Abstract
Despite a central role in angiosperm reproduction, few gametophyte-specific genes and promoters have been isolated, particularly for the inaccessible female gametophyte (embryo sac). Using the Ds-based enhancer-detector line ET253, we have cloned an egg apparatus-specific enhancer (EASE) from Arabidopsis (Arabidopsis thaliana). The genomic region flanking the Ds insertion site was further analyzed by examining its capability to control gusA and GFP reporter gene expression in the embryo sac in a transgenic context. Through analysis of a 5′ and 3′ deletion series in transgenic Arabidopsis, the sequence responsible for egg apparatus-specific expression was delineated to 77 bp. Our data showed that this enhancer is unique in the Arabidopsis genome, is conserved among different accessions, and shows an unusual pattern of sequence variation. This EASE works independently of position and orientation in Arabidopsis but is probably not associated with any nearby gene, suggesting either that it acts over a large distance or that a cryptic element was detected. Embryo-specific ablation in Arabidopsis was achieved by transactivation of a diphtheria toxin gene under the control of the EASE. The potential application of the EASE element and similar control elements as part of an open-source biotechnology toolkit for apomixis is discussed.
In angiosperms, the mature ovule consists of tissues from both the diploid sporophyte and the haploid female gametophyte (megagametophyte or embryo sac; Grossniklaus and Schneitz, 1998; Yang and Sundaresan, 2000; Yadegari and Drews, 2004). It is the site of sexual reproduction, where the transitions between the haploid and diploid generations of the plant life cycle occur.
In a typical angiosperm, during sexual reproduction a single cell in the ovule becomes committed to the gametophytic pathway, differentiates into the megaspore mother cell, and undergoes meiosis. A tetrad of haploid spores is formed but only one of these spores survives. After three mitotic divisions, the embryo sac cellularizes to form an egg cell and two synergids, which form the egg apparatus at the micropylar pole, three antipodals at the chalazal pole, and a binucleate central cell. Fertilization of both the egg cell, which forms the embryo, and the central cell, which gives rise to the endosperm, initiates seed development. The coordinated development of embryo, endosperm, and the surrounding sporophytic tissue eventually produces the mature seed. In some circumstances, asexual reproduction (apomixis) can occur when the sexual life cycle is bypassed and seeds are produced that are genetically identical to their maternal parent (Vielle-Calzada et al., 1996; Grossniklaus, 2001; Koltunow and Grossniklaus, 2003). In apomixis, key steps in sexual reproduction are altered or omitted, such that a seed is formed without meiotic reduction and fertilization, producing a clone of the mother plant. This trait could be of great importance for agricultural applications (Grossniklaus et al., 1998a; Spillane et al., 2004), and, in particular, for hybrid rice (Oryza sativa) production (Jefferson, 1994; Jefferson and Nugroho, 1998).
So far, although significant progress has been made in genetic analyses of female gametophyte development, very little is known about the genes controlling the development and function of the female gametophyte (Grossniklaus and Schneitz, 1998; Yadegari and Drews, 2004). The reasons for the lack of such progress include the inaccessibility of the embryo, the brevity of the developmental stage, and the likely lethality of mutants affecting female gametophyte development.
Application of insertional mutagenesis approaches using heterologous maize (Zea mays) transposons or Agrobacterium-mediated T-DNA insertions has greatly facilitated work on the identification and isolation of novel plant genes that display a mutant phenotype (Pereira, 2000). In particular, marked insertional mutagens make possible the development of screens directed at the isolation of gametophytic mutations, due to the non-Mendelian segregation patterns arising from reduced gametophytic transmission (Moore et al., 1997; Howden et al., 1998; Page and Grossniklaus, 2002). On the other hand, gene trap and enhancer-detector transposons or T-DNA insertions in Arabidopsis (Arabidopsis thaliana; Sundaresan et al., 1995; Topping and Lindsey, 1995; Campisi et al., 1999) are valuable tools that permit identification of genes by their patterns of expression during development and provide direct access to highly specific promoters (Grossniklaus, 2001; Grossniklaus et al., 2003). Enhancer detectors typically rely on a Ds element (DsE) or T-DNA carrying a gusA reporter gene under the control of a minimal promoter. If the DsE or T-DNA inserts in the proximity of regulatory sequences (e.g. enhancers), the minimal promoter is influenced by these cis-regulatory elements and then directs gusA expression in a specific temporal and spatial pattern. This pattern could reflect the expression of a nearby gene controlled by the same regulatory element and thus may allow the identification of genes according to their expression patterns rather than the mutant phenotypes caused by their disruption. The enhancer-detector lines could therefore be extremely useful in detecting genes expressed in specific female gametophyte cells (Grossniklaus et al., 1995, 2003; Grossniklaus, 2001).
In Arabidopsis, large numbers of enhancer-detector lines have been generated by several laboratories over the past decade (Klimyuk et al., 1995; Sundaresan et al., 1995; Campisi et al., 1999; Parinov et al., 1999; Grossniklaus et al., 2003). Some of these lines that show β-glucuronidase (GUS) expression in the developing embryo and/or endosperm after fertilization have been successfully used in the study of the maternal control of early embryo and endosperm development (Vielle-Calzada et al., 2000). Among the enhancer trap lines examined by GUS staining, GUS expression in more than one tissue type seems to predominate (Sundaresan et al., 1995; Springer, 2000; R. Baskar and U. Grossniklaus, unpublished data). This is possibly due to the complex regulation of plant genes, where distinct combinations of regulatory sequences control gene expression in complex patterns during development (Sundaresan et al., 1995). As part of efforts toward the identification of promoters and genes associated with apomixis, this project was initiated to isolate regulatory elements and/or their associated genes involved in female gametophyte development by taking advantage of enhancer-detector line(s) with spatially restricted GUS expression patterns. Here we report the identification of an egg apparatus-specific enhancer (EASE) tagged by Ds in enhancer-detector line ET253, and show that the enhancer element does not regulate a nearby gene. However, we also demonstrate that this enhancer could be a very useful tool in studying and manipulating specific gene expression in the female gametophyte and early embryo development in Arabidopsis.
RESULTS
Egg Apparatus-Specific GUS Expression in the Enhancer-Detector Line ET253
To identify genes expressed in the female gametophyte, a collection of enhancer-detector lines was screened for GUS activity at various stages from cellularization to fertilization. The enhancer-detector lines had been generated in the accession Landsberg erecta (Ler) using the system developed by Sundaresan et al. (1995). Line ET253 shows specific GUS expression in the embryo sac but no GUS activity in other tissues of the plant that were tested. To determine the developmental stages that show GUS expression, some markers associated with approximate stages in floral development (Mansfield and Bowman, 1994) and/or female gametophyte development (Schneitz et al., 1995; Christensen et al., 1997) were used as references.
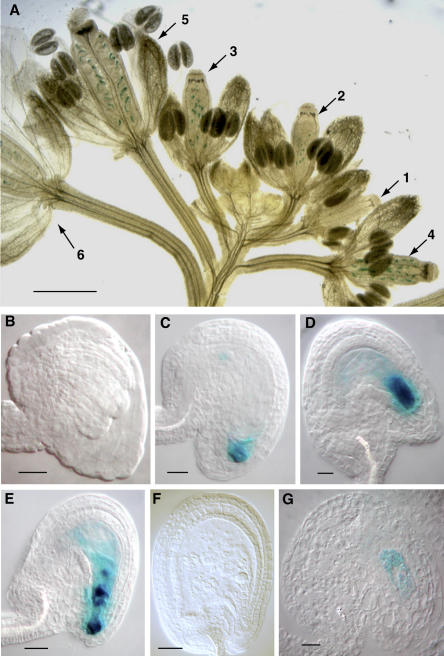
In ET253, GUS activity can be detected from flowers between stage 12 and stage 15. Figure 1A shows a GUS-stained inflorescence with flowers between stage 11 and stage 15 (flowers 1–6). Flower 1 is at floral stage 11, and does not show detectable GUS activity in the ovules (Fig. 1B). GUS activity starts to be detectable at floral stage mid 12 to late 12 (flower 2), where the majority of ovules are at stage 3-IV. The ovules at this stage have the activity restricted to the micropylar end (Fig. 1C), suggesting the GUS gene could be expressed in one or several of the micropylar nuclei. The clear boundary between the stained and nonstained areas in the embryo sac is due to the existence of the large central vacuole. After cellularization (flowers 3 and 4), the embryo sac elongates and the intensity of GUS staining increases while remaining mainly in the synergids and egg cell (egg apparatus; Fig. 1D). GUS activity is very high in the egg apparatus and diffusion of the blue product is often observed in the surrounding sporophytic tissues and the central cell of the female gametophyte. Occasionally, the entire embryo sac shows strong staining (data not shown), suggesting that expression of the reporter gene is activated before cellularization is complete. As this is only observed in a small proportion of the examined ovules (4 out of 75 = 5%), the GUS gene seems typically to be activated in the nuclei of the egg apparatus during cellularization and, most often, this process is completed by the time the reporter gene is fully active such that the product is specifically localized. Egg apparatus-specific staining was confirmed by sectioning of the ovule at this stage (Fig. 1G). It remains highly localized through floral stages 12 and 13. After fertilization (stage 14, flower 5), the zygote shows strong GUS activity (Fig. 1E) but decreasing GUS activity is seen at later stages (stage 15, flower 6) and no GUS expression can be observed in ovules after the globular embryo stage (Fig. 1F). Due to the stability of the GUS protein, the exact stage at which reporter gene expression stops cannot be determined. These observations suggest that GUS expression in ET253 is probably restricted to the late stages of female gametophyte development (starting at cellularization) and persists into early seed development.
Figure 1.
Egg apparatus-specific GUS expression of the enhancer-detector line ET253. A, An inflorescence with flowers at various floral stages from an ET253 plant was subjected to GUS staining. For photography, half of the sepals, petals, and stamens from each numbered flower were removed to reveal the gynoecium. The approximate floral stages for the numbered flowers are as follows: flower 1, stage 11; flower 2, stage mid 12; flower 3, stage late 12; flower 4, stage 13; flower 5, stage 14; and flower 6, stages 15 and 16. Bar = 1 mm. B to F, Optical sections of GUS stained and cleared whole-mount ovules at different stages. After GUS staining, ovules from flowers at different stages were cleared by the modified Hoyer's solution and observed under Nomarski optics. B, An ovule from flower 1 (ovule stages 3-I to 3-II). C, An ovule from flower 2 (ovule stage 3-IV). D, An ovule from flower 3 or 4 (ovule stages 3-V to 3-VI). E, An ovule from flower 5 (ovule stages 4-IV to 4-V). F, An immature seed with embryo at mid globular stage from flower 6 (stage after 4-VI). G, Longitudinal section (2 μm) of a GUS-stained ovule at around the same stage as in D. Bars = 10 μm (B–D and G); 20 μm (E); and 50 μm (F).
The Genomic Region Flanking the DsE Insertion Site in ET253
Since the enhancer-detector line ET253 has only one DsE insertion, which has been confirmed by Southern analysis (data not shown), we tried to identify EASE and/or any associated gene by attempting to amplify both the 5′ and 3′ region flanking the Ds element using thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995) and inverse PCR (Ochman et al., 1993). By using TAIL-PCR, a 170-bp 5′ DsE-flanking fragment was isolated and sequenced (data not shown). Enhancers can often work efficiently at quite a distance from the transcription start site of the gene they control from either upstream or downstream (Maniatis et al., 1987), therefore, we decided to clone longer flanking sequences from both upstream and downstream regions by inverse PCR using primers designed from the 170-bp sequence. We finally obtained 8.5 kb of 5′ and 3 kb of 3′ sequences flanking the DsE in ET253.
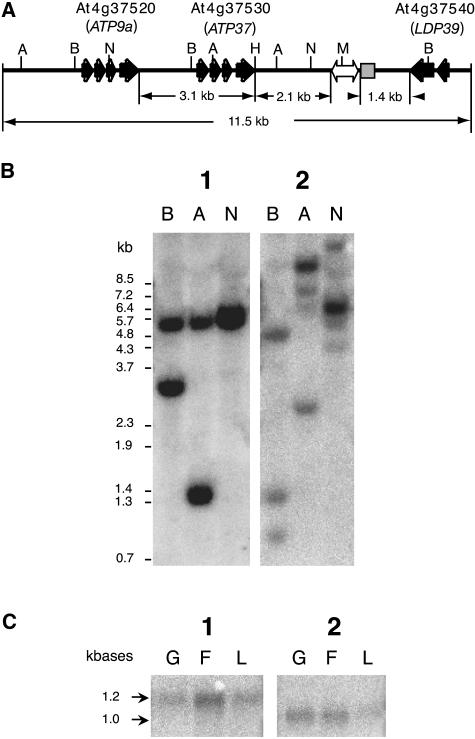
As seen in Figure 2A, sequence analysis using BLAST showed several genes near the DsE insertion site. The upstream region has two peroxidase genes (At4g37520 and At4g37530) identical to the ATP9a gene (GenBank accession no. X98856) and the ATP37 gene (GenBank accession no. AF469928), respectively. The downstream region is a gene (At4g37540) that encodes the Lateral Organ Boundaries (LOB)-domain protein LDP39. At4g37520 and At4g37530 are similarly arranged and share 92% identity. Interestingly, a 165-bp sequence directly downstream of the DsE insertion site (including the 8-bp DsE duplication sequence ATTAAGGC) in ET253 is homologous to many such sequences in all five chromosomes in different locations (more than 30 such sequences by BLAST search; data not shown). This small repetitive sequence is AT rich, having imperfect terminal inverted repeats, and does not seem to have any coding capacity, so it could be a new member of the miniature inverted-repeat transposable elements (MITEs) family (Wessler et al., 1995; Casacuberta et al., 1998). All these genes and/or the MITE-like transposable element were considered as possible candidates for the sequences responsible for the egg apparatus-specific GUS expression pattern.
Figure 2.
Analysis of the genomic region (on chromosome 4) covering the DsE insertion site in ET253. A, Schematic map of the 11.5-kb DsE-flanking region in ET253. The exons and the direction of the three genes are shown in black arrows. The inserted DsE is denoted as the white left-right arrow, and the MITE-like element is shown as a gray box. H, HindIII; M, MfeI. B, Southern-blot analysis of At4g37520, At4g37530, and At4g37540. Genomic DNA from Landsberg erecta (3 μg per lane) was digested with BglII (B), AccI (A), and NcoI plus NheI (N), respectively, and probed with either a 540-bp SpeI/DraI fragment covering the fourth exon of At4g37530 (blot 1) or a 255-bp fragment from the second exon of the LDP39 gene (blot 2). C, Northern-blot analysis of At4g37520, At4g37530, and At4g37540. mRNA was extracted from gynoecium (G), other floral tissues (petal, sepal, and stamen; F), and leaf (L), separated on 1% denaturing agarose gel (0.5 μg per lane), blotted onto nylon membrane, and probed with either the 540-bp SpeI/DraI At4g37530 fragment (blot 1) or the 255-bp At4g37540 fragment (blot 2).
The Two Upstream Peroxidase Genes At4g37520 and At4g37530 and the Downstream Gene At4g37540 Are Not Expressed in a Tissue-Specific Pattern
To determine whether there are more genes with similarity to the two peroxidase genes At4g37520 and At4g37530, Southern analysis was applied to determine their copy number. Using a 540-bp fragment from the fourth exon of At4g37530 as probe, a single band was detected (Fig. 2B, blot 1), showing that the region harboring At4g37520 and At4g37530 is present as a single copy in the genome. Northern blotting was also carried out to determine where the peroxidase genes are expressed. mRNA was prepared from gynoecia, other floral tissues (mainly petals and sepals), and leaves of Ler. Using the same 540-bp fragment from At4g37530 as probe, a band with the size of 1.2 kb, which is the expected size for the mRNA of At4g37520 and At4g37530, could be detected in all tissues tested (Fig. 2C, blot 1). The broad pattern of expression of At4g37520 and At4g37530 is confirmed by the wide range of tissues from which expressed sequence tags (ESTs) have been sequenced (http://www.ncbi.nlm.nih.gov/UniGene). For At4g37520, 66 EST sequences from roots, flowers, green siliques, and seeds have been reported, and for At4g37530, six ESTs from roots and a mixture of developmental stages, respectively. The broad expression patterns of the two peroxidase genes suggest that they are not likely to be associated with egg apparatus-specific expression.
Southern analysis was also carried out to determine the copy number of At4g37540 downstream of the DsE insertion site. A 255-bp fragment from the second exon of the gene was used as probe. As expected for the large LOB-domain gene family, several other family members could be detected with this probe (Fig. 2B, blot 2). Reprobing the northern blot with the same 255-bp probe detected a band of approximately 1 kb (Fig. 2C, blot 2). The size of the band is consistent with the size of the mRNA for the LDP39 gene. mRNA homologous to the LDP39 gene was detected mainly in floral tissues (gynoecium and other floral tissues) and to a lesser extent in leaf tissue, as compared to the two upstream peroxidase genes. The National Center for Biotechnology Information Unigene database reports 10 ESTs for LDP39 from roots and a mixture of developmental stages. These results suggest that the LDP39 gene is not likely to be associated with the egg apparatus-specific expression either.
Identification of a Sequence with the EASE Function
To define the genomic region responsible for the egg apparatus-specific expression, we divided the whole DsE-flanking sequence into several fragments and tested each fragment separately for EASE activity. The two putative promoter regions of At4g37520 and At4g37530 (1.28 kb and 1.2 kb, respectively) and the 2.1-kb region between At4g37530 and the DsE insertion site (Fig. 2A) were tested first. The two putative promoters were fused to the gusA gene in binary vector pCAMBIA1281Z (GenBank accession no. AF234294) to generate pWY-H80 (At4g37530 promoter) and pWY-H84 (At4g37520 promoter). To test the 2.1-kb upstream sequence flanking the DsE insertion site (Fig. 2A), a 2.8-kb segment between the HindIII site at the end of the fourth exon of the At4g37530 and the MfeI site in the gusA gene inside the DsE from ET253 was placed in pCAMBIA1201 (GenBank accession no. AF234293) using the same restriction sites to generate plasmid pWY-F68 (data not shown). In pWY-F68, the structure of the 2.8-kb fragment covering the GUS fusion is exactly the same as the original sequence in ET253. If the EASE were located in this 2.1-kb region, we would expect to obtain a similar pattern of GUS expression when pWY-F68 is introduced into Arabidopsis plants.
The three constructs were then transformed into Ler. In all of the eight pWY-H80-transformed T1 lines, no GUS expression was observed in any of the tissues examined including floral tissues and leaf (root was not included), while six out of 11 pWY-H84-transformed T1 lines showed GUS expression specifically in anthers (mainly in pollen grains; data not shown). However, 15 of 20 pWY-F68-transformed T1 lines revealed GUS expression in the egg apparatus. However, most pWY-F68-transformed lines also showed strong GUS staining in other tissues such as leaves, sepals, petals, and stamen. These results strongly suggest that the 2.1-kb sequence flanking the DsE possesses the EASE activity. The GUS staining observed in other tissues probably reflects the cis-acting effect of the enhancer from the cauliflower mosaic virus (CaMV) 35S promoter controlling the hygromycin resistance gene in the same T-DNA, despite the fact that the 35S promoter controlling the hygromycin resistance gene and the 35S minimal promoter used to detect genomic enhancers are placed in opposite orientation in this construct (W. Yang and R.A. Jefferson, unpublished data).
To narrow down the region responsible for the EASE function, 5′ deletion tests of the 2.1-kb fragment were carried out by using the SpeI and BlpI restriction sites within the fragment, respectively, to generate pWY-J26.2 containing 1.3 kb to the DsE insertion, and pWY-J47.3, containing 318 bp to the DsE (data not shown). The constructs were transformed into Ler. GUS staining of different tissues from the T1 transformants of both pWY-J26.2 and pWY-J47.3 demonstrated that the overall GUS expression pattern of the transformants carrying either of the deletion constructs was very similar to the pattern in plants transformed with pWY-F68. Six of 14 pWY-J26.2-transformed lines and five of eight pWY-J47.3-transformed lines showed clear GUS staining in the embryo sac (data not shown). This indicates that the 318-bp sequence directly flanking the DsE is sufficient to confer GUS expression in the egg apparatus. The background GUS staining pattern in other floral tissues was also similar to the pattern seen in pWY-F68-transformed plants.
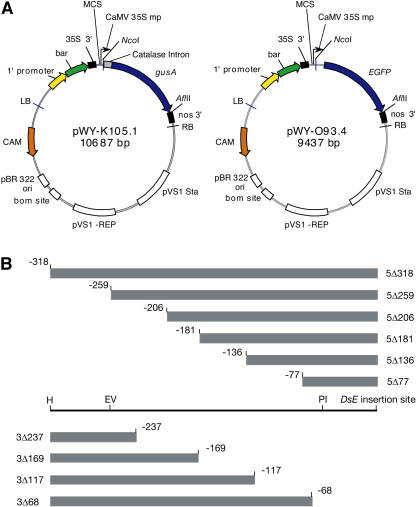
Background GUS expression in pWY-F68, pWY-J26.2, and pWY-J47.3, presumably caused by the cis-acting activity of the enhancers from the full CaMV 35S promoter within the T-DNA, complicates the examination of certain genomic DNA segments with tissue-specific enhancer function, when such segments are tested in pCAMBIA1201 or its derivatives (e.g. pCAMBIA1281Z, etc.). To increase the reliability of our analysis of the candidate tissue-specific enhancers, we constructed new transformation vectors without the CaMV 35S promoter. Figure 3A shows the maps of two new binary vectors pWY-K105.1 and pWY-O93.4. These two vectors use the bar gene under the control of the 1′ promoter as selectable marker in Arabidopsis (Mengiste et al., 1997). The only difference between pWY-K105.1 and pWY-O93.4 is that pWY-K105.1 uses gusA as reporter gene while pWY-O93.4 uses an enhanced green fluorescent protein (EGFP) gene (Yang et al., 1996) as reporter. These two vectors do not show any background expression of the reporter gene (GUS or GFP) when they are transformed into wild-type Arabidopsis (data not shown).
Figure 3.
5′ and 3′ deletion tests of the 318-bp DsE flanking sequence from ET253 for the EASE activity. A, Binary vectors designed for testing enhancer activity of unknown DNA sequence. The two vectors pWY-K105.1 and pWY-O93.4 use bar gene under the control of the 1′ promoter as selectable marker in plant and differ only in the reporter gene. The backbone of these two vectors is the same as pCAMBIA 1201. B, A series of 5′ and 3′ deletion fragments (designated as 5Δ and 3Δ, respectively, from the 318-bp DsE-flanking sequence with the EASE activity) was fused upstream of the CaMV 35S minimal promoter (−1 to −46) in the HindIII site in the multiple cloning site (MCS) in pWY-K105.1. The abbreviated names of the derived constructs are at either side and their full names are given in the text. The numbers above the bars indicate the distance from the DsE insertion site, which is designated as position 0. The Figure is not drawn to scale. H, HindIII; EV, EcoRV; PI, Psp1406I.
A series of further 5′ and 3′ deletions were then designed to dissect the 318-bp sequence to identify the core element responsible for the EASE activity. Figure 3B shows the derivative deletion constructs in detail. The 5′ deletion constructs were named as pWY-5Δ318 (positive control), pWY-5Δ259, pWY-5Δ206, pWY-5Δ181, and pWY-5Δ77, and the 3′ deletion constructs were named as pWY-3Δ237, pWY-3Δ169, pWY-3Δ117, and pWY-3Δ68. Transformation of these constructs into Ler showed that a 77-bp sequence (ccacgatgcaaatatatcgataacgttattaaaaaaagtaaccgcatgatatattctctttcgtatgatattaaggc, GenBank accession no. AX100536) is sufficient to direct the egg apparatus-specific expression of the GUS gene. The deletion of 68 nucleotides from the 3′ end of the 318-bp fragment abolished the EASE function completely (Table I), indicating that the EASE element is within the 77-bp fragment.
Table I.
Egg apparatus-specific activity of 5′ and 3′ deletions in driving GUS expression in Arabidopsis
Arabidopsis tissues including all floral tissues, stem, and leaf from the transformed T1 lines of the 5′ and 3′ deletion constructs were subjected to GUS staining. GUS expression was observed under a dissection microscope. The number of independent transformed T1 lines selected for each deletion construct varies from five to 12. EA, Egg apparatus; –, no GUS expression detected.
| Deletion Constructs
|
GUS Expressed Tissue in Transformed T1 Lines
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| pWY-5Δ318 | – | EA | EA | – | EA | – | EA | EA | – | EA | ||
| pWY-5Δ259 | EA | EA | – | – | EA | – | ||||||
| pWY-5Δ206 | – | EA | – | EA | EA | EA | – | |||||
| pWY-5Δ181 | EA | – | EA | EA | – | EA | EA | – | ||||
| pWY-5Δ136 | EA | EA | – | EA | – | |||||||
| pWY-5Δ77 | EA | EA | EA | – | EA | – | – | EA | EA | – | EA | |
| pWY-3Δ237 | – | – | – | – | – | – | – | – | – | – | – | |
| pWY-3Δ169 | – | – | – | – | – | – | – | – | – | |||
| pWY-3Δ117 | – | – | – | – | – | – | – | – | – | – | ||
| pWY-3Δ68 | – | – | – | – | – | – | – | – | – | – | – | – |
In an attempt to further define the EASE core sequence, the 77-bp sequence was scanned for EASE activity by five 30-bp fragments plus one 27-bp fragment with 20-bp overlapping sequence between the adjacent fragments. However, transformation of these six constructs into wild-type Arabidopsis did not result in any EASE activity (data not shown). This suggests that the EASE core sequence is either bigger than 30 bp or arranged in separate domains.
Basic Features of the EASE Element
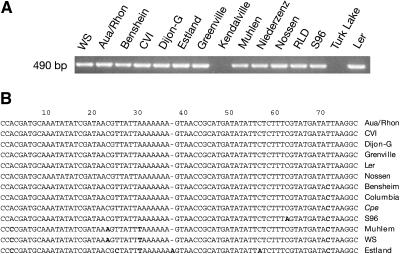
After the identification of the 77-bp sequence responsible for the EASE activity, we know that the EASE is located in a 3.5-kb region (Fig. 2A) where no obvious open reading frame can be found. In addition, it is also clear that the EASE is not part of the MITE-like element described earlier. This raises questions about the origin and the function of this enhancer element in the plant. A Southern blot using the 318-bp HindIII fragment from pWY-5Δ318 as a probe confirmed that the EASE is unique in the Arabidopsis genome (data not shown). To examine the distribution of this enhancer, we surveyed 15 different accessions of Arabidopsis by PCR amplification of the 318-bp EASE-containing sequence plus the 165-bp MITE-like sequence. A 490-bp fragment was PCR amplified from accessions Aua/Rhon, Benshein, Cape Verde Islands, Dijon-G, Estland, Greenville, Ler, Muhlem, Niederzenz, Nossen, RLD, S96, and Wassilewskija but not from accessions Kendalville and Turk Lake (Fig. 4A). The latter indicates that the MITE-like sequence inserted recently, as it is not present in all accessions tested. We sequenced these amplified fragments, aligned the 77-bp EASE sequences together with the EASE sequence from Columbia, and found that they were very conserved (Fig. 4B). Interestingly, the EASE is also present in Cardaminopsis petraca, a close relative of Arabidopsis (Fig. 4B).
Figure 4.
Distribution of the EASE among different accessions of Arabidopsis. A, PCR amplification of the region covering the EASE and the MITE-like element in different accessions. B, Alignment of the 77-bp sequence with EASE activity from different accessions. Sequence variation is shown in bold letters.
To determine whether there is a molecular signature indicative of the selective pressures acting on the EASE, we performed an analysis of nucleotide variation on the 490-bp fragment from the 13 accessions (Fig. 4A) using the DnaSP software (Rozas and Rozas, 1999). The level of nucleotide diversity was estimated as mean pairwise differences (π). The distribution pattern of nucleotide polymorphisms can provide information on the relative role of selection versus genetic drift, which can be evaluated using Tajima's test for selection (Tajima, 1989). In this fragment, although no significant value was obtained for the Tajima test statistic, D, an unusually high level of sequence variation was observed with π = 0.027, which is about 4 to 5 times higher than average. The pattern of nucleotide variation is an unusual one. The 210 bp of the MITE-like element show a π = 0.0070 ± 0.0014, which is close to mean for nuclear genes in Arabidopsis (Shepard and Purugganan, 2003). In contrast, the 77-bp EASE, and the 161-bp sequence flanking it at the 3′ end, have a much higher level of variation, with π = 0.0294 ± 0.0057 and π = 0.0333 ± 0.0029, respectively. This may be due to a high neutral mutation rate in this region or because of selection. As Tajima's test statistic did not provide significant levels for positive selection, we cannot distinguish between these possibilities. However, the presence of the EASE in different accessions eliminated the possibility that it is the result of an accession-specific genomic insertion, deletion, or rearrangement.
To test whether the EASE acts independently of orientation, as expected for an enhancer element, we constructed pWY-5Δ77A, which is the same as pWY-5Δ77 except that the 77-bp EASE sequence is placed in the opposite orientation. This construct was transformed into Ler and 73% of the transformants (11 out of 15) showed egg apparatus-specific GUS expression (data not shown). This result demonstrated that the EASE functions in an orientation-independent fashion.
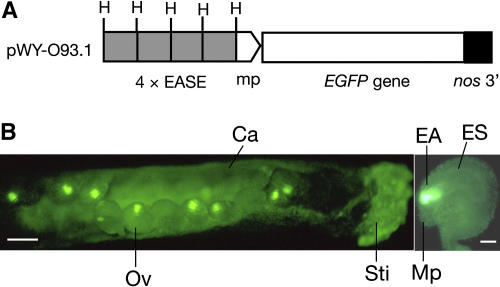
In parallel, we also examined the capability of the EASE to activate GFP gene expression. We inserted four tandem repeats of the 77-bp EASE sequence in to the HindIII site in pWY-O93.4 (Fig. 3A) to generate pWY-O93.1 (Fig. 5A) such that the EGFP gene is under the control of the 35S minimal promoter plus the EASE elements. After transforming Ler with pWY-O93.1, we obtained very strong and specific EGFP expression in the egg apparatus of most transformants (Fig. 5B; five of seven lines examined).
Figure 5.
Specific GFP gene expression under the control of the EASE. A, Schematic map of the EGFP expression construct in pWY-O93.1. The EGFP gene is under the control of the 35S minimal promoter (mp) plus four tandem repeats of the 77-bp EASE sequence (4 × EASE, in gray boxes). H, HindIII; nos 3′, nopaline synthase gene terminator. B, Specific EGFP expression in the egg apparatus of a pWY-O93.1-transformed Arabidopsis plant. The picture on the left shows a gynoecium with ovules expressing EGFP (bar = 100 μm), and the picture on the right displays the location of the expressed EGFP in the egg apparatus in an ovule (bar = 10 μm). Ca, Carpel; EA, egg apparatus; ES, embryo sac; Mp, micropyle; Ov, ovule; Sti, stigma.
Embryo-Specific Ablation under the Control of the EASE Element
We also tested the tissue specificity of the EASE by using a genetic ablation system based on cell-specific expression of the diphtheria toxin A-chain gene (DTA; Thorsness et al., 1993; Day et al., 1995; Nilsson et al., 1998). Since the introduction of the DTA gene directly through the female lineage is presumably lethal, we generated Arabidopsis lines expressing the hybrid Gal4-VP16 transcription activator (Sadowski et al., 1988) under the control of the EASE element plus minimal CaMV 35S promoter (Yang, 2003). Here we name these lines as EASE-Gal4 activator lines. We crossed these activator lines to an Arabidopsis upstream activating sequence (UAS)-DTA target line harboring the DTA gene controlled by an artificial promoter consisting of multiple copies of the UAS recognized by Gal4 and a minimal CaMV 35S promoter (Bougourd et al., 2000) and found that the DTA gene was transactivated specifically in the embryos.
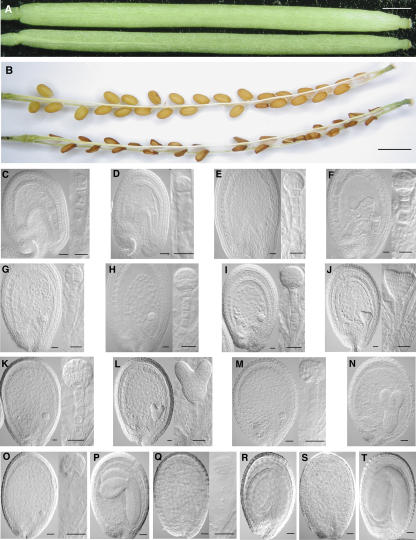
To carry out such ablation test, we crossed flowers from a plant of the target UAS-DTA line with pollen from either an EASE-Gal4 activator line (activator cross) or a wild-type plant (control cross). We observed that silique development following the activator cross and the control cross showed no difference (Fig. 6A). However, F1 seeds from the activator cross were all shriveled, whereas F1 seeds from the control cross were normal (Fig. 6B). Further microscopic observation of the ovule development in the F1 seeds at different stages reveals that, following pollination, the embryo development in the F1 seeds from the activator cross is normal from 1 d after pollination (DAP) to 4 DAP that covers the zygote formation (data not shown), single- or two-terminal cell embryo (Fig. 6C), dermatogen embryo (Fig. 6E), and early globular embryo (Fig. 6G) when compared to the control F1 seeds at same stages (Fig. 6, D, F, and H). The difference of embryo development between the activator cross and the control cross starts to show from 5 DAP. At this time, the embryos of the control F1 seeds have advanced to early heart stage (Fig. 6J) but the embryos of the F1 seeds from the activator cross stay at globular stage (Fig. 6I). From 6 DAP to 12 DAP, the control F1 seeds have their embryos grow from heart stage through to torpedo stage, walking-stick stage, and upturned-U stage toward maturity (Fig. 6, L, N, P, R, and T). In contrast, the F1 seeds from the activator cross go through an embryo degeneration process in the same period. In these seeds, the structure of the globular embryo is maintained up to 8 DAP (Fig. 6, K and M). Embryo degradation then becomes obvious, starting from the embryo proper and hypophyseal cells at 9 DAP (Fig. 6O) to the suspensor cells at 10 DAP (Fig. 6Q). At 12 DAP, the embryos in the F1 seeds from the activator cross are not visible any more. Meanwhile, the endosperm and the seed coat of the F1 seeds from the activator cross develop normally in the whole process as compared to the control F1 seeds. This clearly demonstrated that the DTA gene expression is restricted to the embryos of the F1 seeds defined by the EASE element.
Figure 6.
Embryo-specific ablation by transactivation of the DTA gene under the control of the EASE element. A, Normal-looking siliques produced by an UAS-DTA plant pollinated with either pollen from a wild-type plant (top), or pollen from an EASE-Gal4 activator line (bottom). B, Dissected siliques from the above crosses showing normal (top) and abortive (bottom) F1 seeds, respectively. C to T, Embryo development of the UAS-DTA line following the pollination with pollen from the activator line or a wild-type control plant. Seeds at different stages were dissected from their siliques, cleared in the modified Hoyer's solution (see “Materials and Methods”), and viewed with Nomarski optics. C, E, G, I, K, M, O, Q, and S are seeds from the activator cross at 2, 3, 4, 5, 6, 8, 9, 10, and 12 DAP, respectively. D, F, H, J, L, N, P, R, and T are seeds from the control cross at 2, 3, 4, 5, 6, 8, 9, 10, and 12 DAP, respectively. Close ups of embryos are shown in the insets. Scale bars = 1 mm (A and B); 20 μm (C–L); and 40 μm (M–T).
DISCUSSION
The Endogenous Role of the EASE in the Arabidopsis Genome Remains a Question
We have cloned and characterized an EASE from Arabidopsis using enhancer detection. The sequence conferring the EASE activity was finely mapped to a 77-bp fragment by a 5′ and 3′ deletion analysis. A fine mutation test (Ito et al., 1998) of the 77-bp sequence may be required to determine the core sequence of the EASE. However, our results show that the 77-bp sequence is highly efficient in controlling specific gene expression in the egg apparatus of Arabidopsis.
The sequence bearing the EASE is located at quite a distance from the nearby genes. Without using enhancer detection, it may never have been uncovered. Where does this enhancer originate from and what could be its functions in the genome? BLAST search revealed only three genes in this 11.5-kb region flanking the DsE in ET253: two peroxidase genes, At4g37520 and At4g37530, arranged as a tandem array, and the LDP39 gene (At4g37540). This is consistent with the general detected gene density on chromosome 4, which is 4.6 kb per gene (Arabidopsis Genome Initiative, 2000). As shown in Figure 2A, the DsE inserted in the middle of the region between At4g37530 and At4g37540. According to the orientation of these two genes, the DsE insertion is downstream of both genes. The limited space between these two genes, which is only 3.5 kb from stop codon to stop codon, and the lack of an unambiguous open reading frame, suggest that the 3.5-kb sequence where the DsE inserted is an intergenic region. These facts eliminate the possibility that the EASE is part of a promoter of a gene. The conservation of the EASE among different accessions (Fig. 4B) indicates that the location of this element is not the result of a recent genomic change in a particular accession, although the region shows an unusual distribution pattern of nucleotide diversity. Given its conservation, it is likely that the EASE has a function in the genome but works over a long distance and that the gene controlled by it has not been identified. Enhancer action in trans has been shown to be possible for Drosophila (Chen et al., 2002) and plant (Matzke et al., 2001); the EASE could also act in this fashion. Alternatively, the EASE may only represent a cryptic element that is not involved in the control of gene expression. Given the conservation, specificity, and strength of this element, the latter seems less likely.
One neighbor of EASE is a peroxidase gene. Plant peroxidases play a wide variety of functional roles related to defense, development, lignification, and hormonal signaling (Ostergaard et al., 1998). Some peroxidases are expressed constitutively (Lavid et al., 2001), whereas others are inducible in response to external stress, such as wounding (Kawaoka et al., 1994), pathogen infection (Mohan et al., 1993; Curtis et al., 1997), and salt stress (Botella et al., 1994). Although most peroxidases are expressed in a non-tissue-specific manner, organ- or tissue-specific peroxidases, such as root-specific (Wanapu and Shinmyo, 1996), stem-specific (Omann et al., 1994), and endosperm-specific (Rasmussen et al., 1991) peroxidases, have been reported. In Arabidopsis, more than 40 peroxidase genes are now known, but functional association is complicated due to a general lack of peroxidase substrate specificity (Ostergaard et al., 1998). In our case, since At4g37520 and At4g37530 are similarly arranged based on sequence information, it is difficult to postulate what their respective functions in the genome could be. Although results from RNA-blot analysis suggest that they are not expressed in a tissue-specific manner, it is possible that these two peroxidase genes are expressed in different tissues.
The other neighbor of EASE is LDP39, a gene belonging to a large family comprising several closely related genes as determined by Southern analysis (Fig. 2B). The LOB-domain proteins are characterized by the amino acid motif CX2CX6CX3C defined by its founding member LATERAL ORGAN BOUNDARIES (Shuai et al., 2002). The LDP family has genes distributed on all chromosomes, with the class II members LDP37 (At5g67420), LDP38 (At3g49940), and LDP40 (At1g67100) being most closely related to LDP39. Therefore, the Southern hybridization patterns shown in Figure 2B could represent these genes, with the similarity to LDP39 decreasing in the order of LDP38, LDP37, and LDP40 (data not shown). The mRNA of these genes can be detected in Arabidopsis seedlings 12 d after sowing (Iwakawa et al., 2002), indicating that they are mainly expressed during vegetative growth. Our northern analysis (Fig. 2C) also confirmed that the LDP genes are widely expressed in different vegetative tissues. However, the slight difference of the size between mRNA detected in floral tissues and mRNA detected in leaves also suggests that different LDP genes may be expressed in different tissues. The sizes of mRNA from LDP39, LDP38, and LDP37 are 987, 1,039, and 1,121 bases, respectively. We speculate that the mRNA detected from the floral tissues could be the transcript of the LDP39 gene, but the mRNA detected from the vegetative tissues could be the transcript of the LDP38 or LDP37 gene.
The EASE Is Capable of Driving Gene Expression in the Egg Apparatus and Early Seed Development
Although the endogenous function of the EASE in the Arabidopsis genome is still unknown, our experimental results show that its capability to control gene expression in the egg apparatus and early embryo is clear. Without the minimal promoter, the EASE itself is not functional (data not shown). However, when it is fused to the CaMV 35S minimal promoter, the EASE can drive gusA or GFP gene expression in a very specific and highly efficient manner.
The cell ablation test using the DTA toxin gene under indirect control of the EASE, however, shows that the cell ablation starts in the embryo at the globular stage. This seems to be contrary to the results from the reporter gene expression. If the transactivation of the DTA gene commenced right after fertilization, the cell ablation should have occurred at the zygote formation stage. Since the test was set by hybridization between the EASE-Gal4 activator line and the UAS-DTA line, the delay in cell ablation after cross could be due to delayed activation of the paternal genome during seed development as reported previously by Vielle-Calzada et al. (2000). In their study, they found that in Arabidopsis the paternally inherited allele could become active 3 to 4 d after fertilization. In our case, the late arrest of the embryo development in the activator-crossed F1 seeds is consistent with a predominantly maternal control of seed development and, because of which, transactivation systems only become fully functional at the globular stage (Baroux et al., 2001). In either way crosses are made between the EASE-Gal4 activator line and the UAS-DTA line, the embryo development is arrested at globular stage around 5 d after pollination, which is close to the delayed period of the activation of paternal genome identified by Vielle-Calzada et al. (2000). Once the paternal genome is activated, the DTA gene is then transactivated, resulting in cell death in the embryo. In this way, we specifically ablated the embryo in the hybrid (F1) seeds while the integument and endosperm development remained normal. However, the death of the DTA-expressing F1 seeds is the limitation for this system, which prevents the further analysis of the crossed population. A better system may be needed for examining the timing of the EASE action.
Nevertheless, these experiments clearly demonstrated that the EASE could be used as a tool to study and manipulate gene expression in the female gametophyte and during early embryo development.
Potential Application of the EASE Element in Biotechnology
Regulation of transcription relies at least on two primary DNA components, promoters and enhancers. Enhancers normally consist of sequence-specific transcription factor binding sites that function distal to the transcription initial site in promoters from either an upstream or downstream position (Maniatis et al., 1987; Majumder et al., 1997). At present, little is known about regulatory factors that act during megagametogenesis and the earliest stages of plant embryogenesis. The specificity of the EASE described here is probably associated directly with some particular protein factors, especially if in trans activation is involved. If this is the case, we may be able to design strategies to isolate the EASE-associated protein factor(s) using the EASE as bait in a one-hybrid screen. If this can be achieved, we may be able to discover new genes playing a role in megagametogenesis.
Producing viable embryos in the absence of fertilization (parthenogenesis) is one of the elements of apomixis that must be engineered in order to reach functional apomixis through biotechnology (Jefferson and Bicknell, 1996; Spillane et al., 2004). Recently, several transcription factor genes have been identified which, when misexpressed, lead to somatic embryo formation in seedlings. If this activity could be directed to the egg cell, induction of parthenogenetic development may be possible. To make normal seeds without fertilization, the EASE could be a good candidate for controlling the expression of embryo-inducing genes, because it has no activity at other stages of development and therefore would be unlikely to affect the vegetative development. This strategy could be further combined in the fis class mutants leading to fertilization-independent endosperm development (Grossniklaus et al., 1998b; Luo et al., 1999; Ohad et al., 1999; Köhler et al., 2003; Guitton et al., 2004), for viable seed formation without pollination. However, given that endosperm development is only partial in those mutants, additional issues will have to be addressed and combined for the induction of apomixis.
The use of the EASE sequence and vectors containing it are being made available to the international community as part of an integrated toolkit to develop apomixis in a “protected technology commons.” The EASE element and other components of this technology are accessible under terms consonant with the Biological Open Source (http://www.bios.net) license (Broothaerts et al., 2005), characterized by having no restrictions other than covenants for sharing of improvements, relevant safety information and regulatory data, and for preserving the opportunity for others to freely improve and use the technology.
MATERIALS AND METHODS
PCR Techniques
TAIL-PCR was performed as described by Liu et al. (1995). Nested Ds primers, specific to the 5′ and 3′ ends of the Ds element, were described by Grossniklaus et al. (1998b). Inverse PCR was carried out according to Ochman et al. (1993) by using the GeneAmp XL PCR kit (PE Applied Biosystems).
The two primers ET-TL13-1 (5′-GCTTAGCCTAATATCACAAA-3′) and ET-BP50 (5′-GGCTGTGAATGCTAACCA-3′) were used to amplify the 490-bp fragment covering the 318-bp sequence with EASE activity and the 165-bp MITE-like sequence from genomic DNA prepared from different accessions of Arabidopsis (Arabidopsis thaliana).
Construction of Deletion Constructs
Initial deletions were made using SpeI (1.3 kb from the DsE insertion site) and BlpI (318 bp from the DsE insertion site) digestions of pWY-F68, which were either digested with XbaI/SpeI and self-ligated to generate pWY-J26.2, or digested with HindIII/BlpI, blunt ended by T4 DNA polymerase, and self-ligated to form pWY-J47.3.
For further 5′ deletions of the 318-bp fragment with EASE activity, primers were designed for PCR amplification of a set of five deletion fragments with HindIII site added to both ends for subsequent cloning. Four 3′ deletion fragments were generated in the same fashion. The PCR-amplified deletion fragments were then digested with HindIII and cloned into the HindIII site upstream of the 35S minimal promoter in pWY-K105.1. Orientation of each cloned deletion fragment in pWYK105.1 was determined first by either HindIII/EcoRV digestion for the 3′ deletions or HindIII/Psp1406I digestion for the 5′ deletions, and finally by sequencing.
Transformation of Arabidopsis
Arabidopsis transformation was mainly based on a simplified dip method (Clough and Bent, 1998). For selection of transformants, seeds were selected either on selection medium for hygromycin resistance (Clough and Bent, 1998) with the hygromycin concentration at 30 μg mL−1 (Ye et al., 1999), or on soil for the herbicide Basta resistance (Bechtold et al., 1993) with the Basta concentration at 100 mg L−1 (Mengiste et al., 1997).
Southern and Northern Analyses
Genomic DNA from Arabidopsis was prepared using the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). Total RNA was extracted from gynoecium, other floral tissues (sepal, petal, and stamen), and leaf, using TRIzol reagent (Life Technologies). mRNA was prepared from total RNA using Dynabeads oligo(dT)25 with the Dynabeads mRNA purification kit (Dynal A.S.).
Southern and northern hybridizations were performed essentially according to Sambrook et al. (1989). Probes were prepared by labeling with [32P]dATP using GIGAPRIME DNA labeling kit (Bresatec). Autoradiographic image was taken on Bio-Rad GS-250 Molecular Imager after 4 to 24 h exposure using the Molecular Analyst software version 1.4 (with resolution set at 200 μm).
Histochemical GUS Expression Assay, GFP Detection, and Microscopic Work
Arabidopsis tissues were examined for GUS activity according to the procedure described by Jefferson (1987) with some modification. The samples were immersed in GUS-staining solution (50 mm sodium phosphate, pH 7.0, 10 mm EDTA, 0.1% (v/v) Triton X-100, 2 mm potassium ferrocyanide, 2 mm potassium ferricyanide, 100 μg/mL chloramphenicol, 0.02% (w/v) sodium azide, 1 mg mL−1 X-Gluc), vacuum infiltrated for 5 to 10 min, and incubated at 37°C for overnight. The GUS-stained tissues were then cleared in either a clearing solution (20% [v/v] lactic acid, 20% [v/v] glycerol, 1× phosphate-buffered saline; Vielle-Calzada et al., 2000) for more than 2 h or a modified Hoyer's solution (7.5 g of gum arabic, 100 g of chloral hydrate, 5 mL of glycerin, and 30 mL of water) for at least 5 h. The cleared tissues were observed directly using a Zeiss Axioskop or Leica DMR microscope. Images were recorded by a video/RGB camera attached to Zeiss Axioskop or a Nikon Coolpix 900 digital camera attached to Leica DMR. Cleared whole-mount ovules were observed using differential interference contrast (Nomarski) optics.
For histochemical localization of GUS activity with high resolution, the GUS-stained tissues were fixed in 3% (v/v) glutaraldehyde, embedded in LR White resin, and sectioned on a Reichert Ultracut microtome by C. Miller in the Microscopy Center of the Commonwealth Scientific and Industrial Research Organization. Sections (2 μm) were examined under a Leica DMR microscope and images were taken as mentioned.
Flowers of Arabidopsis with EGFP expression in ovules were collected and placed on a slide for dissection. To view ovules in a whole gyneocium (pistil), a gyneocium was removed from a flower and opened to reveal ovules using a hypodermic needle. Each opened gyneocium was then transferred onto a plate with germination medium. EGFP was examined under a Leica MZ FL III fluorescence stereomicroscope with 470/40 nm excitation filter and 525/50 nm barrier filter. To view individual ovules with higher magnification, ovules were cut out from a carpel with two needles, transferred onto a slide with a drop of 1× phosphate-buffered saline, and then covered with a coverslip. EGFP expression in ovules was monitored with a Leica DMR microscope equipped with the same set of filters for GFP plant fluorescence. Images were taken either on a Nikon Coolpix 900 digital camera or using Fuji chrome 400F film.
Sequence data from this article can be found in the GenBank data library under accession number AX100536.
Acknowledgments
We thank Drs. Andrzej Kilian and Paul Keese for their stimulating discussions on this work and Dr. Marie Connett-Porceddu for critical reading of the manuscript. We are grateful to Professor Michael Purugganan for discussions and comments on nucleotide variation. We also thank Professor Murray Badger and Ms. Prue Kell for help in Arabidopsis transformation, and Dr. Jim Haseloff for providing the UAS-DTA line.
This work was supported in part by the Rockefeller Foundation. W.Y. was supported by a Ph.D. fellowship from the Rockefeller Foundation. U.G. acknowledges the support of the Cold Spring Harbor Laboratory President's Council, the European Molecular Biology Organization, and the Human Frontiers Science Program.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Wei Yang (wei@cambia.org) and Richard A. Jefferson (r.jefferson@cambia.org).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068262.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baroux C, Blanvillain R, Gallois P (2001) Paternally inherited transgenes are down-regulated but retain low activity during early embryogenesis in Arabidopsis. FEBS Lett 509: 11–16 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199 [Google Scholar]
- Botella MA, Quesada MA, Kononowicz AK, Bressan RA, Pliego F, Hasegawa PM, Valpuesta V (1994) Characterization and in situ localization of a salt-induced tomato peroxidase mRNA. Plant Mol Biol 25: 105–114 [DOI] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J (2000) An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J 24: 543–550 [DOI] [PubMed] [Google Scholar]
- Broothaerts W, Mitchell HJ, Weir B, Kaines S, Smith LMA, Yang W, Mayer JE, Roa-Rodriguez C, Jefferson RA (2005) Gene transfer to plants by diverse species of bacteria. Nature 455: 629–633 [DOI] [PubMed] [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T (1999) Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J 17: 699–707 [DOI] [PubMed] [Google Scholar]
- Casacuberta E, Casacuberta JM, Puigdomenech P, Monfort A (1998) Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterisation of the Emigrant family of elements. Plant J 16: 79–85 [DOI] [PubMed] [Google Scholar]
- Chen JL, Huisinga KL, Viering MM, Ou SA, Wu CT, Geyer PK (2002) Enhancer action in trans is permitted throughout the Drosophila genome. Proc Natl Acad Sci USA 99: 3723–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen CA, King EJ, Jordan JR, Drews GN (1997) Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod 10: 49–64 [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Rae AL, Rusu AG, Harrison SJ, Manners JM (1997) A peroxidase gene promoter induced by phytopathogens and methyl jasmonate in transgenic plants. Mol Plant Microbe Interact 10: 326–338 [DOI] [PubMed] [Google Scholar]
- Day CD, Galgoci BF, Irish VF (1995) Genetic ablation of petal and stamen primordia to elucidate cell interactions during floral development. Development 121: 2887–2895 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U (2001) From sexuality to apomixis: molecular and genetic approaches. In Y Savidan, J Carman, T Dresselhaus, eds, Advances in Apomixis Research. International Maize and Wheat Improvement Center (CIMMYT) Press, Mexico, pp 168–211
- Grossniklaus U, Koltunow A, van Lookeren Campagne M (1998. a) A bright future for apomixis. Trends Plant Sci 3: 415–416 [Google Scholar]
- Grossniklaus U, Moore J, Gagliano W (1995) Analysis of Arabidopsis ovule development and megagametogenesis using enhancer detection. In Signaling in Plant Development. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 138
- Grossniklaus U, Moore JM, Brukhin V, Gheyselinck J, Baskar R, Vielle-Calzada JP, Baroux C, Page DR, Spillane C (2003) Engineering of apomixis in crop plants: what can we learn from sexual model systems. In I Vasil, ed, Plant Biotechnology 2002 and Beyond. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 309–314
- Grossniklaus U, Schneitz K (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9: 227–238 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB (1998. b) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of Fertilisation Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131: 2971–2981 [DOI] [PubMed] [Google Scholar]
- Howden R, Park SK, Moore JM, Orme J, Grossniklaus U, Twell D (1998) Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Iwase M, Kodama H, Lavisse P, Komamine A, Nishihama R, Machida Y, Watanabe A (1998) A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Seminarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe Y, Soma T, Ikezaki M, Machida C, et al (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jefferson RA (1994) Apomixis: a social revolution for agriculture. Biotechnology and Development Monitor 19: 14–16
- Jefferson RA, Bicknell R (1996) The potential impacts of apomixis: a molecular genetics approach. In BWS Sobral, ed, The Impact of Plant Molecular Genetics. Birkhäuser, Boston, pp 87–101
- Jefferson RA, Nugroho S (1998) Molecular strategies for hybrid rice: male sterility and apomixis. In SS Virmani, EA Siddiq, K Muralidharan, eds, Advances in Hybrid Rice Technology. International Rice Research Institute, Los Baños, The Philippines, pp 213–234
- Kawaoka A, Kawamoto T, Sekine M, Yoshida K, Takano M, Shinmyo A (1994) A cis-acting element and a trans-acting factor involved in the wound-induced expression of a horseradish peroxidase gene. Plant J 6: 87–97 [DOI] [PubMed] [Google Scholar]
- Klimyuk VI, Nussaume L, Harrison K, Jones JD (1995) Novel GUS expression patterns following transposition of an enhancer trap Ds element in Arabidopsis. Mol Gen Genet 249: 357–365 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003) Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Grossniklaus U (2003) Apomixis: a developmental perspective. Annu Rev Plant Biol 54: 547–574 [DOI] [PubMed] [Google Scholar]
- Lavid N, Schwartz A, Yarden O, Tel-Or E (2001) The involvement of polyphenols and peroxidase activities in heavy-metal accumulation by epidermal glands of the waterlily (Nymphaeaceae). Planta 212: 323–331 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Zhao Z, Kaneko K, DePamphilis ML (1997) Developmental acquisition of enhancer function requires a unique coactivator activity. EMBO J 16: 1721–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Goodbourn S, Fischer JA (1987) Regulation of inducible and tissue-specific gene expression. Science 236: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Bowman JL (1994) Ovules: introduction. In JL Bowman, ed, Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York, pp 299–301
- Matzke M, Mette MF, Jakowitsch J, Kanno T, Moscone EA, van der Winden J, Matzke AJ (2001) A test for transvection in plants: DNA pairing may lead to trans-activation or silencing of complex heteroalleles in tobacco. Genetics 158: 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Amedeo P, Paszkowski J (1997) High-efficiency transformation of Arabidopsis thaliana with a selectable marker gene regulated by the T-DNA 1′ promoter. Plant J 12: 945–948 [DOI] [PubMed] [Google Scholar]
- Mohan R, Bajar AM, Kolattukudy PE (1993) Induction of a tomato anionic peroxidase gene (tap1) by wounding in transgenic tobacco and activation of tap1/GUS and tap2/GUS chimeric gene fusions in transgenic tobacco by wounding and pathogen attack. Plant Mol Biol 21: 341–354 [DOI] [PubMed] [Google Scholar]
- Moore JM, Vielle-Calzada JP, Gagliano W, Grossniklaus U (1997) Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harb Symp Quant Biol 62: 35–47 [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Wu E, Wolfe DS, Weigel D (1998) Genetic ablation of flowers in transgenic Arabidopsis. Plant J 15: 799–804 [DOI] [PubMed] [Google Scholar]
- Ochman H, Ayala FJ, Hartl DL (1993) Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol 218: 309–321 [DOI] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL (1999) Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omann F, Beaulieu N, Tyson H (1994) cDNA sequence and tissue-specific expression of an anionic flax peroxidase. Genome 37: 137–147 [DOI] [PubMed] [Google Scholar]
- Ostergaard L, Pedersen AG, Jespersen HM, Brunak S, Welinder KG (1998) Computational analyses and annotations of the Arabidopsis peroxidase gene family. FEBS Lett 433: 98–102 [DOI] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet 3: 124–136 [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Y, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A (2000) A transgenic perspective on plant functional genomics. Transgenic Res 9: 245–260 [DOI] [PubMed] [Google Scholar]
- Rasmussen SK, Welinder KG, Hejgaard J (1991) cDNA cloning, characterization and expression of an endosperm-specific barley peroxidase. Plant Mol Biol 16: 317–327 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M (1998) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schneitz K, Hulskamp M, Pruitt RE (1995) Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J 7: 731–749 [Google Scholar]
- Shepard KA, Purugganan MD (2003) Molecular population genetics of the Arabidopsis CLAVATA2 region: the genomic scale of variation and selection in a selfing species. Genetics 163: 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Pena CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C, Curtis MD, Grossniklaus U (2004) Apomixis technology development: virgin births in farmers' fields? Nat Biotechnol 22: 687–691 [DOI] [PubMed] [Google Scholar]
- Springer PS (2000) Gene traps: tools for plant development and genomics. Plant Cell 12: 1007–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness MK, Kandasamy MK, Nasrallah ME, Nasrallah JB (1993) Genetic ablation of floral cells in Arabidopsis. Plant Cell 5: 253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping JF, Lindsey K (1995) Insertional mutagenesis and promoter trapping in plants for the isolation of genes and the study of development. Transgenic Res 4: 291–305 [Google Scholar]
- Vielle-Calzada JP, Baskar R, Grossniklaus U (2000) Delayed activation of the paternal genome during seed development. Nature 404: 91–94 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Crane CF, Stelly DM (1996) Apomixis: the asexual revolution. Science 274: 1322–1323 [Google Scholar]
- Wanapu C, Shinmyo A (1996) Cis-regulatory elements of the peroxidase gene in Arabidopsis thaliana involved in root-specific expression and responsiveness to high-salt stress. Ann N Y Acad Sci 782: 107–114 [DOI] [PubMed] [Google Scholar]
- Wessler SR, Bureau TE, White SE (1995) LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev 5: 814–821 [DOI] [PubMed] [Google Scholar]
- Yadegari R, Drews GN (2004) Female gametophyte development. Plant Cell (Suppl) 16: S133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Cheng L, Kain SR (1996) Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res 24: 4592–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W (2003) Molecular characterization of a megagametophyte-specific enhancer in Arabidopsis thaliana. PhD thesis. Australian National University, Canberra, Australia
- Yang WC, Sundaresan V (2000) Genetics of gametophyte biogenesis in Arabidopsis. Curr Opin Plant Biol 3: 53–57 [DOI] [PubMed] [Google Scholar]
- Ye GN, Stone D, Pang SZ, Creely W, Gonzalez K, Hinchee M (1999) Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J 19: 249–257 [DOI] [PubMed] [Google Scholar]