Abstract
The immunosuppressive macrolide rapamycin is used in humans to prevent graft rejection. This drug acts by selectively repressing the translation of proteins that are encoded by an mRNA bearing a 5′-polypyrimidine tract (e.g., ribosomal proteins, elongation factors). The human immunodeficiency virus type 1 (HIV-1) carries a polypyrimidine motif that is located within the tat exon 2. Treatment of human T lymphoid cells with rapamycin resulted in a marked diminution of HIV-1 transcription when infection was performed with luciferase reporter T-tropic and macrophage-tropic viruses. Replication of fully infectious HIV-1 particles was abolished by rapamycin treatment. The rapamycin-mediated inhibitory effect on HIV-1 production was reversed by FK506. The anti-HIV-1 effect of rapamycin was also seen in primary human cells (i.e., peripheral blood lymphocytes) from different healthy donors. Rapamycin was shown to diminish basal HIV-1 long terminal repeat gene expression, and the observed effect of rapamycin on HIV-1 replication seems to be independent of the virus-specific transactivating Tat protein. A constitutive β-actin promoter-based reporter gene vector was unaffected by rapamycin treatment. Kinetic virus infection studies and exposure to reporter viruses pseudotyped with heterologous envelope proteins (i.e., amphotropic murine leukemia virus and vesicular stomatitis virus G) suggested that rapamycin is primarily affecting the life cycle of HIV-1 at a transcriptional level. Northern blot analysis confirmed that this compound is selectively targeting HIV-1 mRNA synthesis.
Human immunodeficiency virus type 1 (HIV-1) has a complex life cycle that begins when the virus binds to a host cell, enters the cell, and, using the host cell's machinery, creates numerous copies of itself within the host cell (28). In an attempt to combat this retroviral infection, several drugs and strategies targeting specific steps of the virus replicative cycle were developed. For instance, anti-CD4 antibodies in association with anti-gp120 antibodies were shown to block HIV-1 infection in human T cells (11), whereas a potent suppression of HIV-1 replication in humans was observed with T-20, a peptide inhibitor of gp41-mediated virus entry (38). In addition, various chemokines (e.g., RANTES, MIP-1α, MIP-1β, and SDF-1) were shown to block the interaction between the gp120/CD4 complex and either the CCR5 or CXCR4 coreceptor on the cell surface, inhibiting the process of viral entry (5, 7, 17, 21, 24, 45, 50, 54).
Other strategies targeting Tat and the viral integrase were also developed in an attempt to fight HIV-1 (40, 51). Interestingly, specific inhibitors of transcriptional factors (NF-κB, NF-AT, and Sp1) were also proposed as effective ways to control HIV-1 replication (4, 23, 32, 66). The current therapy against this retroviral infection, better known as highly active antiretroviral therapy, consists usually in the association of two reverse transcriptase inhibitors (e.g., zidovudine and lamivudine) and a protease inhibitor that blocks the maturation process (43, 55, 73, 74). These potent drugs, however, show strong adverse effects, and their long-term efficiency is severely hampered by the emergence of resistant viruses. Although major advances occurred in the last decade in the management of HIV-1 infection, both less toxic and more effective anti-HIV-1 therapeutics are still needed.
Rapamycin (Rapamune, Sirolimus) is a macrolide exhibiting potent antitumor and immunosuppressive activities (1, 46). This agent has been shown to inhibit T-cell proliferation induced by various stimuli, including, among others, antigen, alloantigen, mitogenic lectins, phorbol ester, and cross-linking of T-cell surface markers with monoclonal antibodies (e.g., CD28) (25, 35-37). The action of rapamycin occurs upon its binding to some specific intracellular proteins called immunophilins. These proteins are divided into two major families: cyclophilins, which bind to cyclosporin A, and FK506-binding proteins (FKBPs), which can interact with both FK506 and rapamycin. Among the FK506-binding proteins, FKBP12 was shown to bind rapamycin, leading to inhibition of FRAP (FK506-binding protein/rapamycin-associated protein, also called mTOR for mammalian target of rapamycin) activity by forming a trimeric complex (FKBP12/rapamycin/FRAP) (16).
FRAP plays an important role in cell physiology by controlling the phosphorylation of both p70s6k and eIF4E repressor (4E-BP1) (10, 72). Activated p70s6k induces the translation of the 5′-polypyrimidine tract mRNA family. The phosphorylation of 4E-BP1 by FRAP abolishes 4E-BP1 suppressive activity on eIF4E, leading to induction of eIF4E-dependent mRNA translation (72). The binding of FRAP to the rapamycin/FKBP12 complex will thus inhibit the translation of mRNAs bearing polypyrimidine tracts at their 5′ termini (34, 44, 48, 70). Included among these 5′-polypyrimidine-containing mRNAs are transcripts encoding ribosomal proteins and elongation factors, i.e., components of the protein synthesis machinery itself. Moreover, polypyrimidine motifs are found in some human viruses, such as the human T-cell leukemia virus type I 5′ long terminal repeat (59). Interestingly, a polypyrimidine tract is present in the tat exon 2 of HIV-1 (64).
Given that a characteristic of the affected mRNAs is the presence of such a polypyrimidine motif, we investigated whether rapamycin could affect HIV-1 replication. We demonstrate here that treatment of established leukemic T-cell lines and primary human cells with rapamycin results in a significant decrease in HIV-1 production. Further experiments indicate that this phenomenon is not due to an effect on the early steps of HIV-1 biology. Reporter gene assays and Northern blot analyses suggest that HIV-1 long terminal repeat-mediated transcriptional activity is targeted by rapamycin. These results suggest that rapamycin acts as an inhibitor of HIV-1 replication in human T cells due to its action on virus-mediated transcriptional activity.
(This work was performed by J.R. and J.-S.P. in partial fulfillment of the Ph.D. degree at the Faculty of Graduate Studies, Department of Medical Biology, Faculty of Medicine, Laval University.)
MATERIALS AND METHODS
Reagents.
Rapamycin was purchased from Calbiochem, while FK506 and zidovudine were obtained from Sigma (St. Louis, Mo.). Trizol was purchased from Life Technologies, Inc. (Grand Island, N.Y.). Recombinant human interleukin-2 (rhIL-2) was kindly supplied by Maurice Gately, Hoffman-La Roche Inc., through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, Rockville, Md.) (41). Cell viability was monitored by performing the MTS [3-(4,5-dimethylthia-zol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, innersalt] assay as described previously (12). Detection of β-galactosidase activity was achieved with the Galacto-Light commercial kit according to the manufacturer's protocol (Applied Biosystems, Bedford, Miss.).
Cells and culture conditions.
The human IL-2-dependent T-lymphoblastoid cell line WE17/10 (75) was provided by the AIDS Repository Reagent Program and was maintained in complete culture medium made of RPMI 1640 supplemented with 20% fetal bovine serum (HyClone Laboratories, Logan, Utah), glutamine (2 mM), penicillin G (100 U/ml), and streptomycin (100 μg/ml), in the presence of rhIL-2 (50 U/ml).
CHCD4 is an established CD4+ T-cell clone that was immortalized with Herpesvirus saimiri (60). These cells were kindly provided by Kunal Saha (Molecular Virology Laboratory, New York, N.Y.). Peripheral blood lymphocytes (PBLs) from healthy donors were isolated by Ficoll-Hypaque density gradient centrifugation and cultured for 48 h at 37°C in complete culture medium containing phytohemagglutinin protein (Sigma) (3 μg/ml) and rhIL-2 (30 U/ml) prior to viral infection. Human T helper cells (CD4+) were negatively isolated from fresh PBLs with the CD4+-T-cell-negative purification kit according to the manufacturer's instructions (Miltenyi Biotec). Briefly, we used an antibody mixture and a magnetic colloid that depletes the cell population of every cell type except CD4+ T lymphocytes upon application to a magnetically charged column.
The human embryonic kidney cell line 293T, expressing the simian virus 40 large T antigen (52), was maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Burlington, Ontario, Canada) supplemented with 10% fetal bovine serum, glutamine (2 mM), penicillin G (100 U/ml), and streptomycin (100 mg/ml). 293T cells were kindly provided by Warner C. Greene (J. Gladstone Institutes, San Francisco, Calif.).
Plasmids.
pNL4-3 is a full-length infectious molecular clone of HIV-1 (2), and pCEP4-Tat is a vector that contains the HIV-1SF2 tat gene ligated to the pCEP4 cytomegalovirus-based expression vector (14). Both molecular constructs were provided by the AIDS Repository Reagent Program. The proviral plasmid pHXB-Luc is a molecular clone of HIV-1 (T-tropic) and was originally derived from pHXB-2D, from which a part of the nef gene was deleted and replaced with the reporter luciferase gene (15). This molecular clone of HIV-1 was kindly supplied by David Baltimore (Massachusetts Institute of Technology, Cambridge, Mass.). pNL4-3-Luc-E−R+ and pcDNA-I/Amp-based expression vectors encoding HIV-1 ADA (R5), JR-FL (R5), BaL (R5), and amphotropic murine leukemia virus full-length envelope proteins were generously provided by Nathaniel R. Landau (The Salk Institute for Biological Studies, La Jolla, Calif.) (24).
pHCMV-G, expressing the broad-host-range vesicular stomatitis virus envelope glycoprotein G under the control of the human cytomegalovirus promoter, has been described previously (76). The pLTRX-LUC plasmid contains a 722-bp fragment (−644 to +78) from HIV-1LAI placed in front of the luciferase reporter gene (61) and was kindly given by Olivier Schwartz (Unité d'Oncologie Virale, Institut Pasteur, Paris, France). The β-galactosidase expression vector pRc/actin LacZ was provided by Michael Karin (University of California, San Diego).
Nucleofection.
WE17/10 and primary CD4+ T lymphocytes were nucleofected with the Nucleofector technology according to the manufacturer's instructions (Amaxa Biosystems). Briefly, WE17/10 and unstimulated primary CD4+ T cells (4 × 106 cells) were harvested, washed once with phosphate-buffered saline, and then resuspended in the human T-cell Nucleofector solution with the indicated plasmids. The cell-DNA mixture was then put into a cuvette, inserted in the Nucleofector, and nucleofected.
Production of virus stocks.
Fully infectious viral entities (NL4-3 backbone) and luciferase-encoding reporter viruses were generated by calcium phosphate cotransfection in 293T cells as described previously (26). Virus-containing supernatants were harvested 40 h posttransfection and frozen at −85°C until used. Virus preparations were quantified with an in-house sensitive double-antibody sandwich enzyme-linked immunosorbent assay (ELISA) specific for the major core viral p24 protein (8).
Virus infection.
Cells were either left untreated or treated with rapamycin (1.1 nM) for 30 min at 37°C before virus infection (see Fig. 1, 3, 4, and 6). Cells were washed once with phosphate-buffered saline, resuspended in complete culture medium (106 cells/ml) containing 20% fetal bovine serum supplemented with rhIL-2 (30 U/ml for PBLs and 50 U/ml for WE17/10), and transferred to 96-well flat-bottomed tissue culture plates (105 cells/well) (Microtest III; Falcon, Becton Dickinson, Lincoln Park, N.J.). Cells were then infected with HIV-1 (10 ng of p24) in the absence or presence or rapamycin (1.1 nM) and kept in culture for 72 h at 37°C. Cells were then lysed, and luciferase activity was monitored with a microplate luminometer as described previously (MLX apparatus from Dynex Technologies, Chantilly, Va.) (13, 26).
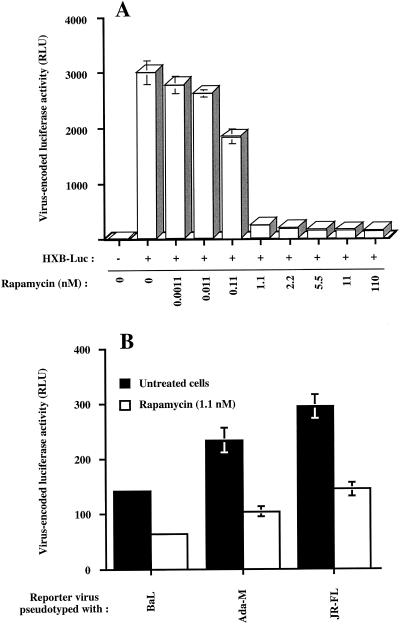
FIG. 1.
Rapamycin inhibits infection of human T lymphoid cells with T-tropic and M-tropic single-cycle reporter virus. (A) WE17/10 cells (105) were initially either left untreated or treated for 30 min with increasing concentrations of rapamycin ranging from 0.0011 to 110 nM. Cells were then infected with HXB-Luc particles (10 ng of p24) in the absence or presence of rapamycin at the indicated concentrations. (B) CHCD4 cells (105) were initially either left untreated or treated for 30 min with rapamycin (1.1 nM). Cells were next infected with luciferase reporter virus pseudotyped with M-tropic envelope proteins from BaL, Ada-M, and JR-FL and either left untreated or treated with rapamycin (1.1 nM). In both instances, cells were kept in culture for a total of 72 h in the absence or presence of the indicated rapamycin concentrations before monitoring luciferase activity. Results shown are the means ± standard deviation of quadruplicate samples and are representative of at least three independent experiments.
In Fig. 2, WE17/10 cells were infected with NL4-3 and left untreated or treated once with rapamycin (1.1 nM). Cells were then left in culture at 37°C, the cell-free supernatants were collected at different time points (3, 6, and 8 days), and virus production was determined with a p24 enzymatic assay. The cells were fed with rhIL-2 (50 U/ml) every 3 days. In Fig. 5, WE17/10 cells were initially infected with HXB-Luc viruses and then either left untreated or treated with rapamycin at different time points postinfection (2, 4, 6, 8, 12, 24, and 48 h). Such cells were kept in culture for 72 h at 37°C before monitoring luciferase activity. In Fig. 7A, WE17/10 cells were first left uninfected or infected with NL4-3 for 72 h to allow a productive infection. Both uninfected and infected cells were then either left untreated or treated with rapamycin for 3 days prior to extraction of total RNA.
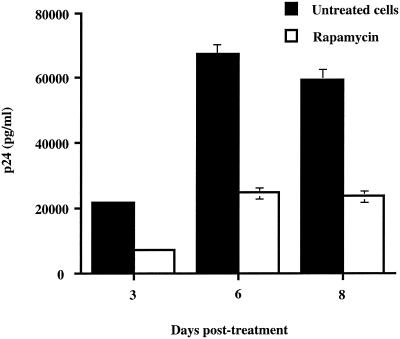
FIG. 2.
Rapamycin abolishes production of fully infectious HIV-1 particles. WE17/10 (106 cells) were infected with fully competent NL4-3 viruses (10 ng of p24), and 6 days later, cells were either left untreated or treated with rapamycin (1.1 nM). Cells were replenished with rhIL-2 every 2 to 3 days. Cell-free supernatants were collected at different time points following infection with HIV-1, and virus production was evaluated with a p24 enzymatic assay. Results shown are the means ± standard deviation of triplicate samples and are representative of at least three independent experiments.
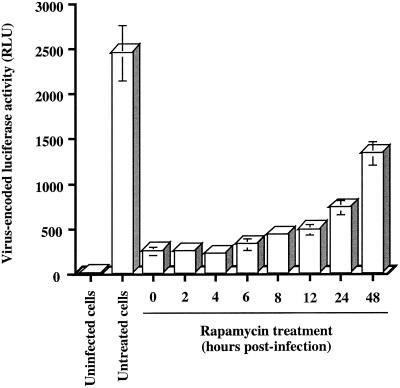
FIG. 5.
Kinetic analysis of rapamycin-mediated diminution of virus-encoded reporter gene activity in human T leukemic cells. WE17/10 cells (105) were infected with HXB-Luc particles (10 ng of p24) and either left untreated or treated with rapamycin (1.1 nM) at the indicated time points after virus infection. Cells were maintained in culture for 72 h, and luciferase activity was assessed as described in Materials and Methods. Results shown are the means ± standard deviation of quadruplicate samples and are representative of at least three independent experiments.
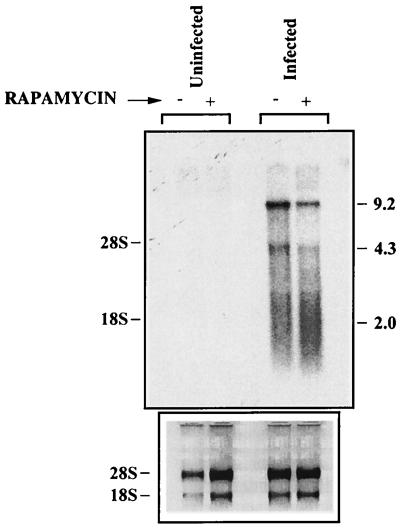
FIG. 7.
HIV-1 transcription is repressed by rapamycin. WE17/10 cells (107) were either left uninfected or infected with whole complete NL4-3 (100 ng of p24) for 6 days. Both infected and uninfected cells were then either left untreated or treated with rapamycin (1.1 nM) for 3 days. Northern blot analysis was next performed on total RNA with an NL4-3 radiolabeled probe. Sizes are shown on the right (in kilobases).
Northern blot analysis.
Total RNA was extracted by the Trizol method (18, 19) from WE17/10 cells. Ten micrograms of total RNA was separated on a formaldehyde-agarose gel (1% agarose, 1× formaldehyde gel buffer morpholinopropanesulfonic acid [MOPS, pH 7.0], 8.3 mM sodium acetate, 1.27 mM EDTA, and 2.2 M formaldehyde). RNA was transferred to Hybond-N nylon membranes (Amersham) by capillary action with 10× SSC (pH 7.0; 1.5 M NaCl, 0.15 M sodium citrate). RNA was fixed to the membrane by UV exposure and hybridized with a radiolabeled probe for NL4-3 at 42°C in 50% formamide-5× SSC-5× Denhardt's solution-0.5% sodium dodecyl sulfate (SDS)-300 μg of denatured fragmented salmon sperm DNA per ml. Probes were radiolabeled with the T7 QuickPrime kit (Pharmacia Biotech). Blots were washed three times (15 min each) in 2× SSC-0.1% SDS at 42°C and three times (15 min each) in 0.1× SSC-0.1% SDS at 42°C and then autoradiographed at −70°C. Laser densitometry was performed with an Alpha Imager 2000 digital imaging and analysis system (Alpha Innotech Corporation, San Leandro, Calif.).
Quantification of viral RNA and virion production.
WE17/10 cells (107 cells in 1-ml total) were infected with NL4-3 (100 ng of p24) for 6 days in RPMI medium supplemented with rhIL-2 (50 U/ml). Culture medium was removed after 3 days and replaced by fresh medium. At day 6 postinfection, cells were transferred in two 25-cm2 culture flasks (10 × 106 cells per flask) and treated with zidovudine (1 μM) for 1 h at 37°C to inhibit reinfection events. Cells were then either left untreated or treated with 1.1 nM rapamycin for 3 days. Cell-free supernatants (100-μl aliquots) and cells (5 × 106) were collected for p24 detection and Northern blot analysis, respectively. Uninfected WE17/10 cells were used as a negative control. Quantitative detection of the main viral core p24 protein was achieved with a p24 assay.
RESULTS
Rapamycin diminished HIV-1 long terminal repeat-driven transcriptional activity in human T lymphoid cells.
The previous demonstration of a polypyrimidine motif in the tat exon 2 (64) prompted us to investigate whether the immunosuppressive drug rapamycin can modulate replication of HIV-1. To this end, the IL-2-dependent CD4/CXCR4-expressing WE17/10 cell line was either left untreated or treated for 30 min with increasing concentrations of rapamycin ranging from 0.0011 to 110 nM. Cells were then infected with recombinant luciferase reporter T-tropic (X4) HIV-1 particles (HXB-Luc) for 72 h in the absence or presence of the indicated rapamycin concentrations. The use of single-cycle reporter viruses enabled us to quantitatively monitor the process of virus infection simply by measuring luciferase activity. As depicted in Fig. 1A, a noticeable decrease in virus-encoded luciferase activity was observed with as little as 0.11 nM rapamycin, whereas maximal inhibition was reached at a concentration of 1.1 nM. Subsequent experiments were thus carried out with rapamycin at a dose of 1.1 nM. Cell viability was not affected by the experimental conditions employed, i.e., exposure of cells to rapamycin for 72 h (data not shown).
We next tested the established T-cell line CHCD4, which expresses both CXCR4 and CCR5 chemokine coreceptors and is thus susceptible to infection with both T-tropic and M-tropic isolates of HIV-1. Cells were either left untreated or treated with rapamycin for 30 min prior to inoculation with reporter viruses pseudotyped with M-tropic envelope protein from BaL, Ada-M, and JR-FL and then kept in culture for 72 in the absence or presence of rapamycin. A decrease in HIV-1 long terminal repeat luciferase activity was still mediated by rapamycin in CHCD4 cells following infection with viruses bearing an R5 phenotype (Fig. 1B). These results suggested that the effect of rapamycin was not cell type specific and was not influenced by virus tropism (i.e., T-tropic or M-tropic).
Virus production is also inhibited by rapamycin treatment.
Our previous results demonstrate that the transcriptional activity of HIV-1 is downregulated by rapamycin. Next, we assessed whether replication of fully competent viruses could also be negatively affected by rapamycin. WE17/10 cells were infected with NL4-3 for 6 days without rapamycin to allow the establishment of a productive infection. Thereafter, cells were treated with zidovudine for 1 h to prevent any reinfection events and then either left untreated or treated with rapamycin for the remaining incubation period (8 days). Treatment of the cells with rapamycin resulted in a significant decrease in virus production at all time points tested following treatment (Fig. 2). Indeed, virus production, as monitored by p24 levels in culture supernatants, was reduced by 68%, 64%, and 60% when tested at days 3, 6, and 8 posttreatment, respectively.
Rapamycin affects basal HIV-1 long terminal repeat expression but not transactivation by Tat and constitutive β-actin promoter.
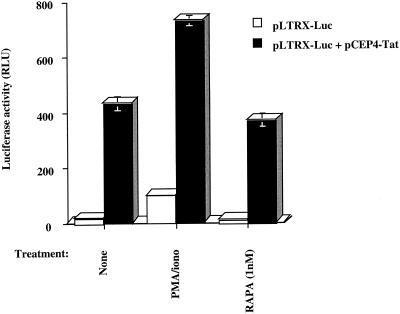
Our results obtained from the experiment with fully infectious viruses showed that rapamycin significantly downregulates HIV-1 production in CD4-expressing T cells. To assess if this effect could be dependent on transactivation of the HIV-1 long terminal repeat by Tat protein, we transiently transfected freshly purified primary CD4+ T cells with pLTRX-Luc alone or in combination with pCEP4-Tat, a Tat-encoding vector. These transfections were done with the Nucleofector technology, which consists of a specific electrical field that transiently alters cell membranes so that DNA is transported directly into the cell's nucleus. Cells were next either left untreated or treated for 24 h with a phorbol myristate acetate/ionophore combination as a positive control (20 ng/ml and 1 μM, respectively) or rapamycin (1.1 nM). We first observed that basal long terminal repeat expression was downregulated by 32% following rapamycin treatment (Fig. 3, 18.2 versus 12.4 relative light units [RLU]). As expected, cotransfection of a Tat-encoding vector led to a significant enhancement in long terminal repeat activity. The Tat-dependent induction of HIV-1 long terminal repeat-driven reporter gene activity was less affected by treatment with rapamycin than basal long terminal repeat activity (14% decrease, 431.8 versus 370.3 RLU).
FIG. 3.
Rapamycin inhibits basal HIV-1 long terminal repeat activity and minimally affects Tat-mediated long terminal repeat transactivation. Freshly isolated primary human CD4+ T lymphocytes (105) were transiently transfected with pLTRX-Luc or cotransfected with pLTRX-Luc and pCEP4-Tat. Cells were then either left untreated or treated with phorbol myristate acetate/ionophore (PMA/iono, 20 ng/ml and 1 μM, respectively) or rapamycin (RAPA, 1.1 nM). Cells were incubated at 37°C for 24 h and then lysed for monitoring luciferase activity. Results shown are the means ± standard deviation of quadruplicate samples and are representative of at least three independent experiments.
In an attempt to evaluate if the observed effect of rapamycin on HIV-1 replication could be the reflection of cell events rather than a specific effect on HIV-1, we transfected WE17/10 cells with a β-galactosidase vector driven by the constitutive β-actin promoter (pRc/actin LacZ), and cells were next either left untreated or treated for 72 h with rapamycin (1.1 nM). No significant downregulation of β-galactosidase activity was observed following treatment of WE17/10 cells with rapamycin (data not shown).
Rapamycin acts via the FK506-binding protein family.
It is well documented that the effect of rapamycin is mediated through FRAP upon its binding to FKBP12. In an attempt to demonstrate that the observed inhibition of HIV-1 replication by rapamycin is specific to its binding to FKBP12, the immunosuppressant drug FK506, which was also shown to bind to FKBP12, was used to compete the binding of rapamycin to FKBP12.
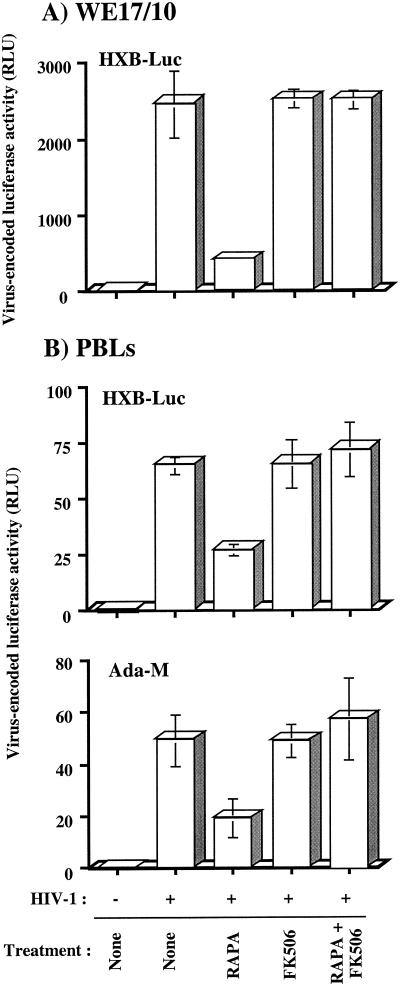
WE17/10 cells were initially either left untreated or treated with rapamycin or FK506 alone or with both agents for 30 min prior to infection. Cells were then infected with HXB-Luc and kept in culture in the absence or presence of rapamycin, FK506, or both agents for 72 h. Again, a significant diminution (about 80%) of reporter gene activity was seen following treatment of cells with rapamycin, and FK506 alone had no effect on HIV-1 long terminal repeat-driven luciferase activity (Fig. 4A). Such rapamycin-dependent diminution of HIV-1 reporter gene expression was totally abolished when FK506 was used in combination with rapamycin. These results suggest that the inhibitory effect of rapamycin on HIV-1 replication was linked to its capacity to bind to FKBP12.
FIG. 4.
Rapamycin inhibits virus transcription in WE17/10 and primary human cells in an FK506-dependent manner. (A) WE17/10 cells (105) were either left untreated or treated for 30 min with rapamycin (RAPA, 1.1 nM), FK506 (100 ng/ml), or both rapamycin and FK506 simultaneously. Cells were then either left uninfected or infected with HXB-Luc particles (10 ng of p24). (B) Freshly isolated PBLs fromthree healthy donors were kept in culture for 48 h in the presence of phytohemagglutinin protein (3 μg/ml) and rhIL-2 (50 U/ml). PBLs (105) were then either left untreated or treated for 30 min with rapamycin (1.1 nM), FK506 (100 ng/ml), or both rapamycin and FK506 simultaneously. Cells were then either left uninfected or infected with HXB-Luc particles (10 ng of p24) or Ada-M-pseudotyped viruses. Cells were incubated at 37°C for 72 h and then lysed for monitoring luciferase activity. Data from only one representative donor are presented. Results shown are the means ± standard deviation of quadruplicate samples and are representative of at least three independent experiments.
The physiological significance of our findings was tested with freshly isolated primary human cells as targets and reporter viruses pseudotyped with T-tropic and M-tropic envelope proteins. PBLs from three healthy volunteers were initially either left untreated or treated with rapamycin, FK506, or the combination of rapamycin and FK506 for 30 min prior to infection with similar amounts of T-tropic and M-tropic HIV-1 pseudotypes. Cells were then kept in culture for 72 h in the presence or absence of rapamycin, FK506, or both agents. Again, rapamycin was found to decrease HIV-1 long terminal repeat-dependent reporter gene activity at various levels depending on the donor tested. Figure 4B presents data from a single donor that are representative of the two other donors tested. It is of interest that replication of both T-tropic and M-tropic HIV-1 pseudotypes was similarly affected by rapamycin treatment. This series of experiments indicated that the observed inhibitory effect of rapamycin on HIV-1 replication is not an epiphenomenon, since similar findings were made in primary human cells from different donors and were not influenced by the tropism of the virus.
Downstream event in HIV-1 replicative cycle is likely target of rapamycin.
In an attempt to shed light on the mechanism through which rapamycin negatively affects the biology of HIV-1, kinetic studies of virus infection were performed to determine the optimal incubation period to achieve maximal inhibition of HIV-1 infection. To this end, WE17/10 cells were again infected with T-tropic luciferase reporter viruses and then either left untreated or treated with rapamycin (1.1 nM) at different time points following virus infection (0, 2, 4, 6, 8, 12, 24, and 48 h). Cells were kept in culture for 72 h prior to monitoring luciferase activity. Our results showed that HIV-1 long terminal repeat-dependent reporter gene expression was reduced by close to 90% when rapamycin was added 2, 4, and 6 h following inoculation of WE17/10 cells with HXB-Luc particles (Fig. 5). A significant decrease in virus-encoded gene expression (i.e., 50%) was still observed when rapamycin was added as late as 48 h after virus infection. This series of experiments suggested that the mechanism of action of rapamycin is not directed toward the initial steps in the HIV-1 life cycle (i.e., attachment and entry).
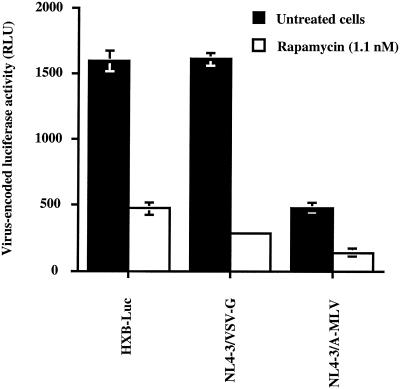
To further substantiate the notion that rapamycin is not modulating the initial steps of HIV-1 replication, CHCD4 and WE17/10 cells were infected with pseudotypes bearing heterologous Env glycoproteins. Luciferase-encoding HIV-1 particles were pseudotyped with vesicular stomatitis virus envelope glycoprotein G and amphotropic murine leukemia virus Env to produce viral entities that could enter target cells independently of CD4 and chemokine coreceptors. Previous works have indicated that vesicular stomatitis virus envelope glycoprotein G-pseudotyped viruses penetrate the cell by endocytosis (42), whereas virions pseudotyped with amphotropic murine leukemia virus Env enter the cell upon an interaction with Pit2, a sodium-dependent phosphate symporter (67). Rapamycin was found to efficiently diminish infection of CHCD4 cells with the two pseudotyped viruses (Fig. 6). Similar observations were made when WE17/10 cells were used as target cells (data not shown). Altogether, the results from these experiments support the idea that rapamycin has no effect on the process of virus attachment or entry but is rather targeting a subsequent step(s) in the HIV-1 life cycle, i.e., reverse transcription, integration, and/or provirus transcription.
FIG. 6.
Rapamycin abolishes infection with amphotropic murine leukemia virus (A-MLV)- and vesicular stomatitis virus envelope glycoprotein G (VSV-G)-pseudotyped HIV-1 particles. CHCD4 cells (105) were infected with HXB-Luc and HIV-1 pseudotypes (10 ng of p24) and either left untreated or treated with rapamycin (1.1 nM). Cells were kept in culture for a total of 72 h before monitoring luciferase activity. Results shown are the means ± standard deviation of quadruplicate samples and are representative of at least three independent experiments.
Downregulation of HIV-1 expression by rapamycin is due to an effect at the transcriptional level.
Northern blot analysis was next performed with a DNA probe which detects three specific viral RNA species, the full-length unspliced 9.2-kb mRNA, the intermediate 4.3-kb mRNAs, and the small multiply spliced 2-kb mRNAs. In this series of investigations, WE17/10 cells were either left uninfected or infected with fully infectious HIV-1NL4-3 particles for 6 days and next treated with rapamycin for an additional 3 days. Total virus-encoding RNA was finally analyzed with a radiolabeled probe specific for NL4-3. As shown in Fig. 7, the level of viral RNAs was diminished by rapamycin treatment. The 9.2- and 4.3-kb mRNAs were quantified with an Alpha Imager 2000 digital imaging and analysis system. The arbitrary units obtained showed that both the 9.2-kb and 4.3-kb mRNA signals were diminished by rapamycin (73,625 versus 38,000 for the 9.2-kb mRNA and 35,625 versus 28,625 for the 4.3-kb mRNA). Normalization of our data to the 28S rRNA indicated that the 9.2-kb mRNAs were reduced by 47.9%, whereas the 4.3-kb mRNAs were diminished by 18.9% upon treatment with rapamycin (Table 1). The rRNA was not affected by rapamycin treatment, suggesting a certain specificity of the mechanism of action of this immunosuppressive agent.
TABLE 1.
Quantification of the 9.2-kb and 4.3-kb HIV-1 mRNAs
| RNA | Amt (arbitrary units)
|
% Inhibition | % Inhibition normalized to 28S rRNA intensity | |
|---|---|---|---|---|
| Untreated | Rapamycin-treated | |||
| 9.2-kb mRNA | 73,625 | 38,000 | 48.4 | 47.9 |
| 4.3-kb mRNA | 35,625 | 28,625 | 19.6 | 18.9 |
| 28S rRNA | 626,076 | 619,938 | 0.98 | |
DISCUSSION
The immunosuppressive drugs rapamycin and FK506 interfere with signal transduction pathways required for T-cell activation and growth and are currently used to prevent graft rejection in humans. Although both rapamycin and FK506 bind to the same intracellular receptor, FKBP12, they have distinct mechanisms of action. The FK506/FKBP12 complex binds to and inhibits calcineurin, a Ca2+-dependent serine-threonine phosphatase known to be a critical component of the T-cell receptor-linked signal transduction pathway leading to cytokine gene transcription (20, 49), whereas the rapamycin/FKBP12 complex associates with a recently defined target protein termed FRAP (3, 47). In addition, it was demonstrated that rapamycin selectively represses translation of a subset of mRNAs bearing a 5′-polypyrimidine motif that include, among others, those encoding ribosomal proteins and elongation factors. This observation, coupled with the presence of a suboptimal 5′-polypyrimidine tracts that is required for viral tat mRNA production (33, 62-64, 68), led us to test the ability of rapamycin to modulate replication of HIV-1.
Our results first demonstrated that rapamycin was effective at inhibiting infection of human T lymphoid cells with luciferase reporter viruses. To the best of our knowledge, this is the first demonstration that rapamycin can affect the biology of HIV-1. Similar observations made in primary human cells, i.e., PBLs, confirm the consistency of this phenomenon and provide a physiological significance for such findings. Several avenues of investigation were explored to identify the mechanism(s) responsible for the rapamycin-mediated abrogation of HIV-1 promoter activity. We initially defined that diminution of expression of virus-mediated reporter gene occurs through the FKBP12 pathway, since the presence of FK506, which also binds FKBP12, completely abolished the inhibitory effect of rapamycin. Data from a time course experiment revealed that rapamycin added as late as 48 h after infection interfered with virus-dependent reporter gene activity, suggesting that rapamycin is not affecting the process of HIV-1 attachment and entry. This postulate was confirmed by experiments with amphotropic murine leukemia virus- and vesicular stomatitis virus envelope glycoprotein G-pseudotyped HIV-1 particles. Finally, Northern blot analysis suggested that rapamycin is most likely acting at the transcriptional level, since viral RNA was diminished upon treatment of human T lymphoid cells with rapamycin. However, it cannot be excluded that the stability of viral RNA could also be affected by rapamycin, as it has been reported for IL-2 and granulocyte-macrophage colony-stimulating factor mRNAs (30, 78). Pulse-chase experiments will be required to assess this possibility. Further experiments will also be necessary to rule out any effect of rapamycin on either reverse transcription or integration of viral cDNA into the host cell genome.
The involvement of Tat in the observed rapamycin-dependent inhibitory effect on HIV-1 replication was evaluated because of previous studies showing that the primary transcript of Tat bears a polypyrimidine motif (33, 62-64, 68). In addition to Tat, other proteins are known to be necessary to achieve efficient transcriptional activation of HIV-1 (e.g., cyclin T1 and CDK9) (reviewed in references 58 and 69). Thus, proteins with which Tat associates within the Tat-binding region of the long terminal repeat represent additional targets for rapamycin that could indirectly affect HIV-1 transcription. Surprisingly, transfection experiments suggested that Tat and its associated cellular proteins are less affected by rapamycin than basal HIV-1 long terminal repeat-dependent activity. Additional studies are needed to clarify the precise mechanism(s) through which rapamycin exerts its inhibitory effect on HIV-1.
Our results do not allow us to completely eliminate the possibility that rapamycin-mediated diminution of HIV-1 replication is due to a nonspecific effect on the cellular transcriptional machinery. A recent work by Grolleau and colleagues showed that many mRNAs are repressed in rapamycin-treated T cells (29). For instance, β-actin mRNAs were abolished by rapamycin in addition to many growth control genes, suggesting that our results could be partly the consequence of nonspecific cellular events. However, it should be stressed that the present work is in sharp contrast with such findings, since we found that β-actin was not affected by rapamycin. Such a discrepancy can be related to differences in the experimental settings, e.g., the cell line tested (WE17/10 in the present study versus Jurkat in Grolleau's study) and the doses of rapamycin used (20-fold more in Grolleau's work).
Host-derived human cyclophilins A (CypA) and B bind specifically to the HIV-1 Gag polyprotein p55gag in vitro (9). The Gag-CypA interaction has been shown to be essential for the life cycle of HIV-1 (27), and virus infectivity is finely tuned by CypA expression level (77). Interestingly, the association of CypA with HIV-1 particles is inhibited by the immunosuppressive agent cyclosporin A (71). In addition to having a direct effect on HIV-1 Gag by affecting the maturation process of HIV-1 (65), cyclosporin A was shown to selectively inhibit HIV-1 replication but not HIV-2 or simian immunodeficiency virus (6, 71). More relevant to the present study, rapamycin was found to repress CypA (29). It can thus be proposed that HIV-1 maturation and infectivity can be negatively affected by rapamycin treatment through a modulatory effect on CypA expression.
Data from in vitro experimental studies demonstrate that rapamycin is not toxic for T cells even at a concentration of 110 nM (39). Rapamycin was reported to arrest the growth of murine IL-2-dependent CTLL-2 cells near the G1/S-phase boundary when used at a concentration as low as 1 nM (47). Results from clinical studies indicate that rapamycin is rapidly absorbed following its administration, with a mean time to peak concentration of approximately 1 h after a single dose in healthy human subjects and approximately 2 h after multiple oral doses in renal transplant recipients (http://www.fda.gov/ohrms/dockets/ac/02/briefing/3832b1_03_FDA-RapamuneLabel.htm).
The systemic availability of rapamycin was estimated to be approximately 14% after an oral administration of this compound. The means of maximal concentrations in renal transplant patients with multiple oral doses of 2 and 5 mg of rapamycin were 12.2 ± 6.8 and 37.4 ± 21 nM, respectively. In such patients, rapamycin was administered 4 h after cyclosporin A. The therapeutic trough levels with a full dose of cyclosporin A are 9.8 and 18.6 nM following a daily administration of rapamycin of 2 and 5 mg, respectively. In healthy human subjects, the mean maximal concentration is 85.6 ±20.0 nM following a single 15-mg oral dose of rapamycin.
Our observation indicating that HIV-1 replication is inhibited by 1 nM rapamycin reveals some physiological significance when considering such previous in vitro and in vivo studies. It should be added that in vivo studies also reported that rapamycin acts as a substrate for both the cytochrome P450 IIIA4 enzyme system and P-glycoprotein. Consequently, it is not surprising to find that several drug-drug interactions occur in patients undergoing rapamycin therapy. For example, diltiazem, verapamil, and erythromycin were shown to increase rapamycin levels, while carbamazepine, phenobarbital, and phenytoin were shown to diminish serum levels of rapamycin. Potential interactions with drugs currently in use for the treatment of HIV-1-infected individuals are also possible, considering that protease inhibitors are primarily metabolized by the cytochrome P450 IIIA4 enzyme system.
In summary, our findings indicate that rapamycin can potently inhibit HIV-1 replication in both human T lymphoid (i.e., WE17/10 and CHCD4) and primary mononuclear cells (i.e., PBLs). Although such an inhibition is not as potent as the one observed with highly active antiretroviral therapy, such an immune-based therapeutic intervention may represent a valuable complement to highly active antiretroviral therapy for treating HIV-1 infection. A recent study in which cyclosporin A was used in combination with highly active antiretroviral therapy demonstrated that a rapid shutdown of T-cell activation in the early phases of primary HIV-1 infection could have long-term beneficial effects and establish a more favorable immunologic set point that affects the ultimate pattern and rate of disease progression (56). Results from other studies with the immunosuppressive agent mycophenolate also suggested that immune-based therapeutic interventions could be an interesting avenue for the treatment of HIV-1 infection (22, 31, 53, 57).
Further studies are required to define the utility of using rapamycin in combination with current highly active antiretroviral therapy. A detailed understanding of the rapamycin-mediated signaling pathway is warranted to identify the various intracellular components playing a role in the anti-HIV-1 properties of this potent immunosuppressive drug.
Acknowledgments
J.R. and J.-S.P. contributed equally to this work.
We thank Corinne Barat for critical reading of the manuscript and helpful discussions.
This study was supported by grants to M.J.T. from the Canadian Institutes of Health Research (CIHR) HIV/AIDS Research Program (HOP-14438, HOP-15575, and MOP-37781). J.R. is the recipient of a CIHR Doctoral Award, and M.J.T. holds a Tier 1 Canada Research Chair in Human Immuno-Retrovirology.
REFERENCES
- 1.Abraham, R. T., and G. J. Wiederrecht. 1996. Immunopharmacology of rapamycin. Annu. Rev. Immunol. 14:483-510. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, A., H. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albers, M. W., R. T. Williams, E. J. Brown, A. Tanaka, F. L. Hall, and S. L. Schreiber. 1993. FK506-binding proteins-rapamycin inhibits a cyclin-dependent kinase activity and a cyclin D1-Cdk association in early G1 of an osteosarcoma cell line. J. Biol. Chem. 268:22825-22829. [PubMed] [Google Scholar]
- 4.Bardini, G., M. C. Re, G. Rosti, and A. R. Belardinelli. 1991. HIV infection and bone-marrow transplantation. Lancet 337:1163-1164. [DOI] [PubMed] [Google Scholar]
- 5.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11:S3-S16. [PubMed] [Google Scholar]
- 6.Billich, A., F. Hammerschmid, P. Peichl, R. Wenger, G. Zenke, V. Quesniaux, and B. Rosenwirth. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 69:2451-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleul, C. C., M. Farzan, H. Choe, C. Parolin, I. Clark-Lewis, J. Sodroski, and T. A. Springer. 1996. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382:829-833. [DOI] [PubMed] [Google Scholar]
- 8.Bounou, S., J. E. Leclerc, and M. J. Tremblay. 2002. The presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4+ T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol. 76:1004-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporin A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 11.Burkly, L., N. Mulrey, R. Blumenthal, and D. S. Dimitrov. 1995. Synergistic inhibition of human immunodeficiency virus type 1 envelope glycoprotein-mediated cell fusion and infection by an antibody to CD4 domain 2 in combination with anti-gp120 antibodies. J. Virol. 69:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttke, T., J. McCubrey, and T. Owen. 1993. Use of an aqueous soluble tetrazolium/formazan assay to measure viability and proliferation of lymphokine-dependent cell lines. J. Immunol. Methods 157:233-240. [DOI] [PubMed] [Google Scholar]
- 13.Cantin, R., J.-F. Fortin, G. Lamontagne, and M. Tremblay. 1997. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J. Virol. 71:1922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715-728. [DOI] [PubMed] [Google Scholar]
- 15.Chen, B., K. Saksela, R. Andino, and D. Baltimore. 1994. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J. Virol. 62:654-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu, M. I., H. Katz, and V. Berlin. 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91:12574-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. Ponath, L. Wu, C. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 18.Chomczynski, P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532-536. [PubMed] [Google Scholar]
- 19.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 20.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357:695-697. [DOI] [PubMed] [Google Scholar]
- 21.Cocchi, F., A. L. DeVico, A. Garzino-Demo, S. K. Arya, R. C. Gallo, and P. Lusso. 1995. Identification of RANTES, MIP-1α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811-1815. [DOI] [PubMed] [Google Scholar]
- 22.Coull, J. J., D. Turner, T. Melby, M. R. Betts, R. Lanier, and D. M. Margolis. 2001. A pilot study of the use of mycophenolate mofetil as a component of therapy for multidrug-resistant HIV-1 infection. J. Acquired Immune Defic. Syndr. 26:423-434. [DOI] [PubMed] [Google Scholar]
- 23.Daelemans, D., A. M. Vandamme, and E. De Clercq. 1999. Human immunodeficiency virus gene regulation as a target for antiviral chemotherapy. Antivir. Chem. Chemother. 10:1-14. [DOI] [PubMed] [Google Scholar]
- 24.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. Sutton, C. Hill, C. David, S. Peiper, T. Schall, D. Littman, and N. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 25.Dumont, F. J., M. J. Staruch, S. L. Koprak, M. R. Melino, and N. H. Sigal. 1990. Distinct mechanisms of suppression of murine T-cell activation by the related macrolides FK-506 and rapamycin. J. Immunol. 144:251-258. [PubMed] [Google Scholar]
- 26.Fortin, J.-F., R. Cantin, G. Lamontagne, and M. Tremblay. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J. Virol. 71:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franke, E. K., and J. Luban. 1996. Inhibition of HIV-1 replication by cyclosporin A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222:279-282. [DOI] [PubMed] [Google Scholar]
- 28.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 29.Grolleau, A., J. Bowman, B. Pradet-Balade, E. Puravs, S. Hanash, J. A. Garcia-Sanz, and L. Beretta. 2002. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 277:22175-22184. [DOI] [PubMed] [Google Scholar]
- 30.Hanke, J. H., L. N. Nichols, and M. E. Coon. 1992. FK506 and rapamycin selectively enhance degradation of IL-2 and GM-CSF mRNA. Lymphokine Cytokine Res. 11:221-231. [PubMed] [Google Scholar]
- 31.Hossain, M. M., J. J. Coull, G. L. Drusano, and D. M. Margolis. 2002. Dose proportional inhibition of HIV-1 replication by mycophenolic acid and synergistic inhibition in combination with abacavir, didanosine, and tenofovir. Antiviral Res. 55:41-52. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson, S. K., R. Y. Calne, and T. G. Wreghitt. 1991. Outcome of HIV infection in transplant patient on cyclosporin. Lancet 337:794.. [DOI] [PubMed] [Google Scholar]
- 33.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29:464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jefferies, H. B., C. Reinhard, S. C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA 91:4441-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahan, B. D., S. Gibbons, N. Tejpal, S. M. Stepkowski, and T. C. Chou. 1991. Synergistic interactions of cyclosporin and rapamycin to inhibit immune performances of normal human peripheral blood lymphocytes in vitro. Transplantation 51:232-239. [DOI] [PubMed] [Google Scholar]
- 36.Kay, J. E., and C. R. Benzie. 1989. T lymphocyte activation through the C28 pathway is insensitive to inhibition by the immunosuppressive drug FK-506. Immunol. Lett. 23:155-159. [DOI] [PubMed] [Google Scholar]
- 37.Kay, J. E., L. Kromwel, S. E. Doe, and M. Denyer. 1991. Inhibition of T and B lymphocyte proliferation by rapamycin. Immunology 72:544-549. [PMC free article] [PubMed] [Google Scholar]
- 38.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 39.Kimball, P. M., R. H. Kerman, and B. D. Kahan. 1991. Production of synergistic but nonidentical mechanisms of immunosuppression by rapamycin and cyclosporin. Transplantation 51:486-490. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura, Y., T. Ishikawa, N. Okui, N. Kobayashi, T. Kanda, T. Shimada, K. Miyake, and K. Yoshiike. 1999. Inhibition of replication of HIV-1 at both early and late stages of the viral life cycle by single-chain antibody against viral integrase. J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 20:105-114. [DOI] [PubMed] [Google Scholar]
- 41.Lahm, H., and S. Stein. 1985. Characterization of recombinant human interleukin-2 with micromethods. J. Chromatogr. 326:357-361. [DOI] [PubMed] [Google Scholar]
- 42.Matlin, K. S., H. Reggio, A. Helenius, and K. Simons. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156:609-631. [DOI] [PubMed] [Google Scholar]
- 43.Matsushita, S. 2000. Current status and future issues in the treatment of HIV-1 infection. Int. J. Hematol. 72:20-27. [PubMed] [Google Scholar]
- 44.Mendez, R., M. G. Myers, Jr., M. F. White, and R. E. Rhoads. 1996. Stimulation of protein synthesis, eukaryotic translation initiation factor 4E phosphorylation, and PHAS-I phosphorylation by insulin requires insulin receptor substrate 1 and phosphatidylinositol 3-kinase. Mol. Cell. Biol. 16:2857-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michael, N. L., and J. P. Moore. 1999. HIV-1 entry inhibitors: evading the issue. Nat. Med. 5:740-742. [DOI] [PubMed] [Google Scholar]
- 46.Molnar-Kimber, K. L. 1996. Mechanism of action of rapamycin (Sirolimus, Rapamune). Transplant. Proc. 28:964-969. [PubMed] [Google Scholar]
- 47.Morice, W. G., G. J. Brunn, G. Wiederrecht, J. J. Siekierka, and R. T. Abraham. 1993. Rapamycin-induced inhibition of p34cdc2 kinase activation is associated with G1/S-phase growth arrest in T lymphocytes. J. Biol. Chem. 268:3734-3738. [PubMed] [Google Scholar]
- 48.Nielsen, F. C., L. Ostergaard, J. Nielsen, and J. Christiansen. 1995. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature 377:358-362. [DOI] [PubMed] [Google Scholar]
- 49.O'Keefe, S. J., J. Tamura, R. L. Kincaid, M. J. Tocci, and E. A. O'Neill. 1992. FK-506- and cyclosporin A-sensitive activation of the interleukin-2 promoter by calcineurin. Nature 357:692-694. [DOI] [PubMed] [Google Scholar]
- 50.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J.-L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J.-M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetsher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 51.Okamoto, H., T. P. Cujec, B. M. Peterlin, and T. Okamoto. 2000. HIV-1 replication is inhibited by a pseudo-substrate peptide that blocks Tat transactivation. Virology 270:337-344. [DOI] [PubMed] [Google Scholar]
- 52.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Press, N., G. Kimel, M. Harris, B. Yip, K. J. Craib, and J. S. Montaner. 2002. Case series assessing the safety of mycophenolate as part of multidrug rescue treatment regimens. HIV Clin. Trials 3:17-20. [DOI] [PubMed] [Google Scholar]
- 54.Proudfoot, A. E., T. N. Wells, and P. R. Clapham. 1999. Chemokine receptors—future therapeutic targets for HIV? Biochem. Pharmacol. 57:451-463. [DOI] [PubMed] [Google Scholar]
- 55.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 56.Rizzardi, G. P., A. Harari, B. Capiluppi, G. Tambussi, K. Ellefsen, D. Ciuffreda, P. Champagne, P. A. Bart, J. P. Chave, A. Lazzarin, and G. Pantaleo. 2002. Treatment of primary HIV-1 infection with cyclosporin A coupled with highly active antiretroviral therapy. J. Clin. Investig. 109:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rizzardi, G. P., A. Lazzarin, and G. Pantaleo. 2002. Potential role of immune modulation in the effective long-term control of HIV-1 infection. J. Biol. Regul. Homeost. Agents 16:83-90. [PubMed] [Google Scholar]
- 58.Romano, G., M. Kasten, G. De Falco, P. Micheli, K. Khalili, and A. Giordano. 1999. Regulatory functions of Cdk9 and of cyclin T1 in HIV tat transactivation pathway gene expression. J. Cell. Biochem. 75:357-368. [PubMed] [Google Scholar]
- 59.Rose, N., and A. Lever. 2001. The rapamycin sensitivity of human T-cell leukaemia virus type I-induced T-cell proliferation is mediated independently of the polypyrimidine motifs in the 5′ long terminal repeat. J. Gen. Virol. 82:435-439. [DOI] [PubMed] [Google Scholar]
- 60.Saha, K., M. Caruso, and D. J. Volsky. 1997. Human immunodeficiency virus type 1 (HIV-1) infection of herpesvirus saimiri-immortalized human CD4-positive T lymphoblastoid cells: evidence of enhanced HIV-1 replication and cytopathic effects caused by endogenous interferon-γ. Virology 231:1-9. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz, O., J. L. Virelizier, L. Montagnier, and U. Hazan. 1990. A microtransfection method with the luciferase-encoding reporter gene for the assay of human immunodeficiency virus long terminal repeat promoter activity. Gene 88:197-205. [DOI] [PubMed] [Google Scholar]
- 62.Si, Z., B. A. Amendt, and C. M. Stoltzfus. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15:4597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 68:3071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Streblow, D. N., M. Kitabwalla, M. Malkovsky, and C. D. Pauza. 1998. Cyclophilin a modulates processing of human immunodeficiency virus type 1 p55Gag: mechanism for antiviral effects of cyclosporin A. Virology 245:197-202. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki, Y. J., B. B. Aggarwal, and L. Packer. 1992. α-Lipoic acid is a potent inhibitor of NF-κB activation in human T cells. Biochem. Biophys. Res. Commun. 189:1709-1715. [DOI] [PubMed] [Google Scholar]
- 67.Tailor, C. S., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tange, T. O., and J. Kjems. 2001. Sf2/asf binds to a splicing enhancer in the third hiv-1 tat exon and stimulates u2af binding independently of the rs domain. J. Mol. Biol. 312:649-662. [DOI] [PubMed] [Google Scholar]
- 69.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 70.Terada, N., H. R. Patel, K. Takase, K. Kohno, A. C. Nairn, and E. W. Gelfand. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. USA 91:11477-11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 72.Thomas, G., and M. N. Hall. 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9:782-787. [DOI] [PubMed] [Google Scholar]
- 73.Vandamme, A. M., K. Van Vaerenbergh, and E. De Clercq. 1998. Anti-human immunodeficiency virus drug combination strategies. Antivir. Chem. Chemother. 9:187-203. [DOI] [PubMed] [Google Scholar]
- 74.Weller, I. V., and I. G. Williams. 2001. ABC of AIDS Antiretroviral drugs. Br. Med. J. 322:1410-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willard-Gallo, K., F. Van de Keere, and R. Kettmann. 1990. A specific defect in CD3 γ-chain gene transcription results in loss of T-cell receptor/CD3 expression late after human immunodeficiency virus of a CD4+ T-cell line. Proc. Natl. Acad. Sci. USA 87:6713-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yee, J. K., A. Miyanohara, P. Laporte, K. Bouic, B. J. C., and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 72:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng, X. X., T. B. Strom, and A. W. Steele. 1994. Quantitative comparison of rapamycin and cyclosporin effects on cytokine gene expression studied by reverse transcriptase-competitive polymerase chain reaction. Transplantation 58:87-92. [PubMed] [Google Scholar]