe Interact 17: 62–69 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2002) Jasmonates and octadecanoids—signals in plant stress response and development. In K Moldave, ed, Progress in Nucleic Acid Research and Molecular Biology, Vol 72. Academic Press, New York, pp 165–221 [DOI] [PubMed]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217–224 [DOI] [PubMed] [Google Scholar]
- Xu X, Vreugdenhil D, van Lammeren A (1998) Cell division and cell enlargement during potato tuber formation. J Exp Bot 49: 573–582 [Google Scholar]
- Ziegler J, Stenzel I, Hause B, Maucher H, Hamberg M, Grimm R, Ganal M, Wasternack C (2000) Molecular cloning of allene oxide cyclase: the enzyme establishing the stereochemistry of octadecanoids and jasmonates. J Biol Chem 275: 19132–19138 [DOI] [PubMed] [Google Scholar]
- Ziegler J, Wasternack C, Hamberg M (1999) On the specificity of allene oxide cyclase. Lipids 34: 1005–1015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.