Abstract
Rhodopseudomonas palustris, an α-proteobacterium, carries out three of the chemical reactions that support life on this planet: the conversion of sunlight to chemical-potential energy; the absorption of carbon dioxide, which it converts to cellular material; and the fixation of atmospheric nitrogen into ammonia. Insight into the transcription-regulatory network that coordinates these processes is fundamental to understanding the biology of this versatile bacterium. With this goal in mind, we predicted regulatory signals genomewide, using a two-step phylogenetic-footprinting and clustering process that we had developed previously. In the first step, 4,963 putative transcription factor binding sites, upstream of 2,044 genes and operons, were identified using cross-species Gibbs sampling. Bayesian motif clustering was then employed to group the cross-species motifs into regulons. We have identified 101 putative regulons in R. palustris, including 8 that are of particular interest: a photosynthetic regulon, a flagellar regulon, an organic hydroperoxide resistance regulon, the LexA regulon, and four regulons related to nitrogen metabolism (FixK2, NnrR, NtrC, and σ54). In some cases, clustering allowed us to assign functions to proteins that previously had been annotated with only putative functions; we have identified RPA0828 as the organic hydroperoxide resistance regulator and RPA1026 as a cell cycle methylase. In addition to predicting regulons, we identified a novel inverted repeat that likely forms a highly conserved stem-loop and that occurs downstream of over 100 genes.
The alpha division of the proteobacteria is defined by a conserved 16S rRNA gene sequence and insertions/deletions in conserved protein sequences (20). Despite being phylogenetically related, the α-proteobacteria have no defining phenotypic characteristics. Among the photosynthetic α-proteobacteria (i.e., purple bacteria) are Rhodobacter sphaeroides, Rhodospirillum rubrum, and Rhodopseudomonas palustris. These metabolically adaptable species are found in a variety of niches, including marine environments, freshwater sediments, and soil, and they have the ability to photosynthesize, fix carbon dioxide, and fix nitrogen. Other members of the α-proteobacteria live in association with eukaryotes, both plants and animals. Plant-associated species include Bradyrhizobium japonicum, a soybean symbiote, and Agrobacterium tumefaciens, the causative agent of crown gall disease. Animal pathogens include Bartonella henselae, Rickettsia prowazekii, and Brucella suis, the causative agents of cat scratch disease, typhus fever, and porcine brucellosis, respectively. At the time of this study, complete genome sequence data are available for 13 α-proteobacterial species, and partial genome sequence data are available for an additional 16 species. Given the wealth of sequence data available and the metabolic and environmental diversity of these bacterial species, the α-proteobacteria constitute an excellent target for a comparative genomics study.
The purple photosynthetic bacterium R. palustris exhibits a highly flexible metabolic lifestyle, reflecting the diversity present in the α-proteobacteria, and was therefore an obvious choice as the reference species for this comparative study. In addition to its ability to photosynthesize, fix carbon dioxide, and fix nitrogen, R. palustris can degrade a variety of organic aromatic compounds, including those found in plant matter and industrial waste (10, 21). R. palustris is able to respire both aerobically and anaerobically. Under anaerobic conditions in the presence of light, the bacterium forms stacked membrane structures that house the photosynthetic machinery and generate energy (51). It can produce hydrogen using a nitrogenase-dependent mechanism, thus making it a potential bioenergy source (2). The R. palustris genome encodes many alternate or redundant systems, including four complete LH2 (light-harvesting) complexes, two NADH-dependent dehydrogenases, and three nitrogenases (24). R. palustris and other α-proteobacterial species, like Caulobacter crescentus and Hyphomonas neptunium, share an asymmetric cell division phenotype, wherein cell division is initiated only by surface-adherent cells, resulting in an adherent cell and a motile cell (22). It has been suggested that the immobilized cells may be exploited in the fabrication of biocatalysts (24). The potential applications in energy production, bioremediation, and biocatalysis make R. palustris a bacterial species of considerable environmental and biotechnological interest. In order to realize that potential, it is important that we first understand the regulation of R. palustris' complement of metabolic pathways.
A consequence of R. palustris' metabolic flexibility is the need for the organism to have an extensive transcription-regulatory network. Cellular processes, such as nitrogen fixation and carbon dioxide fixation, are energy intensive; thus, the enzymes for these pathways must be regulated with respect to nutrient availability and other environmental factors (for reviews, see references 7, 8, and 43). For example, it is most efficient for the bacterium to regulate the synthesis of its photosynthetic machinery with respect to light intensity and wavelength (18). The presence of a sophisticated regulatory network is supported by the large number of annotated transcription-regulatory and signaling proteins (451 genes) encoded in the R. palustris genome (24).
In order to begin to dissect this complex regulatory network, we undertook a comparative study of the α-proteobacteria, focusing on R. palustris. The first goal of this study was to identify transcription factor binding sites (TFBSs) genomewide by phylogenetic footprinting. Phylogenetic footprinting is an approach to discover regulatory motifs in orthologous intergenic regions. The assumption is that among a phylogenetically close group of species, orthologous genes are likely to be regulated by a common transcription factor (TF). Under this assumption, the TFBSs will be conserved, while other noncoding DNA that is not under selective pressure will be free to mutate over time. Thus, the promoter regions upstream of orthologous genes are analyzed for putative regulatory motifs by computationally searching for conserved sequence elements. In the current study, conserved sequences were found using a Gibbs sampling strategy. This approach predicts regulatory motifs de novo without any prior knowledge regarding binding sites and exploits the diversity of the contributing species in order to increase the power of the search for regulatory motifs.
An advantage of using the Gibbs sampler for these studies is that it implements a rigorous Bayesian method to infer the number of sites and their locations (49). This means that not all of the species contributing orthologous promoter data are required to contribute to a motif prediction (detailed discussion and examples can be found at http://bayesweb.wadsworth.org/web_help.PF.html). Accordingly, a TFBS that is present in an intergenic region of the target species and some of the species in the collection may be absent in others. This feature is important, since the selection of species for phylogenetic footprinting is empirical; we require only that the species be phylogenetically related so that they have a significant number of common TFs (30). This requirement was met for the set of α-proteobacteria selected for the current study (Table 1), allowing us to predict regulatory motifs for the promoter regions of 2,044 operons. We have previously demonstrated the power of Gibbs sampling (49) for phylogenetic footprinting within the γ-proteobacteria (29, 30). Specifically, among promoters with experimentally verified TFBSs, ∼74% of the predictions corresponded to the known sites. Furthermore, among the remaining novel predictions were TFBSs predicted upstream of fatty acid biosynthesis genes; these sites were used to affinity purify a previously uncharacterized TF (YijC) that has since been shown to regulate fatty acid biosynthesis in vivo (55).
TABLE 1.
Selected features shared between the α-proteobacterial species used in this study
| Species and description | Characteristics | No. of TFs |
|---|---|---|
| R. palustris; widely distributed | Photosynthesizes | 168a |
| Fixes carbon dioxide | ||
| Fixes nitrogen | ||
| Degrades aromatic compounds | ||
| Asymmetric cell division phenotype | ||
| B. japonicum; soybean symbiote | Fixes nitrogen | 139 |
| C. crescentus; aquatic | Degrades aromatic compounds | 51 |
| Asymmetric cell division phenotype | ||
| B. suis; intracellular animal pathogen | Degrades aromatic compounds | 61 |
| R. sphaeroides; widely distributed | Photosynthesizes | 54b |
| Fixes carbon dioxide | ||
| Fixes nitrogen | ||
| R. rubrum; widely distributed | Photosynthesizes | 69b |
| Fixes carbon dioxide | ||
| Fixes nitrogen | ||
| N. aromaticivorans; widely distributed, opportunistic pathogen | Degrades aromatic compounds | 44b |
| H. neptunium; marine | Asymmetric cell division phenotype | 46b |
Count of TFs identified in each species for the 168 that are found in R. palustris and at least one other species (70 of the 238 R. palustris TFs did not have an ortholog in any of the above species by our criteria).
These counts are from incomplete genomes and are therefore preliminary.
The second goal of this study was to cluster these predicted cis regulatory elements into regulons. Clustering provides a mechanism by which motifs that represent binding sites for the same TF are grouped; this combined evidence improves the reliability of pathway and cognate TF identification. We used a Bayesian clustering algorithm developed previously that was found to accurately cluster Escherichia coli regulatory motifs (38). A total of 101 putative R. palustris regulons were identified, including 8 that are of particular interest.
MATERIALS AND METHODS
Genome sequence data.
The following genome sequences were obtained from the NCBI reference sequence database (ftp://ftp.ncbi.nih.gov/genome/Bacteria/): Rhodopseudomonas palustris CGA009 (NC_005296), Bradyrhizobium japonicum USDA 110 (NC_004463), Brucella suis 1330 (NC_004310 and NC_004311), and Caulobacter crescentus CB15 (NC_002696). Incomplete genome sequence data for Rhodobacter sphaeroides (v 11/9/2003), Rhodospirillum rubrum (v 11/9/2003), and Novosphingobium aromaticivorans (v 11/12/2003) were obtained from the Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). The sequence of Hyphomonas neptunium ATCC 15444 (March 2004) was obtained from The Institute for Genome Research (http://www.tigr.org). R. palustris operon predictions were downloaded from the Department of Energy Joint Genome Institute's site.
Phylogenetic footprinting.
Orthologous intergenic regions were identified as previously described (29). Briefly, TBLASTN (1) was used to identify orthologs of 4,814 R. palustris proteins (excluding 18 of the 4,832 CDS entries in NC_005296 that are pseudogenes) in a database composed of the genome sequence data of the eight species under study. The following criteria were applied in order to select hits corresponding to bona fide orthologs: (i) the expectation value must be <10−20; (ii) the expectation value must be better than the expectation value of the second-best hit in R. palustris; (iii) a hit must start within the first 20 amino acids of the R. palustris query sequence. Upstream orthologous intergenic regions were extracted from the database with a maximum length of 500 bp and a minimum length of 50 bp. In some cases, it was obvious that the start codons for genes in R. palustris were annotated upstream of a more likely 5′ end of the gene (e.g., lexA). To prevent the loss of regulatory signals near the 5′ ends of genes, we reevaluated the start codons using the cross-species BLAST data. In 240 cases, we identified a more likely downstream start codon. The BLAST procedure described above was then rerun with the revised (i.e., shortened) sequences. Potential artifacts arising from the shifting of these start codons were minimized by tracking the revised genes throughout the phylogenetic-footprinting and clustering process.
The recursive Gibbs sampler (Gibbs v 2.06.015) (50) was used to exhaustively search for palindromic and nonpalindromic motifs in each orthologous intergenic data set. A Bayesian segmentation algorithm was used to generate a position-specific background composition model for each sequence in a data set (Unifiedcpp) (26). All models consisted of 16 active columns that were allowed to fragment to a maximum width of 24 columns. For the detection of palindromes, the Gibbs sampler was allowed to choose an even- or odd-width palindromic model, based on the sequence evidence. The sampler was allowed to run for 2,000 iterations, with a plateau period of 200 iterations, and it was reinitialized 40 times using random seeds. The most significant motif found by the recursive Gibbs sampler in a data set was masked, and the sampling procedure was repeated until no motifs were found with an average maximum a posteriori probability (avgMAP) of >1. This average MAP cutoff was chosen empirically. For reference, MAP values of >0 provide a measure of how much more likely the alignment is than the unaligned background. The entire process (sampling and masking) was repeated three times to take advantage of the stochastic nature of Gibbs sampling and to maximize the number of motifs found. Motifs that contained unique sites in R. palustris were then extracted and used for further analysis.
Clustering.
We selected motifs for clustering by first applying a critical-value criterion; palindromic and nonpalindromic motifs were compared to model-specific critical-value criteria derived from random data simulations (30). Coding sequence contamination in the extracted intergenic regions was detected through comparison of the coordinates of each site in a motif to the available genome annotations (R. palustris, B. japonicum, B. suis, and C. crescentus). If more than half of the sites from annotated species overlapped a coding region, the motif was eliminated from clustering. Motifs were also analyzed for the presence of shared sites. If two motifs contained the same site from more than one species, or if motifs contained the same site from R. palustris (which sometimes occurs with divergently transcribed genes), one motif was eliminated from the clustering input.
Clustering was carried out using the Bayesian motif clustering algorithm (BMC v 1.5x) (38). Even-, odd-, and nonpalindromic models were clustered separately using a tuning parameter (q) of 100. This parameter, which affects whether a motif forms a cluster on its own or whether the motif joins an existing cluster, was determined empirically. BMC was run for 10 iterations without fragmentation, followed by 25 iterations with the fragmentation option enabled, to produce the optimal solution. BMC was then allowed to sample for an additional 500 iterations to produce the frequency solution described in this study. All clusters were initially allowed to shift, meaning that motifs could realign with respect to each other within a cluster. The shifting option did not influence the alignment of motifs in palindromic clusters and was turned off. Shifting did, however, improve the alignment of nonpalindromic motifs within a cluster; thus, we allowed nonpalindromic motifs to shift left or right by up to two columns.
Scanning.
Cluster models from the frequency solution were used as input to the Dscan algorithm (Dscan revision 2.3) (34), using either palindromic models (the base counts in paired palindromic columns were summed) or nonpalindromic models, as indicated by the cluster model parity. The complete set of R. palustris intergenic regions was scanned to detect sites that matched the model. Given the input model and database, Dscan uses the approach described by Staden (44) to report sites that match the model above a given level of statistical significance. In this study, we report sites at a P value of <0.05.
RESULTS
Phylogenetic footprinting to identify transcription factor binding sites.
The choice of species to be used for phylogenetic footprinting was governed by two factors: their phylogenetic relatedness and the presence of common metabolic pathways. First, the phylogenetic relatedness of α-proteobacterial species for which a complete or nearly complete genome sequence was available was inferred using the sequences of their 16S rRNA genes. Current alignment algorithms, including Gibbs sampling, do not account for the phylogenetic relatedness (correlation) of the input data, which leads to overestimation of the significance of motifs. Therefore, to achieve sufficient phylogenetic diversity and minimize losses to the power of the cross-species technique due to phylogenetic correlations among the sequence data, we chose R. palustris and seven additional species (Fig. 1), using the technique of Newberg and Lawrence (35). This was done in a “greedy” fashion, so as to maximize the effective number of independent sequences. This means that, although additional genome sequences were available, they were not expected to improve the reliability of predictions in R. palustris and in fact would be likely to skew the results by including highly correlated data. This is why, for instance, only one of the sequenced Brucella species was retained.
FIG. 1.
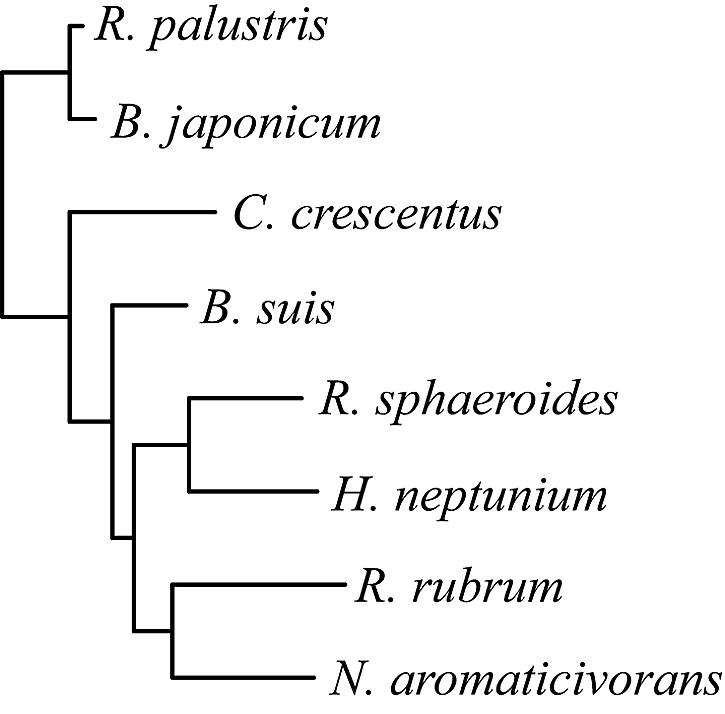
Phylogenetic tree of the α-proteobacterial species used in this study (Rhodopseudomonas palustris, Bradyrhizobium japonicum, Caulobacter crescentus, Brucella suis, Rhodobacter sphaeroides, Hyphomonas neptunium, Rhodospirillum rubrum, and Novosphingobium aromaticivorans) generated using 16S rRNA sequences and the maximum likelihood method (12).
Six of the eight species shown in Fig. 1 approximate independent observations; that is, for each possible species pair comparison, there is no more correlation, measured as the percent identity in the orthologous intergenic regions, than would be expected by chance (∼40% ± 4%). R. palustris and B. japonicum, however, exhibit some correlation in sequence, with an average of 47% ± 8% sequence identity in the intergenic regions. B. japonicum was retained, nevertheless, because the inclusion of a correlated species has been shown to improve reference species predictions in a cross-species study (30), and this low level of correlation can be accounted for in the critical value (P value) calculations (see Materials and Methods). A second factor in our choice of species was the presence of common metabolic pathways and TFs; Table 1 highlights features of each species that are expected to contribute to the analysis of R. palustris. The conservation of TFs across species was examined, and of the 238 R. palustris TFs (24), 168 were found to have an ortholog in at least one of the other seven species.
Using the set of eight species, we compiled orthologous upstream intergenic sequence data sets representing 2,411 R. palustris genes/operons and used them as input to the Gibbs sampler (see Materials and Methods). A total of 4,963 motifs were found for 2,044 of these data sets by phylogenetic footprinting. Statistical-significance criteria were determined using a random data simulation as described by McCue and coworkers (30). By these critical-value criteria, over half of the motifs (2,722) were marginally significant (P < 0.2); of those, 442 were highly significant (P < 0.01). The full set of phylogenetically footprinted motifs, along with significance criteria, can be viewed online (http://bayesweb.wadsworth.org/prokreg.html).
Clustering predicted sites into regulons.
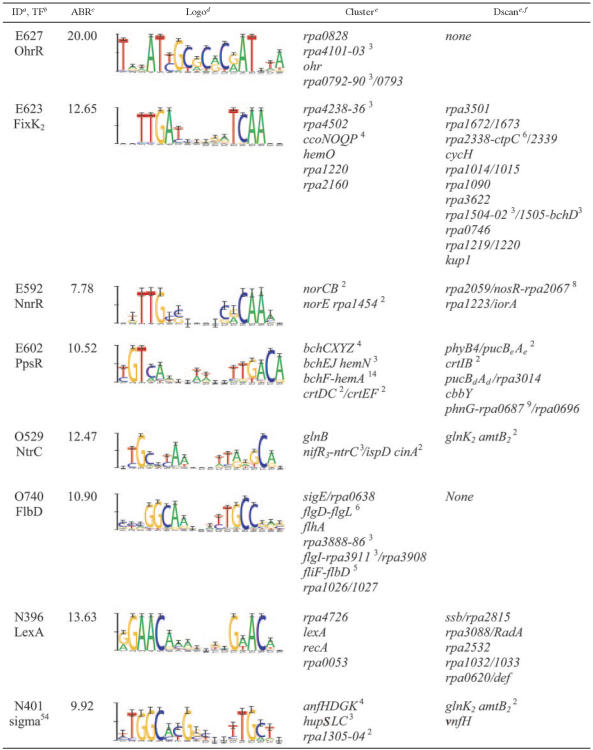
A BMC algorithm (38) was used to infer regulons from the collection of motifs found by phylogenetic footprinting. After applying filtering heuristics (see Materials and Methods) to the 2,722 motifs described above, we identified 1,730 motifs for clustering. At the selected level of statistical significance (P < 0.2), many of these 1,730 motifs were only marginally significant. However, we have found that the noise introduced into the clustering procedure by false-positive motif predictions has little effect on true regulons; specifically, the false-positive motif predictions do not join clusters reproducibly (L. A. McCue, unpublished data). This is supported by the observation that only 472 of the input motifs reproducibly joined a cluster and are reported as members of regulons. Furthermore, we calculate a Bayes ratio as a measure of cluster strength and have found true regulons to rank among the highest scoring. The Bayes ratio is the ratio of the probability of the data belonging in a cluster to the probability of the data existing as separate motifs. Among the 101 clusters identified here, several of the high-scoring clusters are supported by biochemical and genetic data from the literature as bona fide regulons; they are shown, along with sequence logos (41), in Table 2 and are described in detail below.
TABLE 2.
Selected clusters of phylogenetically footprinted motifs
ID, cluster identifier.
Predicted TF.
ABR, average Bayes ratio.
Cluster logo.
Divergently transcribed genes and operons are separated by a slash. Operons are shown as follows. A dash indicates an inclusive range of gene names. Large operons are denoted as a range between the first and last genes. A superscript indicates the number of genes in each operon.
Additional members of the regulon found by Dscan.
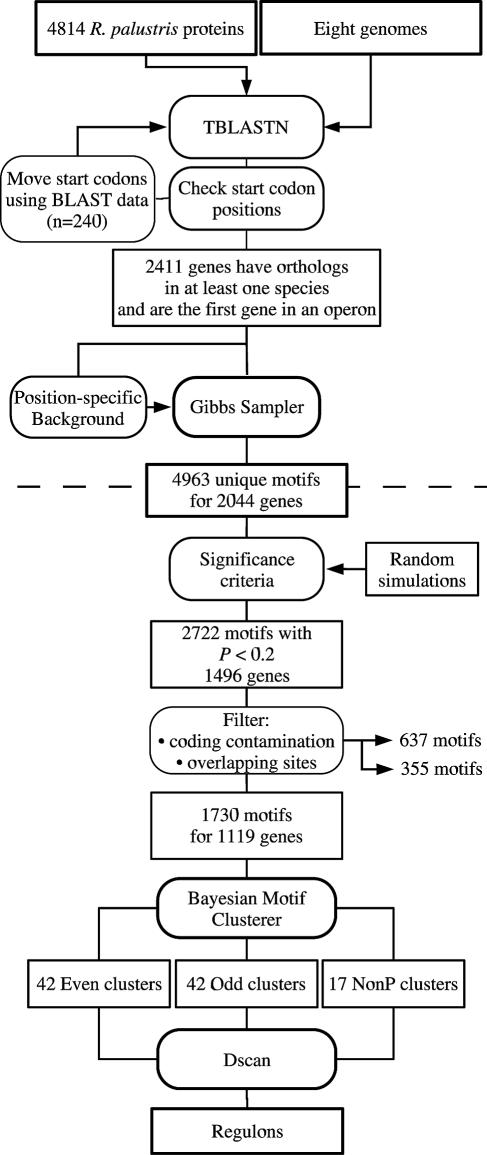
The clusters represent partial regulons, including only those regulatory sites that could be detected by our cross-species approach. For example, R. palustris genes lacking orthologs in other species were never analyzed. It is also the case that the heuristics used to filter out motifs prior to clustering may eliminate some legitimate motifs. One way in which to address this problem is to scan the complete set of intergenic regions for a species, using position weight matrices built from the clusters. This approach often results in the prediction of additional members of a regulon (48). We employed a rigorous statistical algorithm based on Staden's method (44) as implemented by Neuwald and coworkers (34) to scan for additional TFBSs in R. palustris. The algorithm yields a P value for a motif match that represents the probability that a site of equal or greater strength would be found in a random data set of the same size and composition. In scans of the set of all intergenic regions of R. palustris with the cluster matrices, we identified additional members of regulons at a P value of <0.05 (Table 2). Phylogenetic footprinting, clustering, and scanning are summarized in Fig. 2.
FIG. 2.
Flowchart of phylogenetic footprinting, clustering, and scanning. The phylogenetic-footprinting procedure is shown above the dashed line, and the clustering procedure is shown below. Input data and intermediate data sets are shown in boxes. Operations are shown in rounded boxes.
Nomenclature.
The regulon descriptions presented here use the following naming conventions. A motif is defined as a collection of aligned DNA sequences (i.e., putative TFBSs) from phylogenetic footprinting; motifs are simply given the name of the R. palustris gene whose upstream intergenic sequence data were analyzed during motif prediction. A cluster is defined as a collection of aligned motifs. Clusters are referred to as Even, Odd, or NonP depending on whether an even-palindromic, odd-palindromic, or nonpalindromic model best describes them. Each cluster also has a numerical designation that is produced during the clustering process; it is used here solely as a unique identifier (e.g., Even-239). Annotated gene functions were extracted from the published R. palustris CGA009 genome (24). Eight of the clusters have particular relevance to R. palustris biology and are described below.
OhrR cluster.
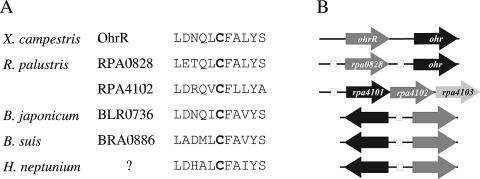
Cluster Even-627 was the highest-scoring cluster by the Bayes ratio criterion. Two of the four genes in the cluster (ohr and rpa4101) are annotated as organic hydroperoxide resistance genes. These two R. palustris genes are homologous to the ohr gene from Xyella fastidiosa, which encodes a thiol-dependent peroxidase that decomposes organic hydroperoxides (5). R. palustris Ohr and RPA4101 are 60% and 45% identical, respectively, to X. fastidiosa Ohr at the protein level and have two conserved cysteines that are required for peroxidase activity. rpa4101 forms an operon with rpa4102 and rpa4103, which, respectively, encode a MarR family TF and a possible glutathione S-transferase. One of the remaining genes in the cluster, rpa0828, is annotated as a MarR family TF.
Organic hydroperoxide resistance genes are regulated by OhrR in Xanthomonas campestris. OhrR is a MarR family TF that senses oxidative stress through a single conserved cysteine residue; this cysteine, when oxidized, disrupts DNA binding (36). An amino acid sequence alignment of X. campestris OhrR, RPA0828, RPA4102, and orthologs from B. japonicum, B. suis, and H. neptunium shows that all contain the critical reactive cysteine surrounded by a number of other well-conserved residues (Fig. 3A). Additionally, it is interesting that the ohr and rpa0828 genes in R. palustris are syntenic with ohr and ohrR in X. campestris. Specifically, the rpa0828 gene is immediately upstream of and on the same strand as ohr, with a regulatory site predicted in each upstream region. In contrast, among B. japonicum, B. suis, and H. neptunium, the ohr orthologs are divergently transcribed from the ohrR orthologs, with a regulatory site located between them (Fig. 3B). These results suggest that RPA0828 is likely the cognate TF of the regulon, although RPA4102 is a probable paralog of RPA0828 and could provide redundancy in the regulatory circuit.
FIG. 3.
Partial alignment of OhrR orthologs. (A) Alignment of the region surrounding the oxidation-sensitive cysteine residue (in boldface) from X. campestris OhrR and its orthologs. Gene names are noted except in the case of the ortholog from H. neptunium, as this species' genome is not yet annotated. (B) Arrangement of the ohr (black arrows) and ohrR (dark-gray arrows) orthologs for the corresponding species in panel A. Motifs found upstream of genes are shown as hatched boxes.
FixK2 and NnrR clusters.
The six motifs that made up cluster Even-623 have a strongly conserved palindromic sequence; when this cluster model was used to scan the database of all R. palustris intergenic regions, 12 additional sites were identified (Table 2). Several of the genes in this expanded regulon have predicted functions related to respiration, specifically, in cytochrome assembly and porphyrin biosynthesis. The motif model for this cluster (TTGA-N6-TCAA) resembles the known binding sequence for the E. coli TF, Fnr (TTGAT-N4-ATCAA) (17), a global regulator of anaerobic respiration. In B. japonicum, nitrogen fixation genes and genes for anaerobic or microaerobic metabolism, including the types of the genes found in this cluster, are controlled by a Crp/Fnr family TF, FixK2 (14, 33), which is regulated by the FixLJ two-component system. It is likely that R. palustris employs a regulatory circuit similar to the B. japonicum FixLJ-FixK2 cascade and that this cluster corresponds to the R. palustris FixK2 regulon. This assertion is supported by the presence of an intact FixLJ two-component system in R. palustris. In addition, when the E. coli Fnr protein sequence is used to search the R. palustris proteome, FixK2 is returned as the top hit and shows 68% identity in the helix-turn-helix region, supporting the observed similarity in binding sites. The presence of three TFs in the cluster (rpa2339, rpa1015, and rpa1090) may thus indicate the existence of downstream regulatory circuits and possible interconnections with other pathways.
Another Crp/Fnr family TF in B. japonicum, NnrR, is regulated as part of the FixLJ-FixK2 cascade and controls genes involved in response to nitric oxide and related free radicals (31). Predicted motifs upstream of the nitric oxide-regulated norCB and norE-rpa1454 operons formed a separate cluster (Even-592), with an average Bayes ratio of 7.779, and are likely regulated by NnrR. This nitric oxide regulatory cluster was used as a model for Dscan, detecting the nosRZDFYLX-rpa2067 operon that is involved in nitric oxide uptake. The fact that FixK2 and NnrR are closely related members of the same TF family is reflected by the high degree of similarity between their binding site consensus sequences (Table 2).
PpsR cluster.
Cluster Even-602 is made up of genes involved in pigment synthesis and photocenter assembly. When the cross-species motif was used as a model for Dscan, several additional motif sites were predicted, including sites upstream of two LH2 complexes (pucBeAe and pucBdAd) and an additional pigment synthesis gene, crtI. One interesting member of the expanded regulon is cbbY, which encodes a haloacid dehalogenase-like hydrolase thought to be involved in the Calvin-Benson-Bassham cycle (15). However, statistically significant sites were not found upstream of any additional genes of the Calvin-Benson-Bassham cycle.
Transcription of the photosynthetic genes in R. sphaeroides (37) and in the unusual photosynthetic Bradyrhizobium sp. strain ORS278 (16, 23) is controlled by the product(s) of the ppsR gene(s). Both R. palustris and Bradyrhizobium sp. strain ORS278 have two forms of PpsR (encoded by ppsR1 and ppsR2), while R. sphaeroides has only one (encoded by ppsR). The binding site consensus for PpsR from R. sphaeroides 2.4.1 and Rhodobacter capsulatus (3), TGT-N10-ACA, matches the sequence logo for this cluster, suggesting that one (or both) of the R. palustris PpsR isoforms is likely the cognate TF for this regulon.
NtrC cluster.
Cluster Odd-529 is made up of two nitrogen-regulated genes, glnB and nifR3. The motifs upstream of these two genes are present in all eight species, indicating that they are part of a regulatory circuit that is highly conserved among the α-proteobacteria. The glnB gene encodes a PII nitrogen-regulatory protein and is located upstream of the glutamine synthase gene, glnA. The nifR3 gene forms an operon with ntrBC, which encodes a two-component system. When the cluster model was used to scan the complete set of R. palustris intergenic regions, an additional site was found upstream of a second PII nitrogen regulatory gene, glnK2.
Given that the TF NtrC is a member of this regulon (in the nifR3-ntrBC operon), it was an obvious candidate for the cognate, autoregulatory TF. There are three lines of evidence that support this hypothesis. The first is that NtrC sites have been found in the promoters of both glnB and nifR3 in R. capsulatus (4). Secondly, two closely spaced copies of the NtrC site are found by Dscan in the promoters of glnB, nifR3, and glnK2; this tandem arrangement is a feature of NtrC-regulated promoters, which are cooperatively bound by NtrC octamers (25, 40). Finally, when DNase I-footprinted NtrC sites from R. capsulatus (4, 27) are aligned, the resulting sequences have a pattern similar to that shown in Table 2 for this cluster, specifically, conserved GC pairs separated by an 11-bp AT-rich region.
FlbD cluster.
Cluster Odd-740 is made up of motifs associated with flagellar synthesis genes, as well as a putative adenine methyltransferase. Since DNA methylation and flagellar assembly are both regulated in a cell-cycle dependent manner in C. crescentus (28), it seemed likely that RPA1026 (the adenine methyltransferase) could have a cell cycle-regulatory function. In fact, a BLAST search identified rpa1026 as an ortholog of ccrMI, the cell cycle methylase of C. crescentus. The ccrM gene is widely distributed among the α-proteobacteria and has been shown to be essential for the viability of C. crescentus (45). Orthology with ccrMI and membership in the flagellar cluster, combined with evidence that ccrMI is under the control of a class II flagellar promoter in C. crescentus (46), support the hypothesis that RPA1026 is part of the flagellar cluster and has a cell cycle-regulatory function. The likely cognate TF for this cluster is FlbD, which is encoded in the fliFG-rpa1266-fliY-flbD operon and is known to be a negative regulator of its own transcription. In addition, the fliF motif found by phylogenetic footprinting and included in this cluster, contains a C. crescentus site (GGTAAATCCTGCC) that has been shown to be bound by FlbD (32).
LexA cluster.
Cluster NonP-396 contains motifs upstream of both recA and lexA from seven of the eight species used for phylogenetic footprinting. These genes encode proteins involved in the SOS pathway for repairing DNA damage (52). When we used this cluster motif as a model to scan the entire set of R. palustris intergenic regions, we identified five additional statistically significant sites, including sites upstream of ssb and RadA, which are also involved in responding to DNA damage. The sequence logo for this cluster matches the previously reported nonpalindromic LexA binding motif described for R. palustris and other α-proteobacteria (9, 13).
Sigma54 cluster.
Cluster NonP-401 is made up primarily of motifs occurring upstream of genes involved in nitrogen and hydrogen metabolism. The anfHDGK operon encodes the alternate nitrogenase, and the hupSLC operon encodes a hydrogenase. When this cluster motif was used as a model for Dscan, a significant site was found upstream of vnfH, which encodes a vanadium-dependent nitrogenase subunit. Since the predicted regulon included subunits for two of the three R. palustris nitrogenases, as well as a nitrogen-regulatory protein (glnK2), we compared the logo for the cluster with the motifs for known nitrogen regulators. The results suggest that this motif is bound by the alternative sigma factor σ54, which is known to regulate many genes involved in nitrogen assimilation and metabolism (54).
A novel inverted repeat.
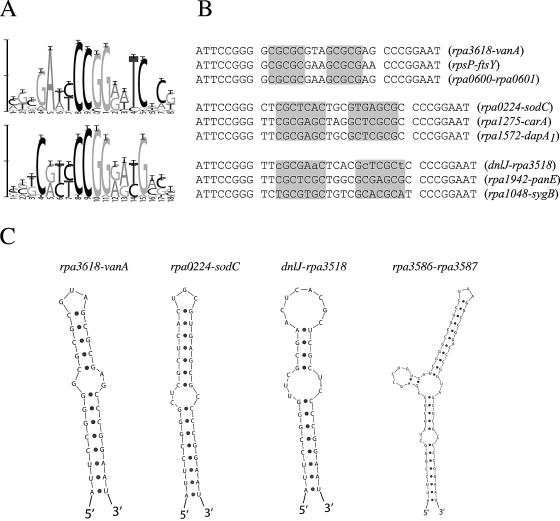
Two high-scoring (by the Bayes ratio criterion) Even clusters had a strongly conserved central CCGG sequence and were composed largely of motifs consisting of sites from only R. palustris and B. japonicum (Fig. 4). When used in Dscan, both of the cluster models identified dozens of intergenic regions in R. palustris with significant matches. Examination of the sequences in these clusters revealed that they were part of a conserved inverted repeat of the form ATTCCGGG-N10-52-CCCGGAAT. The 8-bp ends are identical in all copies of the repeat and are separated by an unconserved, but typically palindromic, region of variable length. The genome sequence of R. palustris was analyzed using simple pattern matching for occurrences of this repeat pattern. Repeats were found almost exclusively in intergenic regions at the 3′ ends of gene sequences. Specifically, these repeats were found with high frequency in intergenic regions between genes transcribed in the same direction (n = 124; 69 intergenic regions) and intergenic regions between convergently transcribed genes (n = 107; 63 intergenic regions) but occurred rarely in intergenic regions separating divergently transcribed genes (n = 5) or in coding regions (n = 7). The other species in this study were analyzed for the presence of the repeats. The B. japonicum genome was found to have a similar density of this repeat, with a similar preference for intergenic regions at the 3′ ends of genes. The other six species do not carry any copies of this repeat, consistent with our observation that these species did not contribute to the motifs making up the clusters represented in Fig. 4A.
FIG. 4.
Features of an R. palustris inverted repeat. (A) Two sequence logos (41) from clusters composed primarily of inverted repeats. (B) Aligned examples of repeats that are 32, 36, and 37 bp in total length from R. palustris intergenic regions. Gene names refer to the flanking genes. Internal palindromic regions are shown in gray boxes (exceptions are in lowercase). The CCGG sequence contributing to the central region of the logos in panel A is found in the flanking inverted repeats. (C) Predicted secondary structures (lowest free energy and most probable) for four repeats having different total lengths. The first three structures correspond to R. palustris sequences labeled in panel B. The fourth structure is an example of one of the largest repeat sequences (68 bp) detected. All structures were inferred using Sfold (6).
The composition of the R. palustris repeat was examined in greater detail. The length of the region separating the 8-bp bounding sequences varied from 10 to 52 bases; however, the distribution of lengths was not uniform. Total repeat lengths of 32 bases, 36 bases, and 37 bases were the most common, accounting for 71% of the repeats. The central variable regions were analyzed using the palindrome tool from EMBOSS (39); 89% of the sequences were found to be palindromic. Randomized data were also analyzed, and fewer than 10% of these sequences were found to be palindromic when the same methodology was used. Thus, conservation of the central region may be based on secondary structure (i.e., hairpin formation) rather than primary sequence. This is supported by the observation that many of the repeats examined are predicted to fold into a stem-loop structure composed of a perfectly conserved 8-bp helix, followed by a bulge and then a variable-sequence hairpin (Fig. 4C).
DISCUSSION
In the present work, we have described a cross-species analysis of transcription regulation in R. palustris. In previous phylogenetic-footprinting studies, using E. coli as the reference species and several additional γ-proteobacteria, we were able to compare our findings to a large number of DNase I-footprinted TFBSs from the literature (29, 30). Since little information is available regarding experimentally identified TFBSs in R. palustris, we focused on clustering the motifs to infer regulons, rather than investigating motifs individually. When the γ-proteobacterial phylogenetic-footprinting data were clustered (38), the majority of clusters with an average Bayes ratio of ≥8 were identified as regulons that had been described in the literature. Among 101 R. palustris clusters, 63 had an average Bayes ratio of ≥8. The clustering results described here, however, differ from the γ-proteobacterial results in that we chose motifs for clustering using a liberal cutoff for statistical significance (P < 0.2), instead of the more stringent cutoff (P < 0.05) used previously (38), and allowed the clustering algorithm to identify those motifs that reproducibly formed clusters instead of identifying a single near-optimal clustering solution. Although the two studies are not directly comparable, we expect that many of the 63 high-scoring (average Bayes ratio, ≥8) clusters from this study merit investigation. All 101 reproducible clusters, as well as the individual motifs with their associated statistical significances, are available at http://bayesweb.wadsworth.org/prokreg.html.
The comparative-genomics approach.
Some caveats associated with phylogenetic-footprinting approaches should be considered, particularly when choosing targets for experimental validation. During either phylogenetic footprinting or scanning of intergenic regions for additional regulon members, TFBSs may be predicted between divergently transcribed genes. Frequently, it is not possible to determine which of the divergently transcribed genes is regulated by the TFBS(s). This is the case during phylogenetic footprinting, when synteny of the two genes in question is conserved among several of the species included in the data set. In addition, when intergenic regions of R. palustris are scanned, the identity of the regulated gene(s) cannot be inferred. In cases where the identity of the regulated gene is unclear, both divergently transcribed genes were listed in Table 2. Another current limitation of the phylogenetic-footprinting approach used here is that it does not reliably detect riboswitches or attenuators; the implications of this are discussed below, in the context of the methionine biosynthesis genes. Finally, phylogenetic footprinting addresses only the regulation of genes that have an ortholog in at least one of the other species, have a similar operon structure, and are regulated by a TF found in at least one of the other species.
Rather than focus on individual motifs from phylogenetic footprinting, we clustered the footprinted motifs into putative regulons. This allowed us to make inferences about their regulatory roles. In addition, putative regulons were expanded by scanning R. palustris intergenic regions. The results presented in this report highlight three advantages of this approach: (i) the ability to make cis-trans connections, (ii) the ability to discriminate between members of a TF family, and (iii) the ability to make predictions in the absence of prior information.
Tan et al. (47) have shown that the presence of autoregulatory sites in the promoters of TFs provides key information to link a particular motif to its cognate TF (i.e., cis-trans connections). In addition, a network analysis of E. coli transcription regulation has shown that ∼50% of TFs are autoregulated (42). In the clusters described in Results, four of the eight contained sites upstream of the likely cognate TF (RPA0828/RPA4102, NtrC, FlbD, and LexA). While there is evidence in the literature of autoregulatory sites for NtrC, FlbD, and LexA among the α-proteobacteria, the connection between RPA0828/RPA4102 and the organic hydroperoxide resistance gene cluster was made de novo by phylogenetic footprinting and clustering.
The R. palustris genome is predicted to encode 15 Crp/Fnr family TFs (24). Some of these family members likely bind to similar TFBSs due to homology in their DNA-binding domains. It was therefore possible that motif clustering could produce “mixed” regulons containing motifs upstream of genes regulated by two or more closely related TFs. It was known from clustering E. coli motifs (38) that the BMC algorithm could correctly separate motifs bound by Crp and Fnr, which have similar binding site consensus sequences. In this study, the separation of FixK2- and NnrR-regulated genes into distinct clusters further demonstrates the ability of the BMC algorithm to detect subtle differences between motifs. Nevertheless, we cannot exclude the possibility that some of our clusters may be mixed regulons.
An advantage of phylogenetic footprinting is that no prior knowledge of the regulatory network is required. This is useful, since regulatory-network information from other species can, in some cases, be misleading. For instance, the motifs that make up the R. palustris LexA cluster identified in this study are upstream of genes likely involved in the SOS response (lexA, recA, and ssb), as they are in E. coli. However, the LexA binding motif identified here, de novo, is distinct from that of E. coli. Specifically, in the α-proteobacteria, LexA binds to a completely different site (GTTC-N7-GTTC) than that of the γ-proteobacteria (CTG-N10-CAG) (13).
We also detected a novel repeat element in R. palustris and B. japonicum with features reminiscent of both a transcriptional terminator and a mobile DNA element. Specifically, the repeat occurs preferentially at the 3′ ends of gene sequences, suggesting a role in terminating transcription, but also has perfectly conserved flanking sequences like those observed for mobile DNA elements (e.g., a transposon). Given the nonuniform distribution of the repeat in the R. palustris genome, we hypothesize that it may be involved in a novel type of transcription termination or perhaps mRNA stability.
Regulation in the α-proteobacteria compared to the γ-proteobacteria.
Our results illustrate some differences in transcription regulation between the γ-proteobacteria and α-proteobacteria. For example, there were a number of regulons that we might have expected to detect, based on our previous experience with phylogenetic footprinting and clustering in the γ-proteobacteria and a general knowledge of prokaryotic gene regulation. However, important amino acid biosynthetic regulatory clusters, such as those for methionine, tyrosine, and tryptophan, were noticeably absent. In fact, orthologs of the E. coli TFs that regulate these pathways (metJ, tyrR, and trpR) are missing in R. palustris, despite the presence of complete metabolic pathways for the synthesis of these amino acids. Because the motif predictions described here were made based on sequence conservation alone, our cross-species method does not require any similarity between the E. coli and R. palustris regulatory networks. However, it is instructive to consider how the transcriptional regulation of these pathways differs in these two species and why regulons, perhaps unique to the α-proteobacteria, were not predicted de novo by phylogenetic footprinting and clustering.
In E. coli, the metA, metC, metE, metF, and metJ genes are regulated by the TF MetJ. Orthologs of the methionine synthesis genes (metA, metC, metE, and metF) are present in R. palustris, but a metJ ortholog is not present. In addition, no common motif is found upstream of the four biosynthesis genes, as would be expected if a single TF regulated them. The metE gene, however, has eight predicted sites that are conserved across four of the species in our study. Similarly, metF has seven distinct sites predicted that are conserved across seven or eight species and that cover 142 bp of the 410 bp in the R. palustris metF intergenic region. These data indicate extensive regions of conservation in the upstream regions of these genes, beyond that which might be expected for straightforward TF-mediated regulation. We found that seven of the eight sites upstream of metE in R. palustris overlap a predicted riboswitch for cobalamine (Rfam) (19). A riboswitch has not been predicted upstream of metF; however, given the limited data available for the prediction of riboswitches, this is perhaps not surprising. These data indicate that the R. palustris methionine biosynthesis pathway may be at least partially controlled by riboswitches, as it is in Bacillus subtilis (11, 53), highlighting the need to extend the phylogenetic-footprinting technique to more efficiently detect riboswitches.
In E. coli, aromatic amino acid biosynthesis is coordinated by TrpR and TyrR. The trpLEDCBA operon encodes proteins involved in tryptophan biosynthesis and is regulated by the TF TrpR and by attenuation. The orthologous genes in R. palustris are separated into three operons: trpG (a trpE ortholog), rpa2889-trpDC-maoC-rpa2893, and trpFBA. Few motifs were found cross-species for these operons, and no motifs were found when the three upstream regions from R. palustris alone were analyzed (data not shown). Similarly, genes encoding the proteins of the tyrosine biosynthesis pathway had few motifs predicted cross-species. The absence of TrpR and TyrR orthologs, as well as the lack of unique motif predictions in the intergenic regions controlling the tryptophan and tyrosine biosynthesis pathways, suggests that the regulation of these pathways has diverged from the E. coli paradigm and may not depend on a classical TF-mediated mechanism.
Conclusions.
The emphasis of this work was on improving our understanding of R. palustris biology. Specifically, we sought to describe the regulatory network of this metabolically versatile species and to make functional predictions that will be of use to the R. palustris research community. This paper provides a description of the most striking patterns that emerged from our genome scale study. From a molecular-biology perspective, many of the predictions made in this study lead directly to hypotheses which can be tested experimentally. For example, interruption of rpa0828 and rpa4102, the putative ohrR orthologs, is predicted to result in constitutive expression of the organic hydroperoxide resistance genes. Interruption of rpa1026, the ccrM ortholog, is likely to have strong effects on cell cycle control and could even be a lethal mutation, as it is in C. crescentus. Our results suggest that the inverted repeat described in this study serves a regulatory role that may involve a stem-loop structure; the first step in investigating this hypothesis is to determine whether the repeat is transcribed, perhaps using nucleic acid hybridization. From a computational-biology perspective, this work demonstrates the applicability of our methods to nonmodel organisms; it illustrates the importance of not only developing computational methods, but also applying them to pertinent bacterial species.
Acknowledgments
We thank the Computational Molecular Biology and Statistics Core Facility at the Wadsworth Center, Lee Newberg for assistance with species selection, William Thompson for assistance with the Gibbs sampler, Michael Palumbo for assistance with the BMC software, and Caroline Harwood for comments on the manuscript. We also thank The Institute for Genome Research and Joint Genome Institute for making sequence data available before completion.
This research was supported by Department of Energy grants DE-FG02-01ER63204 and DE-FG02-04ER63942 to C.L. and L.A.M. and NIH grant R01HG01257 to C.L.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, M. J., J. M. Rocha, J. Tramper, and R. H. Wijffels. 2001. Acetate as a carbon source for hydrogen production by photosynthetic bacteria. J. Biotechnol. 85:25-33. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary, M., and S. Kaplan. 2000. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 28:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen, P. J., W. C. Bowman, D. F. Hartnett, S. C. Reilly, and R. G. Kranz. 1998. Translational activation by an NtrC enhancer-binding protein. J. Mol. Biol. 278:903-914. [DOI] [PubMed] [Google Scholar]
- 5.Cussiol, J. R., S. V. Alves, M. A. de Oliveira, and L. E. Netto. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570-11578. [DOI] [PubMed] [Google Scholar]
- 6.Ding, Y., C. Y. Chan, and C. E. Lawrence. 2004. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 32:W135-W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon, R., and D. Kahn. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621-631. [DOI] [PubMed] [Google Scholar]
- 8.Dubbs, J. M., and F. R. Tabita. 2004. Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28:353-376. [DOI] [PubMed] [Google Scholar]
- 9.Dumay, V., M. Inui, and H. Yukawa. 1999. Molecular analysis of the recA gene and SOS box of the purple non-sulfur bacterium Rhodopseudomonas palustris no. 7. Microbiology 145:1275-1285. [DOI] [PubMed] [Google Scholar]
- 10.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epshtein, V., A. S. Mironov, and E. Nudler. 2003. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. USA 100:5052-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez de Henestrosa, A. R., J. Cune, G. Mazon, B. L. Dubbels, D. A. Bazylinski, and J. Barbe. 2003. Characterization of a new LexA binding motif in the marine magnetotactic bacterium strain MC-1. J. Bacteriol. 185:4471-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, J. L., and F. R. Tabita. 1997. Analysis of the cbbXYZ operon in Rhodobacter sphaeroides. J. Bacteriol. 179:663-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraud, E., L. Hannibal, J. Fardoux, A. Vermeglio, and B. Dreyfus. 2000. Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc. Natl. Acad. Sci. USA 97:14795-14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19:125-137. [DOI] [PubMed] [Google Scholar]
- 18.Gregor, J., and G. Klug. 1999. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol. Lett. 179:1-9. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones, S., A. Bateman, M. Marshall, A. Khanna, and S. R. Eddy. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, R. S. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367-402. [DOI] [PubMed] [Google Scholar]
- 21.Harwood, C. S., and J. Gibson. 1988. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl. Environ Microbiol. 54:712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt, J. G. 1994. Anoxygenic phototrophic bacteria, p. 359. In J. G. Holt (ed.), Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 23.Jaubert, M., S. Zappa, J. Fardoux, J. M. Adriano, L. Hannibal, S. Elsen, J. Lavergne, A. Vermeglio, E. Giraud, and D. Pignol. 2004. Light and redox control of photosynthesis gene expression in Bradyrhizobium: dual roles of two PpsR. J. Biol. Chem. 279:44407-44416. [DOI] [PubMed] [Google Scholar]
- 24.Larimer, F. W., P. Chain, L. Hauser, J. Lamerdin, S. Malfatti, L. Do, M. L. Land, D. A. Pelletier, J. T. Beatty, A. S. Lang, F. R. Tabita, J. L. Gibson, T. E. Hanson, C. Bobst, J. L. Torres, C. Peres, F. H. Harrison, J. Gibson, and C. S. Harwood. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat. Biotechnol. 22:55-61. [DOI] [PubMed] [Google Scholar]
- 25.Lilja, A. E., J. R. Jenssen, and J. D. Kahn. 2004. Geometric and dynamic requirements for DNA looping, wrapping and unwrapping in the activation of E. coli glnAp2 transcription by NtrC. J. Mol. Biol. 342:467-478. [DOI] [PubMed] [Google Scholar]
- 26.Liu, J. S., and C. E. Lawrence. 1999. Bayesian inference on biopolymer models. Bioinformatics 15:38-52. [DOI] [PubMed] [Google Scholar]
- 27.Masepohl, B., B. Kaiser, N. Isakovic, C. L. Richard, R. G. Kranz, and W. Klipp. 2001. Urea utilization in the phototrophic bacterium Rhodobacter capsulatus is regulated by the transcriptional activator NtrC. J. Bacteriol. 183:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAdams, H. H., and L. Shapiro. 2003. A bacterial cell-cycle regulatory network operating in time and space. Science 301:1874-1877. [DOI] [PubMed] [Google Scholar]
- 29.McCue, L., W. Thompson, C. Carmack, M. P. Ryan, J. S. Liu, V. Derbyshire, and C. E. Lawrence. 2001. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res. 29:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCue, L. A., W. Thompson, C. S. Carmack, and C. E. Lawrence. 2002. Factors influencing the identification of transcription factor binding sites by cross-species comparison. Genome Res. 12:1523-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mesa, S., E. J. Bedmar, A. Chanfon, H. Hennecke, and H. M. Fischer. 2003. Bradyrhizobium japonicum NnrR, a denitrification regulator, expands the FixLJ-FixK2 regulatory cascade. J. Bacteriol. 185:3978-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullin, D. A., S. M. Van Way, C. A. Blankenship, and A. H. Mullin. 1994. FlbD has a DNA-binding activity near its carboxy terminus that recognizes ftr sequences involved in positive and negative regulation of flagellar gene transcription in Caulobacter crescentus. J. Bacteriol. 176:5971-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nellen-Anthamatten, D., P. Rossi, O. Preisig, I. Kullik, M. Babst, H. M. Fischer, and H. Hennecke. 1998. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J. Bacteriol. 180:5251-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuwald, A. F., J. S. Liu, and C. E. Lawrence. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4:1618-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newberg, L. A., and C. E. Lawrence. 2004. Mammalian genomes ease location of human DNA functional segments but not their description. Stat. Appl. Genet. Mol. Biol. 3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panmanee, W., P. Vattanaviboon, W. Eiamphungporn, W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45:1647-1654. [DOI] [PubMed] [Google Scholar]
- 37.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin, Z. S., L. A. McCue, W. Thompson, L. Mayerhofer, C. E. Lawrence, and J. S. Liu. 2003. Identification of co-regulated genes through Bayesian clustering of predicted regulatory binding sites. Nat. Biotechnol. 21:435-439. [DOI] [PubMed] [Google Scholar]
- 39.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 40.Rippe, K., N. Mucke, and A. Schulz. 1998. Association states of the transcription activator protein NtrC from E. coli determined by analytical ultracentrifugation. J. Mol. Biol. 278:915-933. [DOI] [PubMed] [Google Scholar]
- 41.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen-Orr, S. S., R. Milo, S. Mangan, and U. Alon. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 31:64-68. [DOI] [PubMed] [Google Scholar]
- 43.Shively, J. M., G. van Keulen, and W. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 44.Staden, R. 1989. Methods for calculating the probabilities of finding patterns in sequences. Comput. Appl. Biosci. 5:89-96. [DOI] [PubMed] [Google Scholar]
- 45.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. USA 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, K., L. A. McCue, and G. D. Stormo. 2005. Making connections between novel transcription factors and their DNA motifs. Genome Res. 15:312-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, K., G. Moreno-Hagelsieb, J. Collado-Vides, and G. D. Stormo. 2001. A comparative genomics approach to prediction of new members of regulons. Genome Res. 11:566-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, W., L. A. McCue, and C. E. Lawrence. 2005. Using the Gibbs Motif Sampler to find conserved domains in DNA and protein sequences, p. 2.8.1-2.8.42. In A. D. Baxevanis and D. B. Davison (ed.), Current protocols in bioinformatics. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 50.Thompson, W., E. C. Rouchka, and C. E. Lawrence. 2003. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 31:3580-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varga, A. R., and L. A. Staehelin. 1983. Spatial differentiation in photosynthetic and non-photosynthetic membranes of Rhodopseudomonas palustris. J. Bacteriol. 154:1414-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker, G. C. 1996. The SOS response in Escherichia coli. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 53.Winkler, W. C., A. Nahvi, N. Sudarsan, J. E. Barrick, and R. R. Breaker. 2003. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10:701-707. [DOI] [PubMed] [Google Scholar]
- 54.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Y. M., H. Marrakchi, and C. O. Rock. 2002. The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 277:15558-15565. [DOI] [PubMed] [Google Scholar]