Abstract
Bamboo mosaic virus (BaMV) has a single-stranded positive-sense RNA genome with a 5′-cap structure and a 3′ poly(A) tail. Deleting the internal loop that contains the putative polyadenylation signal (AAUAAA) in the 3′ untranslated region (UTR) of BaMV genomic RNA appeared to diminish coat protein accumulation to 2% (C. P. Cheng and C. H. Tsai, J. Mol. Biol. 288:555-565, 1999). To investigate the function of the AAUAAA motif, mutations were introduced into an infectious BaMV cDNA at each residue except the first nucleotide. After transfection of Nicotiana benthamiana protoplasts with RNA transcript, the accumulations of viral coat protein and RNAs were determined. Based on the results, three different categories could be deduced for the mutants. Category 1 includes two mutants expressing levels of the viral products similar to those of the wild-type virus. Six mutations in category 2 led to decreased to similar levels of both minus-strand and genomic RNAs. Category 3 includes the remaining seven mutations that also bring about decreases in both minus- and plus-strand RNA levels, with more significant effects on genomic RNA accumulation. Mutant transcripts from each category were used to infect N. benthamiana plants, from which viral particles were isolated. The genomic RNAs of mutants in category 3 were found to have shorter poly(A) tails. Taken together, the results suggest that the AAUAAA motif in the 3′ UTR of BaMV genomic RNA is involved not only in the formation of the poly(A) tail of the plus-strand RNA, but also in minus-strand RNA synthesis.
There are several interesting structural features in the genomes of positive-sense RNA viruses. The 5′ terminus of the viral RNA genome can be a methylated cap structure, as in the eukaryotic mRNAs; a genome-linked viral protein known as Vpg; or just an ordinary nucleotide. A tRNA-like structure, a poly(A) tail, or a non-tRNA-like structure heteropolymeric sequence has been identified at the 3′ end of the viral RNA genome (10). The viral RNA genome can serve as mRNA for viral protein translation from its 5′ terminus and as a template for minus-strand RNA synthesis from its 3′ terminus.
cis-acting sequences, such as the promoters for minus-strand RNA, plus-strand genomic RNA, and subgenomic RNA synthesis, have been identified in several RNA viruses (7, 11, 32, 38, 39). Additional cis elements required for the assembly of replication complexes have been shown in the Brome mosaic virus system expressed in yeast (35). It has been suggested that the replication complexes assembled on the plus-strand templates for minus-strand RNA synthesis could be recycled and modified to recognize the minus-strand templates for plus-strand genomic and subgenomic RNA synthesis (2).
Moreover, the 3′ untranslated region (UTR) of the viral RNA genome could fold into secondary or tertiary structures, acting as cis-acting elements to be recognized by the replicase complex to initiate minus-strand RNA synthesis (10). The genome of a large portion of RNA viruses ends with a poly(A) tail whose length is within the typical range of the eukaryotic mRNAs (20, 47). The poly(A) tail in mRNA has been proposed to be involved in RNA stability (45, 46), translation efficiency (12, 19), and RNA transport from the nucleus to the cytoplasm (3, 4). Longer poly(A) tails appear to be associated with higher infection efficiencies for several viruses, including White clover mosaic virus (WClMV) (15) and Bamboo mosaic virus (BaMV) (43). The poly(A) tail of Bovine coronavirus RNA was shown to be required for viral RNA to interact with host proteins for RNA replication (40). However, some species of viral RNA transcripts without any adenylate residue at their 3′ ends were infectious, and the poly(A) tail was restored in the case of WClMV (15), Sindbis virus (17), and clover yellow vein potyvirus (41).
BaMV has a single-stranded, positive-sense RNA genome with a 5′ m7GpppG structure and a 3′ poly(A) tail (28). The sequence of the entire 6,366-nucleotide (nt) genome [excluding the 3′ poly(A) tail] has been determined (28, 48). Like other potexviral genomes, five open reading frames (ORF1 to -5) encoding 155-, 28-, 13-, 6-, and 25-kDa polypeptides, respectively, were identified. The 155-kDa polypeptide can be synthesized directly from the genomic RNA in an in vitro rabbit reticulocyte lysate (29) and is involved in viral RNA capping (26, 27) and replication (25).
The 3′ UTR of the BaMV RNA has been suggested to fold into a series of stem-loops, including a tertiary pseudoknot structure, by enzymatic and chemical structural probing (5, 43). Two major subgenomic RNAs 2.0 and 1.0 kb in length share the same 3′ terminus (29). A highly conserved potexviral hexamer motif (ACNUAA) and a putative polyadenylation signal (AAUAAA) are located in the apical and internal loops of domain D, respectively, in this region (5, 28, 33). Extensive mutations, including substitutions, deletions, and insertions, were introduced to dissect the structural requirements in domain D for RNA replication (5). One of the mutants, BaMV-O/ΔIL, with a deletion of the putative polyadenylation motif, accumulates coat protein at 2% of the wild-type levels in the protoplasts of Nicotiana benthamiana, likely due to the loss of the conserved AAUAAA sequence.
In this study, the importance of the AAUAAA motif in BaMV genomic RNA replication was investigated. Transcripts derived from the wild-type sequence and its 15 single-point substitution mutants were synthesized in vitro and used to inoculate N. benthamiana protoplasts. Effects on the accumulation of the viral coat protein and RNAs were examined.
MATERIALS AND METHODS
Materials.
Nicotiana benthamiana plants were grown in a controlled growth chamber at 28°C. T7 RNA polymerase, restriction enzymes, and m7GpppG cap analogue were from New England BioLabs (UK) Ltd.; T7 DNA polymerase (Sequenase) and Thermus aquaticus (Taq) DNA polymerase were from Promega (Madison, WI); cellulase was from Yakult Housha Co. Ltd. (Japan); and pectinase was from CalBiochem.
Mutant construction.
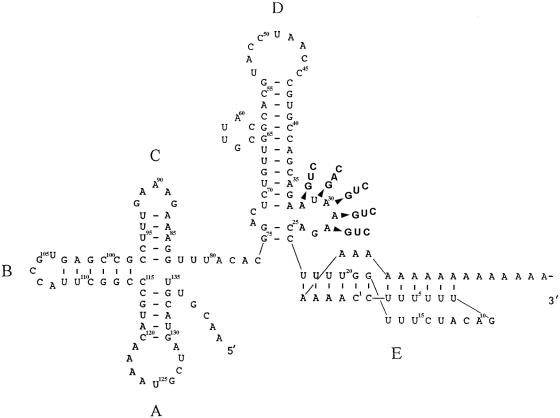
A PCR-based mutagenesis strategy was used to introduce mutations into the conserved AAUAAA sequence in the full-length infectious cDNA clone pBaMV40A (6) using five degenerate primers with three different nucleotides at positions 2 to 6 of the AAUAAA sequence. The sequences of the primers are as follows: BaA32 [5′GCCAGCAGA(G/T/C)TAAAGACCTT3′], BaU31 [5′GCCAGCAGAA(G/A/C)AAAGACCTTT3′], BaA30 [5′GCCAGCAGAAT(G/T/C)AAGACCTTTT3′], BaA29 [5′GCCAGCAGAATA(G/T/C)AGACCTTTTG3′], and BaA28 [5′GCCAGCAGAATAA(G/T/C)GACCTTTTGG3′]. The boldface letters correspond to the substituted residues at each position. Mutants were generated by a two-step PCR (37) involving initial amplification of a 150-bp fragment with the mutagenic primers described above and a downstream primer, M13 universal (5′GTTTTCCCAGTCACGAC3′). The amplified 150-bp fragments were used in the second PCR step as megaprimers, together with the 5′ primer, BaMV + 5910 (5′CCAAACCGACGTTCGCCA3′), to synthesize a 559-bp DNA fragment. The amplified PCR products were cloned into the pGEM-T Easy vector-System I (Promega), and the sequence of each mutant was verified before being subcloned into pBaMV40A using the 5′ NruI (5964) and 3′ BamHI sites. These mutants were designated pBa-A32G, -A32U, -A32C, -U31G, -U31A, -U31C, -A30G, -A30U, -A30C, -A29G, -A29U, -A29C, -A28G, -A28U, and -A28C [nucleotides are numbered from the 3′-end cytosine abutting the poly(A) tail] (Fig. 1).
FIG. 1.
Tertiary folding of domains ABC, D, and E in the 3′ UTR of Bamboo mosaic virus RNA (5, 43). Nucleotides are numbered from the 3′-end cytosine just upstream of the poly(A) tail.
In vitro transcription and inoculation of protoplasts.
Prior to in vitro transcription, pBaMV40A and its derivatives were linearized with BamHI. Transcription reactions were carried out at 37°C for 2 h in a 50-μl reaction mixture containing 100 U of T7 RNA polymerase, 40 mM Tris-HCl, pH 8.0, 2 mM spermidine, 8 mM MgCl2, 10 mM dithiothreitol, 0.5 mM GTP, 1 mM ATP/UTP/CTP, 2 mM m7GpppG cap analogue, and 5 μg of the BamHI-linearized DNA template. Protoplasts (4 × 105 cells) prepared from N. benthamiana leaves were inoculated with 5 μg of the transcripts and incubated for 48 h at 25°C under constant light as described previously (43).
Northern and Western blotting analyses.
For Northern blotting analysis, total RNAs were extracted from the inoculated protoplasts (2 × 105 cells), glyoxalated, electrophoresed through a 1% agarose gel, and transferred to a membrane (Zeta-Probe; Bio-Rad) as described previously (42). The hybridization probes were 0.6- and 0.7-kb 32P-labeled RNA transcripts derived from HindIII-linearized pBaMV-O/SB2.6 (18) and BglI-linearized pBaMV-O (43), complementary to the 3′ ends of the plus and minus strands of BaMV RNA, respectively. The banding signals were scanned and quantified using a phosphorimager (Fujifilm BAS 1500). Total protein harvested from the inoculated protoplasts (2 × 105 cells) was separated on a 14% polyacrylamide gel containing 0.1% sodium dodecyl sulfate (SDS) and electroblotted onto a nitrocellulose membrane (PROTRAN BA 85; Schleicher & Schuell). The levels of coat protein were detected by Western blotting, using an anti-BaMV capsid protein serum as the primary antibody, a horseradish peroxidase-labeled secondary antibody, and the chromogenic substrate 4-chloro-1-naphthol as described previously (43). Results were quantitated by scanning densitometry (Bioimage Intelligent Quantifier).
In vitro translation.
In vitro translation was performed in the TnT Coupled Transcription/Translation System (Promega). The reaction was carried out in a 12.5-μl reaction mixture containing 2 μg plasmid DNA in nuclease-treated rabbit reticulocyte lysate (Promega) or wheat germ extract (Promega) supplemented with all amino acids except methionine. Proteins were labeled by the incorporation of 5 μCi of l-[35S]methionine (1,000 Ci/mmol; 10 mCi/ml; Amersham) in the reaction. Incubations were performed at 28°C for 1 h and terminated by the addition of Laemmli sample buffer. The translation products were resolved by 8% SDS-polyacrylamide gel electrophoresis (PAGE). Gels were fixed, dried, and analyzed by a phosphorimager (Fujifilm BAS 1500).
Characterization of the poly(A) tail of BaMV RNA.
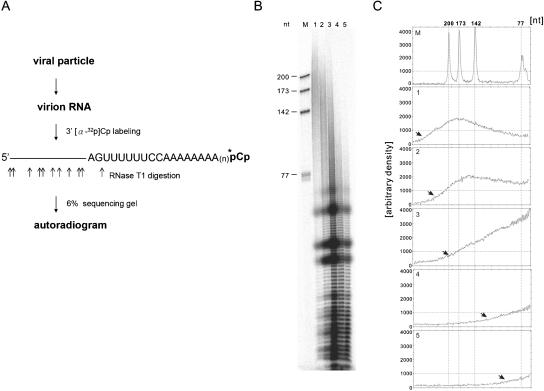
Viral particles were isolated from plants infected with BaMV-S or its derivatives 4 days postinfection (p.i.) by polyethylene glycol precipitation according to the method of Lane (24) and subsequently subjected to RNA purification by two phenol-chloroform extractions, followed by an ethanol precipitation. Viral RNAs, after further purification through oligo(dT)25-coupled paramagnetic beads (Dynal A.S., Oslo, Norway), were labeled with [5′-32P]cytidine 3′,5′-bis(phosphate) ([α-32P]Cp) using T4 RNA ligase, resuspended in urea-containing loading buffer, denatured in boiling water for 90 s, and cleaved with RNase T1 (120 units) at 55°C for 30 min. The cleaved RNA fragments were resolved on 6% sequencing gels.
RESULTS AND DISCUSSION
It has been generally accepted that once a positive-sense RNA virus infects its host cells, the virus-encoded RNA-dependent RNA polymerase (RdRp) is translated and possibly interacts with a host factor(s) (23) to form a functional replicase complex (22). This complex can recognize the 3′ UTR of genomic RNA to initiate minus-strand RNA synthesis (23) to generate the template for genomic and subgenomic RNA syntheses. For RNA with a 3′ poly(A) tail, the polyadenylation step becomes the final process for the maturation of progeny RNAs. The conserved polyadenylation signal, AAUAAA, in mRNAs of eukaryotic cells (16, 34) has also been found in the 3′ UTR of the potexvirus genomic RNA (15).
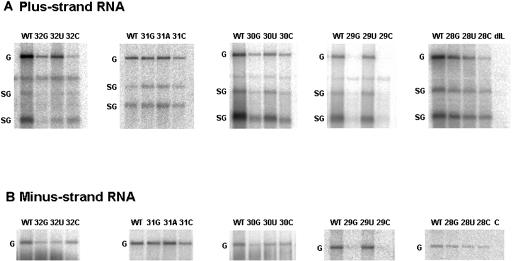
We had demonstrated that a deletion of the internal loop in stem D in the 3′ UTR of BaMV RNA containing the putative polyadenylation signal drastically reduced the accumulation of viral RNA products (5). In order to investigate systematically the function of the putative polyadenylation A1A2U3A4A5A6 motif in BaMV RNA, residues from 2 to 6 were replaced by the other three nucleotides. Transcripts derived from wild-type and mutant plasmids were transfected into N. benthamiana protoplasts. Total protein was extracted after a 48-h incubation and subjected to Western blot analysis (Fig. 2). Northern blot analysis of total RNAs extracted at the same time point, using a probe complementary to the 3′ end of BaMV genomic RNA, showed that the genomic RNA of about 6.4 kb and two subgenomic RNAs about 2 and 1 kb in length could be detected (Fig. 3A). With the 5′-end plus-strand RNA probe, the minus-strand RNAs of 6.4 kb could be detected (Fig. 3B). The levels of the plus- and minus-strand RNAs were quantified based on their relative densities. The level of minus-strand RNA accumulation was used to indicate the efficiency of the early processes of viral RNA replication. The ratio of the plus-strand/minus-strand RNAs was used to deduce the efficiency of genomic RNA accumulation.
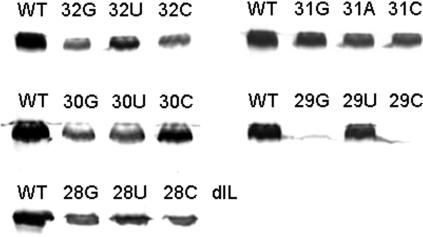
FIG. 2.
Amplification of BaMV40A and its derivatives in N. benthamiana protoplasts. Representative experiments that contributed to the quantitative data in Table 1 are shown. Protoplasts (4 × 105 cells) inoculated with 5 μg of transcripts of 15 single-substitution mutants at nt 2 to 6 of the AAUAAA sequence (32G/U/C, 31G/A/C, 30G/U/C, 29G/U/C, and 28G/U/C) and an internal loop deletion mutant (dIL) were harvested at 48 hr postinoculation. Total-protein extracts were separated on a 14% SDS-polyacrylamide gel, blotted, and probed with an anti-BaMV coat protein serum. The blot was developed using horseradish peroxidase-linked secondary antibody and 4-chloro-1-naphthol color reagent. The identity of the mutant is indicated at the top of each lane. WT, wild type.
FIG. 3.
Amplification of BaMV40A and its derivatives in N. benthamiana protoplasts. Representative experiments that contributed to the quantitative data in Table 1 are shown. (A) Northern blot analysis of the plus-strand genomic (G; 6.4 kb) and two subgenomic (SG; 2.0 and 1.0 kb) RNAs. RNAs were probed with a 32P-labeled RNA transcript complementary to a 0.6-kb region at the 3′ end of genomic RNA. (B) Northern blot analysis of the minus-sense antigenomic (G; 6.4 kb) RNAs. RNAs were probed with a 32P-labeled RNA transcript complementary to a 0.7-kb area at the 3′ end of minus-sense RNA. Lane dIL represents the mutant with a deletion at the 3′ side of the internal loop, and lane c represents 1 ng of virion RNA loaded as a negative control. The other mutants are indicated at the top of each lane. WT, wild type.
Some mutants have no significant effect on viral RNA accumulation.
Mutants BaU31G and BaU31A accumulated 85 and 71% of the wild-type level of coat protein (100%), respectively, and at least 87% of the wild-type level of plus- and minus-sense RNAs at 2 days p.i. The ratio of plus/minus RNAs was close to the wild-type level. These results suggested that mutations at the third position (the only uridine residue of the motif) do not lead to any significant interference with viral RNA accumulation in protoplasts. These mutants are classified in category 1 (Table 1).
TABLE 1.
Levels of accumulation of viral products of BaMV and its derivatives in protoplasts of N. benthamiana
| Mutant | Coat proteina | Genomic RNAb | Subgenomic RNA (2 kb) | Subgenomic RNA (1 kb) | Minus-sense RNA | Plus/minusc | Categoryd |
|---|---|---|---|---|---|---|---|
| Wild type | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| BaA32G | 0.36 ± 0.05 | 0.18 ± 0.08 | 0.33 ± 0.02 | 0.19 ± 0.04 | 0.29 ± 0.04 | 0.62 | 3 |
| BaA32U | 0.70 ± 0.15 | 0.29 ± 0.16 | 0.34 ± 0.16 | 0.22 ± 0.05 | 0.31 ± 0.03 | 0.92 | 2 |
| BaA32C | 0.32 ± 0.14 | 0.39 ± 0.32 | 0.29 ± 0.09 | 0.31 ± 0.17 | 0.49 ± 0.29 | 0.81 | 2 |
| BaU31G | 0.85 ± 0.06 | 0.87 ± 0.22 | 0.77 ± 0.36 | 0.73 ± 0.30 | 0.88 ± 0.18 | 0.99 | 1 |
| BaU31A | 0.71 ± 0.12 | 0.88 ± 0.20 | 1.14 ± 0.33 | 1.13 ± 0.17 | 1.16 ± 0.41 | 0.76 | 1 |
| BaU31C | 0.66 ± 0.11 | 0.43 ± 0.27 | 0.72 ± 0.27 | 0.89 ± 0.26 | 0.51 ± 0.07 | 0.85 | 2 |
| BaA30G | 0.43 ± 0.05 | 0.19 ± 0.10 | 0.32 ± 0.19 | 0.39 ± 0.22 | 0.27 ± 0.17 | 0.70 | 3 |
| BaA30U | 0.52 ± 0.07 | 0.22 ± 0.12 | 0.71 ± 0.38 | 0.62 ± 0.30 | 0.54 ± 0.33 | 0.41 | 3 |
| BaA30C | 0.60 ± 0.09 | 0.22 ± 0.08 | 0.47 ± 0.18 | 0.50 ± 0.13 | 0.39 ± 0.15 | 0.56 | 3 |
| BaA29G | 0.14 ± 0.06 | 0.05 ± 0.02 | 0.15 ± 0.06 | 0.12 ± 0.05 | 0.11 ± 0.04 | 0.49 | 3 |
| BaA29U | 0.56 ± 0.02 | 0.36 ± 0.18 | 0.37 ± 0.19 | 0.31 ± 0.06 | 0.50 ± 0.19 | 0.72 | 3 |
| BaA29C | 0.13 ± 0.03 | 0.04 ± 0.01 | 0.15 ± 0.07 | 0.09 ± 0.07 | 0.10 ± 0.07 | 0.35 | 3 |
| BaA28G | 0.67 ± 0.12 | 0.48 ± 0.25 | 0.68 ± 0.09 | 0.54 ± 0.33 | 0.45 ± 0.19 | 1.07 | 2 |
| BaA28U | 0.86 ± 0.18 | 0.52 ± 0.15 | 0.93 ± 0.17 | 0.70 ± 0.31 | 0.54 ± 0.09 | 0.96 | 2 |
| BaA28C | 0.65 ± 0.16 | 0.37 ± 0.08 | 0.51 ± 0.25 | 0.65 ± 0.27 | 0.34 ± 0.32 | 1.09 | 2 |
Coat protein levels determined by Western blotting compared to that of wild type; data taken from an average of at least three independent runs of protoplast inoculation, and each inoculation was subjected at least two times to Western blot analysis.
BaMV RNA levels determined in Northern blots.
Plus/minus determined from the levels of genomic RNA over the levels of minus-sense RNA.
Category 1, no effect; category 2, effects on minus-strand RNA accumulation but not on plus-strand RNA accumulation; category 3, effects on minus- and plus-strand RNA accumulation.
Some mutants have defects mainly in minus-strand RNA accumulation.
Six mutants, BaA32U, BaA32C, BaU31C, BaA28G, BaA28U, and BaA28C, showed 30 to 50% accumulation levels of the wild-type genomic RNA. The accumulation levels of the minus-strand RNA were also about 30 to 50% of that of the wild type. The ratio of the plus/minus RNAs was close to that of the wild type. These numbers imply that for these mutants, the plus-strand RNA accumulation is dependent on the amounts of minus-strand RNA synthesized. Mutants with a defect in minus-strand RNA synthesis but not in plus-strand RNA synthesis would accumulate smaller amounts of the minus-strand RNA than the wild type, leading to the same plus/minus ratio. These results also suggest that these positions could be parts of the promoter sequence for minus-strand RNA synthesis. Thus, we classified these mutants in category 2, whose members have a defect mainly in minus-strand RNA synthesis.
Some mutants have defects in both minus- and plus-strand RNA accumulation.
Minus-strand RNAs of seven mutants, namely, BaA32G, BaA30G, BaA30U, BaA30C, BaA29G, BaA29U, and BaA29C, showed 10 to 54% of the wild-type accumulation levels, and the plus-strand RNA accumulated to less than 36% of the wild-type level (Table 1). The plus/minus RNA ratios of these mutants were less than 72% that of the wild type. Therefore, these mutants appear to have defects in the accumulation of both plus- and minus-strand RNAs, and they are grouped in category 3 (Table 1).
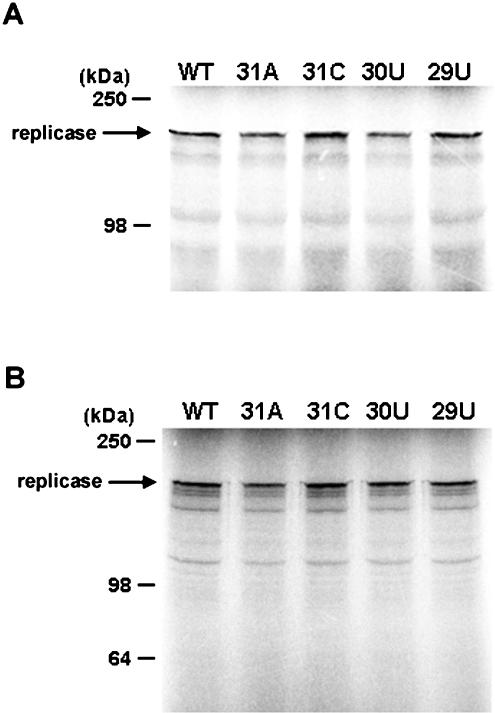
Among the 15 mutants tested, only two showed accumulation levels of viral products similar to those of the wild type. Thirteen mutants showed defects in minus-strand RNA accumulation, with seven of them having extra defects in plus-strand RNA accumulation, indicating that most mutations lead to lower levels of minus-strand RNA accumulation. The lower levels of minus-strand accumulation are unlikely to be due to defects at the translation level, since RNAs of the wild type and mutants from each category can be translated to similar levels in rabbit reticulocyte lysate or wheat germ extract in vitro (Fig. 4). Therefore, we propose that most nucleotides in the AAUAAA motif are involved in the initiation of minus-strand RNA synthesis. These observations have been reported in Potato virus X, where mutations in the 3′ UTR affect the accumulation of both plus- and minus-strand RNAs (33). Furthermore, the mutations at the polyadenylatioin motif also produced less plus-strand RNA accumulation.
FIG. 4.
In vitro translation assay of pBaMV40A and its derivatives. In vitro translation was performed in rabbit reticulocyte lysate (A) or wheat germ extract (B) with plasmid template DNAs of pBaMV40A and its derivatives. The products were separated on 8% SDS-PAGE and analyzed by a phosphorimager (Fujifilm BAS 1500). The molecular masses of the markers are indicated on the left. The arrow indicates the position of the replicase. WT, wild type.
Mutants that have defects in plus-strand RNA accumulation have shorter poly(A) tails.
Mutants in category 3 failed to accumulate plus-strand RNA efficiently, most likely due to altered stability of the genomic RNA. It is possible that mutations of the AAUAAA motif alter the efficiency of the polyadenylation reaction and lead to RNA instability, or these mutations may reduce the stability and leave less time for polyadenylation. To investigate this possibility, the lengths of the poly(A) tails of the mutants from each category were examined. The poly(A) tail of the wild-type genomic RNA can reach up to 300 As (Fig. 5, lane 1). Mutant BaU31A in category 1 and mutant BaU31C in category 2 appeared to have up to 200 As at the 3′ end (Fig. 5, lanes 2 and 3, respectively). Mutants BaA30U and BaA29U in category 3 seemed to have much shorter poly(A) tails of less than 100 As (Fig. 5, lanes 4 and 5). These results indicate that mutations in the AAUAAA motif in the 3′ UTR of BaMV RNA could influence the efficiency of the polyadenylation reaction. It has been reported that deadenylation is a key regulation step in the mRNA surveillance system. The mRNAs with shorter poly(A) tails might have less protection by the poly(A) binding protein from deadynlases or poly(A) ribonucleases and thus shorter half-lives than those with longer poly(A) tails (1, 30, 31, 44). If that is the case, as shown in our mutants, then mutants with shorter poly(A) tails would lead to more efficient turnover and less efficient RNA accumulation in plants.
FIG. 5.
Analysis of the RNA 3′ poly(A) tails of BaMV and its derivatives. (A) Diagram showing the strategy for analyzing the length of the poly(A) tail. Virion RNAs were purified and labeled with [α-32P]Cp using T4 RNA ligase. The labeled RNAs were resuspended in urea-containing loading buffer, denatured, and cleaved with RNase T1. The cleaved RNA fragments were resolved on a 6% sequencing gel. (B) Autoradiogram of the RNase T1-digested 3′-end-labeled RNA fragments. Lane M, size markers indicated on the left of the gel; lane 1, BaMV RNA; lane 2, BaU31A RNA from category 1; lane 3, BaU31C RNA from category 2; lanes 4 and 5, BaA30U and BaA29U RNAs, respectively, from category 3. (C) Length distribution of panel B determined by densitometer scanning, accompanied by size markers. The arrows indicate the sizes of the longest RNA fragments detected.
Since the infection cycle of BaMV and the other members of the same family has not been reported to be associated with the nucleus, the polyadenylation step is presumed to proceed in the cytoplasm. However, cytoplasmic distribution of the poly(A) polymerase has not been reported in plants. Cytoplasmic polyadenylation has been reported mainly in animal systems, with the early developmental stages in many species depending on maternally inherited mRNAs because of quiescent transcription (36). The poly(A) lengths of these dormant or masked RNAs are the key to regulating levels of gene expression. Two cis elements in the 3′ UTRs of the mRNAs are essential for cytoplasmic RNA polyadenylation: the hexanucleotide motif, specifically the near-ubiquitous AAUAAA sequence, and the U-rich cytoplasmic polyadenylation element that often resides about 20 nt 5′ of the hexamer motif (8, 36). At least three trans factors are required for the process of cytoplasmic polyadenylation, the cytoplasmic polyadenylation element binding protein, the hexanucleotide binding protein, and the poly(A) polymerase (9, 13, 21, 36). Although these trans factors have not been reported in plant cells, the potexvrial RNAs do have the hexanucleotides motif (AAUAAA) and several plausible U-rich elements upstream (Fig. 1). Therefore, we cannot rule out the possibility that the polyadenylation of viral RNAs is carried out by host-encoded proteins in the cytoplasm. Alternatively, the polyadenylation of viral RNA is directed by the core of the viral RdRp complex, which has been reported to have nontemplate terminal transferase activity (14). It is probably the most efficient method for virus RNA replication, having RdRp complete plus-strand RNA synthesis as a polymerase and immediately switch into a terminal transferase to synthesize its poly(A) tail.
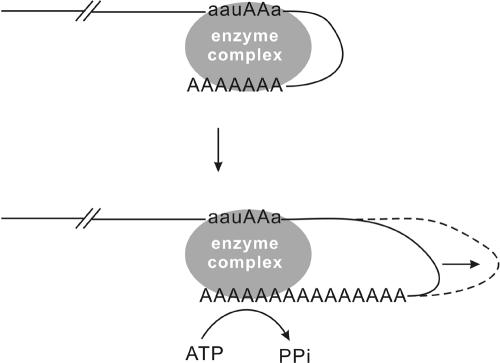
In this study, 15 single-nucleotide substitutions were used to study the function of the putative polyadenylation signal, A1A2U3A4A5A6, of BaMV RNA. U3 (the only uridylate residue) apparently is less important in viral RNA accumulation in protoplasts and plants. Conversely, A2, A4, A5, and A6 are likely involved in minus-strand RNA accumulation. Moreover, substitutions at A4 and A5 appeared to affect polyadenylation efficiency (Fig. 6). Similarly, reductions of local lesions on cowpea plants and in the length of the poly(A) tail from 200 to 300 As to 100 to 200 As were observed in WClMV when its A4 was changed to G4 (15). The A1A2U3A4A5A6 motif in BaMV RNA is sensitive to a change from A4 to U4. Viral particles containing RNA genomes with shorter poly(A) tails can be isolated only at 4 days p.i. while still retaining the mutation. At 7 days p.i., reversion mutants with poly(A) tails of wild-type length were detected in the viral RNAs isolated from virus particles. Overall, we conclude that the AAUAAA sequence in the 3′ UTR of BaMV RNA is not only involved in minus-strand RNA accumulation, but also plays a role in plus-strand RNA accumulation, possibly by regulating the polyadenylation reaction.
FIG. 6.
Cartoon representation of the BaMV RNA polyadenylation model. The two adenylate residues of the aauAAa motif involved in polyadenylation are in capital letters. The oval enzyme complex is responsible for BaMV RNA polyadenylation. The interactions between the aauAAa motif and the enzyme complex could regulate the length of the poly(A) tail.
Acknowledgments
This work was supported by the National Science Council through research grants NSC 91-2311-B-005-036 and 92-2311-B-005-014.
REFERENCES
- 1.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castagnetti, S., and A. Ephrussi. 2003. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development 130:835-843. [DOI] [PubMed] [Google Scholar]
- 4.Chekanova, J. A., and D. A. Belostotsky. 2003. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA 9:1476-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, C. P., and C. H. Tsai. 1999. Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. J. Mol. Biol. 288:555-565. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, W. W., Y. H. Hsu, and C. H. Tsai. 2002. Specificity analysis of the conserved hexanucleotides for the replication of bamboo mosaic potexvirus RNA. Virus Res. 83:159-167. [DOI] [PubMed] [Google Scholar]
- 7.Deiman, B. A., A. K. Koenen, P. W. Verlaan, and C. W. Pleij. 1998. Minimal template requirements for initiation of minus-strand synthesis in vitro by the RNA-dependent RNA polymerase of turnip yellow mosaic virus. J. Virol. 72:3965-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.deMoor, C. H., and J. D. Richter. 1997. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 17:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson, K. S., S. R. Thompson, N. K. Gray, and M. Wickens. 2001. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 276:41810-41816. [DOI] [PubMed] [Google Scholar]
- 10.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 11.Duggal, R., F. Lahser, and T. Hall. 1994. cis-acting sequences in the replication of plant viruses with plus-sense RNA genomes. Annu. Rev. Phytopathol. 32:287-309. [Google Scholar]
- 12.Galli, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translation efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 13.Gebauer, F., and J. D. Richter. 1995. Cloning and characterization of a Xenopus poly(A) polymerase. Mol. Cell. Biol. 15:1422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, H., and A. E. Simon. 2000. Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc. Natl. Acad. Sci. USA 97:12451-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilford, P. J., D. L. Beck, and R. L. S. Forster. 1991. Influence of the poly(A) tail and putative polyadenylation signal on the infectivity of white clover mosaic potexvirus. Virology 182:61-67. [DOI] [PubMed] [Google Scholar]
- 16.Guo, Z., and F. Sherman. 1996. 3′-End-forming signals of yeast mRNA. Trends Biochem. Sci. 21:477-481. [DOI] [PubMed] [Google Scholar]
- 17.Hill, K. R., M. Hajjou, J. Y. Hu, and R. Raju. 1997. RNA-RNA recombination in Sindbis virus: roles of the 3′ conserved motif, poly(A) tail, and nonviral sequences of template RNAs in polymerase recognition and template switching. J. Virol. 71:2693-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, C. Y., and C. H. Tsai. 1998. Evolution of bamboo mosaic virus in a nonsystemic host results in mutations in the helicase-like domain that cause reduced RNA accumulation. Virus Res. 58:127-136. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, R. J., and N. Standart. 1990. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell 62:15-24. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson, S. J., and S. W. Peltz. 1996. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells Annu. Rev. Biochem. 65:693-739. [DOI] [PubMed] [Google Scholar]
- 21.Juge, F., S. Zaessinger, C. Temme, E. Wahle, and M. Simonelig. 2002. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 23:6603-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 23.Lai, M. M. 1998. Cellular factor in the transcription and replication of viral RNA genomes: a paralleled to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 24.Lane, L. 1986. Propagation and purification of RNA plant viruses. Methods Enzymol. 118:687-696. [Google Scholar]
- 25.Li, Y. I., Y. M. Cheng, Y. L. Huang, C. H. Tsai, Y. H. Hsu, and M. Meng. 1998. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of bamboo mosaic virus. J. Virol. 72:10093-10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y. I., Y. J. Chen, Y. H. Hsu, and M. Meng. 2001. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. J. Virol. 75:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Y. I., T. W. Shih, Y. H. Hsu, Y. L. Huang, and M. Meng. 2001. The helicase-like domain of plant potexvirus replicase participates in formation of RNA 5′-triphosphatase activity. J. Virol. 75:12114-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, N. S., B. Y. Lin, W. W. Lo, C. C. Hu, T. Y. Chow, and Y. H. Hsu. 1994. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. J. Gen. Virol. 75:2513-2518. [DOI] [PubMed] [Google Scholar]
- 29.Lin, N. S., F. Z. Lin, T. Y. Huang, and Y. H. Hsu. 1992. Genome properties of bamboo mosaic virus. Phytopathology 82:731-734. [Google Scholar]
- 30.Mitchell, P., and D. Tollervey. 2000. mRNA stability in eucaryotes. Curr. Opin. Genet. Dev. 10:193-198. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, P., and D. Tollervey. 2001. mRNA turnover. Curr. Opin. Cell Biol. 13:320-325. [DOI] [PubMed] [Google Scholar]
- 32.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pillai-Nair, N., K. H. Kim, and C. Hemenway. 2003. cis-acting regulatory elements in the potato virus X 3′ non-translated region differentially affect minus-strand and plus-strand RNA accumulation. J. Mol. Biol. 326:701-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot, N. J., and G. G. Brownlee. 1976. 3′ Non-coding region sequences in eukaryotic messenger RNA. Nature 263:211-214. [DOI] [PubMed] [Google Scholar]
- 35.Quadt, R., M. Ishikawa, M. Janda, and P. Ahlquist. 1995. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc. Natl. Acad. Sci. USA 92:4892-4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter, J. D. 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 63:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 38.Singh, R. N., and T. W. Dreher. 1997. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology 233:430-439. [DOI] [PubMed] [Google Scholar]
- 39.Song, C., and A. E. Simon. 1995. Requirement of a 3′-terminal stem-loop in in vitro transcription by an RNA-dependent RNA polymerase. J. Mol. Biol. 254:6-14. [DOI] [PubMed] [Google Scholar]
- 40.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tacahashi, Y., and I. Uyeda. 1999. Restoration of the 3′ end of potyvirus RNA derived from poly(A)-deficient infectious cDNA clones. Virology 265:147-152. [DOI] [PubMed] [Google Scholar]
- 42.Tsai, C. H., and T. W. Dreher. 1991. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J. Virol. 65:3060-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai, C. H., C. P. Cheng, C. W. Peng, B. Y. Lin, N. S. Lin, and Y. H. Hsu. 1999. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. J. Virol. 73:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Hoof, A., and R. Parker. 2002. Messenger RNA degradation: beginning at the end. Curr. Biol. 12:R285-R287. [DOI] [PubMed] [Google Scholar]
- 45.Vassalli, J.-D., J. Huarte, D. Belin, P. Gubler, A. Vassalli, M. L. O'Connell, L. A. Parton, R. R. Rickles, and S. Strickland. 1989. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 3:2163-2171. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, E. A., G. M. Gilmartin, and J. R. Nevins. 1991. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 10:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, L., T. Ueda, and J. Messing. 1995. The formation of mRNA 3′-ends in plants. Plant J. 8:323-329. [DOI] [PubMed] [Google Scholar]
- 48.Yang, C. C., J. S. Liu, C. P. Lin, and N. S. Lin. 1997. Nucleotide sequence and phylogenetic analysis of a bamboo mosaic potexvirus isolate from common bamboo (Bambusa vulgaris McClure). Bot. Bull. Acad. Sin. 38:77-84. [Google Scholar]