Abstract
In Saccharomyces cerevisiae, ASH1 mRNA is localized to the tip of daughter cells during anaphase of the cell cycle. ASH1 mRNA localization is dependent on four cis-acting localization elements as well as Myo4p, She2p, and She3p. Myo4p, She2p, and She3p are hypothesized to form a heterotrimeric protein complex that directly transports ASH1 mRNA to daughter cells. She2p is an RNA-binding protein that directly interacts with ASH1 cis-acting localization elements and associates with She3p. Here we report the identification of seven She2p mutants—N36S, R43A, R44A, R52A, R52K, R63A, and R63K—that result in the delocalization of ASH1 mRNA. These mutants are defective for RNA-binding activity but retain the ability to interact with She3p, indicating that a functional She2p RNA-binding domain is not a prerequisite for association with She3p. Furthermore, the nuclear/cytoplasmic distribution for the N36S and R63K She2p mutants is not altered, indicating that nuclear/cytoplasmic trafficking of She2p is independent of RNA-binding activity. Using the N36S and R63K She2p mutants, we observed that in the absence of She2p RNA-binding activity, neither Myo4p nor She3p is asymmetrically sorted to daughter cells. However, in the absence of She2p, Myo4p and She3p can be asymmetrically segregated to daughter cells by artificially tethering mRNA to She3p, implying that the transport and/or anchoring of the Myo4p/She3p complex is dependent on the presence of associated mRNA.
Keywords: SHE1/MYO4, SHE2, SHE3, IST2, Saccharomyces cerevisiae, mRNA localization, myosin, RNA-binding protein
INTRODUCTION
Generating cellular polarity requires the asymmetric sorting of specific proteins to distinct regions of a cell. RNA localization is one pathway by which proteins can be asymmetrically sorted, and numerous eukaryotic organisms rely on mRNA localization for the intracellular and intercellular sorting of proteins (Bashirullah et al. 1998; Palacios and St. Johnston 2001; Kloc et al. 2002). The initial event in mRNA localization is the identification of specific cis-acting sequences in the mRNA, termed zipcodes, by trans-acting mRNA-binding proteins (Kislauskis and Singer 1992; Bashirullah et al. 1998; Palacios and St. Johnston 2001; Kloc et al. 2002). Following specific recognition of the cis-acting element, mRNA localization can be achieved through a number of mechanisms: directed transport, generalized degradation with localized protection, and entrapment/anchoring of RNA at the site of localization (Bashirullah et al. 1998; Palacios and St. Johnston 2001; Kloc et al. 2002).
The sorting of Ash1p to daughter cell nuclei in Saccharomyces cerevisiae is a model system for investigating the asymmetric segregation of a cell-fate determinant by mRNA localization (Long et al. 1997; Takizawa et al. 1997). Asymmetric sorting of Ash1p results from transport and anchoring of ASH1 mRNA to the cortex of daughter cells during anaphase of the cell cycle (Long et al. 1997; Takizawa et al. 1997). In daughter cell nuclei, Ash1p functions as a specific transcriptional repressor of HO, restricting mating-type switching to mother cells (Bobola et al. 1996; Jansen et al. 1996; Sil and Herskowitz 1996; Maxon and Herskowitz 2001).
ASH1 mRNA contains four cis-acting localization elements: E1, E2A, E2B, and E3 (Chartrand et al. 1999; Gonzalez et al. 1999). Each of these cis-acting elements can independently localize a heterologous reporter mRNA to daughter cells (Chartrand et al. 1999; Gonzalez et al. 1999). In higher eukaryotic organisms, most cis-acting elements reside in the 3′-untranslated region (3′-UTR) of the mRNA (Bashirullah et al. 1998; Palacios and St. Johnston 2001; Kloc et al. 2002). The ASH1 E3 mRNA localization element is located predominantly in the 3′-UTR, whereas elements E1, E2A, and E2B reside within the ASH1 open reading frame (ORF; Long et al. 1997; Takizawa et al. 1997; Chartrand et al. 1999; Gonzalez et al. 1999). The sequence as well as the secondary structure for these elements appear crucial for mRNA localization activity (Chartrand et al. 1999; Gonzalez et al. 1999; Chartrand et al. 2002). The position of the cis-acting localization elements is not critical for ASH1 mRNA localization but apparently regulates the translation of Ash1p until its mRNA is properly localized (Chartrand et al. 2002).
In addition to the cis-acting localization elements, ASH1 mRNA localization requires numerous trans-acting factors: KHD1, LOC1, BUD6/AIP3, SHE1–5, as well as the actin cytoskeleton (Long et al. 1997; Takizawa et al. 1997; Beach and Bloom 2001; Long et al. 2001; Irie et al. 2002). SHE1/MYO4, SHE2, and SHE3 appear to have the most direct role in ASH1 mRNA localization. She1p was previously identified as Myo4p, a type V nonprocessive myosin that directly transports the ASH1 mRNA ribonucleoprotein particle to the bud tip and colocalizes with ASH1 mRNA at the distal tip of the bud (Haarer et al. 1994; Jansen et al. 1996; Bertrand et al. 1998; Beach et al. 1999; Münchow et al. 1999; Reck-Peterson et al. 2001). The C-terminal tail of Myo4p interacts with a domain in the N terminus of She3p (Böhl et al. 2000). The Myo4p/She3p complex does not associate with ASH1 mRNA independent of She2p (Münchow et al. 1999; Takizawa and Vale 2000). Furthermore, asymmetric sorting of Myo4p to daughter cells requires She2p (Münchow et al. 1999).
She2p directly and specifically interacts with each of the ASH1 cis-acting localization elements (Böhl et al. 2000; Long et al. 2000). Furthermore, She2p directly interacts with a domain in the C terminus of She3p (Böhl et al. 2000; Long et al. 2000). However, unlike Myo4p and She3p, which colocalize with ASH1 mRNA at the bud tip, She2p, when expressed at endogenous levels, is uniformly distributed throughout the cell (Böhl et al. 2000). These observations have led to the following model. The RNA-binding protein, She2p, recruits the Myo4p/She3p transport complex to ASH1 mRNA through interaction with She3p, and once the Myo4p/She3p/She2p/ASH1 mRNA ribonucleoprotein particle is assembled, it is competent for localization to daughter cells (Böhl et al. 2000; Long et al. 2000).
Apparently, She2p is a novel RNA-binding protein, because homology searches have not identified any known RNA-binding motifs within She2p, and the molecular details for the Myo4p/She3p/She2p/mRNA interactions remained to be characterized. Consequently, to study the assembly and transport of this complex in the absence of bound mRNA and to further characterize She2p RNA-binding activity, we sought to identify and characterize novel mutants of She2p specifically defective for RNA-binding activity. Using these mutants, we could investigate the various protein–protein interactions required for ASH1 mRNA localization in the absence of She2p RNA-binding activity. Here we report the identification of seven novel mutations in SHE2, resulting in amino acid substitutions N36S, R43A, R44A, R52A, R52K, R63A, and R63K, which are defective for mRNA localization and RNA-binding activity. The nuclear/cytoplasmic distribution for She2p mutants N36S and R63K is not altered, and both mutants retain the ability to interact with She3p, demonstrating that in vivo the She2p/She3p interaction is independent of associated mRNA. Furthermore, though all the protein–protein interactions required for mRNA localization are detected with She2p mutants N36S and R63K, the absence of She2p RNA-binding activity affects the sorting of Myo4p and She3p to daughter cells. However, the sorting of Myo4p and She3p to daughter cells in the absence of She2p can be rescued when RNA is artificially tethered to She3p. Consequently, this work provides new insight into the mechanistic details of mRNA localization in S. cerevisiae.
RESULTS
Identification of She2p point mutants defective for ASH1 mRNA localization
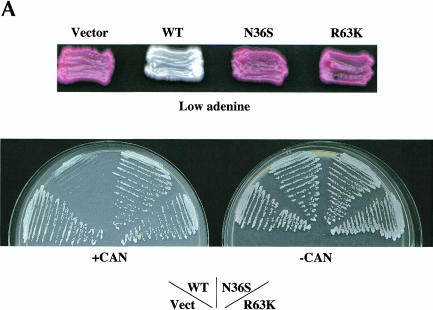
We isolated mutations in She2p defective for ASH1 mRNA localization by adapting a genetic selection/screen that monitors the asymmetric regulation of the HO promoter (Bobola et al. 1996; Jansen et al. 1996). Yeast strain YLM851 is deleted of SHE2 and contains ADE2 and CAN1 regulated by the HO promoter. In the absence of ADE2 expression, this strain appears red on media containing a low concentration of adenine. If CAN1 is expressed, the growth of this strain is sensitive to the arginine analog canavanine. When this strain was transformed with the vector plasmid YCplac111, the cells appeared red on media containing a low concentration of adenine and were insensitive to canavanine, indicating that both ADE2 and CAN1 expression were repressed in mother and daughter cells by the symmetric segregation of Ash1p (Fig. 1A ▶). In contrast, when this strain was transformed with a plasmid containing wild-type SHE2, the cells appeared white and were sensitive to canavanine because of the asymmetric regulation of ADE2 and CAN1, respectively (Fig. 1A ▶).
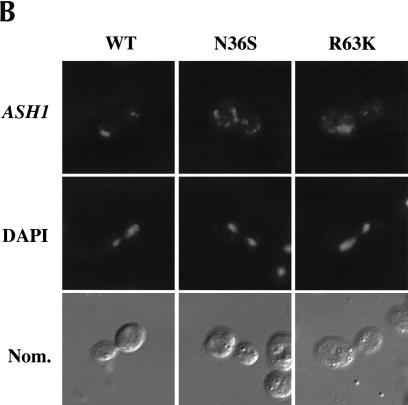
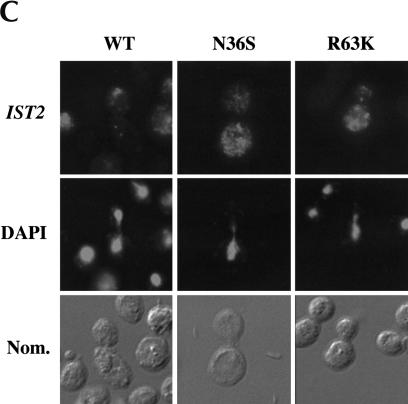
FIGURE 1.
ASH1 mRNA and IST2 mRNA are delocalized in cells expressing She2p mutants N36S and R63K. (A) Strain YLM851 was transformed with plasmids YCplac111 (vector), pRL199 (She2p-wt), pRL322 (She2p-R63K), or pRL324 (She2p-N36S). Transformants were patched on low adenine media devoid of leucine or streaked for single colonies on media devoid of leucine and arginine in the (+) presence and (−) absence of canavanine. (B) Strain YLM777 was transformed with plasmid C3431 (YEplac195/ASH1) and pRL199, pRL322, or pRL324. Transformants were grown in media devoid of leucine and uracil. Cells were processed for in situ hybridization using Cy3-conjugated complementary DNA probes for ASH1 mRNA. Representative images for in situ hybridization, DAPI staining, and (Nom.) Nomarski optics are shown. (C) Strain YLM777 was transformed with plasmid pRL277 [pHZ18(A)-IST2] and pRL199, pRL322, or pRL324. Cells were processed for in situ hybridization using Cy3-conjugated complementary DNA probes for lacZ-IST2 mRNA.
To identify mutants of She2p defective for ASH1 mRNA localization, yeast strain YLM851 was transformed with a randomly mutagenized plasmid pool of SHE2-myc6. Canavanine-resistant transformants that displayed the red adenine phenotype were chosen for further characterization. Mutant candidates were screened for full-length expression of She2p-myc6 by Western blot, and the DNA sequence was determined for candidates that expressed full-length protein. Here we report the characterization of two She2p mutants, N36S and R63K. Both mutants displayed the red adenine phenotype and were insensitive to canavanine (Fig. 1A ▶). Additional She2p mutants isolated from this selection/screen are listed in Supplemental Table S1, at http://www.mcw.edu/microbiology/rml/rna2003support.html.
The growth assays described above are indirect assays for the localization of ASH1 mRNA and Ash1p sorting. To directly observe the distribution of ASH1 mRNA in cells expressing the N36S and R63K She2p mutants, fluorescent in situ hybridization (FISH) was performed for ASH1 mRNA (Fig. 1B ▶). Compared with wild-type She2p, ASH1 mRNA was delocalized in cells expressing either the N36S or R63K She2p mutants (Fig. 1B ▶; Table 1 ▶). From the plate assays and FISH results, we conclude that ASH1 mRNA localization and Ash1p sorting are defective in cells expressing She2p mutants N36S and R63K.
TABLE 1.
ASH1 mRNA localization in cells expressing She2p mutants
| Plasmid/She2pa | ASH1 mRNA localizationb |
| pRL199/She2-wt | 48% |
| pRL324/She2p-N36S | 0% |
| pRL745/She2p-R43K | 54% |
| pRL740/She2p-R43A | 18% |
| pRL746/She2p-R44K | 52% |
| pRL741/She2p-R44A | 2% |
| pRL747/She2p-R49K | 46% |
| pRL742/She2p-R49A | 24% |
| pRL748/She2p-R52K | 0% |
| pRL743/She2p-R52A | 2% |
| pRL322/She2p-R63K | 0% |
| pRL744/She2p-R63A | 0% |
aThe indicated plasmids were transformed into strain YLM777 along with plasmid C3431 (YEplac195/ASH1).
bFifty anaphase cells with signal for ASH1 mRNA were counted. The percent of cells with localized ASH1 mRNA is indicated.
In S. cerevisiae, at least one other mRNA, IST2, localizes to daughter cells dependent on She2p (Takizawa et al. 2000). Consequently, we investigated if She2p mutants N36S and R63K also affected IST2 mRNA localization (Fig. 1C ▶). Identical to the ASH1 mRNA results, IST2 mRNA localization was defective in cells expressing either the N36S or R63K mutant, demonstrating that these two She2p mutants are defective for a common activity shared by the ASH1 and IST2 mRNA localization pathways.
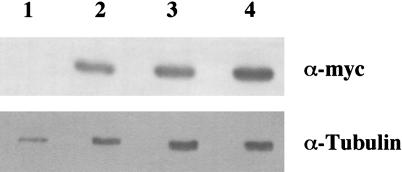
Altering the intracellular concentration of She2p could affect the localization of ASH1 and IST2 mRNA. Specifically, the N36S and R63K amino acid substitutions could reduce or increase the steady-state level of She2p, resulting in the delocalization of mRNA. The steady-state level for wild-type and mutant She2ps was determined by Western blot (Fig. 2 ▶). When normalized to the tubulin loading control, there was no apparent difference in the steady-state level of the N36S (Fig. 2 ▶, lane 3) and R63K (Fig. 2 ▶, lane 4) She2p mutants compared with the wild-type control (Fig. 2 ▶, lane 2). Therefore, this result implies that delocalization of mRNA in cells expressing either the N36S or R63K She2p mutants is not a result of differences in the intracellular concentration of She2p. Rather, we hypothesized that the N36S and R63K She2p amino acid substitutions affect She2p RNA-binding activity, the She2p–She3p interaction, or both.
FIGURE 2.
She2p amino acid substitutions N36S and R63K do not affect the steady-state level of She2p. Yeast strain YLM777 was transformed with plasmids (lane 1) YCplac111 (vector), (lane 2) pRL199 (She2p-wt), (lane 3) pRL324 (She2p-N36S), or (lane 4) pRL322 (She2p-R63K). Transformants were grown in media devoid of leucine, and cell lysates were prepared. Cell lysates were used for Western blotting with anti-myc monoclonal antibody or anti-α-tubulin antibody.
She2p mutants N36S and R63K are able to directly interact with She3p
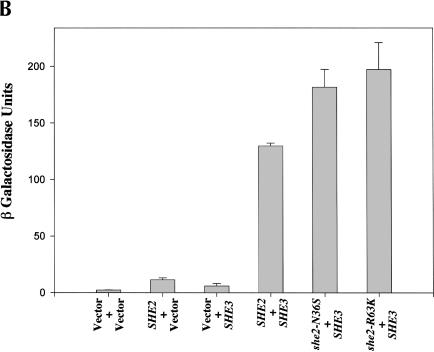
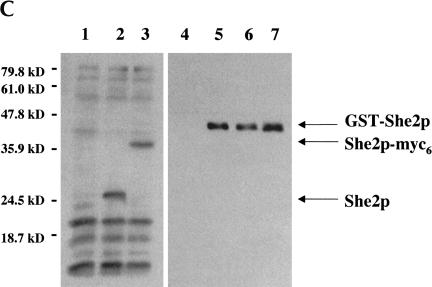
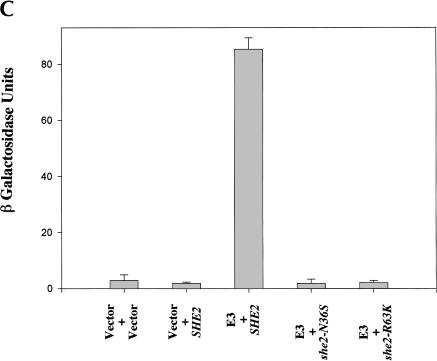
Proper ASH1 mRNA localization apparently requires She2p to interact with She3p (Böhl et al. 2000; Long et al. 2000). Therefore, we compared the ability of wild-type and mutant She2p to interact with She3p using an in vitro GST pull-down assay (Fig. 3A ▶). GST (Fig. 3A ▶, lane 1) alone was unable to pull down She3p, whereas wild-type GST-She2p (Fig. 3A ▶, lane 2) as well as GST-She2p mutants N36S (Fig. 3A ▶, lane 3) and R63K (Fig. 3A ▶, lane 4) were able to precipitate She3p. We further studied the She2p/She3p interaction in vivo by two-hybrid analysis. She2p mutants N36S and R63K expressed She3p-dependent lacZ activity at levels comparable to wild-type She2p (Fig. 3B ▶). These results demonstrate that She2p amino acid substitutions N36S and R63K do not alter the ability of She2p to directly interact with She3p in vitro and in vivo, implying that the overall structure of the N36S and R63K She2p mutants apparently is not altered by these two substitutions.
FIGURE 3.
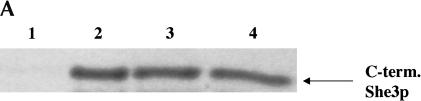
She2p mutants N36S and R36K are able to interact with She3p in vitro and in vivo. (A) In vitro GST pull-down assays were performed with 1 μg of (lane 1) GST, (lane 2) GST-She2p-wt, (lane 3) GST-She2p-N36S, or (lane 4) GST-She2p-R63K incubated with a C-terminal domain of She3p prepared by coupled in vitro transcription/translation. (B) She2p mutants N36S and R63K are able to interact with She3p by the two-hybrid assay. Strain PJ69-4a was transformed with the following combinations of plasmids: pGBDU-c1 (vector)/pACT2 (vector), pRL418 (pGBDU-c1-She2p-wt)/pACT2, pGBDU-c1/pEP13 (pACT2-She3p), pRL418/pEP13, pRL419 (pGBDU-c1-She2p-N36S)/pEP13, and pRL431 (pGBDU-c1-She2p-R63K)/pEP13. Transformants were grown in liquid culture and processed for β-galactosidase assays. (C) Characterization of the anti-She2p rabbit antiserum. Extracts were prepared from yeast strains YLM777 transformed with (lane 1) YCplac111 (vector), (lane 2) pRL453 (YCplac111-She2p), and (lane 3) pRL199 (YCplac111-She2p-myc6). Transformants of YLM777 were grown in synthetic media devoid of leucine. Lysates were prepared and used for Western blotting with anti-She2p rabbit antiserum. In addition, 5 ng of purified (lane 4) GST, (lane 5) GST-She2p-wt, (lane 6) GST-She2p-N36S, and (lane 7) GST-She2p-R63K were analyzed by Western blot with anti-She2p rabbit antiserum. The bands corresponding to She2p, She2-myc6, and GST-She2p are indicated by the arrows. (D) She2p mutants N36S and R63K associate with full-length endogenous She3p in vivo. Yeast strain YLM1320 was transformed with the following combinations of plasmids: (lane 1) YCplac22 (vector)/pRL453 (YCplac111-She2p-wt), (lane 2) pRL460 (YCplac22-She3p-myc6)/pRL453, (lane 3) pRL460/YCplac111 (vector), (lane 4) pRL460/pRL462 (YCplac111-She2p-N36S), or (lane 5) pRL460/pRL463 (YCplac111-She2p-R63K). Transformants were grown in synthetic media devoid of leucine as well as tryptophan, and lysates were prepared. An aliquot of the total lysate was analyzed by Western blot for She3p-myc6 and She2p (total). She3p-myc6 was immunoprecipitated from the lysate using anti-myc antibody, and the immunoprecipitates were analyzed by Western blot for the presence of She3p-myc6 and She2p (IP). She2p was detected in the immunoprecipitates using anti-She2 rabbit antiserum. Subsequently, the Western blot was stripped and reprobed with anti-myc to detect She3p-myc6.
We further characterized the interaction between the She2p mutants and She3p under physiological conditions. For these experiments, rabbit She2p antiserum was generated (Fig. 3C ▶). Although some cross-reactivity was observed with all yeast lysates tested (Fig. 3C ▶, lanes 1–3), this antiserum recognized a protein corresponding to the predicted molecular mass, 28.3 kD, for She2p in lysates containing endogenous levels of She2p (Fig. 3C ▶, lane 2), and this protein was absent in extracts prepared from cells deleted of She2p (Fig. 3C ▶, lane 1). This result indicated that the antiserum was capable of detecting She2p.
To conclusively demonstrate that the antiserum was detecting She2p, we performed two additional experiments. First, we probed yeast extracts prepared from a she2Δ strain expressing a She2p-myc6 construct (Fig. 3C ▶, lane 3). We did not observe the 28.3-kD form of She2p in this extract. Rather, because of the presence of the myc6 tag, we observed a slower-migrating form of She2p that was not detected in either of the other yeast lysates tested. In the second experiment, recombinant GST-She2p fusion proteins were purified from Escherichia coli, separated by SDS-PAGE, and detected by Western blot with the She2p antiserum. We observed that the antiserum had the ability to detect a protein corresponding to the molecular mass of wild-type GST-She2p (Fig. 3C ▶, lane 5), GST-She2p N36S (Fig. 3C ▶, lane 6), and GST-She2p R63K (Fig. 3C ▶, lane 7), and this protein was absent from the GST negative control (Fig. 3C ▶, lane 4). The sum of these results indicates that the antiserum has the ability to detect She2p in yeast lysates as well as recombinant She2p purified from E. coli.
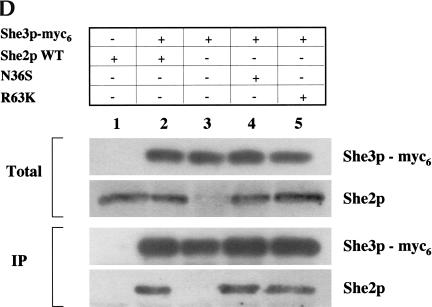
To study the interaction between full-length She3p-myc6 and mutant She2p, a coimmunoprecipitation experiment was performed. She3p-myc6 was immunoprecipitated, and coimmunoprecipitating proteins were analyzed for She2p by Western blot with the She2p antiserum (Fig. 3D ▶). Wild-type She2p (Fig. 3D ▶, lane 2) as well as She2p mutants N36S (Fig. 3D ▶, lane 4) and R63K (Fig. 3D ▶, lane 5) coimmunoprecipitated with She3p, indicating that under physiological conditions She2p amino acid substitutions N36S and R63K do not affect the ability of She2p to interact with She3p.
She2p mutants N36S and R63K are defective for association with ASH1 and IST2 mRNA
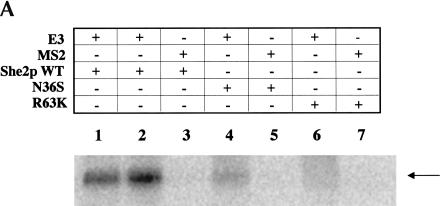
Because the She2p/She3p interaction was apparently not altered for the N36S and R63K She2p mutants, we suspected that these two mutants could be defective for RNA association. To explore this possibility, we first investigated the ability of the She2p mutants to interact with RNA by UV cross-linking (Fig. 4A ▶; Long et al. 2000). As previously observed (Long et al. 2000), a UV cross-link product corresponding to wild-type GST-She2p and the ASH1 E3 localization element was detected (Fig. 4A ▶, lanes 1,2). Furthermore, to demonstrate that the She2p RNA-binding activity is specific for the E3 localization element, we performed the UV cross-linking assay with MS2 RNA. The MS2 RNA, like E3, forms a stem–loop structure (Valegard et al. 1997). However, we observed no UV cross-link product corresponding to an She2p/MS2 RNA complex (Fig. 4A ▶, lane 3). From this result we can conclude that in vitro She2p RNA-binding activity is apparently specific for the E3 stem–loop structure.
FIGURE 4.
She2p mutants N36S and R63K are defective for ASH1 and IST2 mRNA-binding activity. (A) The UV cross-linking assay was performed by incubating (lane 1) 0.25 μg of GST-She2p-wt, (lanes 2,3) 0.5 μg of GST-She2p-wt, (lanes 4,5) 5.0 μg of GST-She2p-N36S, or (lanes 6,7) 5.0 μg of GST-She2p-R63K with 1 ng of (lanes 1,2,4,6) 32P-labeled E3 RNA or (lanes 3,5,7) MS2 RNA. The arrow indicates the position of the GST-She2p/RNA complex. (B) Apparent gel shift of She2p with E3 RNA is not caused by RNA–protein complex formation. (Lane 1) Trace (1 nM) labeled E3 RNA combined with (lane 2) mobility shift buffer (MSB) produces an apparent mobility shift in the absence of protein. Addition of (lanes 3–6) 125 nM She2p with (lanes 4–6) increasing molar equivalents of unlabeled E3 competitor RNA results in an increase, not a decrease, in the amount of the shifted species, and is consistent with the formation of a base-paired RNA duplex, not an RNA–protein complex. (C) She2p RNA-binding activity for mutants N36S and R63K as measured by three-hybrid assay. Strain YLM585 was transformed with the following combinations of plasmids: pIIIA-MS2–2(vector)/pGAD-c1 (vector), pIIIA-MS2-2/pRL445 (pGAD-c1-Shep-wt), pRL80 (pIIIA-MS2-2-E3)/pRL445, pRL80/pRL441 (pGAD-c1-She2p-N36S), and pRL80/pRL442 (pGAD-c1-She2p-R63K). Transformants were grown in liquid culture and processed for β-galactosidase assay. (D) She2p RNA-binding activity for She2p mutants N36S and R63K as measured by IP/RT-PCR. Yeast strain YLM777 was transformed with plasmid C3431 (YEplac195/ASH1) and plasmid (lanes 1,2) pRL199 (She2-myc6-wt), (lane 3) pRL324 (She2-myc6-N36S), or (lane 4) pRL322 (She2p-myc6-R63K). Transformants were grown in synthetic media devoid of leucine and uracil; lysates were prepared and immunoprecipitations were performed with anti-myc. Immunoprecipitations were subsequently used for RT-PCR reactions with primers specific for ASH1 mRNA or IST2 mRNA. RT-PCR reactions corresponding to an aliquot of the lysates prior to immunoprecipitation (total) are shown as well as reactions corresponding to the immunoprecipitation reactions with and without RT (+RT and −RT). Also, an aliquot of the immunoprecipitation was analyzed by Western blot using anti-myc.
Using the UV cross-linking assay, we investigated the in vitro RNA-binding activity for She2p mutants N36S and R63K. This experiment was performed identically to that for wild-type She2p with the exception that 10–20-fold more mutant She2p was incubated with the radiolabeled RNAs. We detected a faint UV cross-link product for both She2p N36S/E3 (Fig. 4A ▶, lane 4) and She2p R63K/E3 (Fig. 4A ▶, lane 6). Furthermore, we investigated if the RNA-binding specificity might be altered for She2p mutants N36S and R63K. Identically to wild-type She2p, we observed no UV cross-link product corresponding to She2p N36S/MS2 (Fig. 4A ▶, lane 5) or She2p R63K/MS2 (Fig. 4A ▶, lane 7). These results imply that these two amino acid substitutions apparently do not affect the specificity of RNA-binding activity, but greatly impair the She2p/E3 ASH1 mRNA association, most likely because of a reduced affinity for the RNA substrate.
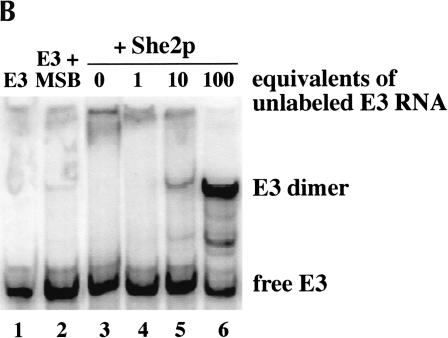
Given these observations, we sought to determine the relative binding affinities of wild-type She2p and the two She2p mutants for the E3 localization element. Using untagged She2p and in vitro transcribed RNA, we performed an extensive series of RNA-binding studies using a quantitative electrophoretic mobility shift assay (EMSA). However, despite considerable effort, we have been unable to identify a mobility shift that conclusively corresponds to direct and specific binding of She2p to either E1 or E3 RNA. Previously, an electrophoretic mobility shift was observed with GST-She2p and E3 (Böhl et al. 2000), but we now believe this shifted species represents an alternative RNA conformation rather than She2p:E3 binary complex. Notably, this alternative RNA conformation was further stimulated in a self-competition experiment, in which increasing amounts of unlabeled E3 RNA are titrated into a solution containing a trace amount of labeled E3 RNA and She2p (Fig. 4B ▶, lanes 3–6). Normally, unlabeled competitor RNA decreases the intensity of the shifted band through competition for protein binding. However, addition of competitor increases the amount of shifted species (Fig. 4B ▶), indicating that the shifted band is comprised of a dimeric E3 RNA. A similar result was also obtained for the E1 localization element (data not shown). Consequently, because of the inability to detect an She2p:E3 complex by electrophoretic mobility shift assay, we have been unable to determine the relative RNA-binding affinities for purified She2 and the She2p mutants. We conclude that weak She2p interactions with RNA can be trapped by UV cross-linking, but there is no evidence for a high-affinity binary complex between She2p and RNA.
To further explore the UV cross-linking results, the association between the She2p mutants and the E3 element was monitored in vivo by three-hybrid analysis. In the three-hybrid assay, the She2p RNA interaction was specific for the ASH1 E3 cis-acting element (Fig. 4C ▶; data not shown) and was independent of Myo4p and She3p (data not shown). Furthermore, the level of lacZ expression was at least 10-fold lower for the N36S and R63K She2p mutants when compared with wild-type She2p (Fig. 4C ▶). Therefore, these results are consistent with the in vitro UV cross-linking data demonstrating that She2p mutants N36S and R63K are defective for interaction with E3 ASH1 mRNA.
The experiments described above monitor She2p association with a single cis-acting localization element, E3, which is one of four cis-acting localization elements found in ASH1 mRNA. It therefore remained a formal possibility that She2p mutants N36S and R63K could remain associated with full-length ASH1 mRNA in vivo. Consequently, we investigated if She2p mutants N36S and R63K could associate with full-length ASH1 mRNA using an IP/RT-PCR assay (Fig. 4D ▶). Compared with wild-type She2p (Fig. 4D ▶, lane 2), we observed a reduction in the amount of the ASH1-specific IP/RT-PCR product coprecipitating with She2p mutants N36S (Fig. 4D ▶, lane 3) and R63K (Fig. 4D ▶, lane 4). Together, these results indicate that She2p mutants N36S and R63K are severely impaired for association with ASH1 mRNA compared with wild-type She2p. Furthermore, because these mutants retain the ability to interact with She3p in vivo, we can conclude that She2p association with RNA is not required for interaction with She3p.
Given that both ASH1 and IST2 mRNA localization were defective in cells expressing either She2p N36S or R63K mutants, we investigated if these mutants were also defective for association with IST2 mRNA (Fig. 4D ▶). Using the IP/RT-PCR assay, IST2 mRNA coprecipitated with wild-type She2p (Fig. 4D ▶, lane 2) but not with She2p mutants N36S (Fig. 4D ▶, lane 2) or R63K (Fig. 4D ▶, lane 4). Therefore, we conclude that She2p mutants N36S and R63K are unable to associate with either ASH1 mRNA or IST2 mRNA.
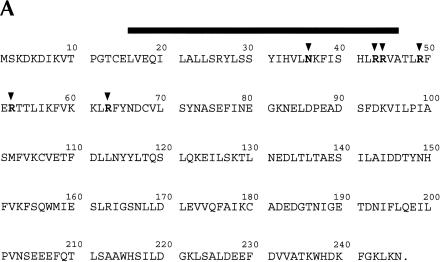
She2p RNA-binding activity is also dependent on arginine residues 43, 44, and 52
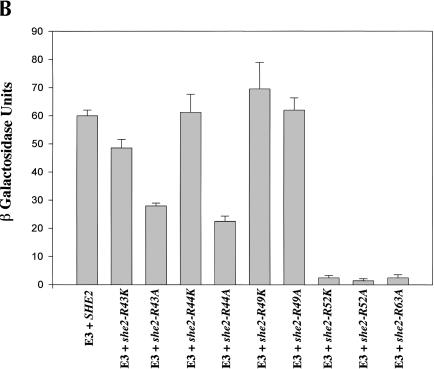
The observation that R63 is important for She2p RNA-binding activity provided a potential clue for defining an RNA-binding motif within She2p. Several RNA-binding motifs, the arginine-rich motif and the arg–gly–gly (RGG) box, contain arginine residues that are critical for RNA-binding activity. Five of the seven arginines in She2p reside in a relatively arginine-rich region encompassing amino acids 43–63 (Fig. 5A ▶). To determine whether these arginines are important for She2p RNA-binding activity, we individually converted R43, R44, R49, and R52 to alanine or lysine, and arginine residue 63 to alanine. The RNA-binding activity for each mutant was determined by three-hybrid analysis (Fig. 5B ▶). When compared with wild-type She2p, we observed that lacZ expression was reduced at least 10-fold for She2p mutants R52A, R52K, and R63A and twofold to threefold for She2p mutants R43A and R44A. We observed very little, if any, difference in the level of lacZ expression for She2p mutants R43K, R44K, R49A, and R49K compared with the wild-type control. Importantly, each of these mutants retains the ability to interact with She3p (data not shown). These results demonstrate that She2p residues R52 and R63 are critical for She2p RNA-binding activity; R43 and R44 are less important for She2p RNA-binding activity; and R49 apparently has very little, if any, role in She2p RNA-binding activity. Analogous to arginine 63, a positive charge at position 52 is not sufficient for She2p RNA-binding activity because the R52K She2p mutant is defective for RNA-binding activity. These observations imply that arginine residues 52 and 63 participate in RNA binding in a manner that cannot be mimicked by lysine residues.
FIGURE 5.
R43, R44, and R52 are required for She2p association with ASH1 mRNA. (A) Amino acid sequence for She2p with amino acids N36, R43, R44, R49, R52, and R63 indicated by the black arrowheads. The black rectangle indicates the region of She2p corresponding to the predicted leucine zipper motif. (B) She2p RNA-binding activity for mutants R43K, R43A, R44K, R44A, R49K, R49A, R52K, R52A, and R63A as measured by three-hybrid assay. Strain YLM585 was transformed with the following combinations of plasmids: pRL80 (pIIIA-MS2-2-E3)/pRL445 (pGAD-c1-She2p-wt), pRL80/pRL552 (pGAD-c1-She2p-R43K), pRL80/pRL669 (pGAD-c1-She2p-R43A), pRL80/pRL553 (pGAD-c1-She2p-R44K), pRL80/pRL670 (pGAD-c1-She2p-R44A), pRL80/pRL554 (pGAD-c1-She2p-R49K), pRL80/pRL671 (pGAD-c1-She2p-R49A), pRL80/pRL555 (pGAD-c1-She2p-R52K), pRL80/pRL672 (pGAD-c1-She2p-R52A), and pRL80/pRL673 (pGAD-c1-She2p-R63A). Transformants were grown in liquid culture and processed for β-galactosidase assays.
We next wanted to investigate whether She2p mutants that reduced the expression of lacZ in the three-hybrid assay also affected ASH1 mRNA localization (Table 1 ▶). We observed that ASH1 mRNA localization was reduced to varying degrees in cells expressing She2p mutants R43A, R44A, R52K, R52A, and R63A. Collectively, these results indicate a trend between reduced She2p RNA-binding activity and the ability to localize ASH1 mRNA. However, it is not a strict correlation because She2p mutants R43A and R44A have very similar, if not identical, RNA-binding activity (Fig. 5B ▶), whereas the R44A mutant is apparently more impaired for ASH1 RNA localization activity than the R43A mutant (Table 1 ▶). Furthermore, although we observed no reduction in lacZ expression in the two- and three-hybrid assays for She2p mutant R49A (Fig. 5B ▶; data not shown), we observed a modest reduction in the ability of this She2p mutant to localize ASH1 mRNA (Table 1 ▶). Therefore, although the two- and three-hybrid assays can reveal deficiencies in She2p protein–protein interactions and She2p RNA-binding activity, these assays may not completely recapitulate the in vivo physiology for She2p in ASH1 mRNA localization.
Amino acid substitutions N36S and R63K do not affect the nuclear/cytoplasmic distribution of She2p
In summary, we have investigated all of the known characteristics associated with She2p and have determined that amino acid substitutions N36S and R63K specifically affect only She2p RNA-binding activity. Using these two mutants of She2p, we were poised to investigate mechanistic details of ASH1 mRNA localization that were previously addressed only by deletion of SHE2 or large truncations in She2p.
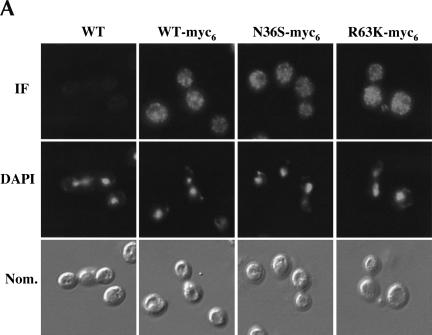
Under normal physiological conditions, Myo4p and She3p colocalize with ASH1 mRNA at the cortex of the bud and are excluded from the nucleus (Jansen et al. 1996; Bertrand et al. 1998; Münchow et al. 1999). In contrast, under identical conditions, She2p is not excluded from the nucleus and is uniformly distributed between mother and daughter cells (Jansen et al. 1996; Bertrand et al. 1998; Böhl et al. 2000). Furthermore, it has recently been shown that an N-terminal truncation of She2p, ΔN70, which affects both RNA-binding activity and the ability to interact with She3p, accumulates in the nucleus (Kruse et al. 2002). Using our She2p point mutants, we were able to specifically address if nuclear accumulation of She2p is a result of the loss of RNA-binding activity, the loss of interaction with She3p, or both. Immunofluorescence analysis revealed that the intracellular distribution of She2p mutants N36S and R63K was no different from that of wild-type She2p (Fig. 6A ▶), indicating that the absence of RNA-binding activity has no effect on the intracellular distribution of She2p.
FIGURE 6.
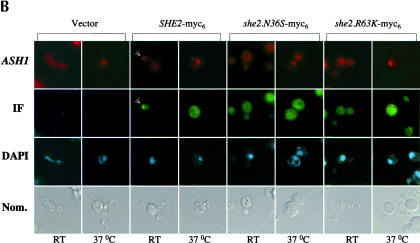
(A) The intracellular distribution of She2p mutants N36S and R63K is not altered. Representative images for the intracellular distribution of She2p-myc6. Yeast strain YLM777 was transformed with pRL453 (control), pRL199 (She2p-myc6-wt), pRL324 (She2p-myc6-N36S), or pRL322 (She2p-myc6-R63K). Transformants were grown in synthetic media minus leucine; subsequently, the cells were fixed and processed for immunofluorescence using anti-myc. (B) She2p does not accumulate in the nucleus in the absence of mRNA export. Yeast strain YLM1769 was transformed with the following combinations of plasmids: pRL718 (pESC-URA-ASH1)/YCplac22 (vector), pRL718/pRL676 (YCplac22-She2p-myc6-wt), pRL718/pRL677 (YCplac22-She2p-myc6-N36S), and pRL718/pRL678 (YCplac22-She2p-myc6-R63K). Transformants were grown at the permissive and nonpermissive temperatures as described in Materials and Methods, Temperature Shift Experiment. For immunofluorescence, anti-myc antibody was used and ASH1 mRNA was detected by FISH.
It remained a formal possibility that in vivo She2p mutants N36S and R63K retain a sufficiently low level of RNA-binding activity that results in a wild-type distribution of She2p. Mex67p is required for nuclear export of poly(A)+ RNA (Hurt et al. 2000), and it has recently been shown that when ASH1 mRNA is overexpressed, She2p accumulates in the nuclei of mex67 cells shifted to a nonpermissive temperature (Kruse et al. 2003). We reasoned that if She2p mutants N36S and R63K contain low levels of RNA-binding activity, then these two mutants should also accumulate in the nuclei of mex67 cells shifted to the nonpermissive temperature. The intracellular distribution of both wild-type and mutant She2p was determined by immunofluorescence, and the subcellular location of ASH1 mRNA was simultaneously detected by FISH in mex67 cells grown at permissive and nonpermissive temperatures (Fig. 6B ▶). As expected, we observed ASH1 mRNA (Fig. 6B ▶) and poly(A)+ RNA (data not shown) in the cytoplasm of mex67 cells grown at the permissive temperature and in the nuclei of mex67 cells shifted to the nonpermissive temperature. Furthermore, in mex67 cells expressing wild-type She2p grown at the permissive temperature, we observed localized ASH1 mRNA and a fraction of wild-type She2p colocalized with ASH1 mRNA at the bud tip (Fig. 6B ▶), consistent with previous observations (Böhl et al. 2000). In mex67 cells expressing She2p RNA-binding mutants, we did not observe localized ASH1 mRNA, and no She2p was observed at the bud tip (Fig. 6B ▶). At the nonpermissive temperature, we observed ASH1 mRNA accumulating in nuclei. In contrast to ASH1 mRNA, we did not observe nuclear accumulation of wild-type She2p in mex67 cells grown at the nonpermissive temperatures (Fig. 6B ▶). Our observation is not consistent with those previously reported (Kruse et al. 2002). Furthermore, the intracellular trafficking of She2p between the nucleus and cytoplasm in mex67 cells is not altered in the absence of She2p RNA-binding activity (Fig. 6B ▶). These results support the conclusions that nuclear export of She2p is independent of mRNA transport to the cytoplasm and that subcellular trafficking of She2p between the nucleus and cytoplasm is not dependent on RNA-binding activity.
She2p RNA-binding activity is required for proper asymmetric sorting of Myo4p and She3p
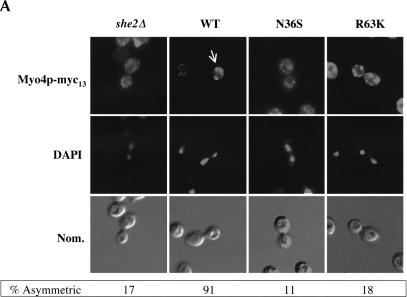
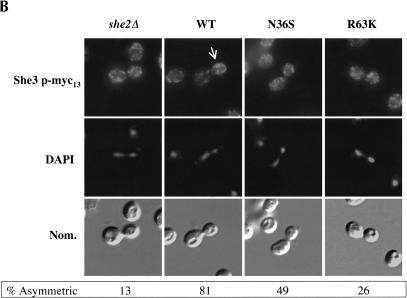
Myo4p and She3p are asymmetrically sorted to daughter cells (Jansen et al. 1996; Bertrand et al. 1998; Münchow et al. 1999). Sorting of Myo4p to daughter cells during anaphase is dependent on She2p (Münchow et al. 1999). However, because this observation was made with a strain deleted of SHE2, it remained to be determined if proper Myo4p sorting to daughter cells is dependent on She2p RNA-binding activity, the She2p–She3p interaction, or both. We investigated by immunofluorescence if She2p RNA-binding activity is required for proper asymmetric sorting of Myo4p and She3p to daughter cells. The intracellular distribution of Myo4p-myc13 was determined in yeast cells expressing either wild-type She2p or She2p mutants N36S and R63K (Fig. 7A ▶). As previously reported, Myo4p-myc13 was symmetrically distributed in she2 deletion cells compared with the wild-type control. We further observed that the intracellular distribution of Myo4p-myc13 was altered in cells expressing either the N36S or R63K She2p mutant. Analogous to Myo4p-myc13, the intracellular distribution of She3p-myc13 was altered in cells expressing either the N36S or R63K She2p mutant compared with the wild-type control (Fig. 7B ▶). Although the penetrance of the She3p sorting phenotype for the N36S She2p mutant was not as high as for the R63K mutant, the sorting of She3p in cells expressing the N36S mutant was defective compared with the wild-type control. The difference in penetrance of She3p sorting phenotype may be due to subtle differences in residual RNA association of the two mutants (Fig. 4D ▶). Regardless, we conclude that correct sorting of Myo4p and She3p to daughter cells is defective in cells expressing the She2p RNA interaction mutants.
FIGURE 7.
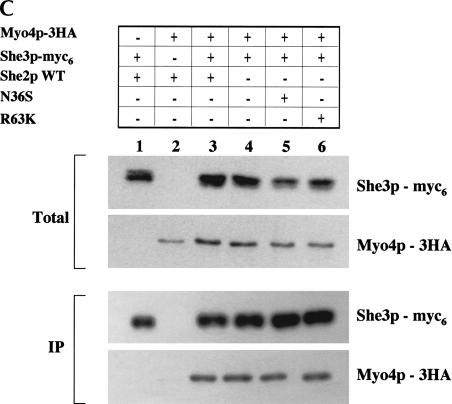
The asymmetric distribution of Myo4p and She3p in anaphase cells is dependent on She2p mRNA-binding activity, but formation of the Myo4p/She3p complex is independent of She2p RNA-binding activity. (A) Representative images for the intracellular distribution of Myo4p-myc13 in cells devoid of She2p, expressing wild-type She2p, She2p mutant N36S, and She2p mutant R63K. Strain YLM1309 was transformed with plasmid YCplac111 (vector), pRL453 (YCplac111-She2p-wt), pRL462 (YCplac111-She2p-N36S), or pRL463 (YCplac111-She2p-R63K). Transformants were grown in synthetic media minus leucine; subsequently, the cells were fixed and processed for immunofluorescence using anti-myc. Then 100 anaphase cells with signal corresponding to Myo4p were counted; below each panel, the percentage of anaphase cells with asymmetrically sorted Myo4p is indicated. (B) Representative images for the intracellular distribution of She3p-myc13 in cells devoid of She2p, expressing wild-type She2p, She2p mutant N36S, or She2p mutant R63K. Strain YLM1368 was transformed and processed for immunofluorescence identically to strain YLM1309 described above. Then 100 anaphase cells with signal corresponding to She3p were counted; below each panel, the percentage of anaphase cells with asymmetrically sorted She3p is indicated. (C) The Myo4p/She3p complex can form in the absence of She2p RNA-binding activity. Strain YLM921 was transformed with plasmids (lane 1) pRL460 (YCplac22-She3p-myc6)/pRL453 (YCplac111-She2p-wt). Yeast strain YLM1320 was transformed with the following combinations of plasmids: (lane 2) vector (YCplac22)/pRL453, (lane 3) pRL460/pRL453, (lane 4) pRL460/YCplac111 (vector), (lane 5) pRL460/pRL462 (YCplac111-She2p-N36S), or (lane 6) pRL460/pRL463 (YCplac111-She2p-R63K). Transformants were grown in synthetic media minus leucine and tryptophan. Cells were harvested, lysates were prepared, and an aliquot of the total lysate was saved for Western blotting. She3p-myc6 was immunoprecipitated from the remaining portion of each lysate with anti-myc. The yeast lysates (total) and immunoprecipitates were analyzed by Western blot for the presence of She3p-myc6 and Myo4p-3HA.
A simple explanation for the symmetric sorting of Myo4p and She3p in cells expressing She2p RNA-binding mutants N36S and R63K is that the formation of the Myo4p/She3p complex might be defective. Therefore, we investigated if Myo4p-3HA and She3p-myc6 are capable of interacting in vivo in the absence of She2p RNA-binding activity (Fig. 7C ▶). She3p-myc6 was immunoprecipitated from yeast lysates, and the presence of coprecipitating Myo4p-3HA was determined by Western blot. As expected, Myo4p-3HA coprecipitated dependent on She3p-myc6 (Fig. 7C ▶, cf. lanes 1–3). Furthermore, Myo4p-3HA and She3p-myc6 were capable of interacting in cells expressing either the N36S (Fig. 7C ▶, lane 5) or R63K (Fig. 7C ▶, lane 6) She2p mutants or in the complete absence of She2p (Fig. 7C ▶, lane 4). These results indicate that the interaction between Myo4p and She3p is independent of She2p and She2p RNA-binding activity and are consistent with previously cited results (Takizawa and Vale 2000). We conclude that association of RNA with She2p affects Myo4p/She3p localization by promoting proper sorting and not by directly affecting complex formation. However, we can conclude that She2p, but not associated RNA, is dispensable for tight polarization of Myo4p to daughter cells, because tethering RNA to She3p (Long et al. 2000) rescues asymmetric sorting of Myo4p and She3p to daughter cells (data not shown).
DISCUSSION
Previous studies demonstrated that She2p directly interacts with She3p and is a novel RNA-binding protein that directly and specifically interacts with each of the ASH1 mRNA cis-acting localization elements (Böhl et al. 2000; Long et al. 2000). However, She2p does not exhibit any obvious characteristics of known RNA-binding motifs, and the details of the She2p/She3p in vivo interaction remained largely uncharacterized. To begin a molecular dissection of She2p function, we sought to identify novel alleles of SHE2 that delocalize ASH1 mRNA by specifically altering She2p RNA-binding activity or the She2p/She3p interaction, but not both. Initially, we identified two individual amino acid substitutions in She2p, N36S and R63K, that result in the delocalization of ASH1 mRNA. These two mutants retain the ability to interact with She3p as determined by three independent assays: GST pull-down reactions, two-hybrid analysis, and coimmunopreciptation experiments (Fig. 3 ▶). However, these two mutants are compromised for association with ASH1 mRNA as determined by three different assays: UV cross-linking, three-hybrid analysis, and IP/RT-PCR (Fig. 4 ▶). From our observations, we conclude that in vivo the She2p/She3p interaction is independent of a functional She2p mRNA-binding domain.
Interestingly, our in vitro work using purified components indicates that She2p may bind ASH1 mRNA too weakly to be measured by EMSA (Fig. 4B ▶), but it interacts long enough to be photochemically trapped by UV cross-linking (Fig. 4A ▶). The UV cross-linking results are not dependent on the GST portion of GST-She2p, because we observe a specific cross-link product with untagged She2p (data not shown). These results are consistent with a model in which the She2p/ASH1 mRNA interaction is stabilized by other cellular factors as part of a cooperatively assembled ribonucleoprotein complex. In support of this model, the U snRNA nuclear export complex involves cooperative interactions between the cap-binding complex (CBC), phosphorylated adaptor for RNA export (PHAX), and capped RNA for the nuclear export of U snRNA (Ohno et al. 2000). Alternatively, She2p association with RNA may be regulated by posttranslational modification. The RNA-binding activity of AKIP1, abscisic-acid-activated-protein-kinase-interacting protein, for dehydrin mRNA is positively regulated by phosphorylation (Li et al. 2002). Although She2p is phosphorylated in vivo (data not shown), the functional significance of She2p phosphorylation with respect to RNA-binding activity and RNA localization remains to be determined.
How might N36 and R63 function in She2p RNA-binding activity? Both of these amino acids could be located within a domain that directly contacts RNA. Alternatively, these amino acid substitutions could affect the She2p:RNA interaction through disruption of protein–protein contacts required for RNA binding. The region of She2p containing amino acids 16–46 is predicted to form a leucine zipper motif (Johnson and McKnight 1989). Several proteins require oligomerization through leucine zipper domains for RNA-binding activity (McAfee et al. 1996a,b; Wu et al. 1998; Tan et al. 2001). We propose that She2p RNA-binding activity requires oligomerization dependent on the leucine zipper motif and substitution of serine for asparagine at position 36 alters the structure of the leucine zipper motif, preventing dimerization of She2p. In support of this, She2p has been observed to interact with itself by two-hybrid analysis (Ito et al. 2001). However, definitive biochemical proof for dimerization of She2p is lacking.
R63 might be directly involved with She2p RNA-binding activity, because several RNA-binding motifs, including the RNA recognition motif (RRM), the arginine-rich motif (ARM), the RGG box, and the double-stranded RNA-binding motif (dsRBD), all contain conserved arginine residues (Burd and Dreyfuss 1994). Inspection of the She2p amino acid sequence revealed that five out of seven arginine residues are clustered between amino acids 43 and 63, indicating that this region of She2p may represent an ARM motif. Mutational analysis of She2p revealed that arginine residues 52 and 63 are critical for She2p association with RNA (Figs. 4 ▶, 5B ▶). Our finding that She2p mutants R52K and R63K are defective for association with ASH1 mRNA indicates that a positive charge at either of these positions is not sufficient for She2p RNA-binding activity. Because arginine has more atoms capable of forming hydrogen bonds with RNA compared with lysine (Calnan et al. 1991; Burd and Dreyfuss 1994), we propose that R52 and R63 make specific interactions that cannot be mimicked by lysine residues. In contrast, a positive charge at positions 43 and 44 is apparently sufficient for She2p association with RNA because amino acid substitutions R43K and R44K have no apparent affect on She2p RNA-binding activity, whereas alanine substitutions are not tolerated at these positions (Fig. 5B ▶). Possibly, a positive charge at R43 and R44 increases nonspecific affinity for RNA and could facilitate the search for high-affinity binding sites requiring amino acids R52 and R63.
As expected, from the two- and three-hybrid results for She2p mutants R43A, R44A, R52K, R52A, and R63A (Fig. 5B ▶; data not shown), we observed that ASH1 mRNA was delocalized in cells expressing these mutants (Table 1 ▶). Unexpectedly, we observed a modest reduction for ASH1 mRNA localization in cells expressing She2p mutant R49A (Table 1 ▶), even though this mutant is capable of interacting with She3p and associating with RNA (Fig. 5B ▶; data not shown). It is possible that the sensitivity of the two- and three-hybrid assays is not sufficient to detect subtle differences in She2p interactions that could result in delocalization of ASH1 mRNA. However, if these two assays accurately reflect the ability of She2p to interact with She3p and associate with RNA localization elements, the inability of She2p mutant R49A to localize ASH1 mRNA implies that She2p may have an additional unidentified function in the ASH1 mRNA localization pathway.
Recently, an N-terminal deletion mutant of She2p, ΔN70, that lacks RNA-binding activity was observed to accumulate in the nucleus, and wild-type She2p was observed to accumulate in nuclei only when poly(A)+ mRNA nuclear export is inhibited (Kruse et al. 2002). These two observations led Kruse and colleagues to propose that She2p RNA-binding activity is required for She2p nuclear export. Consequently, if nuclear export of She2p is dependent on mRNA-binding activity, we would predict amino acid substitutions N36S and R63K should result in nuclear retention of these two She2p mutants. However, in wild-type cells, the nuclear/cytoplasmic distribution of She2p mutants N36S and R63K is indistinguishable from wild-type She2p (Fig. 6A ▶). Furthermore, in an mex67 strain, we did not observe nuclear accumulation of wild-type She2p or either She2p mutant even after confirming accumulation of ASH1 mRNA and poly(A)+ RNA in the nucleus of mex67 cells grown at the nonpermissive temperature (Fig. 6B ▶; data not shown). Therefore, we favor the idea that nuclear export of She2p is independent of RNA-binding activity. How might these differing observations be explained?
With respect to the mex67 experiments, subtle differences in the expression level of She2p could explain the discrepancy between the observed results. The SHE2 construct used in this report contains the wild-type SHE2 promoter and 3′-UTR with insertion of the myc6 tag immediately before the She2p termination codon. Kruse and coworkers placed a myc9 tag into the SHE2 genomic locus by homologous recombination, which results in the replacement of the wild-type 3′-UTR. It is known that the 3′-UTR can influence the level of expression of the corresponding protein. Consequently, we propose that the different 3′-UTRs affect the level of She2p expression. Whereas the construct used by Kruse and coworkers was integrated at the SHE2 locus, ensuring only a single copy of She2p-myc per cell, our She2p-myc6 was harbored on a centromere (CEN)-containing plasmid. Although this difference between the two experiments might alter the level of expression of She2p, we observe very little, if any, difference in expression level between endogenous She2p and She2p expressed from a CEN plasmid (data not shown).
Besides a difference in expression levels between the N36S and R63K She2p mutants and ΔN70, other intrinsic properties of these proteins could affect the steady-state distribution of these molecules. For example, if the nuclear import rate of the N36S and R63K are slower than ΔN70, the N36S and R63K RNA-binding mutants could appear uniformly distributed throughout the cell. However, we favor the hypothesis that a unique property of ΔN70 leads to nuclear accumulation of this protein. Because ΔN70 is defective for ASH1 mRNA binding activity and for association with She3p, we hypothesize that ΔN70 more effectively interacts with nuclear proteins, resulting in nuclear accumulation (Kruse et al. 2002). In support of this proposal, two-hybrid experiments indicate that She2p can interact with several nuclear proteins, including Hmo1p, Tye7p, and a putative protein encoded by the yeast ORF YJL048c (Uetz et al. 2000; Ito et al. 2001; R.M. Long, unpubl.). We predict that ΔN70 retains the ability to interact with these nuclear proteins.
By identifying mutations in She2p that specifically affect RNA-binding activity, we were able to address a mechanistic detail required for ASH1 mRNA localization. Given that She2p is required for the asymmetric sorting of Myo4p to daughter cells, proper sorting of Myo4p and She3p to daughter cells could be solely dependent on the formation of the heterotrimeric Myo4p/She3p/She2p complex and independent of any associated mRNA (Münchow et al. 1999). The in vivo data presented here demonstrate that the protein–protein interactions required for the formation of the Myo4p/She3p/She2p complex are detectable in cells expressing the She2p RNA-binding mutants N36S or R63K (Fig. 7 ▶). However, even though the protein–protein interactions are maintained, Myo4p and She3p are not properly sorted to daughter cells during anaphase of the cell cycle (Fig. 7 ▶). Consequently, polarization of Myo4p and She3p to daughter cells is dependent on the presence of associated mRNA. While our manuscript was in preparation, an article reaching a similar conclusion was published using different experimental approaches (Kruse et al. 2002).
These observations are extremely informative toward understanding the mechanism of mRNA localization. Even though the protein–protein interactions required for ASH1 mRNA localization are maintained in the absence of She2p RNA-binding activity, the lack of associated mRNA apparently restricts the transport and/or anchoring of the protein complex. In the absence of associated mRNA, we hypothesize that the Myo4p/She3p complex may not properly anchor at the cortex of daughter cells, because in small budded cells we can detect Myo4p and She3p sorting to daughter cells (Münchow et al. 1999; data not shown). We propose that the Myo4p/She3p complex contains or interacts with a molecular sensor that is sensitive to the presence of mRNA. This sensor could be a novel protein at the bud cortex that anchors the ribonucleoprotein complex when it arrives at its destination. The sensing mechanism apparently is not directly attributed to She2p, because we have previously demonstrated that RNA can be localized to daughter cells when artificially tethered to She3p (Long et al. 2000). Furthermore, both Myo4p and She3p are asymmetrically sorted to daughter cells in the absence of She2p when mRNA is artificially tethered to She3p (Kruse et al. 2002; data not shown). The suggestion that the transport of the She3p/Myo4p complex is sensitive to the presence/absence of mRNA cargo is supported by reports that transport by additional molecular motors is regulated by the cargo (for review, see Karcher et al. 2002). With respect to RNA transport and localization, subtle changes in RNP cargo affect the efficiency of motor transport in Drosophila embryos (Bullock et al. 2003). Regulating the transport of molecular motors by sensing the appropriate cargo further illustrates that eukaryotic cells have developed sophisticated mechanisms for regulating complex cellular processes.
MATERIALS AND METHODS
Yeast strains, media, and plasmids
The yeast strains used in this study are listed in Table 2 ▶. The plasmids used in this study are listed in Table 3 ▶; they were constructed using standard molecular techniques (Sambrook et al. 1989). Details for plasmid constructions are available upon request. Yeast cells were transformed using lithium acetate (Ito et al. 1983) and were grown either in rich media or in defined synthetic media lacking the nutrients indicated (Rose et al. 1990). As described previously, gene deletions were created with PCR products generated from plasmid pUG6 (Güldener et al. 1996). Silent deletions were subsequently created using plasmid pSH47 (Güldener et al. 1996). For tagging endogenous yeast genes with the myc13 or 3HA epitopes, DNA cassettes containing the respective epitope were generated by PCR with gene-specific primers (Longtine et al. 1998).
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Source |
| YBZ-1 | MATa,ade2, his3-200, leu2-3, 112, trp1-1, ura3-52, LYS2::(LexAop)-lacZ, LexA-MS2-MS2 coat (N55K) | Zhang et al. 1997 |
| PJ69-4a | MATahis3-200, leu2-3, 112, trp1-90, ura3-52, gal4, gal80, GAL2-ADE2, LYS2::GAL1-HIS3, met2::GAL7-lacZ | James et al. 1996 |
| K699 | MATa, ade2-1, his3-11, leu2-3,112, ura3, trp1-1, ho, can1-100 | Jansen et al. 1996 |
| K4452 | MATα, ade2-1, leu2-3, trp1-1, ura3, HO-ADE2, HO-CAN1 | Jansen et al. 1996 |
| RS453 | MATa, ade2-1, his3-11,15, leu2-3,112, trp1-1, ura3-52, can1-100, GAL+ | Segref et al. 1997 |
| YLM585a | MATa, ade2, his3-200, leu2-3,112, trp1-1, ura3-52, LYS2::(LexAop)-lacZ, LexA-MS2-MS2 coat (N55K), she2::kanMX | Long et al. 2000 |
| YLM777b | MATa, ade2-1, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, she2Δ silent | This study |
| YLM851c | MATα, ade2-1, leu2-3, trp1-1, ura3, HO-ADE2, HO-CAN, she2::kanMX | This study |
| YLM921b | MATa, ade2-1, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, she2Δ silent, she3::kanMX | This study |
| YLM946b | MATa, ade2-1, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, trp1Δ silent, SHE3-myc13:TRP1 | This study |
| YLM1309b | MATa, ade2-1, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, she2Δ silent, SHE1-myc13:kanMX6 | This study |
| YLM1320b | MATa, ade2-1, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, she2Δ silent, she3Δ silent, SHE1-3HA: kanMX6 | This study |
| YLM1368b | MATa, ade2-, his3-11, leu2-3,112, trp1-1, ura3, ho, can1-100, trp1Δ silent, SHE3-myc13:TRP1, she2::kanMX | This study |
| YLM1769d | MATa, ade2, his3, leu2, trp1, ura3, mex67::HIS3, she2::kanMX, pUN100-mex67-5 | This study |
aYBZ-1 genetic background.
bK699 genetic background.
cK4452 genetic background.
dRS453 genetic background.
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Source |
| pACT2 | Yeast vector for expressing Gal4p activation domain fusion proteins | Li et al. 1994 |
| pET15b | E. coli vector for generating His6-tagged fusion proteins | Novagen |
| pESC-URA | Yeast multicopy plasmid marked with URA3 for galactose-regulated expression | Stratagene |
| pGAD-c1 | Yeast vector for expression of Gal4p activation domain fusion proteins | James et al. 1996 |
| pGBDU-c1 | Yeast vector for expression of Gal4p DNA-binding domain fusion proteins | James et al. 1996 |
| pGEX-2T | E. coli vector for generating GST-tagged fusion proteins | Pharmacia |
| pHZ18(A) | Yeast plasmid containing galactose-inducible lacZ | Long et al. 1995 |
| pHMTc-She2p | E. coli expression vector for His6-MBP-TEV-She2p | This work |
| YCplac22 | Yeast single-copy shuttle plasmid marked with TRP1 | Gietz and Sugino 1998 |
| YCplac111 | Yeast single-copy shuttle plasmid marked with LEU2 | Gietz and Sugino 1998 |
| YEplac112 | Yeast multicopy shuttle plasmid marked with TRP1 | Gietz and Sugino 1998 |
| C3431 | YEplac195 containing ASH1 | Long et al. 1997 |
| pGEM-4Z | Vector for in vitro transcription | Promega |
| pUG6 | Plasmid for generating KAN disruption cassettes by PCR | Güldener et al. 1996 |
| pSH47 | Plasmid containing a galactose-inducible cre recombinase construct | Güldener et al. 1996 |
| pIIIA/MS2-2 | Three-hybrid vector for the expression of MS2-fusion RNAs | Zhang et al. 1997 |
| pEP13 | pACT2 containing entire She3p open reading frame | Long et al. 2000 |
| pRL80 | pIIIA/MS2-2 containing the 127-nt SmaI E3 ASH1 localization element | Long et al. 2000 |
| pRL104 | pET15b containing a C-terminal domain of She3p | This work |
| pRL168 | pGEM-3Z containing element E3 (118 nt) | Long et al. 2000 |
| pRL172 | pGEX-2T containing the open reading frame corresponding to She2p | Long et al. 2000 |
| pRL199 | YCplac111 containing She2p-myc6 | This work |
| pRL266 | pET15b containing the open reading frame corresponding to She2p | This work |
| pRL270 | Identical to pRL172 except carrying the R63K amino acid substitution in She2p | This work |
| pRL273 | Identical to pRL172 except carrying the N36S amino acid substitution in She2p | This work |
| pRL277 | pHZ18(A)/IST2 2098-3141 | This work |
| pRL322 | Identical to pRL199 except carrying an R63K amino acid substitution in She2p | This work |
| pRL324 | Identical to pRL199 except carrying an N36S amino acid substitution in She2p | This work |
| pRL418 | pGBDU-c1 containing wild-type She2p | This work |
| pRL419 | pGBDU-c1 containing N36S She2p | This work |
| pRL431 | pGBDU-c1 containing R63K She2p | This work |
| pRL441 | pGAD-c1 containing N36S She2p | This work |
| pRL442 | pGAD-c1 containing R63K She2p | This work |
| pRL445 | pGAD-c1 containing wild-type She2p | This work |
| pRL453 | YCplac111 containing wild-type She2p | This work |
| pRL460 | YCplac22 containing She3p-myc6 | This work |
| pRL462 | YCplac111 containing N36S She2p | This work |
| pRL463 | YCplac111 containing R63K She2p | This work |
| pRL552 | pGAD-c1 containing R43K She2p | This work |
| pRL553 | pGAD-c1 containing R44K She2p | This work |
| pRL554 | pGAD-c1 containing R49K She2p | This work |
| pRL555 | pGAD-c1 containing R52K She2p | This work |
| pRL669 | pGAD-c1 containing R43A She2p | This work |
| pRL670 | pGAD-c1 containing R44A She2p | This work |
| pRL671 | pGAD-c1 containing R49A She2p | This work |
| pRL672 | pGAD-c1 containing R52A She2p | This work |
| pRL673 | pGAD-c1 containing R63A She2p | This work |
| pRL676 | YCplac22 containing wild-type She2p-myc6 | This work |
| pRL677 | YCplac22 containing N36S She2p-myc6 | This work |
| pRL678 | YCplac22 containing R63K She2p-myc6 | This work |
| pRL679 | pGEM-4Z containing MS2 cloned from pIIIA/MS2-2 | This work |
| pRL718 | pESC-URA containing ASH1 | This work |
| pRL740 | YCplac111 containing R43A She2p-myc6 | This work |
| pRL741 | YCplac111 containing R44A She2p-myc6 | This work |
| pRL742 | YCplac111 containing R49A She2p-myc6 | This work |
| pRL743 | YCplac111 containing R52A She2p-myc6 | This work |
| pRL744 | YCplac111 containing R63A She2p-myc6 | This work |
| pRL745 | YCplac111 containing R43K She2p-myc6 | This work |
| pRL746 | YCplac111 containing R44K She2p-myc6 | This work |
| pRL747 | YCplac111 containing R49K She2p-myc6 | This work |
| pRL748 | YCplac111 containing R52K She2p-myc6 | This work |
Genetic selection/screen for She2p point mutations
To isolate She2p point mutants defective for ASH1 mRNA localization, plasmid pRL199 was initially propagated in E. coli strain mutD5, which contains a defect in the 3′ → 5′ exonuclease activity of the polymerase III holoenzyme (Echols et al. 1983; Schaaper 1988). The randomly mutagenized pool of plasmid was transformed into yeast strain YLM851. All yeast transformants were initially selected using defined synthetic media lacking leucine. Yeast transformants were recovered from minus leucine plates, and 500 colony-forming units were replated onto synthetic media containing 0.03% canavanine but lacking arginine and leucine. Canavanine-resistant colonies were isolated and screened for the red adenine phenotype. Cell lysates were prepared from colonies displaying the correct phenotypes, and cells expressing full-length mutant She2ps were identified by Western blot using anti-myc monoclonal antibody 9E10 (Roche), goat anti-mouse conjugated with horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories, Inc.), and SuperSignal West Pico Chemiluminescent Substrate (Pierce). The plasmids corresponding to the full-length mutant She2p clones were recovered from yeast, and the DNA was sequenced to determine the She2p mutation.
Temperature shift experiment
With few modifications, the mex67 temperature shift experiment was performed as described previously (Kruse et al. 2002). Yeast cells were grown to exponential phase overnight at 25°C in synthetic media lacking uracil and tryptophan containing 2% raffinose. The cells were harvested by centrifugation and resuspended in YEP media containing 2% galactose. The cells were grown at 25°C for an additional 3 h in YEP galactose media to induce expression of ASH1 mRNA. Subsequently, half of the culture was shifted to the nonpermissive temperature, 37°C, by addition of an equal volume of 49°C YEP galactose media, and incubation was continued at 37°C for an additional 30 min. An equal volume of 25°C YEP galactose media was added to the remaining half of the 25°C YEP galactose culture, and incubation was continued at 25°C for an additional 30 min. Following the 30-min incubation, cells were harvested, fixed, and processed for in situ hybridization and immunofluorescence.
Two- and three-hybrid analysis
For two-hybrid analysis, yeast strain PJ69–4a (James et al. 1996) was used, and for three-hybrid (SenGupta et al. 1996) analysis, yeast strain YLM585 was used. Transformants were grown in synthetic media devoid of leucine and uracil. Quantitative β-galactosidase expression levels were determined by liquid assay using O-nitrophenyl-β-D-galactopyranoside (ONPG; Guarente 1983). Cells were grown overnight in 50 mL of synthetic liquid media to mid-log phase and harvested by centrifugation. The cell pellets were resuspended in 5 mL of buffer Z (60 mM Na2HPO4, 40 mM NaHPO4, 10 mM KCl, 1 mM MgSO4, and 50 mM β-mercaptoethanol, adjusted to pH 7.0), and the OD600 was determined in triplicate. An aliquot of the cell suspension was analyzed in triplicate for β-galactosidase activity in 1 mL of 60 mM Na2HPO4, 40 mM NaHPO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, 0.0025% SDS, and 5% chloroform by vortexing the cell suspension for 10–15 sec, followed by equilibration at 30°C for 15 min and incubation with 0.67 mg/mL ONPG at 30°C for 30 min. Reactions were terminated with 300 mM Na2CO3 and centrifuged for 5 min at 1100g. The OD420 and OD550 of the supernatant were determined. β-Galactosidase activity was calculated using the formula U = 1000 × [(OD420) − (1.75 × OD550)]/(time in min.) (volume of aliquot of cell suspension used in assay in milliliters) (OD600). The reported β-galactosidase expression levels represent the average of three independent experiments.
Immunoprecipitation/RT-PCR analysis
With few modifications, immunoprecipitation of She2p-myc and detection of associated mRNAs was performed essentially as described previously (Irie et al. 2002). Briefly, exponentially growing yeast cultures corresponding to 3 × 108 cells were harvested by centrifugation, and the cells were disrupted with glass beads in 300 μL of breaking buffer: 25 mM HEPES-KOH (pH 7.5), 150 mM KCl, 2 mM MgCl2 200 U/mL RNasin (Promega), 0.1% NP-40, 1 mM DTT, 0.2 mg/mL heparin, 0.1 μg/mL chymostatin, 2 μg/mL aprotinin, 1 μg/mL pepstatin, 0.5 μg/mL leupeptin, and 0.01 μg/mL benzamidine. The cell lysates were cleared by centrifugation at 4000g at 4°C for 10 min, and immunoprecipitation was performed at 4°C for 1 h using anti-myc monoclonal antibody 9E10. Anti-myc immune complexes were purified by incubation with Protein-A–agarose (Pierce) at 4°C for 1 h. Protein-A–agarose complexes were recovered by centrifugation and washed three times with 500 μL of wash buffer: 25 mM HEPES-KOH (pH 7.5), 150 mM KCl, and 2 mM MgCl2. Protein–RNA complexes were eluted from protein-A–agarose by incubation at 65°C for 10 min in 100 μL of elution buffer: 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, and 1% SDS. A small aliquot of the eluted material was solubilized with Laemmli buffer and analyzed by Western blot. RNA was extracted from the remainder of the immunoprecipitate using phenol/chloroform/isoamyl alcohol, and the RNA was precipitated with ethanol. The RNA pellet was resuspended in 20 μL of nuclease-free water and analyzed with the Access/RT-PCR kit (Promega) using primers specific for either ASH1 mRNA or IST2 mRNA. RT-PCR reactions were performed as described by the manufacturer for 25 cycles. Under these conditions, the RT-PCR experiments were in the linear range of the assay.
Coimmunoprecipitation assays
Exponentially growing cultures corresponding to 3 × 108 yeast cells were harvested, and soluble lysates were prepared as described in Supplemental materials. Protein complexes were purified by the addition of 7.5 μg of anti-myc 9E10 monoclonal antibody. The antibody was incubated with the lysate at 4°C for 1 h. The protein–antibody complexes were subsequently purified using Protein-A–agarose (Pierce). The matrix was washed four times with 500 μL of wash buffer: 50 mM HEPES-KOH (pH 7.3), 50 mM potassium acetate, 2 mM EDTA, 0.1% Triton X-100, and 5% glycerol. The bound proteins were eluted by boiling in Laemmli buffer. Equivalent amounts of cell extract (total fraction) and precipitated sample (IP fraction) were separated by SDS-PAGE and analyzed by Western blot. Proteins containing the 3HA epitope were detected using anti-HA monoclonal antibody 16B12 (Babco).
UV cross-linking and electrophoretic mobility shift assays
UV cross-linking reactions were performed with 0.25 μg, 0.5 μg, and 5.0 μg of recombinant protein and 1 ng of 32P-labeled E3 RNA (Long et al. 2000). Recombinant protein and RNA were incubated at 30°C for 10 min in 20 mM HEPES (pH 7.9), 33 mM KCl, 3.2 mM MgCl2, 6% glycerol, and 4 mM dithiothreitol. Subsequently, reaction mixtures were UV cross-linked at 4°C for 20 min using a 254-nm UV lamp (Spectroline, EF-180C) at a distance of 9 cm, followed by treatment with 1.0 mg/mL RNase A at 37°C for 15 min. Reactions were analyzed by SDS-PAGE and autoradiography.
For electrophoretic mobility shift assays, E3 RNA was folded by heating to 90°C for 2 min and immediately transferring the samples to ice. Successful folding of E3 RNA was monitored by comparing the mobility of prefolded and folded samples by gel shift (data not shown). Trace (1 nM) 32P-labeled E3 RNA and unlabeled competitor E3 RNA were combined in a buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 125 μg/mL acetylated BSA (Invitrogen), and 0.01% igepal CA-630 prior to adding purified She2p to 125 nM. Reactions were equilibrated at 30°C for 2.5 h and loaded onto a running 8% (29:1) native gel. Samples were electrophoresed for 2 h at 250 V, dried, and subjected to PhosphorImager analysis.
Pull-down assays between GST-She2p and in vitro transcribed and translated She3p
Radiolabeled She3p was produced by coupled in vitro transcription–translation using plasmid pRL104 with the TnT Coupled Reticulocyte Lysate Kit (Promega). The reaction was assembled using 35S-methionine according to the manufacturer’s directions. An aliquot of the transcription–translation reaction was incubated with 1 μg of GST or 1 μg of GST-She2p (wild type or mutant) bound to glutathione Sepharose. The binding reaction was performed at room temperature for 90 min in 500 μL of binding buffer: 50 mM HEPES-KOH (pH 7.3), 20 mM potassium acetate, 2 mM EDTA, 0.1% Triton X-100, and 5% glycerol. The matrix was recovered by centrifugation and washed four times with 500 μL of binding buffer. The bound proteins were eluted by boiling in Laemmli buffer and separated by SDS-PAGE, and She3p was visualized by autoradiography.
Supplemental Materials
Experimental protocols for soluble lysate preparation, production of anti-She2p rabbit serum, She2p recombinant protein expression and purification, in vitro transcription, in situ hybridization, and immunofluorescence as well as in vitro mutagenesis are available at http://www.mcw.edu/microbiology/rml/rna2003support.html.
Acknowledgments
We thank P. Traktman, J. Barbieri, R. Misra, C.R. Urbinati, and members of the Long lab for helpful discussions and critical reading of the manuscript. We thank the laboratory of R.-P. Jansen for generously providing yeast strains and plasmids, as well as the laboratory of D. Frank for generously providing anti-tubulin antibody. Work in the R.M.L. laboratory is supported by NIH grant GM60392, the Pew Scholars Program in the Biomedical Sciences, and a March of Dimes Basil O’Connor Starter Scholar Research Award. Work in the J.R.W. laboratory is supported by NIH grant GM53320 to J.R.W. and by Ruth L. Kirschstein National Research Service Award GM65718 from the NIH to K.A.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5120803.
REFERENCES
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 1998. RNA localization in development. Annu. Rev. Biochem. 67: 335–394. [DOI] [PubMed] [Google Scholar]
- Beach, D.L. and Bloom, K. 2001. ASH1 mRNA localization in three acts. Mol. Biol. Cell 12: 2567–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, D.L., Salmon, E.D., and Bloom, K. 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9: 569–578. [DOI] [PubMed] [Google Scholar]
- Bertrand, E., Chartrand, P., Schaefer, M., Shenoy, S.M., Singer, R.H., and Long, R.M. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Bobola, N., Jansen, R.-P., Shin, T.H., and Nasmyth, K. 1996. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 84: 699–709. [DOI] [PubMed] [Google Scholar]
- Böhl, F., Kruse, C., Frank, A., Ferring, D., and Jansen, R.-P. 2000. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19: 5514–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock, S.L., Zicha, D., and Ish-Horowicz, D. 2003. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 22: 2484–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of function of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Calnan, B.J., Tidor, B., Biancalana, S., Hudson, D., and Frankel, A.D. 1991. Arginine-mediated RNA recognition: The arginine fork. Science 252: 1167–1171. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.H., Singer, R.H., and Long, R.M. 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9: 333–336. [DOI] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.H., Huttelmaier, S., Donato, D., and Singer, R.H. 2002. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell 10: 1319–1330. [DOI] [PubMed] [Google Scholar]
- Clackson, T., Güssow, D., and Jones, P.T. 1991. In PCR, a practical approach (eds. M.J. McPherson et al.), Chapter 12, pp. 187–214. IRL Press, Oxford, UK.
- Echols, H., Lu, C., and Burgers, P.M.J. 1983. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. 80: 2189–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D. and Sugino, A. 1998. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Gonzalez, I., Buonomo, S.B., Nasmyth, K., and von Ahsen, U. 1999. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9: 337–340. [DOI] [PubMed] [Google Scholar]
- Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101: 181–191. [DOI] [PubMed] [Google Scholar]
- Güldener, U., Heck, S., Fiedler, T., Beinhauer, J., and Hegemann, J.H. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer, B.K., Petzold, A., Lillie, S.H., and Brown, S.S. 1994. Identification of MYO4, a second class V myosin gene in yeast. J. Cell Sci. 107: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Hurt, E., Sträβer, K., Segref, A., Bailer, S., Schlaich, N., Presutti, C., Tollervey, D., and Jansen, R.-P. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275: 8361–8368. [DOI] [PubMed] [Google Scholar]
- Irie, K., Tadauchi, T., Takizawa, P.A., Vale, R.D., Matsumoto, K., and Herskowitz, I. 2002. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 21: 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukudua, Y., Murata, K., and Kimura, A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bact. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R.-P., Dowser, C., Michaelis, C., Galova, M., and Nasmyth, K. 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell 84: 687–697. [DOI] [PubMed] [Google Scholar]
- Johnson, P.F. and McKnight, S.L. 1989. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 58: 799–839. [DOI] [PubMed] [Google Scholar]
- Karcher, R.L., Deacon, S.W., and Gelfand, V.I. 2002. Motor–cargo interactions: The key to transport specificity. Trends Cell Biol. 12: 21–27. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E.H. and Singer, R.H. 1992. Determinants of mRNA localization. Curr. Opin. Cell Biol. 4: 975–978. [DOI] [PubMed] [Google Scholar]
- Kloc, M., Zearfoss, N.R., and Etkin, L.D. 2002. Mechanisms of subcellular mRNA localization. Cell 108: 533–544. [DOI] [PubMed] [Google Scholar]
- Kruse, C., Jaedicke, A., Beaudouin, J., Böhl, F., Ferring, D., Güttler, T., Ellenberg, J., and Jansen, R.-P. 2002. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 159: 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Elledge, S.J., Peterson, C.A., Bales, E.S., and Legerski, R.J. 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc. Natl. Acad. Sci. 91: 5012–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Kinoshita, T., Pandey, S., Ng, C.K.-Y., Gypi, S.P., Shimazake, K.I., and Assmann, S.M. 2002. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418: 793–797. [DOI] [PubMed] [Google Scholar]
- Long, R.M., Singer, R.H., Meng, X.-H., Gonzalez, I., Nasmyth, K., and Jansen, R.-P. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383– 387. [DOI] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Lorimer, E., Singer, R.H., and Chartrand, P. 2000. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 19: 6592–6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.M., Gu, W., Meng, X.-H., Gonsalvez, G., Singer, R.H., and Chartrand, P. 2001. An exclusively nuclear RNA binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie III, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953– 961. [DOI] [PubMed] [Google Scholar]
- Maxon, M.E. and Herskowitz, I. 2001. Ash1p is a site-specific DNA-binding protein that actively represses transcription. Proc. Natl. Acad. Sci. 98: 1495–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAfee, J.G., Shahied-Milam, L., Soltaninassab, S.R., and LeStourgeon, W.M. 1996a. A major determinant of hnRNP C protein binding to RNA is a novel bZIP-like RNA binding domain. RNA 2: 1139–1152. [PMC free article] [PubMed] [Google Scholar]
- McAfee, J.G., Soltaninassab, S.R., Lindsay, M.E., and LeStourgeon, W.M. 1996b. Proteins C1 and C2 of heterogeneous nuclear ribonucleoprotein complexes bind RNA in a highly cooperative fashion: Support for their contiguous deposition on pre-mRNA during transcription. Biochemistry 35: 1212–1222. [DOI] [PubMed] [Google Scholar]
- Münchow, S., Sauter, C., and Jansen, R.-P. 1999. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J. Cell Sci. 112: 1511–1518. [DOI] [PubMed] [Google Scholar]
- Ohno, M., Segref, A., Bachi, A., Wilm, M., and Mattaj, I.W. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198. [DOI] [PubMed] [Google Scholar]
- Palacios, I.M. and St. Johnston, D. 2001. Getting the message across: The intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17: 569–614. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Tyska, M.J., Novick, P.J., and Mooseker, M.S. 2001. The yeast class V myosins, myo2p and myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153: 1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. 1990. Methods in yeast genetics. A laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaaper, R.M. 1988. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: Role of DNA mismatch repair. Proc. Natl. Acad. Sci. 85: 8126–8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Lührmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M.P. 1996. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Nat. Acad. Sci. 93: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil, A. and Herskowitz, I. 1996. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84: 711–722. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A. and Vale, R.D. 2000. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. 97: 5273–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., Sil, A., Swedlow, J.R., Herskowitz, I., and Vale, R.D. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389: 90–93. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., DeRisi, J.L., Wilhelm, J.E., and Vale, R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290: 341–344. [DOI] [PubMed] [Google Scholar]
- Tan, J.H., Kajiwara, Y., Shahied, L., Li, J., McAfee, J.G., and LeStourgeon, W.M. 2001. The bZIP-like motif of hnRNP C directs the nuclear accumulation of pre-mRNA and lethality in yeast. J. Mol. Biol. 305: 829–838. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, C., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627. [DOI] [PubMed] [Google Scholar]
- Valegard, K., Murray, J., Stonehouse, N., van den Worm, S., Stockley, P., and Liljas, L. 1997. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein–RNA interactions. J. Mol. Biol. 270: 724–738. [DOI] [PubMed] [Google Scholar]
- Wu, W.-Z., Xu, L., and Hecht, N.B. 1998. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 26: 1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., Gallegos, M., Puoti, A., Durkin, E., Field, S., Kimble, J., and Wickens, M.P. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484. [DOI] [PubMed] [Google Scholar]