Abstract
The mouse hepatitis virus (MHV) genome's 3′ untranslated region contains cis-acting sequences necessary for replication. Studies of MHV and other coronaviruses have indicated a role for RNA secondary and tertiary elements in replication. Previous work in our laboratory has identified four proteins which form ribonucleoprotein complexes with the 3′-terminal 42 nucleotides [3′(+)42] of the MHV genome. Defective interfering (DI) RNA replication assays have demonstrated a role for the 3′(+)42 host protein binding element in the MHV life cycle. Using gel mobility shift RNase T1 protection assays and secondary structure modeling, we have characterized a possible role for RNA secondary structure in host protein binding to the 3′-terminal 42-nucleotide element. Additionally we have identified a role for the 3′-terminal 42-nucleotide host protein binding element in RNA replication and transcription using DI RNA replication assays and targeted recombination and by directly constructing mutants in this protein binding element using a recently described MHV reverse genetic system. DI RNA replication assays demonstrated that mutations in the 3′(+)42 host protein binding element had a deleterious effect on the accumulation of DI RNA. When the identical mutations were directly inserted into the MHV genome, most mutant genomes were viable but formed smaller plaques than the wild-type parent virus. One mutant was not viable. This mutant directed the synthesis of genome-sized negative-sense RNA approximately as efficiently as the wild type did but had a defect in subgenomic mRNA synthesis. These results point to a potential role for sequences at the extreme 3′ end of the MHV genome in subgenomic RNA synthesis.
Mouse hepatitis virus (MHV) is a positive-sense single-strand RNA virus of the order Nidovirales. MHV is a prototypical coronavirus; its genome is 32 kilobases in length and contains cis-acting elements responsible for viral mRNA transcription and genome replication (1, 6, 7, 11, 16, 19, 34). The genome also serves as an mRNA which contains two overlapping reading frames translated by an RNA frameshifting mechanism to produce a large polypeptide, which is autocatalytically proteolyzed into 16 different polypeptides, many of which contain or are predicted to contain a variety of enzymatic activities (31). These include RNA-dependent RNA polymerase, helicase, papain-like protease, picornavirus 3C-like protease, methyltransferase, and two different nuclease activities, all of which are likely required for viral replication. Coronavirus-infected cells contain five to eight subgenomic mRNAs, each of which is made up of two noncontiguous segments, an approximately 70-nucleotide (nt) leader RNA identical to the corresponding 5′ end of the genome, which is joined to the downstream body of the mRNA (20, 28). Current thinking is that discontinuous transcription during negative-strand synthesis results in the production of a nested set of subgenomic negative-strand RNAs containing antileader RNAs that function as the sole templates for subgenomic mRNA synthesis (2, 26). Genome replication is thought to occur through the production of a negative-sense full-length RNA followed by synthesis of genomic RNA which is packaged and released as fully infectious virions.
Most studies of coronavirus cis-acting replication signals have utilized defective interfering (DI) RNAs as model replicons due to the large size of coronavirus genomes. DIssE is a 2.2-kb, naturally occurring DI RNA which was isolated by serial passage of MHV-JHM (19). DIssE is composed of three regions from the MHV-JHM genome. Region I is from nt 1 to 864, region II is derived from a 784-nt internal region 3.3 to 4 kb from the 5′ end of the genome, and region III is derived from the 601 3′-terminal nucleotides (10). Studies of DIssE deletion mutants demonstrated that both the 5′-terminal 474 nt and the 3′-terminal 436 nt are necessary for replication, as is a 58-nt element derived from region II (1, 11, 19). Studies of various MHV-A59 DI RNAs have indicated that the requirement for the region II element is specific for MHV-JHM DI RNAs (11, 25). Spagnolo and Hogue demonstrated that at least five A residues in the poly(A) tail are required for replication and poly(A) tail formation (29). Relatively few studies have attempted to dissect particular functions in genome replication for the various cis-acting sequences. Lin et al. demonstrated that DI RNAs in which the last 55 nt had been deleted were unable to serve as templates for negative-strand RNA synthesis when transfected into MHV-infected cells (16), thus implicating these sequences in negative-strand RNA synthesis.
RNA secondary structures within cis-acting sequences often function in viral replication. Computer-aided predictions of secondary structure combined with biochemical probing to detect such structures demonstrated the existence of a 68-nucleotide bulged stem-loop near the 5′ end of the MHV 3′ untranslated region (UTR) (6, 7, 21). Replication studies with DI RNAs containing mutations within this structure demonstrated that this stem-loop was essential for DI RNA replication (6). These studies used targeted recombination to generate viruses containing mutations within this 68-nt bulged stem-loop and demonstrated that both primary and secondary structures were important for viral replication. Immediately downstream of this stem-loop structure is a second stem-loop, first identified in the 3′ UTR of bovine coronavirus, which can form a pseudoknot with the 3′-most segment of the upstream bulged stem-loop (32). The interaction of these two RNA secondary structures is proposed to form a molecular switch that may regulate viral RNA synthesis (4). Mutagenesis of these two structures in the MHV genome supports their existence and function in viral replication. Computer-assisted modeling and enzymatic probing of the secondary structure of the MHV 3′ UTR downstream of the pseudoknot indicated the formation of a complex multiple stem-loop structure in this region of the 3′ UTR (18). Mutagenesis of this structure in the DI RNA DIssE, combined with studies of the ability of these mutant DI RNAs to replicate, supported a role for this RNA secondary structure in MHV replication.
Studies of protein binding to the 3′ UTR identified two host protein binding elements within the 3′-terminal 166 nucleotides (18, 34, 35). Mutation of these protein binding elements decreased host protein binding and decreased the ability of DIssE RNAs carrying these mutations to replicate, suggesting that these protein binding elements were functionally important during MHV replication (17, 34, 35). Recently we identified mitochondrial aconitase and mitochondrial HSP70, as well as HSP60 and HSP40, as the four proteins which bound to the 3′-terminal 42-nucleotide [3′(+)42] element within the 3′ UTR (23, 24). The role of secondary structure of the 3′(+)42 element in binding to mitochondrial aconitase and mitochondrial HSP70, HSP60, and HSP40 and the effect of 3′-end RNA-protein interactions on viral replication have not been studied. Computer-based secondary structure modeling of the 3′(+)42 element suggests the formation of a stem-loop structure. Nuclease probing of RNA secondary structure did not provide any information on the terminal 12 nucleotides but suggested the formation of a mostly single-stranded, bulged stem-loop structure which differs from the predominantly double-stranded model proposed by Yu and Leibowitz (34, 35).
In this report we demonstrate that mutations within the 3′-terminal 42-nt genomic RNA lead to increased and decreased RNP complex formation. Introduction of these same mutations into DI replicons lead to decreases in positive-strand DI RNA replication. Viable viruses with altered plaque morphology were recovered when these mutations were introduced into the MHV genome, with one exception. For one mutation, viable virus could not be recovered due to a defect in subgenomic RNA synthesis.
MATERIALS AND METHODS
Cells, cell lysates, and viruses.
17Cl-1 cells were maintained at 37°C and 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum (DMEM10). L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. L2 cells for targeted recombination experiments were adapted to grow in suspension and maintained at 37°C with 3% CO2 in spinner DMEM supplemented with 10% calf serum. Cytoplasmic lysates for RNA binding assays were prepared from 17Cl-1 cells as previously described (35).
The origin and growth of MHV-A59 have been described previously (17). A temperature-sensitive mutant of MHV-A59, Alb4, was generously provided by Paul S. Masters (12) and used in targeted recombination experiments.
Template preparation and transcription of gel mobility shift RNase T1 protection assay probes.
PCR was carried out on DE25, a cDNA of a naturally occurring DI RNA, DIssE, using primers listed in Table 1. PCR products were then purified by electrophoresis in a 1× Tris-acetate-EDTA, 4% NuSieve GTG agarose gel; the amplified fragment was excised from the gel and recovered using the Qiaquick gel extraction kit. Transcription of radiolabeled RNAs was conducted as described previously (35). Probes were quantified based upon incorporation of [α-32P]UTP, and 1.4 picomoles of labeled RNA was used for each binding reaction. Unlabeled wild-type probe was prepared in the same manner as labeled probe, quantitated by spectrophotometry, and resuspended in diethyl pyrocarbonate-treated water to appropriate concentrations.
TABLE 1.
Primers used for gel mobility shift RNase T1 protection assays, site-directed mutagenesis, and RT-PCR
| Primer name | Sequence, 5′-3′ |
|---|---|
| WTA | TAATACGACTCACTATGGGCGAGAGTAAATGAATGAAGT |
| MT1A | TAATACGACTCACTATGGGCGAGAGTAAACGAATGAAGTTGATCATGGC |
| MT2A | TAATACGACTCACTATGGGCGAGAGTAAAACAATGAAGTTGATCATGG |
| M19C | TAATACGACTCACTATGGGCGAGAGTAAAACTATGAAGT |
| MT5A | TAATACGACTCACTATGGGCGAGAGTAAAacttaGAAGTTGATCATGG |
| ATW17-20 | TAATACGACTCACTATGGGCGAGAGTAAATGAATGAAGTACTACATGGC |
| ATW | GACTATCTTCCAATT |
| U3RA | GTGATTCTTCCAATTGGCCATGATCAACTTC |
| M19CR | GTGATTCTTCCAATTGGCGGAT |
| MD10 | CAATTGGCCATGATCAAC |
| B36SacI | GAAGAGCTCACATCAGG |
| B36HindIII | CCAGTGCCAAGCTTATGC |
| RJATW3LF | GAAGTTGATCATGGCCAATTGGAAGATAGTCA |
| RJATW3LR | TTTTTTTTTTTTTTTTTTTGACTATCTTCCAATTGGCCATGTCAACTTC |
| RJATW5LF | GCCCTAGTAAATGAATGAAGTACTACATGGCCAATTGGAAGAATC |
| RJATW5LR | GATTCTTCAATTGGCCATGTAGTACTTCATTCATTTACTAGGGC |
| RJ19CF | GAATGAAGTTGATCATCGCCAATTGGAAG |
| RJ19CR | GAACCAATTGGCGATGATCAACTTCATTC |
| RJMT3CLF | CCTGAATGCCCTAGTAAAACTATGAAGTTGATCATGGCC |
| RJMT3CLR | GGCCATGATCAACTTCATAGTTTTACTAGGGCATTGCAGG |
| 5666 | GTAGTGCCAGATGGGTTA |
| B36For | TAGTACTCTACCTGGTTTT |
| B36Rev | ATTGCAGGAATAGTACCC |
| B36n-1 | GGCTTCCTATTTACATCC |
| 17TG | TTTTTTTTTTTTTTTTTG |
| GAPDH-1 | GCCAAAAGGGTCATCATCTC |
| GAPDH-1 | GTAGAGGCAGGGATGATGTTC |
| a59Sp6Ng(+) | TCGGCCTCGATGGCCATTTAGGTGACACTATAGATGTCTTTTGTTCCTGGGCAAG |
| A59Ng3′ | TCCGGA(TTT)8TTACACATTAGAGTCATCTTCTAACC |
| B36PACI3 | GCGCGCTTAATTAACGCGCCGGTGCA |
| B36PACI5′ | CCGGCGCGTTAATTAAGCGCGCTGCA |
| A59(+)(14639-14658) | GTGGATACACATCGTTATCG |
| A59(−)(16596-16577) | TACTGTGGTTTATGGTCCTC |
| A59(+)(16038-16059) | ATGAAGTCTACCTTCCATACCC |
| 7077(1-20) | TATAAGAGTGATTGGCGTCC |
| Le(7-23) | AGTGATTGGCGTCCGTA |
| L-internal(26-47) | TACCCTCTCAACTCTAAAACTC |
| 7065a (−)(31288-31270) | CATTGCAGGAATAGTACCC |
| N (−)(29937-29920) | GGCACTCCTTGTCCTTCT |
| S (−)(24672-24654) | TGGGTTGCAGATGAAAGGT |
| S (−)(24284-24265) | GCACCTGATGGCGTACTTGT |
Gel mobility shift RNase T1 protection assays.
The gel mobility shift RNase T1 protection assays were carried out as described by Yu and Leibowitz (34). Radiolabeled probe was incubated with 17Cl-1 cell lysate for 20 minutes followed by digestion with 0.006125 units of RNase T1 per 1.4 pmol of RNA for 20 minutes at 22°C. All reactions were carried out with unlabeled wild-type competitor, and reactions with nonspecific tRNA competitor at 10, 25, 50, and 100× molar concentrations were carried out in parallel. The complete reaction mixture was loaded onto a 1× Tris-buffered EDTA, 6% nondenaturing polyacrylamide gel and electrophoresed for 4.5 h. Gels were transferred to Whatman 3CHR paper and dried at 80°C under vacuum for 3 h and then exposed to Kodak Biomax MS film and quantitated with a phosphoimager. Six gel shift assays of each mutant were used to generate an average binding efficiency, with wild type always set to 100%.
Computer models.
The Mfold RNA version 3.0 folding algorithm was used to predict RNA secondary structure (36). The following parameters were used for all secondary structure modeling and are in keeping with previous work in our lab (34): no folding constraints, RNA sequence = linear, temperature fixed by the program at 37°C, ionic conditions fixed by the program at 1 M NaCl with no divalent cations, percent suboptimality = 5, upper band = 50, default = window, maximum distance between paired bases set, output at high resolution, format = bases, base number frequency = default, structure rotation angle = 0, and structure annotation by p-num.
UV cross-linking assays.
UV cross-linking of RNA binding proteins to radiolabeled probes was performed as described previously (35). Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the labeled proteins were visualized by autoradiography.
Oligonucleotide primers.
The oligonucleotide primers used in this work are shown in Table 1. For many of the primers, the position of the primer relative to the MHV-A59 genome, GenBank accession no. NC001846, is given in parentheses as part of the primer name.
Site-directed mutagenesis and pB36 mutant construction.
Paul S. Masters kindly provided the plasmid pB36, which contains a cDNA representing the B36 synthetic DI RNA (7). To ensure that only mutations in the 3′(+)42 host protein binding element were introduced into pB36, a subcloning strategy to create a mutagenesis template was adapted. The B36 HindIII-SacI 178-nucleotide fragment was generated as a mutagenesis template by PCR and TA cloning using primers B36SacI and B36HindIII and Invitrogen's 2.1 TA cloning kit to produce plasmid 2-1.
The MT3C, M19C, ATW, ATW5′, and ATW3′ mutations were introduced into plasmid 2-1 using Stratagene's QuickChange site-directed mutagenesis kit with primers listed in Table 1. After the procedure for mutagenesis was carried out, potential mutant 2-1 plasmids were screened for their intended mutations. Plasmids containing the desired mutations were used to introduce these mutations into pB36 by restriction fragment exchange using the SacI and HindIII restriction sites. The presence of the mutations and the absence of other changes were verified by sequencing the mutant pB36 plasmids using primer 5666 (5′-GTAGTGCCAGATGGGTTA).
In vitro transcription of DI RNAs.
DI RNAs for targeted recombination were generated by linearizing pB36 wild type and mutants with NsiI followed by gel purification. Transcription reactions were carried out using Ambion's mMessage mMachine kit following the manufacturer's protocol. Each transcription reaction was phenol:chloroform:isoamyl alcohol (25:24:1) extracted twice, chloroform extracted twice, ethanol precipitated, redissolved in diethylpyrocarbonate-treated water, and quantitated by spectrophotometry.
Targeted recombination.
Targeted recombination was carried out as described by Koetzner et al. (12). L2 cells grown in suspension were electroporated with 5 μg of DI RNA and then infected with Alb4. Virus stocks were prepared and heat inactivated at 40°C for 24 h, and representative heat-resistant viruses were plaque purified. Isolated virus was incubated until syncytia involved 90 to 95% of a 17Cl-1 monolayer. The medium was drawn off the monolayer and stored at −70°C, the monolayer was washed once with Dulbecco’s phosphate-buffered saline, and total cellular RNA was isolated using the RNeasy Mini kit (QIAGEN).
Reverse transcription PCR (RT-PCR) and sequencing of isolated virus.
RT reactions were carried out using the primer 17TG following Invitrogen's protocol for SuperScript II RNase H− reverse transcriptase. PCR was carried out on 1 microliter of RT product using the primers B36For and B36Rev, which flank the 87-nucleotide deletion present in Alb4 (21). PCR products were resolved on 2% agarose gels run in parallel to φX174 HaeIII markers. Viruses in which the 87-nucleotide deletion was repaired by a recombination event were identified by the presence of a 542-bp fragment compared to the 455-bp fragment observed when Alb4 was used as template. Recombinant viruses were subjected to further analysis to determine if they carried the desired mutations. A second PCR was carried out using the B36For and 17TG primers and 17TG-primed cDNA under the conditions used for screening. The amplified fragment was purified by gel electrophoresis and subjected to automated sequencing using the primer 5666.
B36 DI replication assays.
The method established by Masters' lab was used with modifications (21). 17Cl-1 and L2 cells were grown as monolayers until confluent. 17Cl-1 cells were washed, trypsinized, centrifuged, resuspended in DMEM10, and seeded onto six-well cluster plates 2 h prior to electroporation of L2 cells. Just prior to electroporation of the L2 cells the medium was changed to DMEM supplemented with 2% serum. To prepare L2 cells for electroporation, a confluent flask of cells was trypsinized, resuspended in DMEM containing 10% fetal bovine serum, and after centrifugation resuspended in DMEM containing 2% fetal bovine serum (DMEM2) at a concentration of 1.0 × 106 cells/ml. For each DI RNA 1.0 × 106 L2 cells were infected in suspension with MHV-A59 at a multiplicity of infection (MOI) of 3 for 2 h at 37°C. After 2 h of infection L2 cells were centrifuged at 1,000 × g, resuspended in 10 ml Ca2+- and Mg2+-free phosphate-buffered saline, and centrifuged again at 1,000 × g. This wash procedure was repeated two times. For each DI RNA 1.0 × 106 L2 cells were electroporated with 5 μg of DI RNA, or without RNA for the mock-transfected control, at maximum resistance, 0.8 kV and 25 μF. Two pulses were delivered 5 seconds apart. The electroporated cells were resuspended in 1 ml of DMEM2 and overlaid onto the 17Cl-1 cells.
The overlaid-infected-electroporated cells were incubated until syncytia consumed roughly 65% of the monolayer, usually 8 to 10 h postinfection. At this point the DMEM2 was removed and replaced with 2 ml phosphate-free DMEM with 2.5 μg/ml actinomycin D and incubated at 37°C for 20 min. At the end of 20 min the medium was replaced with phosphate-free medium containing 2.5 μg/ml actinomycin D, 2% dialyzed serum, and 0.5 mCi of 32PO4 and incubated at 37°C until syncytia enveloped 90 to 95% of the monolayer, usually 14 to 16 h postinfection. RNA was extracted with QIAGEN′s RNeasy kit. Three to six μg of RNA was denatured in RNA loading buffer and resolved by electrophoresis in 1% agarose gels containing formaldehyde as described previously (35). The 18S rRNA bands in ethidium bromide-stained gels were quantitated by densitometry to normalize the phosphorimager signal to total RNA loaded per sample. B36 DI RNA was set at 100%, and replicating DI RNAs were also adjusted to account for small differences in quantification and loading based on rRNA staining. Gels were photographed under UV light, fixed in 80% methanol, dried, and exposed to film. Bands located between RNAs 6 and 7, which represent replicating DI RNAs, were quantified with Molecular Dynamics Phosphorimager and Storm 8.2 software; three experiments for each set of mutants were used to generate an average.
RT-PCR and sequencing of replicating DI RNAs.
For each DI RNA evaluated, 1 μg of total RNA cell extracted from electroporated cells underwent reverse transcription with 2 pmol of primer B36n-1 using Invitrogen's Superscript II RT following the manufacturer's protocol. B36n-1 is specific to the negative-strand B36 DI RNA. This ensured that only replicated DI B36 would be detected. No-RT, no-RNA, and A59 controls were included in the RT step. The no-RT control used RNA extracted from the B36 DI RNA-transfected culture to ensure that residual digested transcription template DNA was eliminated and not amplified by PCR. RNA extracted from A59 infected-untransfected cells was used as a control to show specificity of the primers for B36. PCRs for each DI RNA were carried out for 31 cycles of 95°C for 30 seconds, 35°C for 60 seconds, and 72°C for 90 seconds, followed by a 15-minute extension at 72°C using 200 nM primers B36n-1 and 17TG and 1 μl of RT product as template in a 50-μl volume. PCR products were electrophoresed on a 1.0% agarose gel, stained with ethidium bromide, photographed, excised, and sequenced using primer 5666.
Introduction of mutations into MHV-A59.
The reverse genetic system described by Yount et al. (33) was used to recover wild-type MHV-A59 (known as MHV-A59 1000) and mutant viruses carrying the MT3C, M19C, ATW3′, ATW5′, and ATW mutations described above. Restriction fragment exchange was used to move the 3′(+)42 mutations from pB36 into the reverse genetic system plasmid G. To generate plasmids that contained compatible restriction sites, 5 micrograms of DI B36 mutants MT3C, M19C, ATW, ATW5′, and ATW3′ was digested with NsiI, and linearized plasmids were gel purified. An adaptor containing a PacI site was prepared by annealing 100 nmol of oligonucleotides B36PacI3′ and B36PacI5′. The adaptor was ligated into NsiI-cut pB36, and transformants containing the PacI site were identified by restriction digestion and verified by sequencing. These modified pB36 plasmids were digested with NruI and PacI, and the178-nucleotide NruI-PacI fragments were isolated. Plasmid G was similarly digested with NruI and PacI and ligated with the pB36-derived NruI-PacI fragment. The identities of G plasmids carrying the MT3C, M19C, ATW, ATW5′, and ATW3′ mutations were verified by sequencing with primer 5666. The MHV cDNAs were excised from mutant and wild-type G plasmids and ligated to MHV-A59 cDNAs A to F to generate cDNAs corresponding to the entire MHV genome under the control of a T7 promoter. Genome-sized RNAs were transcribed in vitro. Transcripts corresponding to the N gene coding sequence were also transcribed in vitro as described by Yount et al. (33). A mixture of genome and N gene transcripts was electroporated as described elsewhere to recover infectious virus (33).
After electroporation of the transcription reaction mixtures into MHV receptor-expressing BHK cells, the cells were overlaid onto a T-75 flask freshly seeded with 2 × 106 DBT cells. Cultures were incubated for 2 to 3 days. The cells were then frozen at −80°C, sonicated, and clarified by low-speed centrifugation. Viruses were plaque cloned on DBT or L2 cells. Individual plaques were isolated and inoculated onto a 70% confluent monolayer of 17Cl-1 cells until cytopathic effect enveloped 70 to 85% of the monolayer. At this point the medium was removed and stored at −70°C.
RNA was extracted and purified from the monolayer of infected 17Cl-1 cells using the RNeasy RNA extraction kit (QIAGEN). Reverse transcription was carried out using the primer 17TG. RT reactions were carried out using 1 μg of total RNA and 500 ng of primer 17TG; following incubation the RT was heat inactivated at 70°C for 15 min. A fragment containing the last 590 nucleotides of the genome was amplified by PCR using the B36For and 17TG primers under the conditions described above. Amplified fragments were sequenced with the primer 5666.
One-step growth curves.
17Cl-1 cells were grown in 96-well plates, and replicate wells were infected with MHV-A59 1000 or mutant virus at an MOI of three. Samples were frozen at 2, 4, 6, 8, 10, 12, 16, and 24 h postinfection, and virus production was quantitated by plaque assay. Triplicate samples were obtained for all time points.
Detection of negative-strand RNA and subgenomic RNAs by RT-PCR.
RT-PCR was employed to detect negative-strand genomic RNA and subgenomic mRNAs that might be expressed after electroporation of mutant viral genomes which failed to give rise to viable virus in multiple independent experiments. To enhance the sensitivity of the assays and to be certain that electroporated DNA was not detected, the standard procedure used to recover virus described above was slightly modified. After transcription of MHV genome RNA and prior to electroporation, transcription reaction mixtures were digested with 2 U of RNase-free DNase I (Ambion) for 20 min at 37°C. After electroporation, transfected BHK-R cells were plated directly in T-25 tissue culture flasks in the absence of DBT cells. After 4 h and 8 h of incubation at 37°C or 34°C, cells were harvested and total RNA was isolated using the RNeasy kit (QIAGEN). For one experiment total RNA was extracted at 8, 12, and 24 h after electroporation. One microgram of total RNA in a 10-μl volume was treated with 2 U of RNase-free DNase I for 30 min at 37°C; the DNase I was then inactivated at 65°C for 20 min. RT-PCRs to detect negative-strand genome-sized RNA and subgenomic RNAs were performed using SuperScript II RNase H− reverse transcriptase according to the manufacturer's protocol. To detect genome-sized negative-strand RNA, oligonucleotide A59(+)(14639-14658) was used as the RT primer for cDNA synthesis. This was followed by nested PCR. In the first PCR oligonucleotide A59(+)(14639-14658) was the forward primer, while oligonucleotide A59(−)(16596-16577) was used as the reverse primer. The primer pair for the second PCR was oligonucleotide A59(+)(16038-16059) and oligonucleotide A59(−)(16596-16577). A nested RT-PCR protocol was also used to detect negative-strand RNA complementary to subgenomic mRNA7. One microgram of total RNA was treated with DNase I, and cDNA synthesis was primed with oligonucleotide 7077(1-20) as described earlier. In the first PCR, oligonucleotide Le(7-23) and oligonucleotide A59(−)(31288-31270) were used as the forward and reverse primers, respectively. After 30 PCR cycles, 1 microliter of the first PCR product was used as template for the second nested PCR of 40 cycles. The forward and reverse primers were oligonucleotide L-internal(26-47) and oligonucleotide N(−)(29937-29925), respectively. Nested RT-PCR was also used to detect subgenomic mRNA7. Oligonucleotide 7065a(−)(31288-31270) was used for cDNA synthesis during the RT reaction, and then oligonucleotides Le(7-23) and 7065a(−)(31288-31270) were used for the first 30 cycles of PCR. For the subsequent nested PCR, 1 microliter of this PCR product was mixed with oligonucleotides L-internal(26-47) and N(−)(29937-29925). A similar nested RT-PCR protocol was also used to detect negative-strand RNA complementary to subgenomic mRNA3 (S gene). One microgram of total RNA was treated with DNase I, and cDNA synthesis was primed with oligonucleotide 7077(1-20) as described above. In the first PCR oligonucleotide Le(7-23) and oligonucleotide S(−)(24672-24654) were used as forward and reverse primers, respectively, for 30 cycles of PCR. One microliter of the first PCR product was mixed with oligonucleotide L-internal(26-47) and oligonucleotide S(−)(24284-24265) and subjected to another 40 cycles of amplification. PCR mixtures were electrophoresed in 1.5% agarose gels and ethidium bromide stained. Parallel reactions in which reverse transcriptase was omitted from the cDNA step were always performed to ensure that the PCRs did not detect residual DNA transcription templates which entered the cells during electroporation. Nested RT-PCR was also used to detect subgenomic mRNA3. The RT primer was oligonucleotide S(−)(24672-24654). Nested PCR was performed with the same primers and conditions used for detecting negative-sense RNA3.
RESULTS
Gel mobility shift RNase T1 protection assays and RNA secondary structure predictions.
Previous work in our laboratory defined two host protein binding elements located at positions 154 to 129 upstream of the poly(A) tail and the 3′-terminal 42 nucleotides of the 3′ UTR (18, 34, 35). A sequence comparison of the two elements revealed a shared 11-nt motif, UGARNGAAGUU. Secondary structure analysis using Mfold 2.0 predicted that the 3′-terminal 42-nt host protein binding element formed a bulged stem-loop structure with the 11-nt motif forming part of the right portion of the bulged stem (35). Reexamination of the secondary structure with a more recent version of Mfold (version 3.0) predicted a different secondary structure. Mfold 3.0 predicted that nucleotides 27 to 42 [all nucleotides herein are referred to by their position relative to the 3′ terminus of the MHV genome, with the first nucleotide upstream of the poly(A) tail designated as nucleotide 1] were single stranded with the remaining nucleotides forming a bulged stem-loop structure (Fig. 1). Nuclease structural probing was more consistent with the 42-nt structure folding into the Mfold-predicted structure shown here, rather than the published model based on a longer 166-nt RNA (17). Liu et al. (17) were able to obtain nuclease digestion data on nucleotides 13 to 42. Comparison of the predicted secondary structure of the wild-type 3′(+)42 model (Fig. 1) with the nuclease digestion of the 166-nt RNA indicated that the 42-nucleotide model matches the digestion data in 28 of 30 positions. When the Mfold 3.0-predicted secondary structure of the 166-nt RNA was compared to the same digestion data, only 18 of 30 nucleotides matched (17).
FIG. 1.
Gel mobility shift RNase T1 protection assays for the mutant MT3C and secondary structure models of the 3′(+)42 host protein binding element mutants. Mutant nucleotides are shaded. The dark triangle represents increasing concentration of wild-type competitor RNA, and the open triangle indicates an increasing concentration of nonspecific tRNA competitor. The arrow indicates the RNP complexes which were quantitated (Table 2).
To investigate the role of RNA secondary structure as a determinant of protein binding activity, the wild-type secondary structure predicted by Yu and Leibowitz was used as a basis for designing mutations within the 42-nucleotide protein binding element. Initially five mutations were created within this genome fragment (MT1A, MT2A, MT3C, MT5A, and MD10; Table 2). All of these mutations were located within the 11-nucleotide motif previously implicated in protein binding activity, with the exception of MD10, which is a 10-nt deletion from the 3′ end. Two of these mutations were predicted by Mfold to have no effect on RNA secondary structure, and three were predicted to produce major changes in secondary structure. The effect of these mutations on the RNA binding activity of the 42-nt protein binding element was then examined in a series of gel mobility shift RNase T1 protection assays. In our gel mobility shift RNase T1 protection assays, the complex of interest designated by the arrow in Fig. 1 represented the largest RNP complex formed that could be resolved within the gel and was quantified by phosphoimager and compared with the wild type. The bands at the top of the gel are in the well and are most likely nonspecific complexes that could not enter the gel, while the faster-migrating complexes probably represent RNP complexes which lack one or more of the four proteins composing the complete complex, as described by Nanda et al. (23).
TABLE 2.
Summary of RNA protein binding data
| Probe | Sequenceb | % Binding | SD | Predicted effect on second structure |
|---|---|---|---|---|
| Wild type | 5′ AGUAAAUGAAUGAAGUUGAUC | 100 | NAa | NAa |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| MT1A | 5′ AGUAAAaGAAUGAAGUUGAUC | 68.5 | 11.3 | None |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| MT2A | 5′ AGUAAAacAAUGAAGUUGAUC | 64 | 21.7 | None |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| MT3C | 5′ AGUAAAacuAUGAAGUUGAUC | 6.3 | 6.1 | Disrupts |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| MT5A | 5′ AGUAAAacuuaGAAGUUGAUC | 25.2 | 5.5 | Disrupts |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| MD10 | 5′ AGUAAAUGAAUGAAGUUGAUC | 44.1 | 2.5 | Disrupts |
| AUGGCCAAUUG | ||||
| M19C | 5′ AGUAAAacuAUGAAGUUGAUC | 74.1 | 8.6 | Restores |
| AUcGCCAAUUGGAAGAAUCAC | ||||
| 19C | 5′ AGUAAAUGAAUGAAGUUGAUC | 73.5 | 25.4 | None |
| AUcGCCAAUUGGAAGAAUCAC | ||||
| ATW5′ | 5′ AGUAAAUGAAUGAAGUacuaC | 7.2 | 8.7 | Disrupts |
| AUGGCCAAUUGGAAGAAUCAC | ||||
| ATW3′ | 5′ AGUAAAUGAAUGAAGUUGAUC | 400 | 84.4 | Disrupts |
| AUGGCCAAUUGGAAGAuaguC | ||||
| ATW | 5′ AGUAAAUGAAUGAAGUacuaC | 185.8 | 34.7 | Restores |
| AUGGCCAAUUGGAAGAuaguC |
NA, not applicable.
Mutations are indicated by lowercase type.
RNAs which contained mutations predicted to maintain RNA secondary structure, MT1A and MT2A, had protein binding activity decreased to approximately 65% of that observed with wild-type RNA. RNAs that contained mutations predicted to disrupt secondary structure, like MT3C, MT5A, and MD10, disrupted protein binding activity to 6 to 40% of the wild-type value. The results are summarized in Table 2. Mutant MT3C is identical to mutant mB4 described by Yu and Leibowitz (34). The decrease in binding was expected based on previous studies but does not determine the contribution of secondary structure in RNP complex formation.
To further explore the role of RNA secondary structure in host protein binding, Mfold was used to identify a series of compensatory mutations that are predicted to restore the wild-type secondary structure. Compensatory mutation effects on host protein binding were determined by gel mobility shift RNase T1 protection assays. A representative mobility shift RNase T1 protection assay for mutant MT3C is shown in Fig. 1. MT3C had the greatest predicted change in secondary structure and the least amount of binding activity compared to wild-type virus (Fig. 1). The predicted compensatory G-to-C change at position 19 in the MT3C mutant not only restored wild-type RNA secondary structure (Fig. 1, mutant M19C) but dramatically restored protein binding activity of the substrate to about 74% of the wild-type value (Table 2). RNA carrying only the G-to-C mutation at position 19 (mutant 19C, Fig. 1 and Table 2) is predicted to maintain the wild-type secondary structure and has a protein binding activity which is approximately 74% of that achieved by wild-type RNA. These data suggest that wild-type secondary structure, and to a lesser extent sequence context, may play a role in the ability of the 3′(+)42 host protein binding element to bind to host proteins.
Because the updated version of Mfold predicted a different secondary structure for the wild-type sequence reported by Yu and Leibowitz (35), another panel of mutations were designed to disrupt interactions between nucleotides 23 to 26 and nucleotides 2 to 5. Nucleotide 26U is the last nucleotide composing the 11-nt motif. The ATW5′ mutation (Table 2) is predicted to disrupt RNA secondary structure (Fig. 1) with the only base pairing occurring between nucleotides 10 to 11 and 16 to 17 with a small loop formed by nucleotides 12 to 15. Gel mobility shift RNase T1 protection assays demonstrated that ATW5′ exhibited 7.2% as much protein binding activity as wild-type sequence. The ATW3′ mutation (Table 2) was predicted to produce an RNA secondary structure similar, but not identical, to that predicted for the wild-type sequence. Gel mobility shift RNase protection assays demonstrated that the ATW3′ probe has a fourfold increase in protein binding activity relative to the wild-type RNA probe. The double mutant ATW contains both the ATW5′ and ATW3′ mutations; ATW is predicted to restore the wild-type secondary structure and binds 1.85 times as well as wild type. Taken together, the results support the hypothesis that both primary and secondary structure play a role in host protein binding to the 3′(+)42 host protein binding element.
Since two of the mutant probes had increased protein binding activity relative to wild-type RNA, we performed a UV cross-linking assay to determine if proteins of identical molecular weights were bound by wild-type, ATW3′, and ATW RNAs. After RNA complexes were allowed to form in solution, the RNAs were cross-linked to their protein binding partners and the labeled proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described in Materials and Methods. The proteins that formed complexes with the mutant and wild-type probes were identical in molecular weight. Moreover, the relative amount of the four labeled proteins was similar in wild-type and mutant complexes except that the overall signal intensity of each sample paralleled that observed in the gel mobility shift RNase T1 protection assays for the wild-type, ATW3′, and ATW RNAs (data not shown).
DI RNA replication assays.
To investigate the effect of these mutations on MHV replication, we first employed a series of DI RNA replication assays. The M19C, MT3C, ATW5′, ATW3′, and ATW mutations were introduced into a plasmid containing a cDNA corresponding to the B36 DI RNA (12). We chose the B36 DI RNA as our model replicon because it also would allow us to attempt to introduce these mutations into MHV-A59 by targeted recombination (12, 21). For simplicity, replication assays were performed in two groups, the first with the mutants involving the 11-nt host protein binding motif, MT3C and M19C. The MT3C mutation in B36 is identical to the mB4 mutation in DIssE described by Yu and Leibowitz, which replicated poorly and readily recombined with wild-type genome sequences (34). We chose to explore this mutation in the context of the B36 DI RNA because it had the greatest effect on RNP formation and was predicted to cause a drastic change in RNA secondary structure. M19C was derived from MT3C and encoded a second compensatory change that largely restored protein binding activity as well as the predicted secondary structure of the RNA, establishing a possible connection between secondary structure and RNP complex formation. Cells were infected with MHV-A59; wild-type B36, MT3C, M19C, or DIssE (an MHV-JHM-derived DI RNA which replicates to higher levels than B36) was electroporated into the infected cells; and the cultures were metabolically labeled with [32P]orthophosphate in the presence of actinomycin D as described in Materials and Methods. After the labeling period RNA was extracted and the MHV-specific RNAs were resolved by electrophoresis (Fig. 2). Three complete experiments were used to determine an average DI RNA replication efficiency. A complete experiment was defined as one in which DI B36 RNA and DIssE RNA both replicated to detectable levels regardless of how well the mutant DI RNAs replicated and time constants for all electroporations were 0.8, indicating that electroporation conditions were optimal. MHV-A59-infected cells transfected with mutants MT3C and M19C synthesized positive-strand DI RNA with varying efficiency compared to wild-type DI B36, as detected by metabolic labeling. Quantitation by phosphorimager analysis revealed that MT3C replicated 78% as well as B36 and M19C replicated 69% as efficiently as B36. In order to determine if replicated DI RNAs contained the intended mutations, RT-PCR for negative-strand RNA was carried out for DI RNAs MT3C and M19C. The negative strand was amplified to avoid interference by transfected positive-strand DI RNAs. Sequence analysis revealed that the mutant DI RNA had undergone recombination events that restored the wild-type sequence and that the repaired, replication-competent DI RNAs became the dominant genotype detected in the culture, making it impossible to measure levels of mutant DI RNA replication; any mutant DI RNA that could replicate did so well below our detection level. Recombination was not entirely unexpected, since recombination to restore wild-type sequence is a common occurrence in DI RNA replication experiments (6, 17, 34, 35).
FIG. 2.
Replication assays for DI RNAs B36, MT3C, and M19C. (A) Representative electropherogram of MHV-infected cells which were mock, DIssE, B36, MT3C, and M19C electroporated as indicated. Brightness and contrast have been adjusted to enhance the visibility of the B36, MT3C, and M19C bands using Adobe Photoshop 6.0. The arrow indicates the position of bands which were used to quantitate DI RNA replication. (B) Average replication efficiency for three experiments using mutants MT3C and M19C. DIssE replicated an average of 1.6-fold more efficiently than wild-type B36 in these experiments.
The second group consisted of the mutants based upon the more recent Mfold data, two of which, ATW and ATW3′, formed RNP complexes at higher levels than wild-type probe did in gel mobility shift RNase T1 protection assays. The replication efficiency of ATW, ATW5′, and ATW3′ was determined in a similar set of experiments. In contrast to the first set of mutants, ATW, ATW5′, and ATW3′ did not replicate nearly as efficiently as DI B36 and were generally undetectable by metabolic labeling (Fig. 3). Sequencing was carried out by RT-PCR for negative-strand DI RNA followed by sequencing of the PCR products to determine if the intended mutations had been maintained. For two of the three complete experiments obtained, the mutations were maintained in ATW, ATW5′, and ATW3′. In one of those two experiments ATW3′ replicated at 4% of B36 levels and was undetectable in the other, while ATW and ATW5′ were undetectable by phosphor-imager analysis in both experiments.
FIG. 3.
Replication assays for DI RNAs B36, ATW, ATW5′, and ATW3′. (A) Representative electropherogram of MHV-infected cells which were mock electroporated or DIssE, ATW, B36, ATW5′, and ATW3′ electroporated as indicated. The arrow indicates the position of bands which were used to quantitate DI RNA replication. (B) Summation of the DI RNA replication data for the two experiments in which mutant DI RNAs did not recombine.
Sequencing data from the third experiment indicated a mixed population of DI RNAs. Some of the mutant nucleotides reverted to wild type within the DI RNA population, while part of the population had either a mutant or a wild-type base in a particular position. Sequencing was repeated for these DI RNAs with identical results. The data are summarized in Table 3. The results from this experiment may point to particular nucleotides within and outside the 11-nt motif that play a role in replication and present an opportunity for further study of the 3′-terminal 42-nucleotide host protein binding element. DI RNA replication assays for mutants ATW, ATW3′, and ATW5′ are consistent with the formation of an RNP complex at the 3′-terminal 42 nt of the MHV genome playing an important role in replication.
TABLE 3.
Sequence variation during DI RNA replication assays with ATW5′, ATW3′, and ATW DI RNAs
| Assay | Nucleotide at position:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 26 | 25 | 24 | 23 | 5 | 4 | 3 | 2 | |
| Wild type | U | G | A | U | A | U | C | A |
| ATW | A | C | U | A | U | A | G | U |
| ATW recovered | A/U | C | U | A/U | U/A | U | G/C | U |
| ATW5′ | A | C | U | A | A | U | C | A |
| ATW5′ recovered | A/U | C | U | A/U | A | U | C | A |
| ATW3′ | U | G | A | U | U | A | G | U |
| ATW3′ recovered | U | G | A/U | U | U/A | U | C | U |
Isolation and phenotypic properties of MHV-A59 mutants.
Targeted recombination using B36 RNAs carrying the M19C, MT3C, ATW5′, ATW3′, and ATW mutations as the donor and Alb4 as the MHV-A59 recipient was carried out as described in Materials and Methods. A total of 87 individual plaques were screened, and 28 of these plaques were demonstrated to be recombinants (they repaired the deletion present in the Alb4 N gene) by RT-PCR. All 28 recombinant plaques were sequenced and found to be wild type; this indicated that an even number of recombination events had occurred and that under these conditions the introduced mutations were not recovered.
To evaluate the role of the 3′(+)42 host protein binding element in MHV replication in the context of a whole genome, we utilized the reverse genetic system recently developed in the Baric lab (33). Mutants MT3C, M19C, ATW5′, ATW3′, and ATW were introduced into the G fragment which encompasses the 3′ 8.7 kb of the MHV genome by introducing a PacI site downstream of the poly(A) tail in the pB36 constructs and then performing restriction fragment exchanges. All constructs were confirmed by sequencing. cDNAs representing the entire MHV-A59 genome with either the wild-type sequence or the MT3C, M19C, ATW5′, ATW3′, and ATW mutations were constructed by ligation of the G fragments to fragments A to F as described previously (33). Mutant and wild-type MHV genomes were transcribed and electroporated into cells as described by Yount et al. (33). After incubation for 72 h, or until cytopathic effect (syncytia) encompassed roughly 70% of the cell monolayer, whichever was first, potential mutant virus was harvested and plaque purified. For all mutants generated except ATW, viable virus was isolated and sequenced and found to carry the intended mutation. Syncytium formation became apparent for all viruses except the ATW mutant by 24 h after electroporation. Multiple independent experiments failed to recover virus containing the ATW mutation. Interestingly, ATW encodes both the ATW3′ and ATW5′ mutation sets. These included experiments in which the electroporated cells were incubated at 34°C and 40°C.
The plaque phenotypes of the MT3C, M19C, ATW5′, and ATW3′ viruses were different (Fig. 4) and are summarized in Table 4. In L2 cells wild-type MHV-A59 has a relatively large (2.4- to 3.38-mm-diameter) plaque size which varied from experiment to experiment. All of the mutants had somewhat reduced plaque sizes relative to wild-type MHV-A59 assayed in the same experiment. Virus carrying the MT3C mutation produced plaques 46.4% (1.57 mm) of the diameter of those formed by wild-type virus, while virus carrying the M19C mutation had a plaque size only slightly smaller than that of wild-type MHV-A59. Viruses carrying the ATW5′ or ATW3′ mutation produced plaques approximately one-third the diameter of wild-type plaques. One-step growth curves were performed with all of these mutants to further characterize their ability to replicate (Fig. 5). At an MOI of 3, all viruses had growth kinetics similar to those of wild-type virus. Metabolic labeling experiments with [32P]orthophosphate in the presence of actinomycin D analyzed by denaturing gel electrophoresis demonstrated that cells infected with these mutant viruses synthesized similar amounts of genomic and subgenomic MHV-specific RNAs (data not shown).
FIG. 4.
Average plaque sizes for viable 3′ 42-nucleotide mutant viruses generated with the MHV infectious clone. Plaque sizes were measured after monolayers were stained with crystal violet. The monolayer and a millimeter ruler were digitally photographed and projected. The projected plaque sizes and the degree of enlargement were determined with a millimeter ruler, and the actual plaque sizes were calculated.
TABLE 4.
Summary of relative plaque sizes of mutants
| Expt no. | Virus | Viable | Plaque size (mm) | Relative plaque size |
|---|---|---|---|---|
| 1 | A59 1000 | Yes | 3.38 ± 0.50 | 1.0 |
| MT3C | Yes | 1.57 ± 0.20 | 0.46 | |
| 2 | A59 1000 | Yes | 2.40 ± 0.05 | 1.0 |
| M19C | Yes | 2.15 ± 0.05 | 0.90 | |
| ATW5′ | Yes | 0.80 ± 0.04 | 0.33 | |
| 3 | A59 1000 | Yes | 3.40 ± 0.05 | 1.0 |
| ATW3′ | Yes | 1.14 ± 0.04 | 0.37 | |
| ATW | No | NAa | NAa |
NA, not applicable.
FIG. 5.
Growth curves for MHV mutants. Replicate cultures in 96-well plates were infected at an MOI of 3 and harvested at the times shown, and titers were determined by plaque assays. Triplicate wells were assayed for each time point. Error bars represent the standard errors of the means.
Since we could not recover viable ATW mutant virus and the DI RNA replication assays with this mutant indicated a defect in viral RNA synthesis, we employed a nested RT-PCR assay (shown schematically in Fig. 6A) to determine if genome-size negative-strand RNA was synthesized in cells electroporated with MHV-A59 genomic RNA containing this mutation. As shown in Fig. 6B, cells electroporated with ATW and wild-type MHV RNA yielded similar amounts of a 559-base-pair fragment in this assay (lanes 3 and 5). When reverse transcriptase was omitted from the RT step (lanes 2 and 4), no PCR product of the expected size was obtained from electroporated cells, demonstrating that any template DNA was sufficiently degraded by DNase digestion so that it could not be amplified. To more accurately define the relative amounts of genome-sized negative-strand RNA present in the electroporated cells, we performed a second series of nested RT-PCRs using various dilutions of the initial RT product. Over this series of dilutions of cDNA, there is no noticeable difference between the amount of ATW and that of wild-type PCR products. This result, although not quantitative, indicates that the amounts of full-length negative-strand RNA in cells electroporated with these two RNA genomes are roughly similar and that the replication defect lies in another step of the viral replication cycle.
FIG. 6.
The defect in mutant ATW is not at the level of genome-length negative-strand RNA synthesis. (A) Schematic drawing of the assay used to detect negative-strand genome-sized RNA and the relative locations of the primers within the genome. (B) RT-PCR of RNA extracted from wild-type (WT)- and ATW mutant-electroporated cells at 8 hours after electroporation. A 1-kbp ladder was used as a molecular size marker. (C) RT-PCR assay done with the indicated dilutions of the RT product. For each dilution the wild-type sample is on the left and the ATW mutant sample is on the right.
To determine if negative-strand RNA complementary to subgenomic mRNAs was synthesized, we employed a second nested RT-PCR assay utilizing an RT primer, oligonucleotide 7077(1-20), that corresponded to the first 20 nucleotides of the leader sequence present at the 5′ end of all the MHV mRNAs and the genome (Fig. 7A and 8A). To detect negative-sense RNA complementary to mRNA7, encoding the N gene, nested PCR was carried out using primer sets that correspond to leader-specific primers [Le(7-23) and L-internal(26-47)] and primers within the N coding sequence. As shown in Fig. 7C, RNA extracted from cells electroporated with ATW or wild-type MHV RNA at 4 and 8 h after electroporation (prior to cell fusion in cultures electroporated with wild-type RNA) yielded roughly similar amounts of a 330-base-pair fragment in this assay. Sequencing of the amplicon showed that it corresponded to the targeted region and that the amplicon carried the leader sequence fused to the body of the N gene. When reverse transcriptase was omitted from the RT step, no PCR product of the expected size was obtained from electroporated cells (data not shown). A similar result was obtained when RNA was extracted from cells at later times (12, 16, and 24 h) after electroporation (data not shown). Real-time RT-PCR assays were performed using the same nested PCR strategy to obtain a more quantitative comparison in the amount of negative-sense RNA7 present. These assays failed to demonstrate consistent differences between cultures electroporated with wild-type and those electroporated with ATW RNA, even at late times when cultures electroporated with the wild type had undergone considerable cell fusion. To investigate if mRNA transcription was impaired for the ATW mutant, a nested RT-PCR assay specific for subgenomic mRNA7 was used (Fig. 7A). As shown in Fig. 7B, RNA extracted from cells electroporated with ATW or wild-type MHV at 4 and 8 h after electroporation yielded roughly equal amounts of the expected 330-base-pair amplicon in this assay. Samples extracted at later times after electroporation also failed to demonstrate an obvious difference in the quantities of mRNA7 present (not shown). When reverse transcriptase was omitted from the RT step, no PCR product of the expected size was obtained from electroporated cells (data not shown). Somewhat surprisingly, real-time RT-PCR assays (data not shown) failed to demonstrate consistent differences in mRNA7 levels at either early times (4 and 8 h) or late times (12, 16, and 24 h) after electroporation, indicating that similar amounts of mRNA7 were present in cells electroporated with ATW and those electroporated with wild-type genomes.
FIG. 7.
Nested RT-PCR analyses for RNA7. (A) Schematic representation of the RT-nested PCR strategy and primers used to detect negative- and positive-sense subgenomic RNA7. The relative sizes of the leader sequence in RNA7 and the body of RNA7 are not to scale. The position of each primer in the MHV-A59 genome, GenBank accession no. NC0001846, is given in parentheses as part of the primer name. Primer sequences are listed in Table 1. (B) Representative RT-nested PCR for positive-strand subgenomic RNA7. The arrow indicates the specifically amplified RNA7 fragment. The sizes of the marker DNAs in base pairs are shown to the left of the electropherogram. (C) Representative RT-nested PCR for negative-strand subgenomic RNA7. The arrow indicates the specifically amplified RNA7 fragment. The sizes of the marker DNAs in base pairs are shown to the left of the electropherogram.
FIG. 8.
Cells electroporated with ATW genomes do not synthesize RNA3. (A) Schematic representation of the primers used to detect both positive- and negative-strand subgenomic RNA3. (B) Representative experiment for positive- and negative-strand subgenomic RNA3 synthesis at 4 and 8 hours postelectroporation. The sizes of the marker DNAs in base pairs are shown to the left of the electropherogram.
The data for mRNA7 are inconsistent with the ATW phenotype; no syncytia are observed in cells electroporated with ATW genomes, leading us to believe that there must be a defect in subgenomic mRNA synthesis. One possible explanation for the nested RT-PCR result is recombination between the in vitro-transcribed N coding sequence RNA lacking leader and 3′ UTR sequences with genomic sequences yielding mRNA7 and its negative-strand complement.
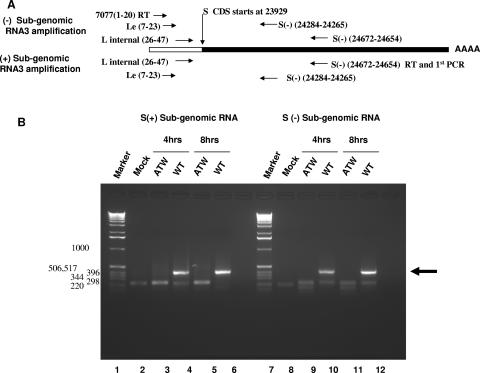
RNA3, which encodes the S protein responsible for cell fusion, was chosen as a target for RT-PCR to further examine the effect of the ATW mutation on subgenomic RNA synthesis (Fig. 8). A nested-set RT-PCR approach similar to that described for mRNA7 was employed (Fig. 8A). With RNA3 as a target it became obvious that negative- and positive-strand subgenomic RNA synthesis was impaired (Fig. 8B). Nested RT-PCR analysis of RNA extracted from cultures electroporated with wild-type genome (lanes 4 and 6) at 4 and 8 h postelectroporation amplified a DNA fragment of the expected 401-bp size, indicated by the arrow in Fig. 8B, whereas cultures electroporated with ATW genomes (lanes 3 and 5) did not yield a fragment of the expected size. The approximately 220-bp fragment observed in the ATW lanes was also present in the mock-electroporated sample (lane 2), indicating that this likely represents nonspecific amplification of a host RNA. These results indicate that no mRNA3 was produced in ATW- electroporated samples, while mRNA3 is abundant in wild-type-electroporated samples. A similar nested RT-PCR experiment was performed to determine if negative-strand subgenomic RNA3 was present. At 4 and 8 h postelectroporation no negative-strand ATW RNA3 could be detected (lanes 9 and 11), although negative-sense RNA3 was easily detected in cells electroporated with wild-type genome (lanes 10 and 12). The origin of the smaller (approximately 200- to 220-bp) fragments amplified in mock, ATW, and wild-type samples was not investigated further. As subgenomic RNA abundance is regulated by 3′-end proximity and transcription regulatory sequence regulatory domains, a similar experimental approach was used to investigate mRNA6, encoding the M gene, and identical results were obtained. Neither positive- nor negative-sense RNA corresponding to RNA6 was detected in cultures electroporated with ATW genomes (data not shown).
DISCUSSION
Using gel mobility shift RNase T1 protection assays, we have demonstrated a possible role for RNA secondary structure for host protein binding to the MHV 3′ 42-nucleotide element. Previous work in our lab has demonstrated that an 11-nt motif was important for RNA binding and DI RNA replication (17, 18, 34, 35). We have also identified the host proteins that bind specifically to this region as mitochondrial aconitase and mitochondrial HSP70, HSP60, and HSP40 (23, 24). Gel mobility shift RNase T1 protection assays with probes containing mutations within the 11-nt motif of the 3′ 42-nucleotide element demonstrated an overall decrease in host protein binding compared to wild-type probe. Two mutants, MT3C and MT5A, showed dramatic changes in computer-predicted RNA secondary structure compared to wild-type sequence and had much lower protein binding activities, 6.3 and 25.2% of wild-type values, respectively. Probe M19C, which contained compensatory changes that restored the wild-type predicted secondary structure, also restored host protein binding activity to 74.1% of the wild-type level. The 19G-to-C mutation alone modestly decreased host protein binding to 73.5% of the wild-type level but did not alter computer-predicted RNA secondary structure. These probes provide support for the hypothesis that proper RNA folding is essential for host protein binding to the 3′ 42-nucleotide element, although additional biochemical studies are needed to prove the existence of the predicted structures.
A second set of mutant probes was developed based on the Mfold 3.0 predictions of the wild-type 3′ 42-nucleotide element. Probes ATW, ATW3′, and ATW5′ carried mutations in a predicted double-stranded region. Computer predictions suggest that probes ATW3′ and ATW5′ would have altered RNA secondary structures while the compensatory mutant ATW should have restored the wild-type structure. ATW5′, which is predicted to be mostly single stranded, had little protein binding activity (7.2%), while probe ATW3′, which is predicted to have only a slight alteration in RNA secondary structure, had a protein binding activity fourfold greater than that of the wild type. The double mutant, ATW, is predicted to maintain wild-type secondary structure and formed RNP complexes 1.85 times more efficiently than wild-type probes did. The results with the ATW mutant suggest the possibility that this RNA may fold into two equally stable structures, with one structure binding RNA as efficiently as the ATW3′ probe and the second conformation unable to efficiently form RNP complexes, producing the observed result. The failure to recover ATW mutant virus using the infectious clone underlines the importance of primary structure at the 3′ end of the genome in the MHV life cycle. Other studies have established the importance of host proteins interacting with the MHV genome. Polypyrimidine tract binding protein (PTB) binds to the leader RNA as well as the 5′ end of the genome complement (9, 14). It has recently been demonstrated that overexpression of PTB decreases viral mRNA transcription but not translation (3). PTB also alters RNA secondary structure and may help mediate 5′-to-3′ interactions of the MHV genome through PTB interactions with hnRNP-A1 (8). hnRNP-A1 interacts with MHV transcription regulatory sequences and 3′ UTR; DI mutants with decreased hnRNP-A1 binding also had a decrease in transcription and replication (8, 15) The importance of hnRNP-A1 in MHV transcription and replication is controversial, as cell lines deficient in hnRNP-A1 produce viral RNAs as efficiently as cell lines expressing hnRNP-A1 (27).
Spagnolo and Hogue characterized an interaction between the poly(A) tail of DI RNAs and poly(A) binding protein (PABP). PABP is a host cellular protein that is required for translation of eukaryotic mRNAs and is reported to play a vital role in poliovirus replication and transcription by circularization of the poliovirus genome via its interaction with eIF4G (5, 22, 30). It is likely that PABP binding to the MHV 3′ UTR has a similar effect on virus replication as what Spagnolo and Hogue hypothesize (29). Recently Zúñiga et al. developed a model predicated upon a role for 5′ and 3′ genomic interactions in the regulation of subgenomic RNA synthesis (37). They demonstrated that base pairing of transcription-regulating sequences with the leader sequence of the genome directly correlated with the amount of subgenomic RNA produced and that discontinuous transcription occurs during negative-strand synthesis. Their data raise the possibility that specific regions of the 3′ UTR may be required for interactions with the 5′ end of the genome, either directly or through protein-protein interactions, to regulate additional aspects of MHV replication, such as negative-strand synthesis.
Nanda and Leibowitz established the identity of four proteins that specifically interact with the MHV 3′(+)42 host protein binding element as mitochondrial aconitase and mitochondrial HSP70, HSP60, and HSP40 (23, 24). They also established that metabolic regulation of the RNA binding activity of mitochondrial aconitase did influence viral growth, although it's less clear whether these proteins have high or low penetrant effects on virus replication. Mitochondrial aconitase has a cytosolic counterpart that is a conditional RNA binding protein, iron regulatory protein. Increasing iron concentrations in MHV-infected cell cultures increased viral titer at early times postinfection compared to infected cells that did not receive supplemental iron (23, 24), emphasizing the importance of RNP complex formation involving mitochondrial aconitase and the MHV genome.
Our functional studies to determine the role for RNP complexes of the 3′ 42-nucleotide element in viral replication demonstrated that mutations within the 3′ 42-nucleotide host protein element resulted in defects in replication. Targeted recombination experiments to generate viruses containing the MT3C and M19C and ATW, ATW3′, and ATW5′ mutations resulted in the recovery of even-numbered recombinants which restored the wild-type sequence. The data suggest that the mutant viruses which should have been generated by a single or an odd number of crossover events were at a replicative disadvantage compared to the wild-type genomes which were generated by a less-likely even number of crossovers. An alternative strategy that may have yielded a more favorable result would have been to use the feline coronavirus spike protein targeted recombination system (13). Selection by spike protein would have been more stringent and probably reduced the background and even-number recombination events that likely occurred, giving our negative results.
DI RNA replication assays using DI B36 and DI B36 mutants MT3C, M19C, ATW, ATW3′, and ATW5′ were performed. Mutant DI RNAs MT3C and M19C carry mutations within the 11-nt motif previously implicated in protein binding (18, 34, 35). DI mutants MT3C and M19C recombined to restore wild-type sequence in all three experiments evaluated, demonstrating that the mutants could not successfully compete against wild-type recombinant DI RNAs, suggesting the importance of this element in replication. DI RNAs ATW, ATW3′, and ATW5′ replicated poorly. In two experiments in which the mutant sequence was maintained in the DI RNAs (as shown by RT-PCR sequencing of the negative strand), ATW and ATW5′ positive-strand replication was undetectable by metabolic labeling, and ATW3′ replication was barely detectable in one of those experiments.
Comparison of the gel mobility shift RNase T1 protection assays with the DI RNA replication data indicates the importance of proper primary and secondary structures. Mutant MT3C was dramatically reduced in protein binding activity, while mutants M19C and 19C had more modest decreases in host protein binding. When introduced into DI B36 the DI RNAs MT3C and M19C were not able to replicate and only recombined wild-type DI RNA was detected. Although the gel shift data suggest that probe 19C and M19C, a mutation containing the MT3C and 19C mutations, have wild-type secondary structures and near-wild-type binding activity, the M19C mutation did not restore the replicative capacity of the DI RNA, suggesting the importance of wild-type primary structure in the 11-nt host protein binding motif. In contrast the ATW set of mutants suggest the importance of proper secondary structure in host protein binding and replication. The mutant ATW3′ could be detected only in one of two recombination-free experiments and at 4% of the wild-type DI B36 level. The ATW3′ mutants do not lie within the 11-nt element, and secondary structure is disrupted compared to wild type, suggesting that secondary structure is important for replication.
When the MT3C, M19C, ATW, ATW5′, and ATW3′ mutations were introduced into a full-length virus genome, results unfettered by recombination repair of DI RNAs were obtained. Kinetic analysis of the mutant viruses indicates that there is no impairment in the ability of MT3C, M19C, ATW5′, and ATW3′ to replicate in one-step growth curve experiments performed at moderate multiplicities of infection, although more dramatic changes might be noted at lower MOIs. This result is somewhat difficult to reconcile with the negative effects that these mutations had in DI RNA replication assays and underscores some of the difficulties in using DI RNA models for investigating genome replication. The inherent competition of mutant DI RNAs with both helper virus and any wild-type DI RNAs that arise by recombination complicates the interpretation of DI RNA replication assays. Recombination is a stochastic event; thus, DI RNA recombination varies from experiment to experiment and recombination leading to repair of introduced mutations is unlikely to be equivalent among constructs. Recombination probably varies depending upon the fitness of the mutation introduced and transfection efficiency. Recombination is associated with replication, and a slightly more robust mutant DI RNA may repair more quickly, resulting in wild-type DI RNAs. In spite of these limitations, which make interpretation difficult and quantitative comparisons of assays where recombination has repaired a mutation impossible, the inherent competitive nature of the DI RNA assay makes them similar in some respects to assays of competition between wild-type and mutant viruses, which may bring out deficits in replication fitness which are not apparent in one-step growth curve experiments.
Each of the mutant viruses produced plaque sizes smaller than wild-type virus. The cause of the small-plaque phenotype is not clear. Metabolic labeling experiments in cells infected with the wild-type MHV-A59 and MT3C, M19C, ATW5′, and ATW3′ mutant viruses indicated that approximately the same amount of each species of virus-specific RNA is present regardless of the virus examined (J. L. Leibowitz and J. J. Millership, unpublished data). Since plaques are largely created by lysis of syncytial giant cells and syncytium formation is mediated by the amount of S protein on the cell surface, it is possible that the differences in plaque size reflect modest differences in translational efficiency of virus-specific mRNAs carrying these mutations in their 3′ UTRs. This question requires further investigation to be resolved.
Virus carrying the ATW mutation could not be recovered. A previous study by Lin et al. demonstrated that the 3′-terminal 55 nucleotides were necessary for negative-strand synthesis and subsequent DI RNA replication (16), and the ATW mutation falls within this region. Our results, using nested RT-PCR, demonstrate that negative-strand RNAs complementary to genome were produced by the ATW virus. However, it was impossible to determine if positive-strand genomes were produced at early times (before the onset of cell fusion in cultures electroporated with wild-type genomes) after electroporation due to the presence of large amounts of the input genomes which persist up to 24 h postelectroporation even in the absence of viral replication (H. Kang and J. L. Leibowitz, unpublished data). Subgenomic positive- and negative-strand mRNA3 could not be detected in cultures electroporated with ATW genomes (Fig. 8), and similar results were obtained for RNA6 (data not shown). Negative- and positive-strand subgenomic mRNA7 was detected using nested RT-PCR, seemingly contradicting the data obtained for mRNA3 and mRNA6. The PCR product generated by nested RT-PCR of mRNA7 was cloned and sequenced and found to contain the expected leader sequence. The 3′ UTR of RNA7 could not be recovered and sequenced since RT-PCRs extending from the leader to the poly(A) tail failed to yield specific products. The RNA7 products are most likely the result of a recombination event with N coding sequences electroporated with the ATW genome to increase the efficiency of generating recombinant virus. However, we cannot rule out the possibility that ATW genomes support the synthesis of subgenomic RNA7 but not other subgenomic RNAs.
Our data support a role for the 3′ 42-nucleotide host protein binding element in mRNA production. It is possible that the 3′ 42-nucleotide host protein binding element is part of a larger complex of viral RNA viral proteins and host proteins which are involved in transcription through mediation of 5′-to-3′ interactions as recently proposed by Zúñiga et al. (37), who suggest that looped-out structures of the 3′ UTR are involved in subgenomic transcription and replication. Another alternative role for the 42-nt segment is in RNA stability. Proper primary and secondary structure of the viral RNAs may contribute to RNA stability through either interactions with host and viral proteins or intramolecular interactions of the RNA. Forty-two-nucleotide segment sequences other than wild type may have a higher turnover rate leading to alterations in observed phenotype; this possibility requires further examination.
An alternative explanation for our results which cannot be excluded is that the eight point mutations in ATW produce an extensive alteration in RNA structure which disrupts a structure outside this region that is required for mRNA synthesis. However, previous nuclease structural probing in the context of a 166-nt 3′ genomic RNA (17) is most consistent with the 42-nt segment folding into the Mfold-predicted structure shown here, and the ATW mutation is predicted to have the wild-type structure, suggesting that distant effects of this mutation are not likely.
Mutant viruses MT3C, M19C, ATW5′, and ATW3′ demonstrate a weak correlation between plaque size and protein binding activity. Mutant MT3C virus demonstrated a dramatic decrease in host protein binding activity (6.3%) and a small-plaque phenotype (Table 3). The mutant virus M19C has a plaque only slightly smaller than that of wild-type virus, and gel shift assays with the corresponding mutant probe produced RNP complexes 74% as efficiently as wild-type RNA probes. In contrast to mutant M19C, the ATW3′ mutation resulted in RNAs with a 400% increase in protein binding activity compared to wild type and also resulted in a virus with the second smallest plaque size observed in this study, only marginally larger than that seen with the ATW5′ virus. RNA probes containing the ATW5′ mutation had a very low protein binding activity and produced a virus with the smallest plaque size, 33% of that of wild-type MHV-A59. Thus, the mutant with binding activity nearest that of the wild type also had a near-wild-type plaque size, while mutants that had binding activities which differed greatly from that of the wild type had plaque sizes approximately one-third that of the wild type. These data imply that RNP complex formation must be kept within a certain range if it has a role in efficient viral replication. If the data are perceived as showing 100% wild-type binding as the optimal RNP complex, then deviations, in terms of RNP complex formation (in our study measured as binding relative to wild type), above and below that optimum could result in the phenotype demonstrated by these mutants.
The explanation holds for all mutant viruses studied except for mutant ATW. The ATW mutation resulted in a 1.85-fold increase in RNA binding activity and produced a viral genome with an obvious replication defect in that no virus could be recovered. Regardless of RNA binding activity it is evident that ATW primary structure contained too many mutations (eight total point mutations, nucleotides 2 to 5 and 23 to 26) for the production of positive- and negative-strand mRNAs.
The experiments with the ATW, ATW5′, and ATW3′ series of DI RNAs discussed above suggest that the wild-type sequence is required at position 4 but that mutations can be tolerated in positions 2, 24, and 25. This possibility has not yet been tested in the context of the intact virus. Thus, from the data generated in this study we cannot conclude that binding of mitochondrial aconitase and mitochondrial HSP70, HSP60, and HSP40 to the 3′ 42-nt host protein binding element plays an essential role in viral replication, but it may play a role in regulating positive- or negative-strand subgenomic RNA synthesis. Any effects that protein binding to this sequence element had on replication appear to be relatively subtle, since mutants with mutations which produced a very low protein binding activity were viable.
Acknowledgments
This work was supported by in part by NIH grants AI051493 and AI23946, National Multiple Sclerosis Society grant RG2203-B-6, and a generous gift from the Stearman family.
We gratefully acknowledge Paul Masters for providing us with pB36 and the Albany 4 mutant of MHV-A59. We thank Rajesh Miranda, Texas A&M University System Health Science Center College of Medicine, for the use of his real-time PCR instrument and Leena Kumar for advice on real-time RT-PCR.
REFERENCES
- 1.An, S., and S. Makino. 1998. Characterization of coronavirus cis-acting RNA elements and the transcription step affecting its transcription efficiency. Virology 243:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi, K. S., P. Huang, and M. M. C. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. [DOI] [PubMed] [Google Scholar]
- 4.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:582-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, P., and M. M. C. Lai. 2001. Heterogenous nuclear ribonucleoprotein A1 binds to the 3′ untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, P., and M. M. C. Lai. 1999. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering RNA conformation. J. Virol. 73:9110-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, Y., S. Jeong, and S. Makino. 1993. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology 197:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, Y., and S. Makino. 1995. Characterization of a murine coronavirus defective interfering RNA internal cis-acting replication signal. J. Virol. 69:4963-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koetzner, C. A., M. M. Parker, C. S. Ricard, L. S. Sturman, and P. S. Masters. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted recombination. J. Virol. 66:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. M. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, H., P. Huang, S. Park, and M. M. C. Lai. 1999. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 73:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, H., X. Zhang, R. Duncan, L. Comai, and M. M. C. Lai. 1997. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc. Natl. Acad. Sci. USA 94:9544-9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, Y., C.-L. Liao, and M. M. C. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Q., R. F. Johnson, and J. L. Leibowitz. 2001. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 75:12105-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Q., W. Yu, and J. L. Leibowitz. 1997. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology 232:74-85. [DOI] [PubMed] [Google Scholar]
- 19.Makino, S., C. K. Shieh, L. H. Soe, S. R. Weiss, and M. M. C. Lai. 1989. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology 166:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino, S., F. Taguchi, N. Hirano, and K. Fujiwara. 1984. Analysis of genomic and intracellular viral RNAs of small plaque mutants of mouse hepatitis virus, JHM strain. Virology 139:138-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel, Y. M., A. M. Borman, S. Paulous, and K. M. Kean. 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 21:4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanda, S. K., R. F. Johnson, Q. Liu, and J. L. Leibowitz. 2004. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the murine hepatitis virus genome. Arch. Virol. 149:93-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanda, S. K., and J. L. Leibowitz. 2001. Mitochondrial aconitase binds to the 3′ untranslated region of the mouse hepatitis virus genome. J. Virol. 75:3352-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Repass, J. F., and S. Makino. 1999. Importance of the positive-strand RNA secondary structure of a murine coronavirus defective interfering RNA internal replication signal in positive-strand RNA synthesis. J. Virol. 72:7926-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen, X., and P. S. Masters. 2001. Evaluation of the role of heterogeneous nuclear ribonucleoprotein A1 as a host factor in murine coronavirus transcription and genome replication. Proc. Natl. Acad. Sci. USA 98:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shieh, C.-K., L. H. Soe, S. Makino, M. F. Chang, S. A. Stohlman, and M. M. C. Lai. 1987. The 5′ end sequence of the murine coronavirus genome: implications for multiple fusion sites in leader-primed transcription. Virology 156:321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interaction with eIF4G stimulates picornavirus IRES-dependent translation. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 31.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 32.Williams, G. D., R.-Y. Chang, and D. A. Brian. 1999. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 73:8349-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, W., and J. L. Leibowitz. 1995. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology 214:129-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu, W., and J. L. Leibowitz. 1995. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J. Virol. 69:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-47. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology, 1st ed. Kluwer Academic Publishers, San Diego, Calif.
- 37.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]