Abstract
We have previously shown that the novel ATP-dependent chromatin-remodeling complex WINAC is required for the ligand-bound vitamin D receptor (VDR)-mediated transrepression of the 25(OH)D3 1α-hydroxylase (1α(OH)ase) gene. However, the molecular basis for VDR promoter association, which does not involve its binding to specific DNA sequences, remains unclear. To address this issue, we investigated the function of WSTF in terms of the association between WINAC and chromatin for ligand-induced transrepression by VDR. Results of in vitro experiments using chromatin templates showed that the association of unliganded VDR with the promoter required physical interactions between WSTF and both VDR and acetylated histones prior to VDR association with chromatin. The acetylated histone-interacting region of WSTF was mapped to the bromodomain, and a WSTF mutant lacking the bromodomain served as a dominant-negative mutant in terms of ligand-induced transrepression of the 1α(OH)ase gene. Thus, our findings indicate that WINAC associates with chromatin through a physical interaction between the WSTF bromodomain and acetylated his tones, which appears to be indispensable for VDR/promoter association for ligand-induced transrepression of 1α(OH)ase gene expression.
Keywords: acetylated histone, transrepression, VDIR, VDR, WSTF
Introduction
Lipophilic ligands, such as fat-soluble vitamins A and D, as well as thyroid/steroid hormones, are thought to exert their physiological effects through transcriptional control of target genes via cognate nuclear receptors (NRs) (Mangelsdorf et al, 1995). NRs form a gene superfamily, and they act as ligand-inducible activators. A number of coregulator complexes that support ligand-dependent transcription control have been identified, and these complexes can be classified into three categories according to function (Glass and Rosenfeld, 2000). The first coregulator complex class regulates transcriptional control directly, through a physical interaction with general transcription factors and RNA polymerase II (Rachez et al, 1998; Gu et al, 1999). Members of the second coregulator complex class modify histone tails covalently, for example by acetylation, in promoter nucleosomal arrays (Onate et al, 1995; Kamei et al, 1996; Heinzel et al, 1997; Yanagisawa et al, 2002). The major function of the last class of complexes is chromatin remodeling, which involves the ATP-dependent dynamic remodeling of chromatin structure (Ito et al, 1997; Fyodorov and Kadonaga, 2001; Lemon et al, 2001; Narlikar et al, 2002; Kitagawa et al, 2003). Chromatin-remodeling complexes utilize energy from ATP hydrolysis to rearrange nucleosomal arrays in a non-covalent manner. As chromosomal DNA is generally packed as nucleosomal arrays, chromatin-remodeling complexes are thought to render specific promoter regions accessible to other coregulator complex classes and sequence-specific regulators.
Recently, we identified a novel, multifunctional, ATP-dependent chromatin-remodeling complex, designated WINAC, which consists of 13 subunits (Kitagawa et al, 2003). It contains SWI/SNF chromatin-remodeling complex components and DNA replication-related factors. Vitamin D receptor (VDR) interacts with WINAC in a ligand-independent manner through the Williams syndrome transcription factor (WSTF). WSTF belongs to bromodomain adjacent to a zinc-finger motif (BAZ) protein family. Members of this family harbor both a PHD finger and a bromodomain in their C-terminal domain (Jones et al, 2000). As bromodomains have been recently shown to bind acetylated histones, it is possible that WSTF serves as an adaptor protein for acetylated histones, facilitating the association between WINAC and chromatin (Dhalluin et al, 1999; Winston and Allis, 1999; Jacobson et al, 2000; Hassan et al, 2002).
Expression of the 25(OH)D3 1α-hydroxylase (1α(OH)ase) gene, a key enzyme in vitamin D biosynthesis, is negatively regulated by vitamin D (Takeyama et al, 1997; Yoshizawa et al, 1997; Murayama et al, 1998). We recently reported that a bHLH-type activator, VDR-interacting repressor (VDIR), directly binds to the negative vitamin D response element (1αnVDRE) in the human 1α(OH)ase gene promoter, thus activating transcription (Murayama et al, 2004). However, ligand-induced association between VDR and VDIR results in ligand-induced transrepression of 1α(OH)ase gene expression. This occurs through the switching of coregulator complexes from histone acetyltransferase (HAT) coactivator complexes to histone deacetylase (HDAC) corepressors upon VDIR binding to 1αnVDRE (Murayama et al, 2004). However, the molecular basis of VDR targeting to 1αnVDRE in the chromatinized 1α(OH)ase gene promoter remained unclear.
In the present study, we demonstrate, using chromatin templates in vitro, that both ligand-free VDR and WINAC avidly associate with the 1α(OH)ase gene promoter when the promoter region is reconstituted as a nucleosomal array. The WSTF bromodomain in WINAC appears to serve as a chromatin-targeting module that escorts ligand-free VDR to the promoter via a physical interaction with acetylated histones. Thus, our findings show that WINAC associates with chromatin through a physical interaction between the WSTF bromodomain and acetylated histones. This apparently contributes to the association between unliganded VDR and the promoter, resulting in a ligand-induced transrepression of human 1α(OH)ase gene expression.
Results
Involvement of WSTF in ligand-induced transrepression by VDR/VDIR of the 1α(OH)ase gene promoter
It has previously been shown that VDIR activates transcription via specific binding to 1αnVDRE in the promoter (Murayama et al, 2004). The association of liganded VDR with VDIR then inhibits VDIR transactivation function through recruitment of an HDAC corepressor complex, resulting in ligand-induced transrepression (Murayama et al, 2004). Although WSTF was previously shown to be involved in 1α(OH)ase gene regulation (Kitagawa et al, 2003; Kato et al, 2004), the function of WSTF, with respect to ligand-induced VDR/VDIR transrepression, remained unclear.
We employed a transient expression assay using MCF7 cells, which express the 1α(OH)ase gene endogenously, and a luciferase reporter gene plasmid containing two consensus 1αnVDRE sequences recognized by VDIR. These sequences confer negative responsiveness to 1α,25(OH)2D3 in gene repression. 1α,25(OH)2D3 transrepressed transcription of the reporter gene, and this repression was enhanced in the presence of VDR/RXR expression (Figure 1A).
Figure 1.
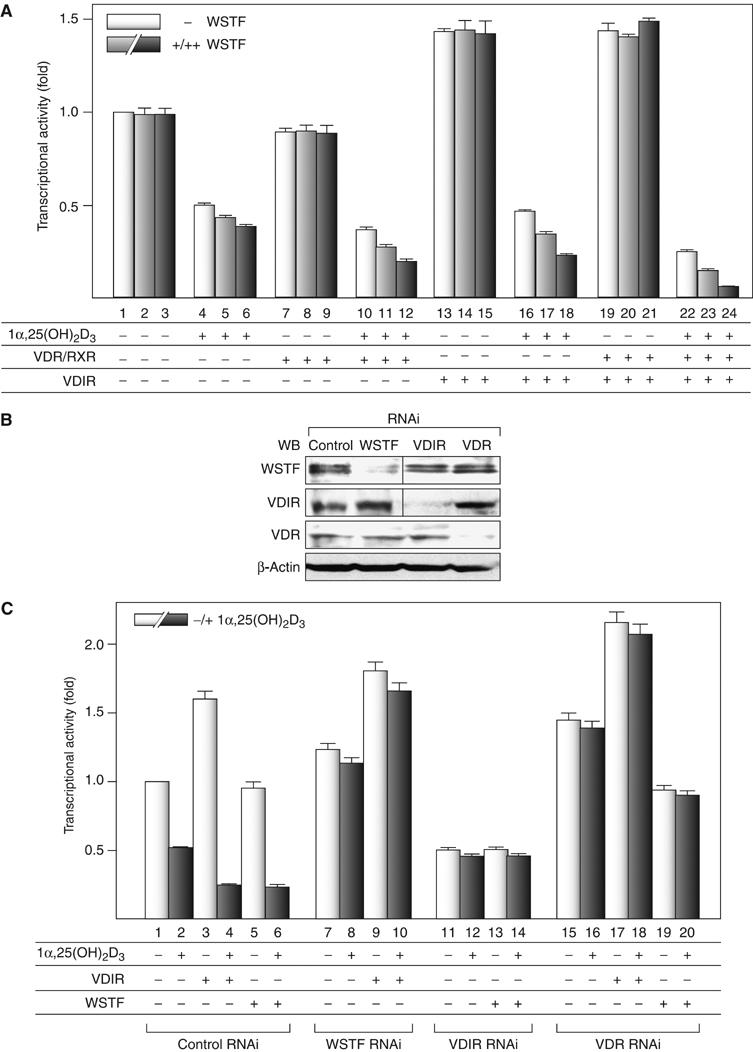
WSTF enhances 1α,25(OH)2D3-induced transrepression of 1α(OH)ase gene expression, but not transactivation by VDIR. (A) Coordinate transrepression of the 1α(OH)ase gene by VDR, WSTF and VDIR in a luciferase reporter assay. MCF7 cells were transfected with a luciferase reporter gene expression vector containing 1αnVDRE (× 2) driven by a TATA promoter (0.4 μg), pML-CMV (2 ng), and either pSG5-rat VDR and pSG5-rat RXRα (0.2 μg each), pcDNA3-VDIR (0.1 μg), pcDNA3-WSTF (0.1[+], 0.3[++] μg), or combinations thereof in the presence or absence of 1α,25(OH)2D3 (10−8 M) (Kitagawa et al, 2003; Murayama et al, 2004). Bars in each graph show the fold change in luciferase activity relative to basal activity obtained in the absence of ligand. All values are means±s.d. for at least three independent experiments. (B) Gene-specific knockdown of WSTF, VDIR or VDR by RNAi was confirmed by Western blots using anti-WSTF, -VDIR, -VDR and β-actin (as a control). Whole-cell extracts were prepared from MCF7 cells transfected with 0.3 μg of double-stranded siRNA and further cultured for 48 h. (C) Effect of gene-specific knockdown of endogenous factors, WSTF, VDIR and VDR on 1α(OH)ase gene expression in a luciferase reporter assay. MCF7 cells were transfected with 0.3 μg of the indicated siRNAs, 48 h after the transfection luciferase reporter gene containing 1α(OH)ase native promoter was transfected again into the cells. Luciferase activity was assessed after 12 h culture in the presence or absence of 1α,25(OH)2D3 (10−8 M).
We further assessed whether endogenous VDR, VDIR and WSTF were responsible for ligand-induced negative responsiveness of the 1α(OH)ase gene, using RNAi in a transient expression assay, in which a reporter gene was driven by the native promoter. The analysis confirmed that RNAi downregulated expression of the target endogenous factors without modulating the expression levels of the other factors (Figure 1B). We confirmed that WSTF-RNAi and VDIR-RNAi are able to abrogate the transrepression function of VDIR and WSTF, respectively, and VDR-RNAi eliminates both of them (Figure 1C). These results show that it is likely that WSTF mediates the ligand-induced transrepression of the 1α(OH)ase gene, together with VDR and VDIR.
WSTF associates with VDIR in a ligand-dependent manner
Based on our findings that WSTF appears to play a role in ligand-induced VDR/VDIR transrepression, we further examined the complex formed by these three factors in MCF-7 cells using an immunoprecipitation assay. As unliganded VDR was reported to associate with NCoR corepressor complex (Glass and Rosenfeld, 2000), the corepressor dissociation of exogenous VDR was observed in response to ligand binding (Figure 2A, lanes 3 and 4). As previously reported (Kitagawa et al, 2003; Kato et al, 2004; Murayama et al, 2004), while VDR associated with WSTF irrespective of 1α,25(OH)2D3 binding, 1α,25(OH)2D3 binding enhanced the interaction between VDR and VDIR (Figure 2A, lanes 3–8). Exogenous WSTF co-immunoprecipitated with exogenous VDIR in a ligand-dependent manner, and with endogenous NCoR corepressor complex components (NCoR, mSin3 and HDAC2) in the presence of 1α,25(OH)2D3 (Figure 2A, lanes 7 and 8). Furthermore, we found that an HDAC inhibitor trichostatin A (TSA)-released HDAC activity was contained in immunoprecipitates of exogenous FLAG-WSTF, and this activity was enhanced in a ligand-dependent manner (Figure 2B). The ligand-dependent association of VDR/WSTF with VDIR was also observed for endogenous proteins in MCF7 cells (Figure 2C). This ligand binding is presumably required for the association between VDR/WSTF and VDIR, and it results in recruitment of an HDAC corepressor complex.
Figure 2.

WSTF interacts with VDIR through 1α,25(OH)2D3-bound VDR. (A) Exogenous WSTF interacted with exogenous VDIR and endogenous corepressors in an 1α,25(OH)2D3-dependent manner in vivo. MCF7 cells were transfected with 0.3 μg of WSTF, VDR and VDIR expression vector. The panels show results of immunoprecipitation with anti-VDR, -VDIR or -FLAG (WSTF) antibodies, followed by Western blot analysis using the indicated antibodies. (B) WSTF associates with HDAC activity in a ligand-dependent manner. MCF7 cells were transfected with pcDNA3 or FLAG-WSTF/pcDNA3 and the extracted cell lysates were then immunoprecipitated with anti-FLAG M2 resin. HDAC activity in the immunoprecipitates was measured by fluorometric detection using an HDAC assay kit. (C) 1α,25(OH)2D3-dependent interaction between endogenous WSTF and VDIR in vivo. MCF7 cells cultured with or without 1α,25(OH)2D3 for 12 h were subjected to immunoprecipitation with anti-WSTF or anti-VDIR antibodies. Immunoprecipitates were Western blotted with specific antibodies as shown on the left. (D) SDS–PAGE gels of a series of GST-fused WSTF deletion mutants (left panel) and recombinant VDR (right panel) were visualized by CBB staining. Recombinant proteins were expressed in Escherichia coli and purified by affinity chromatography. (E) GST pull-down assay. Schematic diagrams of the WSTF deletion mutants used are illustrated. 35S-labeled VDR translated in vitro was incubated with deletion mutants immobilized onto glutathione-Sepharose beads in the presence or absence of 1α,25(OH)2D3 (10−6 M). Bound proteins were resolved by SDS–PAGE followed by autoradiography (upper panel). Autoradiographs show 35S-labeled VDIR, preincubated with (lower panel) or without (middle panel) cold recombinant VDR, bound to the GST-fused mutants immobilized on beads (Murayama et al, 2004).
We tested this hypothesis using an in vitro GST pull-down assay on a series of bacterially expressed GST-fused WSTF mutants (Figure 2D). The WSTF m1 domain (aa 163–576, illustrated as a shaded box above the panel) was found to interact with in vitro-translated VDR, irrespective of 1α,25(OH)2D3 binding (Figure 2E, upper panel). No clear association of VDR with the other regions was detected, even in the presence of 1α,25(OH)2D3. We then assessed the interaction of VDIR with the WSTF mutants. While none of the WSTF regions exhibited physical interaction with VDIR, in the presence of 1α,25(OH)2D3-bound VDR, an association between WSTF and VDIR was detected (Figure 2E, middle and lower panels). Together, these findings suggest that while WSTF interacts with VDR, VDIR is stably recruited only when VDR is liganded.
WSTF mediates 1α(OH)ase gene promoter occupancy of ligand-unbound VDR
To test whether WSTF was recruited to VDIR via liganded VDR in the nuclei of living cells, we performed a chromatin immunoprecipitation (ChIP) assay using endogenous proteins and the native 1α(OH)ase gene promoter. In agreement with previous reports (Kitagawa et al, 2003; Murayama et al, 2004), VDIR was constitutively bound to 1αnVDRE, while the NCoR corepressor complex components were recruited to the promoter 45 min after the addition of 1α,25(OH)2D3 (Figure 3A). SNF2h, an ISWI chromatin-remodeling complex ATPase, was used as a negative control. As WSTF RNAi remarkably attenuated the promoter occupancy of VDR in the absence of ligand, WSTF appeared to facilitate the binding of ligand-unbound VDR to the 1αnVDRE region (Figure 3A). The occupancy of VDR and WSTF was undetectable in the 1α(OH)ase-distal region, confirming promoter-specific binding of the factors. A Re-ChIP assay was performed to verify the formation of the unliganded VDR, WSTF and VDIR complex at 1αnVDRE. Ligand-independent association of VDR and WSTF was observed in the 1αnVDRE region, while clear association of VDR with VDIR required ligand binding (Figure 3B). Finally, to verify a physiological role for VDR in WINAC promoter targeting, isolated primary mouse embryonic fibroblasts (MEFs) derived from VDR knockout mice were used, after confirming the ablation of VDR protein without changes in the expression level of VDIR and WSTF by Western blotting analysis (Figure 3C). In a ChIP assay using VDR−/− MEFs, VDR appeared to mediate the recruitment of these factors to the 1αnVDRE-proximal region (Figure 3D). Based on these findings, it appeared that WSTF requires VDR to target the promoter nucleosomes, and WSTF facilitates the subsequent retention of unliganded VDR. Furthermore, 1α,25(OH)2D3 binding appears to induce association of VDR with VDIR together with an HDAC complex to trigger ligand-induced transrepression.
Figure 3.
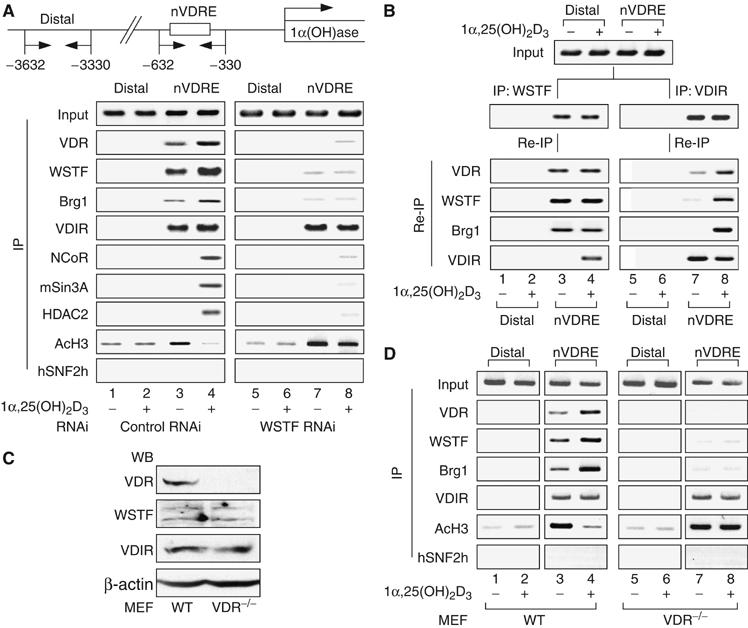
VDR is indispensable for ligand-induced promoter assembly of the WINAC and HDAC corepressor complex. (A) Recruitment of VDR, WSTF, VDIR and other coregulators to the 1α(OH)ase gene promoter in vivo, as shown by ChIP analysis. Soluble chromatin was prepared from MCF7 cells treated with 1α,25(OH)2D3 (10−8 M) for 45 min and immunoprecipitated with the indicated antibodies. Extracted DNA samples were amplified using primer pairs that covered the 1α(OH)ase negative VDRE region (1αnVDRE) (Kitagawa et al, 2003; Murayama et al, 2004) or a distal region (3 kb upstream of 1αnVDRE) as a control. (B) Recruitment of ligand-free VDR/WSTF complexes to the 1α(OH)ase gene promoter, shown by the re-chromatin immunoprecipitation (Re-ChIP) assay. Chromatin prepared from MCF7 cells cultured in the presence or absence of 1α,25(OH)2D3 (10−8 M) for 45 min was subjected to the ChIP procedure with the indicated antibodies and immunoprecipitated using the antibodies as shown on the left. (C) Cessation of VDR expression in VDR−/− MEF cells was confirmed by Western blot analysis. VDR−/− and wild-type (WT) MEF cells were generated from VDR−/− knockout mouse embryos and WT littermates (E 13.5). (D) Effect of VDR disruption in recruitment of WSTF and VDIR to the 1α(OH)ase gene promoter in vivo by ChIP analysis. Soluble chromatin was prepared from VDR−/− and WT MEF cells treated with 1α,25(OH)2D3 (10−8 M) for 45 min and subjected to the ChIP procedure as described in panel A.
Promoter targeting of unliganded VDR via WSTF requires chromatin structures
To clarify the mechanism by which WSTF targets unliganded VDR to the promoter in vitro, we addressed which factors are indispensable for the promoter targeting of unliganded VDR by employing an immobilized DNA/chromatin template recruitment assay. DNA fragments containing either 1αnVDRE (−60 to −615) or the 1α(OH)ase distal region (−3632 to −3032) were end-biotinylated to allow their immobilization onto streptavidin beads. These fragments were reconstituted as highly regular nucleosome arrays using recombinant dNAP1 protein and dACF complexes, along with purified histone octamers from HeLa cells and bacterially expressed histone octamers as unmodified histones for negative control (Figure 4A), as previously reported (Nakagawa et al, 2001; Kitagawa et al, 2003). The expected functions of the chromatin reconstitution factors were verified by supercoiling or standard micrococcal nuclease (MNase) digestion assays (data not shown). Using these factors, the end-biotinylated DNA fragments were correctly reconstituted into nucleosome arrays according to the MNase digestion assay (Figure 4B). As dAcf-1 in the dACF complex is highly homologous to WSTF, chromatin templates were analyzed by Western blots to confirm the absence of dAcf-1 after final washing (Figure 4C).
Figure 4.

Chromatin structures are required to target unliganded VDR to the 1α(OH)ase promoter. (A) SDS–PAGE analysis of purified HeLa histone octamers, recombinant histone octamers, recombinant Drosophila NAP1 (dNAP1) and Drosophila ACF (dACF) complexes. HeLa histone octamers were purified from HeLa nuclear pellets by traditional hydroxylapatite chromatography, as described in Materials and methods. Each component of the recombinant histone octamer, H2A, H2B, H3 and H4, was expressed in an insoluble form in E. coli and extracted with guanidine hydrochloride. Extracted crude proteins were further purified by traditional gel filtration and ion exchange chromatography, as described previously (Luger et al, 1999). Affinity-tagged recombinant dNAP1 and dACF complex components (FLAG-dAcf1 and dISWI) were expressed in Sf9 cells by infection with recombinant baculoviruses and purified by affinity chromatography as described in Materials and methods. (B) Chromatin template containing the 1α(OH)ase gene promoter immobilized to streptavidin beads. Schematic representation of the DNA template containing the 1α(OH)ase gene promoter is illustrated above. Chromatinized template reconstituted in vitro was confirmed using the standard MNase digestion assay. The 123 bp ladder DNA was used as a size marker. (C) Immobilized template was subjected to Western blot analysis with an anti-FLAG antibody. To eliminate possible contamination by recombinant dACF complexes, immunoblotting of the beads using anti-FLAG, acetylated histone H3 and unmodified histone H3 (as a control) antibodies was performed after extensive washing with high-salt buffer. (D) Schematic diagram of the in vitro-immobilized DNA/chromatin template assay. (E) Stabilization of the ligand-free VDR/WSTF complex on the 1α(OH)ase promoter required chromatin structure in vitro. Whole-cell extracts from MCF7 cells stably expressing FLAG-WSTF treated with or without 1α,25(OH)2D3 (10−8 M) were mixed with immobilized templates. The template beads were then concentrated using a magnet and analyzed by Western blotting using the indicated antibodies.
Whole-cell extracts from MCF-7 cells that stably expressed FLAG-tagged WSTF treated with or without 1α,25(OH)2D3 were incubated with either naked or chromatin DNA templates containing 1αnVDRE. Proteins bound to the DNA templates were then analyzed by immunoblotting (Figure 4D). WSTF and VDR bound to naked DNA templates only in the presence of ligand, while VDIR stably associated with naked DNA templates even in the absence of ligand (Figure 4E, lanes 3 and 4). The specific recruitment of WSTF, together with VDR and VDIR, to 1αnVDRE was confirmed by the finding that addition of excessive synthetic 1αnVDRE oligonucleotides blocked recruitment (Figure 4E, lanes 5 and 6). In contrast, for the chromatin templates with HeLa histone octamers, recruitment of WSTF and VDR was ligand-independent (Figure 4E, lanes 7 and 8). However, WSTF and VDR were removed to significant extents from chromatinized templates by the addition of excess 1αnVDRE oligonucleotides (Figure 4E, lanes 9 and 10), indicating a role for DNA-bound VDIR in the stable association of VDR/WSTF with chromatin. This significant association of WSTF and VDR with chromatin template was not seen when the distal 1α(OH)ase gene promoter region was used (Figure 4E, lanes 11 and 12), or when the promoter region with the octamers of the unmodified histones was used (Figure 4E, lanes 13–16). This supported the significance of WSTF function in unliganded VDR recruitment to the native 1α(OH)ase promoter.
WSTF associates with acetylated nucleosomes and facilitates transcriptional repression through its C-terminal region, which contains the bromodomain
As a member of the BAZ protein family, WSTF harbors two characteristic domains, designated the bromodomain and the PHD finger, in its C-terminal region. Because these domains are present in a number of chromatin-remodeling factors (Ito et al, 1997; Dhalluin et al, 1999; Winston and Allis, 1999; Jacobson et al, 2000; Hassan et al, 2002), we speculated that the WSTF bromodomain and the PHD finger play a role in the observed association between WSTF and chromatin. To address whether these domains served as the direct contact site for histone octamers, we performed in vitro histone binding experiments with purified histone octamers from HeLa cells, and GST fusion proteins containing the bromodomain and PHD finger regions. We also prepared anti-FLAG immunoprecipitates having HDAC activity from whole lysates of MCF7 cells with 1α,25(OH)2D3 (Figure 2B), in order to deacetylate the HeLa histone octamers (Figure 5A). The results indicated that the WSTF bromodomain bound to the histone octamers only when histones were preincubated with the anti-FLAG immunoprecipitates and TSA, while the PHD finger bound the octamers only weakly (Figure 5B, even number lanes). Because bromodomains are generally known to serve as a motif responsible for binding acetylated lysine in histone tails, we reasoned that histone acetylation mediates the interaction between the WSTF bromodomain and HeLa histone octamers. This idea was supported by the observed in vitro association between the WSTF bromodomain and histone octamers, which was indeed abrogated when histone octamers were deacetylated using anti-FLAG immunoprecipitates (Figure 5B, odd number lanes).
Figure 5.
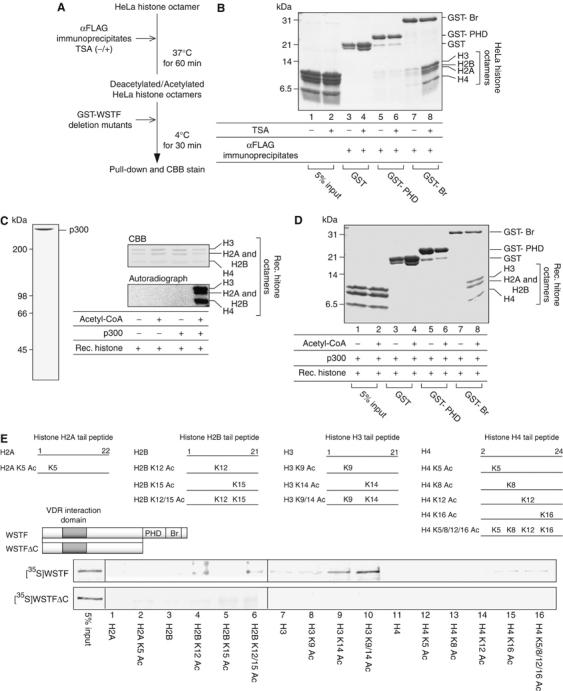
WSTF bromodomains in its C-terminal region interact with acetylated histone octamers. (A) Schematic diagram of a histone binding assay. HDAC immunoprecipitate was prepared using an anti-FLAG antibody from whole-cell extracts of 1α,25(OH)2D3-treated MCF7 cells transiently expressing FLAG-WSTF (see Figure 2B). (B) An in vitro histone binding assay showed that the interaction between WSTF bromodomains and HeLa histone octamers is decreased after deacetylation of the HeLa histone octamers. HeLa histone octamers were incubated with HDAC complexes, mixed with GST-fused WSTF bromodomain and PHD finger regions and then immobilized onto glutathione-Sepharose beads. Bound materials were eluted from the resin and resolved by 18% SDS–PAGE. Proteins were visualized by CBB staining. (C) SDS–PAGE analysis of recombinant p300 and in vitro histone acetylation of recombinant histone octamers. Affinity-tagged recombinant p300 was expressed in Sf9 cells using a baculovirus system and purified by affinity chromatography as described in Materials and methods. Histone octamers were acetylated in vitro by p300 with radiolabeled acetyl-CoA and the gels were visualized by CBB staining following exposure to film. (D) Interaction between recombinant histone octamers and WSTF bromodomains is enhanced after in vitro histone acetylation by p300. Recombinant histone octamers were preincubated with p300 in the absence (upper panel) or presence (lower panel) of acetyl-CoA and subjected to histone binding assay. (E) Site-specific recognition between the WSTF bromodomain and histones with tail modification. Schematic diagrams of the WSTF deletion mutants used are illustrated (upper panel). 35S-labeled WSTF and a WSTFΔC mutant translated in vitro were incubated with a series of acetylated N-terminal histone tails immobilized onto streptavidin beads. Histone tail peptides were tested for WSTF binding (middle panel). Bound WSTF was resolved by SDS–PAGE, followed by autoradiography (lower panel).
To determine whether the histone acetylation was sufficient to promote this interaction, we established an in vitro histone binding assay using recombinant histone octamers. The integrity and HAT activity of purified recombinant p300 were assessed by CBB stain of the protein and in vitro acetylation of recombinant histone octamers (Figure 5C). The WSTF bromodomain bound to recombinant histone octamers only when the histones were acetylated (Figure 5D). We then further analyzed selective recognition of acetylated lysine residues in histone N-terminal tails by WSTF. Among biotin-conjugated tail peptides, WSTF selectively recognized some of the known acetylated lysine residues, histone AcH2BK12, AcH3K14, AcH4K16. However, ablation of the WSTF bromodomain (WSTFΔC) resulted in loss of the WSTF interaction with acetylated histones (Figure 5E). Our results thus suggested that the WSTF C-terminal region selectively associated with post-translationally acetylated lysine residues in the histone tails through the WSTF bromodomain.
Physical interactions between the WSTF C-terminal region and acetylated histones are required for the ligand-induced transrepression of the 1α(OH)ase gene
We next examined the role of WSTF in binding to acetylated histones during VDR-mediated gene repression. A WSTFΔC construct was first assessed in MCF7 cells by immunoprecipitation using anti-FLAG antibodies and Western blotting with anti-acetylated lysine 14 of histone H3 (AcH3K14), anti-acetylated histone H3 (AcH3), anti-Brg1 and anti-VDR antibodies. Both WT WSTF and the WSTFΔC mutant appeared to associate with VDR and Brg1, which is an ATPase subunit of WINAC, as well as the other SWI/SNF chromatin-remodeling complex subtypes. However, the WSTFΔC mutant exhibited no clear association with AcH3 and AcH3K14 (Figure 6A). These findings imply that the highly acetylated state of histones induces a stable association between WSTF and chromatin in vivo.
Figure 6.

The WSTF C-terminal region is indispensable for the promoter targeting of ligand-unbound VDR and for VDR-mediated transrepression of the 1α(OH)ase gene. (A) WSTFΔC shows no binding to acetylated histone H3. MCF7 cells transfected with FLAG-tagged WSTF, FLAG-tagged WSTFΔC or pcDNA3 vector as a control were lysed and subjected to immunoprecipitation with anti-FLAG. Immunoprecipitates were Western blotted with indicated antibodies (lower panel). (B) Histone acetylation-dependent recruitment of WSTF to the 1αnVDRE region. MCF7 cells transfected with FLAG-tagged WSTF or FLAG-tagged WSTFΔC were treated with either 1α,25(OH)2D3 (10−8 M) for 45 min or TSA (10−7 M) for 120 min and then subjected to ChIP analysis. (C) A WSTF mutant with a deleted C-terminal bromodomain and PHD finger (WSTFΔC) exerted a partial dominant-negative effect on the ligand-induced transrepression function of VDR. The amounts of each transfected plasmid are described in Figure 1A.
To further characterize this association, a ChIP assay was performed. TSA treatment potentiated histone H3 and H3K14 acetylation around 1αnVDRE (nVDRE), as expected, with increased recruitment of VDR and WSTF (Figure 6B, left panel). In the distal region of the 1α(OH)ase gene promoter, histone H3 acetylation was partially enhanced by TSA, though WSTF, VDR or VDIR was undetectable. As expected from the previous findings of 1α,25(OH)2D3-induced recruitment of HDAC to 1αnVDRE via VDIR/VDR (Murayama et al, 2004), 1α,25(OH)2D3 treatment caused clear deacetylation of histone H3 around 1αnVDRE. The WSTFΔC mutant was again unable to associate with the 1αnVDRE region, regardless of the hyperacetylated state of histone H3 (Figure 6B, right panel). A reporter assay using the luciferase gene driven by the 1α(OH)ase promoter showed that WSTF expression potentiated the 1α,25(OH)2D3-induced transrepression along with Brg1 (Figure 6C), whereas the WSTFΔC mutant acted as a dominant-negative factor in terms of the ligand-induced transrepression (Figure 6C, black bars). Thus, it appears that WSTF interacts with acetylated histones via the bromodomain in vivo, and that this interaction is indispensable for VDR-mediated transrepression through 1αnVDRE.
Discussion
Role of WINAC in the ligand-dependent corepressor recruitment
Transcription control by NRs encompasses multiple steps with the help of a large number of coregulator complexes (Glass and Rosenfeld, 2000; McKenna and O'Malley, 2002). ATP-dependent chromatin-remodeling complexes are considered to support the promoter-specific recruitment of other coregulator complexes (Emerson, 2002; Narlikar et al, 2002). We have previously reported that the WINAC dysfunction resulted in a failure of proper transcriptional regulation by VDR, possibly because of impairment of coregulator recruitment to VDR target gene promoter. These findings strongly suggested that ATP-dependent chromatin-remodeling activity is indispensable for subsequent coregulator recruitment in response to ligand binding. However, the molecular mechanism in ligand-induced transrepression has not been well understood.
It has been considered that ligand-unbound VDR/RXR on VDRE mainly associates with HDAC complex to actively repress target genes. Reflecting this model, ligand binding led to corepressor dissociation from VDR (Figure 2A). In contrast, we also showed that WINAC assisted promoter recruitment of HDAC corepressor complex in VDIR-mediated transrepression on a negative VDRE (Figure 3A). These results may indicate a difference in the set of associating factors/complexes with unliganded VDR on negative VDREs from positive VDREs on the VDR target gene promoters. Indeed, ligand binding significantly increases the interaction of VDR/WINAC with HDAC complex (Figure 2A and B). A WSTF mutant with a deleted C-terminal region containing bromodomains functions as a dominant-negative mutant in terms of ligand-induced transrepression by VDR (Figure 6C), although this mutant could interact with VDR (Figure 6A). Moreover, this mutant abrogated ligand-dependent histone deacetylation, considering loss of acetylated histone recognition and subsequent HDAC corepressor recruitment. Hence, in addition to ligand-induced transactivation by VDR, WINAC has an important role in VDR-mediated transrepression mechanism. The proposed mechanism of the ligand-induced transrepression by VDR in the present study appears to be dependent on the promoter content, since it is unlikely that all of the VDR target gene promoters for vitamin D-induced transrepression harbor VDIR binding sites. The other mode and mechanism of ligand-induced transrepression might be unrevealed in the other promoters for VDR and the other NRs.
Promoter targeting of VDR requires chromatin structures
In this report, we investigated the role of WINAC in the recruitment of VDR to 1αnVDRE in the 1α(OH)ase gene promoter with consequent ligand-induced transrepression. We showed in vitro that WINAC potentiated association of VDR with 1αnVDRE, irrespective of the ligand binding, when the promoter DNA was configured as a nucleosome array (Figure 4). In contrast, in naked DNA fragments, only liganded VDR was recruited to 1αnVDRE via association with VDIR. Moreover, biochemical mapping experiments demonstrated that the WSTF bromodomain has an interaction surface with histone octamers (Figure 5B and E). These results suggest that WINAC aids promoter occupancy by unliganded VDR, acting as a tether between VDR and promoter nucleosomal arrays. This model was further supported by the in vivo observation that a decrease in WSTF levels due to RNAi expression attenuated VDR retention on the endogenous 1α(OH)ase gene promoter (Figure 3A). As VDR directly interacts with WSTF in a ligand-independent manner, VDR could be targeted to the promoter irrespective of ligand binding through its association with WSTF.
Transition from the transactivation state to the transrepression state
1α(OH)ase gene expression is induced by calciotropic peptide hormones such as parathyroid hormone (PTH) (Brenza et al, 1998; Murayama et al, 1998). It has been previously shown that VDIR is phosphorylated by PKA as a downstream effect of PTH activity, which then leads to HAT p300/CBP complex binding and transactivation (Murayama et al, 2004). The recruited HAT coactivator complex is presumed to acetylate the nucleosomal array around 1αnVDRE, and consequently this acetylation renders the 1α(OH)ase promoter accessible to the 1α,25(OH)2D3-unbound VDR/WINAC complex. Furthermore, the weak interaction between unliganded VDR and VDIR, shown in Figure 3B, uncovered the existence of this transition stage (see Figure 7). In this study, we show that a WSTF mutant with a deletion of the C-terminal region, which includes bromodomains, abolished the retention of the WINAC complex on the acetylated 1α(OH)ase gene promoter. This deletion mutant also functions as a dominant-negative form of WSTF to carry out ligand-induced transrepression (Figure 6). These results indicate that assembly of these factors prior to ligand binding is indispensable to initiate ligand-induced transrepression. Upon VDR binding to 1α,25(OH)2D3, VDR stably associates with VDIR, leading to recruitment of the HDAC corepressor complex with the assistance of WINAC chromatin-remodeling activity. Alternatively, it is also possible to presume that WINAC stabilizes the association of VDR with the HDAC corepressor complex. In any case, ligand binding to VDR results in transcriptional repression of these gene expressions through DNA-bound VDIR and the WINAC complex.
Figure 7.
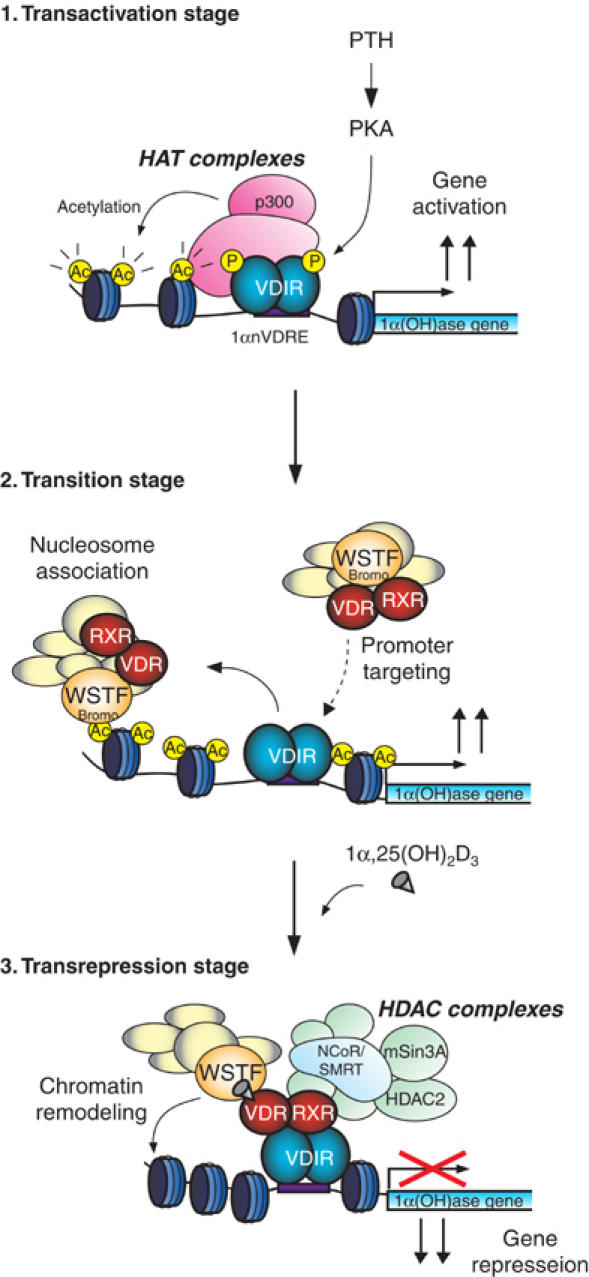
Model demonstrating the role of WINAC in the ligand-induced transrepression function of VDR at the 1α(OH)ase gene promoter. p300 is recruited to VDIR, which was phosphorylated via PKA signaling, and it acetylates the nucleosomes around the 1α(OH)ase gene promoter region (transactivation stage). WINAC, along with VDR, sequentially targets VDIR through interaction between unliganded VDR and VDIR, and is retained on the acetylated promoter via the WSTF bromodomain. VDR becomes receptive to 1α,25(OH)2D3 binding (transition stage). Upon 1α,25(OH)2D3 binding, HDAC corepressor complexes are recruited to the ligand-bound VDR/VDIR complex, and they then deacetylate the nucleosomes. WINAC then exerts its ATP-dependent chromatin-remodeling activity (transrepression stage).
Several pieces of evidence support our finding of a physical interaction between acetylated histones and ATP-dependent chromatin-remodeling complexes. For instance, histone acetylation by Gcn5 has been shown to recruit SWI/SNF complexes during activation of the interferon-β promoter (Agalioti et al, 2000). Likewise, ligand-induced transactivation by RAR/RXR also requires histone acetylation prior to chromatin remodeling by SWI/SNF complexes in vitro (Dilworth et al, 2000). However, all of these studies focused on the role of histone acetylation in transcriptional activation. Thus, we have revealed a novel mechanism by which histone acetylation may be linked with transcriptional repression, even though the acetylation of nucleosomes is generally considered to enhance eukaryotic gene expression.
VDR/WINAC targets VDIR on 1αnVDRE and anchors on the promoter via a physical interaction between the WSTF bromodomain and acetylated histones
In this report, we showed that the WSTF bromodomain physically interacts with acetylated histone octamers in vitro (Figure 5), and that the WSTF bromodomain is indispensable for ligand-induced transrepression by VDR (Figure 6). Physical interaction of acetylated histones with the bromodomains harbored in ATP-dependent chromatin-remodeling complex components is considered to be a critical step in the activation of chromatin, modulating its architecture by rearrangement of nucleosome arrays (Winston and Allis, 1999; Jones et al, 2000). Hassan et al (2002) found that both the SAGA and SWI/SNF complexes are capable of anchoring to promoter nucleosomes through direct contact of acetylated histones with the bromodomains. However, it remained unclear how these chromatin-remodeling complexes selectively discriminate their target chromosomal areas from others through their chromatin recognition modules. To address this point, we have examined two aspects of WINAC promoter targeting. One is the mechanism by which specific chromatin areas are recognized by WSTF through association with sequence-specific regulators, and the other is the preference of WSTF for a specific chromatin condition.
To address the first aspect of WINAC promoter targeting, we investigated the role of VDR by using VDR−/− MEF cells. Our experiments showed in vivo that WSTF recruitment to the 1α(OH)ase promoter was significantly impaired in VDR−/− MEF cells, even though the acetylation level of histones and VDIR occupancy in the promoter were unchanged when compared to WT MEF cells (Figure 3D). These results suggested that the association between the WSTF bromodomain and acetylated nucleosomes itself is not stable enough to anchor the VDR/WINAC complex on the promoters. Indeed, clear VDR/WSTF retention on the chromatin templates required VDIR bound to 1αnVDRE (Figure 4F, lanes 11 and 12). Previous reports have shown that SWI/SNF complexes are able to associate with promoter nucleosomes, but the association was somewhat unstable on an unmodified nucleosomal array (Cote et al, 1998). Therefore, ATP-dependent chromatin-remodeling complexes may require a physical interaction with the sequence-specific regulators to maintain their promoter occupancy. In this respect, VDIR could be considered as a hallmark for targeting of VDR/WINAC to the specific chromosomal areas.
The second aspect of WINAC promoter targeting was addressed by showing a preferential interaction of WSTF with acetylated amino-acid residues in histones. Acetylation of lysines on the histone tails is thought to establish a distinct histone code that directs molecular processes, including gene regulation (Strahl and Allis, 2000). A number of factors have turned out to harbor bromodomains, and these are believed to utilize diverse histone codes to carry out different cellular functions. We found that the WSTF bromodomain preferentially binds to acetylated Lys14 of histone H3 (Figure 5F). This lysine residue is considered to be the best HAT substrate for p300, as a previous study showed that p300 effectively acetylates Lys14 in histone H3 in a nucleosomal context (Schiltz et al, 1999; Lau et al, 2000). Together with our previous findings that p300/CBP is a HAT coactivator for VDIR bound to 1αnVDRE, this suggests that interaction of the WSTF bromodomain with acetylated H3 Lys14 by p300 enables WINAC to discriminate, at least to some extent, the target promoter regions from other nonspecific acetylated regions.
Thus, considering all of these results, we conclude that WINAC facilitates VDR-mediated transrepression of the 1α(OH)ase gene through a physical interaction between the WSTF bromodomain and an acetylated nucleosomal array. This mechanism is likely indispensable for the biological functions of the VDR, such as a negative feedback control in the 1α,25(OH)2D3 biosynthesis pathway.
Materials and methods
Plasmids and RNAi
For transfection studies, the two 1αnVDRE sequences (5′-CAT TTT AGC CCA TTA ACC CAC CTG CCA TCT GCC C-3′) and the 1α(OH)ase promoter (nucleotides −615 to −60 relative to the RNA start site) were inserted into the pGL3-Luciferase vector (Promega) under the control of a TATA promoter. Expression vectors for full-length rat VDR, rat RXR and human FLAG-tagged WSTF were as previously described (Kitagawa et al, 2003). Full-length mouse VDIR cDNA tagged with 6 × His was inserted into pcDNA3 (Invitrogen). cDNA encoding a human WSTF deletion mutant (amino acids 1–1185, WSTFΔC) N-terminally tagged with FLAG was cloned into pcDNA3. A series of human WSTF deletion mutants fused with GST were cloned into pGEX-4T (Pharmacia) (Kitagawa et al, 2003). For an immobilized template recruitment assay, the human 1α(OH)ase promoter distal region (nucleotides −3615 to −3060 relative to the RNA start site) was cloned into the pGL3 vector.
The two short RNA oligomers denatured at 90°C were annealed for 1 h at 60°C (Kitagawa et al, 2003). The RNAi sequences used were as follows: WSTF (5′-GAG UAU GAA GCC CGC UUG GTT-3′ and 5′-CCA AGC GGG CUU CAU ACU CTT-3′); VDR (5′-UCA AUG CUA UGA CCU GUG AUU-3′ and 5′-UCA CAG GUC AUA GCA UUG AUU-3′); VDIR (5′-GAA CCU GAA UCC CAA AGC AUU-3′ and 5′-UGC UUU GGG AUU CAG GUU CUU-3′); lamin A/C Duplex (as a control; Dharmacon) (5′-CUG GAC UUC CAG AAG AAC ATT-3′ and 5′-TTG ACC UGA AGG UCU UCU UGU-3′).
Protein purification
HeLa histone octamers were prepared from HeLa nuclear pellets as previously described (Hassan et al, 2002). HeLa cells (3 l culture) were lysed in lysis buffer (20 mM HEPES, pH 7.5, 0.25 M sucrose, 3 mM MgCl2, 0.2% Nonidet-P40 (NP-40), 3 mM 2-mercaptoethanol, 0.4 mM PMSF, 1 μM pepstatin A and 1 μM leupeptin) by Dounce homogenization with pestle B. After washing the pellet with buffer B (20 mM HEPES, pH 7.5, 3 mM MgCl2, 0.2 mM EGTA, 3 mM 2-mercaptoethanol, 0.4 mM PMSF, 1 μM pepstatin A and 1 μM leupeptin), nuclear proteins were extracted with buffer B containing 0.3 M KCl and 5% glycerol. The nuclear pellets were resuspended in HAP buffer (50 mM sodium phosphate, pH 6.8, 0.6 M NaCl, 1 mM 2-mercaptoethanol and 0.5 mM PMSF). To remove extra DNA fragments and histone H1, 20 g of dry BioGel HTP powder (Bio-Rad) was added to the suspension and the slurries were poured into a column (2.5 × 20 cm). After washing the resin extensively with HAP buffer, HeLa histone octamers were eluted with HAP buffer containing 2.5 M NaCl. Purified histone octamers were concentrated using Centriprep-3 (Amicon).
Recombinant Xenopus histones (H2A, H2B, H3, H4) were expressed and purified as previously described (Luger et al, 1999; Dyer et al, 2004). Recombinant p300, dNAP1 and dACF complexes were prepared essentially as described previously (Ito et al, 1997; Dilworth et al, 2000; Nakagawa et al, 2001). Briefly, His6-tagged p300 and His6-tagged dNAP1 were purified from baculovirus-infected Sf9 cells using Ni-NTA resin (Qiagen), followed by conventional anion exchange chromatography using SOURCE 15Q (Pharmacia) for dNAP1. Recombinant dACF complexes were prepared from Sf9 cells infected with baculovirus vectors encoding dACF complex components, FLAG-tagged dAcf-1 and dISWI, by affinity chromatography using anti-FLAG M2 agarose (Sigma). All proteins were dialyzed against HEG buffer (HEPES, pH 7.6, 1 mM EDTA, 10% glycerol, 0.15 M NaCl, 1 mM DTT and 0.5 mM PMSF), and protein concentrations were evaluated by SDS–PAGE and visualized with CBB R-250, using BSA as a standard.
Establishment and maintenance of WT and VDR−/− MEF cell lines
MEF cell lines were obtained from WT or VDR−/− 13.5-day-old embryos and used at the 10th generation (Ito et al, 2000). The MEF cell lines were replated at a density of 1 × 106 cells on gelatin-coated 10-cm dishes and maintained in DMEM supplemented with 10% FBS at 37°C in 5% CO2.
Immunoprecipitation
After washing MCF7 cells with ice-cold PBS, cells were collected and resuspended in 100 μl lysis buffer (20 mM Tris–HCl, pH 7.9, 1% NP-40, 1 mM EDTA, 150 mM NaCl, 2.5 mM MgCl2, 5% glycerol, 5 mM DTT, 10 μg/ml aprotinin and 1 mM PMSF) containing 0.1% SDS, incubated on ice for 30 min and then centrifuged for 30 min at 12 000 g. After centrifugation, the supernatants were diluted 10 times with lysis buffer and used as MCF7 whole-cell extracts for immunoprecipitation using anti-FLAG (Sigma), anti-VDR (Neomarkers), anti-WSTF (Cell signaling) or anti-VDIR (Santa Cruz) antibodies (Yanagisawa et al, 2002; Kitagawa et al, 2003; Murayama et al, 2004).
ChIP and Re-ChIP assay
ChIP analysis was performed using the ChIP assay kit (Upstates) (Yanagisawa et al, 2002; Kitagawa et al, 2003), according to the manufacturer's instructions. Intact MCF7 cells or transfected cells, and WT or VDR−/− MEF cells were cultured in the presence of ligand. Soluble chromatin prepared from 1 × 106 cells was immunoprecipitated with antibodies against the indicated proteins, including anti-NCoR (Alexis), anti-AcH3 (Upstates) or hSNF2h (Santa Cruz) as a negative control.
Immobilized DNA/chromatin template recruitment assays
Recruitment assays were performed as previously described (Hassan et al, 2002). The 1α(OH)ase promoter or distal fragment was cleaved from the 1α(OH)ase promoter/pGL3 or 1a(OH)ase distal region/pGL3 with MluI and BglII, end-labeled with biotin-14-dATP (Gibco) and gel purified. The fragments (0.2 μg DNA) were then reconstituted as chromatin with purified HeLa histone octamers (0.15 μg), recombinant histone octamers (0.3 μg), purified recombinant dNAP1 (2.5 μg), and purified recombinant dACF complexes (10 ng) and ATP (5 mM; Sigma) as described previously (Kitagawa et al, 2003). Biotinylated DNA or nucleosomal arrays were incubated at room temperature for 1 h with paramagnetic beads coupled to streptavidin (Dynabeads streptavidin, Dynal) in binding buffer (10 mM Tris–HCl, pH 7.9, 0.3 M KCl, 5 mM DTT, 5 mM PMSF, 5% glycerol and 0.25 mg/ml BSA) and washed extensively with binding buffer supplemented with 0.5 M KCl. Whole-cell extracts, prepared from three 75 cm3 dishes of MCF-7 cells stably expressing FLAG-hWSTF treated with or without 10−8 M 1α,25(OH)2D3, were preincubated with 1 μg of circular competitor chromatin for 30 min and then added to the prepared templates and further incubated in lysis buffer for 1 h at 37°C. Either 100 pmol of double-stranded 1αnVDRE-oligo (5′-TAA CCC ACC TGC CAT CTG CCC AGT-3′) as a competitor or 100 pmol of double-stranded DR5-oligo (5′-TAA GGG TTC ACC GAA AGT TCA CTC GCA T-3′) as a control was then added and samples were further incubated for 1 h at 30°C. Subsequently, templates were concentrated using a magnet and washed twice in binding buffer containing 50 mM KCl before being used in Western blot analysis.
In vitro histone acetylation and histone deacetylation
For in vitro histone acetylation experiments, recombinant histone octamers (0.5 μg) were incubated with or without recombinant p300 (200 ng) in HEG buffer. Acetyl-CoA mix (1 μM radiolabeled and 9 μM cold acetyl-CoA) was then added and histone acetylation was carried out at 30°C for 30 min. After stopping the reaction by incubating on ice for 30 min, samples were analyzed on an 18% SDS–PAGE gel, which was visualized with Coomassie staining before immersion in Enhance (NEN) fluorography reagent as the manufacturer's instructions. The gel was then dried and visualized by autoradiography.
For in vitro histone deacetylation, anti-FLAG immunoprecipitates were prepared from whole extracts of MCF7 cells transfected with empty pcDNA3 vector or FLAG-WSTF/pcDNA3 and cultured with or without 10−8 M 1α,25(OH)2D3 for 48 h. HDAC activity of the immunoprecipitates was measured using an HDAC assay kit (Fluorometric detection, Upstate Biotech.) according to the manufacturer's instructions. HeLa histone octamers were deacetylated by incubation with immunoprecipitates containing HDAC activity in the presence or absence of 10−6 M TSA at 37°C for 60 min.
Peptide binding assay
Peptide binding assay was performed as previously reported (Dey et al, 2003). Briefly, 35S-labeled proteins translated in vitro were incubated with 2 μg of biotin-labeled synthetic peptides corresponding to the N-terminal tails of histone H2A, H2B, H3 and H4 (purchased from Upstate, or synthesized from Genemed Synthesis) in a binding buffer (50 mM Tris–HCl (pH 7.5), 15 mM MgCl2, 150 mM NaCl, 0.5 mM DTT and 0.1% NP-40) for 2 h at 4°C, followed by incubation with 20 μl of M-280 streptavidin beads (Dynal). Bound materials were subjected to SDS–PAGE, followed by autoradiography.
Acknowledgments
We thank Dr Timothy J Richmond for kindly providing the xHistone expression vector, Dr JT Kadonaga for the recombinant baculovirus expressing human p300 and Dr K Luger for technical discussions. We also thank Mr Y Mezaki and Dr AP Kouzmenko for technical support, and Ms H Higuchi for manuscript preparation. This work was supported in part by a grant-in-aid for Basic Research Activities for Innovative Biosciences (BRAIN) and priority areas from the Ministry of Education, Science, Sports, and Culture of Japan (to SK).
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103: 667–678 [DOI] [PubMed] [Google Scholar]
- Brenza HL, Kimmel-Jehan C, Jehan F, Shinki T, Wakino S, Anazawa H, Suda T, DeLuca HF (1998) Parathyroid hormone activation of the 25-hydroxyvitamin D3-1alpha-hydroxylase gene promoter. Proc Natl Acad Sci USA 95: 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J, Peterson CL, Workman JL (1998) Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA 95: 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA 100: 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P (2000) ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol Cell 6: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K (2004) Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol 375: 23–44 [DOI] [PubMed] [Google Scholar]
- Emerson BM (2002) Specificity of gene regulation. Cell 109: 267–270 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT (2001) The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106: 523–525 [DOI] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev 14: 121–141 [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell 3: 97–108 [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387: 43–48 [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG (2000) Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell 5: 683–693 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu J, Shimane M (2000) A novel family of bromodomain genes. Genomics 63: 40–45 [DOI] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85: 403–414 [DOI] [PubMed] [Google Scholar]
- Kato S, Fujiki R, Kitagawa H (2004) Vitamin D receptor (VDR) promoter targeting through a novel chromatin remodeling complex. J Steroid Biochem Mol Biol 89-90: 173–178 [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S (2003) The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 113: 905–917 [DOI] [PubMed] [Google Scholar]
- Lau OD, Kundu TK, Soccio RE, Ait-Si-Ali S, Khalil EM, Vassilev A, Wolffe AP, Nakatani Y, Roeder RG, Cole PA (2000) HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol Cell 5: 589–595 [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R (2001) Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414: 924–928 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304: 3–19 [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM (1995) The nuclear receptor superfamily: the second decade. Cell 83: 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108: 465–474 [DOI] [PubMed] [Google Scholar]
- Murayama A, Kim MS, Yanagisawa J, Takeyama KI, Kato S (2004) Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J 23: 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Murayama A, Takeyama K, Kitanaka S, Kodera Y, Hosoya T, Kato S (1998) The promoter of the human 25-hydroxyvitamin D3 1 alpha-hydroxylase gene confers positive and negative responsiveness to PTH, calcitonin, and 1 alpha,25(OH)2D3. Biochem Biophys Res Commun 249: 11–16 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Bulger M, Muramatsu M, Ito T (2001) Multistep chromatin assembly on supercoiled plasmid DNA by nucleosome assembly protein-1 and ATP-utilizing chromatin assembly and remodeling factor. J Biol Chem 276: 27384–27391 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270: 1354–1357 [DOI] [PubMed] [Google Scholar]
- Rachez C, Suldan Z, Ward J, Chang CP, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP (1998) A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev 12: 1787–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem 274: 1189–1192 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Takeyama K, Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S (1997) 25-Hydroxyvitamin D3 1alpha-hydroxylase and vitamin D synthesis. Science 277: 1827–1830 [DOI] [PubMed] [Google Scholar]
- Winston F, Allis CD (1999) The bromodomain: a chromatin-targeting module? Nat Struct Biol 6: 601–604 [DOI] [PubMed] [Google Scholar]
- Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon SB, Cole MD, Tora L, Takahashi N, Kato S (2002) Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol Cell 9: 553–562 [DOI] [PubMed] [Google Scholar]
- Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y, Kawakami T, Arioka K, Sato H, Uchiyama Y, Masushige S, Fukamizu A, Matsumoto T, Kato S (1997) Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet 16: 391–396 [DOI] [PubMed] [Google Scholar]


