Abstract
Yeasts from caves have rarely been examined. We examined yeasts collected from bat guano samples from 20 bat-inhabited limestone and volcanic caves located in 11 prefectures in Japan. Of ∼700 yeast-like colonies, nine Trichosporon species were recovered from 15 caves. Two of these were known species, and the remaining seven are potentially novel species, based on molecular phylogenetic analyses. In addition to Trichosporon species, identifiable strains of eight ascomycetous yeasts and one basidiomycetous yeast were recovered at frequencies of 5 to 35%. Our findings suggest that Trichosporon spp. are the major yeast species in bat guano in Japan and that bat guano is a potentially rich source of previously undescribed yeast species.
Caves usually are nutrient-limited sites that are sequestered from the outside environment and may contain novel, diverse microbial populations. Bat biologists, speleologists, and tourists sometimes develop clinical symptoms, such as fever or cough, after visiting bat-inhabited caves. A pulmonary disease (histoplasmosis) that is commonly contracted in bat-inhabited caves of North and Latin America is caused by Histoplasma capsulatum (3, 10, 20). Histoplasmosis is rare in Japan, and it is generally assumed that most of the cases that do arise are from infections contracted in other countries (7, 18). The excreta of feral birds and animals, including bats, contain medically significant fungi, such as Cryptococcus neoformans and Cryptococcus laurentii (5, 12, 23), and locations that contain large amounts of such excreta are potential sites of human infection. In this presentation, we analyze yeasts from bat guano from bat-inhabited caves in Japan.
Sample collection.
Sixty-two bat guano samples were collected between December 2003 and February 2004 from 20 bat-inhabited caves of speleological interest (1 to 13 samples per cave), located in 11 Japanese prefectures (Table 1).
TABLE 1.
Yeast isolation from bat guano samples in various Japanese caves
| Cave | Location | No. of samples (n = 62) | Isolation of speciesa
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Trichosporon
|
Non-Trichosporon
|
|||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||
| 1.Tateishi-no-ouana (limestone cave) | Fukushima | 2 | X | X | X | |||||||||||||||
| 2.Koumori-ana (limestone cave) | Fukushima | 1 | X | X | ||||||||||||||||
| 3.Medama-ana and Nuke-ana (limestone cave) | Gunma | 2 | X | X | ||||||||||||||||
| 4.Aoiwa-shonyudo (limestone cave) | Yamanashi | 1 | X | |||||||||||||||||
| 5.Kamiza-fuketsu (volcanic cave) | Yamanashi | 2 | X | |||||||||||||||||
| 6.Nippara-shonyudo (limestone cave) | Tokyo | 3 | X | X | X | X | X | X | X | |||||||||||
| 7.Oukubo-kazaana (limestone cave) | Ibaraki | 1 | X | |||||||||||||||||
| 8.Bussekizan-shonyudo (limestone cave) | Saitama | 3 | X | X | X | |||||||||||||||
| 9.Shoumeishi-do (limestone cave) | Mie | 1 | ||||||||||||||||||
| 10.Sazare-do (limestone cave) | Yamaguchi | 2 | X | X | X | |||||||||||||||
| 11.Akiyoshi-do (limestone cave) | Yamaguchi | 13 | X | X | X | X | X | X | ||||||||||||
| 12.Koumori-ana (limestone cave) | Yamaguchi | 5 | X | |||||||||||||||||
| 13.Senbutsu-shonyudo (limestone cave) | Yamaguchi | 7 | X | X | X | X | X | |||||||||||||
| 14.Mejiro-do (limestone cave) | Fukuoka | 3 | X | X | X | |||||||||||||||
| 15.Seiryu-kutsu (limestone cave) | Fukuoka | 6 | X | X | X | X | X | |||||||||||||
| 16.Komusou-ana (limestone cave) | Fukuoka | 1 | ||||||||||||||||||
| 17.Gouya daiichi-do (limestone cave) | Fukuoka | 1 | ||||||||||||||||||
| 18.Kyusen-do (limestone cave) | Kumamoto | 1 | ||||||||||||||||||
| 19.Tsuzurase-do (limestone cave) | Kumamoto | 5 | X | X | X | X | X | |||||||||||||
| 20.Shouryu-do, Daini-do (limestone cave) | Kagoshima | 2 | X | X | ||||||||||||||||
1 to 7, novel Trichosporon species; 8, T. laibachii; 9, T. porosum; 10, Cryptococcus podzolicus; 11, Candida palmioleophila; 12, Debaryomyces hansenii; 13, Hanseniaspora species; 14, C. lusitaniae; 15, Saccharomyces cerevisiae; 16, S. kluyveri; 17, Williopsis calfornica; 18, Zygosaccharomyces florentinus. X indicates that the species was isolated.
Isolation of yeasts.
Approximately 0.5 to 1.0 g of each sample of bat guano was suspended in YM broth (3 g yeast extract, 3 g malt extract, 5 g peptone, 10 g glucose, 20 g agar; Becton Dickinson, Paramus, NJ) that contained 50 μg/ml chloramphenicol (Sankyo, Tokyo, Japan), 400 IU/ml penicillin (Meijiseika, Tokyo, Japan), and 400 IU/ml streptomycin (Meijiseika). Aliquots of 100 μl of the broth supernatants were then inoculated onto YM agar plates (at least 10 plates per sample) that contained the three antibiotics listed above, and the plates were incubated at 27°C until colonies could be seen.
Identification of yeast isolates.
Each yeast isolate was identified with rRNA sequence analysis. Genomic DNA was extracted (11), and the D1/D2 26S rRNA and the internal transcribed spacer (ITS) region, which includes the 5.8S rRNA, were sequenced directly from the PCR products with primer pairs NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) plus NL-4 (5′-GGTCCGTGTTTCAAGACGG-3′) (9) and pITS-F (5′-GTCGTAACAAGGTTAACCTGCGG-3′) plus pITS-R (5′-TCCTCCGCTTATTGATATGC-3′) (17), respectively. The PCR products were sequenced with an ABI 310 DNA sequencer and the Big Dye Terminator Cycle Sequencing Ready kit (Perkin-Elmer Applied Biosystems, Foster City, CA), according to the manufacturer's instructions. Strains with a ≥99% ITS or D1/D2 26S rRNA sequence similarity were defined as conspecific (15, 17). The sequence data were analyzed with the National Center for Biotechnology Information (NCBI; Bethesda, MD) BLAST system (http://www.ncbi.nlm.nih.gov/BLAST/).
Molecular phylogenetic analysis of new species.
DNA sequences were aligned using CLUSTAL W (19). For the neighbor-joining analysis (16), distances between the sequences were calculated using Kimura's two-parameter model (8). A bootstrap analysis was performed using 1,000 replications (4).
Yeasts in bat guano.
We identified approximately 700 yeast-like colonies from the YM agar plates. Trichosporon spp. were found in samples from 15 of the 20 caves.
Trichosporon laibachii and Trichosporon porosum were isolated from 7 and 5 of the 20 caves, respectively. Based on the molecular phylogenetic analysis, seven new Trichosporon spp. (designated species 1 to 7) were recovered from 10 of the caves. In addition to the Trichosporon spp., eight ascomycetous yeasts (Candida palmioleophila, Candida lusitaniae, Debaryomyces hansenii, Hanseniaspora spp., Saccharomyces cerevisiae, Saccharomyces kluyveri, Williopsis californica, and Zygosaccharomyces florentinus) and one basidiomycetous yeast (Cryptococcus podzolicus) were isolated from the guano samples at frequencies that ranged from 5 to 35% for the 20 caves (Table 1).
Phylogenetic analysis of the new Trichosporon species.
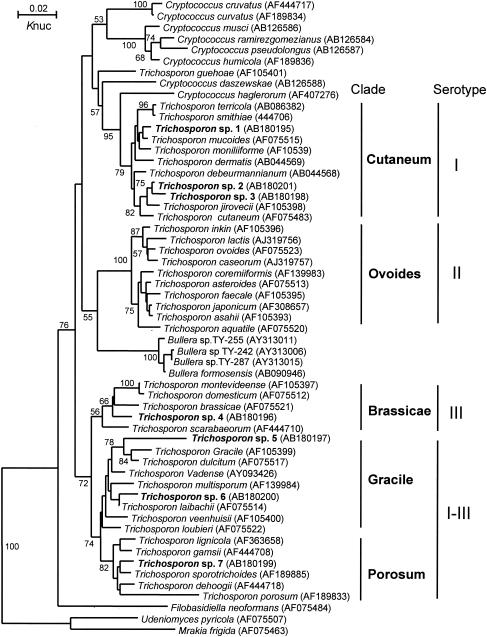
As the level of dissimilarity of the D1/D2 26S rRNA or ITS sequences between Trichosporon species 1 to 7 and the phylogenetically closest species was >1%, we concluded that Trichosporon species 1 to 7 were new species. The new species were not located within a single clade on the tree (Fig. 1) but were distributed across several clades (Gracile, Porosum, Brassicae, and Cutaneum). The Ovoides clade was the only clade for which we did not identify a new species.
FIG. 1.
Phylogenetic trees constructed using the D1/D2 26S rRNA sequences of Trichosporon spp. that were isolated from bat guano samples and from known species of Trichosporon. DDBJ/GenBank accession numbers are indicated in parentheses. The numerals near branch points are confidence limits for 1,000 bootstrap replicates (limits of <50% are not indicated). Knuc, Kimura's parameter (8).
In recent years, research on bio-speleology and microbial diversity has increased. Pathogenic fungi are potential sources of infection and allergy for speleologists. A similar investigation in Italy (13) found the pathogenic yeasts Cryptococcus neoformans and Cryptococcus laurentii in 3 of 25 cavernicolus fungus-inhabited caves. Although C. neoformans colonization of pigeon droppings is well known (23), there are few reports of this microorganism from caves.
We detected nine Trichosporon species, including seven novel species, in bat guano samples from 75% of the caves sampled. It is not known whether the Trichosporon spp. are part of the cutaneous or intestinal microbiota of the bats or if they are introduced into bat guano from other organisms, the soil, or other excreta in the cave. Repeated inhalation of Trichosporon cells may cause summer-type hypersensitivity pneumonitis, with consequent development of cough, dyspnea, and remittent fever (1). These symptoms are similar to those that may develop in speleologists after they visit caves.
Trichosporon spp. have four different serotypes (types I, II, III, and I-III) (6, 14). Each serotype is associated with the occurrence of summer-type hypersensitivity pneumonitis, and there is a correlation between the serotype of inhalation challenge-positive antigens with those of Trichosporon spp. from patients' homes (2). These serotypes also correspond to the phylogenetic clades. Trichosporon serotypes I, II, III, and I-III are associated with species in the Cutaneum, Ovoides, Brassicae, and Gracile-Porosum clades, respectively (Fig. 1).
In addition to the Trichosporon species, we isolated Candida lusitaniae and Debaryomyces hansenii from the collected bat guano. These species have been implicated in opportunistic infections of immunocompromised patients (21, 22). Because bats are widely distributed in the environment, bat guano may mediate the exchange of these pathogens, just as pigeon excreta mediate the exchange of Cryptococcus neoformans, the causal agent of cryptococcosis (23).
In conclusion, our analysis provides new, basic information on potential sources of infection and allergy for speleologists. Japanese bat guano contains several novel yeast species and may be of significant mycological interest.
Acknowledgments
We thank the members of the Speleological Society of Japan and the local and university caving clubs that cooperated in collecting bat guano samples.
This study was supported in part by a Health Science Research Grant for “Research on Emerging and Re-emerging Infectious Diseases” from the Ministry of Health, Labor, and Welfare of Japan.
REFERENCES
- 1.Ando, M., K. Arima, R. Yoneda, and M. Tamura. 1991. Japanese summer-type hypersensitivity pneumonitis. Geographic distribution, home environment, and clinical characteristics of 621 cases. Am. Rev. Respir. Dis. 144:765-769. [DOI] [PubMed] [Google Scholar]
- 2.Ando, M., T. Sakata, K. Yoshida, H. Yamasaki, S. Araki, K. Onoue, and T. Shinoda. 1990. Serotype-related antigen of Trichosporon cutaneum in the induction of summer-type hypersensitivity pneumonitis: correlation between serotype of inhalation challenge-positive antigen and that of the isolates from patients' homes. J. Allergy Clin. Immunol. 85:36-44. [DOI] [PubMed] [Google Scholar]
- 3.Erkens, K., M. Lademann, K. Tintelnot, M. Lafrenz, U. Kaben, and E. C. Reisinger. 2002. Histoplasmosis group disease in bat researchers returning from Cuba. Dtsch. Med. Wochenschr. 127:21-25. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 5.García-Hermoso, D., S. Mathoulin-Pélissier, B. Couprie, O. Ronin, B. Dupont, and F. Dromer. 1997. DNA typing suggests pigeon droppings as a source of pathogenic Cryptococcus neoformans serotype D. J. Clin. Microbiol. 35:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeda, R., M. Yokota, and T. Shinoda. 1996. Serological characterization of Trichosporon cutaneum and related species. Microbiol. Immunol. 40: 813-819. [DOI] [PubMed] [Google Scholar]
- 7.Kamei, K., A. Sano, K. Kikuchi, K. Makimura, M. Niimi, K. Suzuki, Y. Uehara, N. Okabe, K. Nishimura, and M. Miyaji. 2003. The trend of imported mycoses in Japan. J. Infect. Chemother. 9:16-20. [DOI] [PubMed] [Google Scholar]
- 8.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 9.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyon, G. M., A. V. Bravo, A. Espino, M. D. Lindsley, R. E. Gutierrez, I. Rodriguez, A. Corella, F. Carrillo, M. M. McNeil, D. W. Warnock, and R. A. Hajjeh. 2004. Histoplasmosis associated with exploring a bat-inhabited cave in Costa Rica, 1998-1999. Am. J. Trop. Med. Hyg. 70:438-442. [PubMed] [Google Scholar]
- 11.Makimura, K., S. Y. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 12.Mattsson, R., P. D. Haemig, and B. Olsen. 1999. Feral pigeons as carriers of Cryptococcus laurentii, Cryptococcus uniguttulatus and Debaryomyces hansenii. Med. Mycol. 37:367-369. [DOI] [PubMed] [Google Scholar]
- 13.Montagna, M. T., M. P. Santacroce, G. Caggiano, D. Tato, and L. Ajello. 2003. Cavernicolous habitats harbouring Cryptococcus neoformans: results of a speleological survey in Apulia, Italy, 1999-2000. Med. Mycol. 41:451-455. [DOI] [PubMed] [Google Scholar]
- 14.Nishiura, Y., K. Nakagawa-Yoshida, M. Suga, T. Shinoda, E. Guého, and M. Ando. 1997. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J. Med. Vet. Mycol. 35:45-52. [DOI] [PubMed] [Google Scholar]
- 15.Peterson, S. W., and C. P. Kurtzman. 1991. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst. Appl. Microbiol. 14: 124-129. [Google Scholar]
- 16.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 17.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzaki, A., M. Kimura, S. Kimura, K. Shimada, M. Miyaji, and L. Kaufman. 1995. An outbreak of acute pulmonary histoplasmosis among travelers to a bat-inhabited cave in Brazil. Kansenshogaku Zasshi 69:444-449. [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdez, H., and R. A. Salata. 1999. Bat-associated histoplasmosis in returning travelers: case presentation and description of a cluster. J. Travel Med. 6:258-260. [DOI] [PubMed] [Google Scholar]
- 21.Viudes, A., J. Peman, E. Canton, M. Salavert, P. Ubeda, J. L. Lopez-Ribot, and M. Gobernado. 2002. Two cases of fungemia due to Candida lusitaniae and a literature review. Eur. J. Clin. Microbiol. Infect. Dis. 21:294-299. [DOI] [PubMed] [Google Scholar]
- 22.Wong, B., T. E. Kiehn, F. Edwards, E. M. Bernard, R. C. Marcove, E. de Harven, and D. Armstrong. 1982. Bone infection caused by Debaryomyces hansenii in a normal host: a case report. J. Clin. Microbiol. 16:545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto, Y., S. Kohno, H. Koga, H. Kakeya, K. Tomono, M. Kaku, T. Yamazaki, M. Arisawa, and K. Hara. 1995. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J. Clin. Microbiol. 33:3328-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]