Abstract
The carbon and energy metabolisms of a variety of cultured chemolithoautotrophic Epsilonproteobacteria from deep-sea hydrothermal environments were characterized by both enzymatic and genetic analyses. All the Epsilonproteobacteria tested had all three key reductive tricarboxylic acid (rTCA) cycle enzymatic activities—ATP-dependent citrate lyase, pyruvate:ferredoxin oxidoreductase, and 2-oxoglutarate:ferredoxin oxidoreductase—while they had no ribulose 1,5-bisphosphate carboxylase (RubisCO) activity, the key enzyme in the Calvin-Benson cycle. These results paralleled the successful amplification of the key rTCA cycle genes aclB, porAB, and oorAB and the lack of success at amplifying the form I and II RubisCO genes, cbbL and cbbM. The combination of enzymatic and genetic analyses demonstrates that the Epsilonproteobacteria tested use the rTCA cycle for carbon assimilation. The energy metabolisms of deep-sea Epsilonproteobacteria were also well specified by the enzymatic and genetic characterization: hydrogen-oxidizing strains had evident soluble acceptor:methyl viologen hydrogenase activity and hydrogen uptake hydrogenase genes (hyn operon), while sulfur-oxidizing strains lacked both the enzyme activity and the genes. Although the energy metabolism of reduced sulfur compounds was not genetically analyzed and was not fully clarified, sulfur-oxidizing Epsilonproteobacteria showed enzyme activity of a potential sulfite:acceptor oxidoreductase for a direct oxidation pathway to sulfate but no activity of AMP-dependent adenosine 5′-phosphate sulfate reductase for a indirect oxidation pathway. No activity of thiosulfate-oxidizing enzymes was detected. The enzymatic and genetic characteristics described here were consistent with cellular carbon and energy metabolisms and suggest that molecular tools may have great potential for in situ elucidation of the ecophysiological roles of deep-sea Epsilonproteobacteria.
Epsilonproteobacteria include physiologically and phylogenetically diverse members from a variety of habitats (for a review, see reference 40) such as the gastrointestinal tracts of animals (12), sulfurous springs (3, 45), activated sludge (50), oil fields (17), Antarctic Ocean water (5), and deep-sea cold seep sediments (22, 30). Deep-sea hydrothermal systems, however, may host the largest biomass and diversity of Epsilonproteobacteria on earth (39, 53).
Based on recent culture-independent molecular ecological surveys, the predominant occurrence of Epsilonproteobacteria in global deep-sea hydrothermal systems has been demonstrated (10, 20, 31, 36, 42, 43, 54, 55). As indicated in phylogenetic trees, most epsilonproteobacterial subgroups consist only of deep-sea epsilonproteobacterial rRNA gene clones obtained from planktonic, benthic, epilithic, and episymbiotic habitats with relatively low temperatures. More recently, the first endosymbiotic epsilonproteobacterium was discovered in a gastropod Alvinoconcha sp. endemic in deep-sea hydrothermal vents and has been added to the list of uncultivated deep-sea Epsilonproteobacteria (51). Despite the dominance and the expansive phylogenetic diversity of deep-sea Epsilonproteobacteria, their physiological properties and ecological roles remain unclear due to their strong resistance to cultivation.
The first successful isolation of previously uncultivated subgroups of deep-sea Epsilonproteobacteria was that of hydrogen-oxidizing, thermophilic chemolithoautotrophs from nests of tube-dwelling polychaetes in deep-sea hydrothermal environments (1, 7, 34). All these isolates were members of a phylogenetic subgroup (group D Epsilonproteobacteria) (35). Subsequently, a number of deep-sea Epsilonproteobacteria representing nearly all of the previously uncultivated subgroups were isolated from a variety of habitats in geologically and geographically distinct hydrothermal fields (53). Physiological characterizations of these isolates have provided a rough sketch indicating that most of the deep-sea Epsilonproteobacteria are hydrogen- and/or sulfur-oxidizing chemolithoautotrophs, and some phylogenetic subgroups are specified with distinctive physiological characteristics (1, 23, 24, 34, 35, 37-39, 53, 54, 58). For instance, group D contains all strictly anaerobic, hydrogen-oxidizing, sulfur- or nitrate-reducing thermophiles (1, 34, 35, 53, 58), and group A also has facultatively anaerobic, hydrogen-oxidizing thermophiles (38, 56). Groups B, F, and G consist of metabolically versatile mesophiles utilizing various electron donors and acceptors (23, 24, 37, 39).
The enzymatic and genetic characteristics of the carbon and energy metabolisms of deep-sea Epsilonproteobacteria have been investigated much less than their cellular physiological characteristics. Carbon assimilation by way of the reductive tricarboxylic acid (rTCA) cycle was first suggested for the marine epsilonproteobacterium “Arcobacter sulfidicus” (proposed name) based on the absence of ribulose 1,5-phosphate carboxylase (RubisCO) activity and the carbon isotopic fractionation between the cellular carbon and the inorganic carbon source (63). Campbell et al. (8) and Campbell and Cary (9) demonstrated that genes for key enzymes of the rTCA cycle—ATP-dependent citrate lyase (ACL) (aclB for the large subunit), pyruvate:ferredoxin oxidoreductase (porA for the α subunit), and 2-oxoglutarate:ferredoxin oxidoreductase (oorA for the α subunit)—were detected in epsilonproteobacterial DNA and RNA assemblages recovered from symbiotic and epilithic deep-sea hydrothermal vent habitats. Recently, Hügler et al. (21) demonstrated the first enzymatic and genetic evidence of operation of the rTCA cycle as autotrophic CO2 fixation in the epsilonproteobacterial strains Thiomicrospira denitrificans and “Arcobacter sulfidicus,” which are sulfur-oxidizing chemolithoautotrophs within group B isolated from the oxic-anoxic interface in coastal marine habitats. These results strongly suggested that the operative rTCA cycle might serve in a diversity of deep-sea Epsilonproteobacteria and might sustain the predominant primary production in situ. However, the energy metabolisms and the enzymatic identification of the rTCA cycle of cultured deep-sea Epsilonproteobacteria have not been thoroughly investigated. In this study, enzymatic and genetic characterizations of carbon and energy metabolisms were conducted for a variety of deep-sea epsilonproteobacterial isolates whose cellular physiological properties have been well characterized. Representative strains from widespread phylogenetic subgroups and physiological types were chosen which would most likely cover the carbon and energy metabolisms dominating in deep-sea hydrothermal microbial communities.
MATERIALS AND METHODS
Organisms and growth conditions.
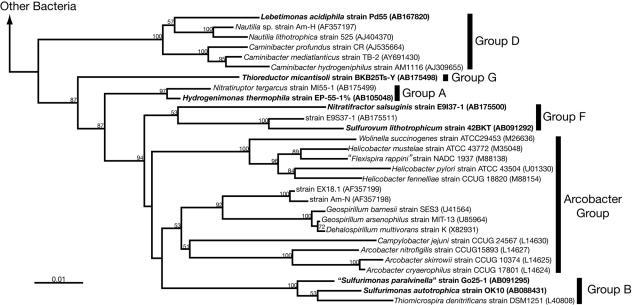
The taxonomic characteristics of the microorganisms used in this study and the cultivation conditions are summarized in Table 1. All the microorganisms were isolated from deep-sea hydrothermal environments, and their cellular physiological properties have been described previously, except for “Sulfurimonas paralvinella” (proposed name) (23, 24, 37-39, 53, 56-58). Although Nitratifractor salsuginis is able to oxidize elemental sulfur to sulfate during growth with molecular hydrogen as the energy source, sulfur does not solely support the growth of these organisms (39). A phylogenetic tree of representative strains of Epsilonproteobacteria inferred from 16S rRNA gene sequences is shown in Fig. 1, indicating that the deep-sea Epsilonproteobacteria examined in this study cover most of the phylogenetic subgroups within Epsilonproteobacteria (Fig. 1).
TABLE 1.
Cellular physiological properties, phylogenetic affiliations, and enzymatic experimental conditions for deep-sea isolates of Epsilon- and Gammaproteobacteria examined in this study
| Characteristic | Hydrogenimonas thermophila strain EP1-55-1% | Sulfurimonas autotrophica strain OK10 | “Sulfurimonas paralvinella” strain GO25 | Lebetimonas acidiphila strain Pd55 | Sulfurovum lithotrophicum strain BKB42 | Nitratifractor salsuginis strain E9137-1 | Thioreductor micantisoli strain BKB25Ts-Y | Thiomicrospira thermophila strain I78 |
|---|---|---|---|---|---|---|---|---|
| Class | Epsilon proteobacteria | Epsilon proteobacteria | Epsilon proteobacteria | Epsilon proteobacteria | Epsilon proteobacteria | Epsilon proteobacteria | Epsilon proteobacteria | Gamma proteobacteria |
| Subgroup or order | Group A | Group B | Group B | Group D | Group F | Group F | Group G | Thiotrichales |
| Place of isolation | Central Indian Ridge chimney | Okinawa Trough hydrothermal sediments | Okinawa Trough, Paralvinella's nest | Mariana Arc vent fluids | Okinawa Trough hydrothermal sediments | Okinawa Trough chimney | Okinawa Trough hydrothermal sediments | Mariana Arc vent fluids |
| Topt for growtha (°C) | 55 | 25 | 30 | 50 | 30 | 37 | 32 | 35 |
| Electron donor | H2 | S0, S2O32−, sulfide | H2, S0 | H2 | S0, S2O32− | H2, S0b | H2 | S0, S2O32−, sulfide |
| Electron acceptor | S0, NO3−, O2 | O2 | NO3−, O2 | S0 | NO3−, O2 | NO3−, O2 | S0, NO3− | O2 |
| Microaerophilic growth | + | + | + | − | + | + | − | + |
| Cultivation conditions | ||||||||
| Temp (°C) | 55 | 25 | 30 | 50 | 30 | 37 | 32 | 35 |
| Electron donor | H2 | S2O32− | H2 | H2 | S2O32− | H2 | H2 | S2O32− |
| Electron acceptor | NO3− | O2 | NO3− | S0 | O2 | NO3− | S0 | O2 |
| Enzyme assay | ||||||||
| Temp (°C) | 55 (45 for RubisCO) | 25 | 30 | 50 (45 for RubisCO) | 30 | 37 | 30 | 37 |
| Reference(s) | 52, 55 | 22, 52 | 52 | 57 | 23, 52 | 37, 52 | 36, 52 | 55 |
Topt, optimum temperature.
*, S0 does not solely support the growth.
FIG. 1.
Phylogenetic tree of representative strains of Epsilonproteobacteria inferred from the 16S rRNA gene sequences by the neighbor-joining method using 1,105 homologous sequence positions for each organism. The phylogenetic tree was constructed as described previously (38). The deep-sea epsilonproteobacterial strains examined in this study are boldfaced. Bootstrap values of >50% for the branches are shown (based on 100 replicates). Bar, 1 substitution per 100 nucleotides. Database accession numbers are given in parentheses.
Cultures were harvested during late-exponential growth by centrifugation under anaerobic conditions, and the cells were washed twice with 100 mM Tris-HCl (pH 7.8), 500 mM NaCl, and 1 mM dithiothreitol (DTT). The cell pellets were preserved at −80°C under anaerobic conditions.
Preparation of cell extracts.
The frozen cell pellets (1 g [wet weight]) were suspended in 3 ml of 100 mM Tris-HCl (pH 7.8) and 1 mM DTT. The cell suspensions were thoroughly disrupted by ultrasonification with acid-washed, autoclaved glass beads (diameter, 425 to 600 μm) on ice and centrifuged at 20,000 × g for 20 min at 4°C. Solid ammonium sulfate was added to the cell extracts (supernatant) to reach 80% saturation. After being stirred for 30 min, the extracts were centrifuged at 20,000 × g for 20 min at 4°C. The pellets were dissolved in 100 mM Tris-HCl (pH 7.8) and 1 mM DTT and dialyzed against the same buffer. All the procedures were performed anaerobically under an N2 gas atmosphere or with N2 gas purging. After ammonium sulfate precipitation, the dialyzed fractions were used as cell extracts for enzyme activity characterization. Protein concentrations were estimated by using the NanoOrange Protein Quantification kit (Molecular Probes, Inc., Eugene, OR) according to the manufacturer's manual.
Enzyme assays.
All the enzyme activities described below were measured in triplicate. Reactions in the absence of the substrate and in the presence of heat-denatured cell extracts were used as the negative controls. Enzyme activity was measured in the same buffer system of 200 mM Tris-HCl (pH 7.8), 2 mM DTT, and 2 mM MgCl2 with substrates, cofactors, and coupling enzymes. Temperatures for enzyme activity measurement are shown in Table 1.
(i) ACL.
ACL, a key enzyme of the rTCA cycle, was assayed spectrophotometrically according to Takeda et al. (59). Reaction mixtures contained 2.5 mM ATP, 10 mM sodium citrate, 0.25 mM NADH, and 10 U ml−1 of malate dehydrogenase (from porcine heart [Sigma, St. Louis, MO] for 25, 30, and 37°C; from Thermus flavus [Sigma] for 50 and 55°C) as a coupling enzyme.
(ii) POR and OGOR.
Pyruvate:acceptor oxidoreductase (POR) and 2-oxoglutarate:acceptor oxidoreductase (OGOR), reversible CO2 assimilation enzymes of the rTCA cycle, were assayed spectrophotometrically according to Shiba et al. (47). Reaction mixtures contained 0.25 mM coenzyme A (CoA), 5 mM methyl viologen (MV), and 10 mM sodium pyruvate for POR or 10 mM sodium oxoglutarate for OGOR.
(iii) ICDH.
Isocitrate dehydrogenase (ICDH), a reversible CO2 assimilation enzyme of the rTCA cycle, was assayed spectrophotometrically according to Shiba et al. (46). Reaction mixtures contained 10 mM sodium oxoglutarate, 10 mM sodium bicarbonate, and 0.25 mM NADH.
(iv) PEPC and PC.
Phosphoenolpyruvate carboxylase (PEPC) and pyruvate carboxylase (PC), anaplerotic (replenishing) CO2 assimilation enzymes related to the rTCA cycle, were assayed spectrophotometrically according to Shiba et al. (46) and Takai et al. (52). Reaction mixtures contained 10 mM sodium bicarbonate, 0.25 mM NADH, 10 mM sodium phosphoenolpyruvate for PEPC or 10 mM sodium pyruvate for PC, and 10 U ml−1 of malate dehydrogenase (from porcine heart [Sigma] for 25, 30, and 37°C; from Thermus flavus [Sigma] for 50 and 55°C) as a coupling enzyme.
(v) RubisCO.
RubisCO, a key CO2 assimilation enzyme of the Calvin-Benson cycle, was assayed by spectrophotometry and high-performance liquid chromatography (HPLC) according to Ezaki et al. (13) and Maeda et al. (32), respectively. For the spectrophotometric assay, reaction mixtures contained 2.5 mM ATP, 10 mM sodium bicarbonate, 0.25 mM NADH, 10 mM sodium ribulose 1,5-bisphosphate, 40 U ml−1 of 3-phosphoglycerate phosphokinase (Sigma), 20 U ml−1 of triose-phosphate isomerase (Sigma), 10 U ml−1 of glyceraldehyde-3-phosphate dehydrogenase (Sigma), and 10 U ml−1 of glycerophosphate dehydrogenase (Sigma) as coupling enzymes. For the HPLC assay, reaction mixtures contained 10 mM sodium bicarbonate and 10 mM sodium ribulose 1,5-bisphosphate. Periodically, reduction of ribulose 1,5-bisphosphate and generation of 3-phosphoglycerate were analyzed by ion chromatography using a Shim-pack IC column (Shimadzu, Kyoto, Japan).
(vi) Hydrogenase.
Soluble hydrogenase, a hydrogen uptake hydrogenase, is derived from cytoplasmic [NiFe] hydrogenase (group III) and/or from membrane-anchored, periplasmically oriented [NiFe] hydrogenase (group I). The activity was assayed spectrophotometrically according to Ishii et al. (25). Reaction mixutures contained 5 mM methyl viologen or 0.25 mM NAD under H2 gas (100%, 1 atm).
(vii) TSO.
Thiosulfate-oxidizing enzymes (TSO), catalyzing the oxidation of thiosulfate to tetrathionate (thiosulfate:acceptor oxidoreductase) or sulfite (rhodanese), were assayed spectrophotometrically according to Tuttle et al. (61) and Singleton and Smith (49). Reaction mixtures contained 10 mM sodium thiosulfate and an acceptor such as 0.25 mM NADH (340 nm), 0.25 mM flavin adenine dinucleotide (420 nm), or 0.25 mM potassium ferricyanide [K3Fe(CN)6] (420 nm).
(viii) APSR and SOR.
Adenosine 5′-phosphate sulfate reductase (APSR) or sulfite oxidoreductase (SOR), a reversible sulfite-oxidizing enzyme of the indirect or direct pathway of sulfite oxidation, was assayed spectrophotometrically according to de Jong et al. (11) or Fritz et al. (16), respectively. Reaction mixtures contained 10 mM sodium sulfite and an acceptor such as 0.25 mM NADH (340 nm), 0.25 mM flavin adenine dinucleotide (420 nm), or 0.25 mM potassium ferricyanide [K3Fe(CN)6] (420 nm) in the presence or absence of 2.5 mM AMP. The difference between the activities in the presence and absence of AMP represented APSR.
Radioisotope labeling experiment.
Cell extracts (final concentration, 1 mg of proteins ml−1) of Hydrogenimonas thermophila and Lebetimonas acidiphila were incubated with radioisotope-labeled sodium bicarbonate (NaH14CO3) (100 μCi ml−1) in a 3-ml reaction mixture containing 200 mM Tris-HCl (pH 7.8), 2 mM DTT, 2 mM MgCl2, 10 mM succinyl-CoA, 10 mM sodium bicarbonate, 0.5 mM NADH, and 5 mM ATP under H2 gas (100%, 1 atm). Reactions were started by addition of the cell extract, reaction mixtures were incubated at 55°C for 5 min, and reactions were stopped by addition of 2 ml of 5 N H2SO4. After the free CO2 was expelled, 20 ml of 100% methanol was added to the mixture. After centrifugation of the mixture, the supernatant (methanol-soluble fraction) was collected and evaporated.
The dry samples were resuspended in 400 μl of distilled, deionized H2O, and 20 μl of each sample was applied to a high-performance thin-layer chromatography (HPTLC) plate (Fertigplatten Kieselgel 60; Merck, Darmstadt, Germany). The plate was developed as follows: first dimension, n-butanol-acetic acid-H2O (6:2:2 [vol/vol/vol]); second dimension, phenol-H2O (75:25 [vol/vol]). After development, organic acids with radioactivity were identified by using a BAS-2000 imaging analyzer (Fuji Film, Tokyo, Japan). Individual organic acids were identified by cochromatography on the HPTLC plate with authentic compounds.
Zymogram of soluble hydrogenases.
In order to check the number of soluble hydrogenases in Epsilonproteobacteria, zymograms of soluble hydrogenase in native polyacrylamide gel electrophoresis (PAGE) were conducted. Cell extracts (100 μg protein) were run in a 4 to 20% (wt/vol) gradient native polyacrylamide gel (Bio-Rad, Hercules, CA). After electrophoresis, the gel was incubated with a reaction mixture containing 200 mM Tris-HCl (pH 7.8), 2 mM DTT, 2 mM MgCl2, and 5 mM methyl viologen under H2 gas (100%, 1 atm) at 37°C. The hydrogenase activity was visualized by reduction of methyl viologen under anaerobic conditions. Kaleidoscope prestained standards (Bio-Rad) were used as a protein molecular weight standard.
Extraction of genomic DNA.
Genomic DNAs of all the Epsilon- and Gammaproteobacteria were prepared from the cell pellets (0.5 g [wet weight]) as described by Marmur and Doty (33).
PCR amplification and sequencing of genes for the rTCA and Calvin-Benson cycles.
PCR amplification of genes for key rTCA cycle enzymes was initially based on previously designed primers (8, 9). For this study, we designed degenerate primers to amplify longer segments of the ACL β subunit gene (aclB), the OGOR gene (oorAB), and the POR gene (porAB) than those described previously. Primer design was based on amino acid alignments for at least three phylogenetically distinct Epsilonproteobacteria. The primers used in this study include aclB275F (5′-TAGAGGATGCRGCTAAWTGGATTGATGA-3′) and aclB1204R (5′-GTTGGGGCCRCCWCKKCKNAC-3′), oorA67F (5′-TTCTTCGCTGGGTAYCCNATHACNCC-3′), oorA177R (5′-CATACCAGCTATYTCRTCYTCCATYTG-3′), oorA345R (5′-CTTGCAGCCTGTNGGMAKNCCNGT-3′), and oorB68R (5′-CCRCANCCCCARCACCA-3′); and porA100F (5′-GCAGCTCGTAGCNTATCCNATHACNCC-3′), porA658F (5′-GCTGCAGARGAAGANTGGCATT-3′), porA900F (5′-GATCAGGTCCTTCAGNCCNTTCCC-3′), porA1101R (5′-RTCICTTYCICCIARACC-3′), and porB230R (5′-TTTTCRAANCCRATRTGSATCCAAG-3′). Primers for amplification of the form I RubisCO gene (cbbL) were modified from previous primers, whereas the form II gene (cbbM) primers were as described previously (9). The cbbL primers include cbbL379F (5′-GACCAGTCGGYAAYGTNTTYGGNTTYAA-3′) and cbbL634R (5′-CTGACCAGTCTNAYRTTYTCRTCRTCYTT-3′).
PCR conditions were as described for aclB (8) with the following changes in annealing temperatures: aclB at 54°C, porA at 49°C, oorA at 52°C, and cbbL at 54°C. All amplifications were performed on a Robocycler 96 (Stratagene) as described previously (8). Not all strains amplified with all primer sets, as outlined in Table 2. Those amplicons with the expected sizes (except for the oorA67F-177R amplicons) were excised from 1% agarose gels (in 0.5× Tris-borate-EDTA) and eluted with a QiaexII DNA elution kit by following the manufacturer's protocol (QIAGEN, Valencia, CA). The amplification products were subsequently sequenced as described previously by using primers to the appropriate gene (8). The oorA67F-177R products were first cloned into a TA-TOPO vector and subsequently sequenced as described previously (8).
TABLE 2.
Summary of PCR amplification for genes involved in the rTCA and Calvin-Benson cycles and for the hyn operon
| Primer pair | Amplification of the following organism (primary energy source) with the indicated primer paira:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hydrogenimonas thermophila (hydrogen) | Sulfurimonas autotrophica (reduced sulfur compounds) | “Sulfurimonas paralvinella” (hydrogen) | Lebetimonas acidiphila (hydrogen) | Sulfurovum lithotrophicum (reduced sulfur compounds) | Nitratifractor salsuginis (hydrogen) | Thioreductor micantisoli (hydrogen) | Thiomicrospira thermophila (reduced sulfur compounds) | |
| rTCA cycle | ||||||||
| aclB275F-aclB1204R | + and S | + and S | + and S | + and S | + and S | + and S | + and S | − |
| porA900F-porA1101R | − | + | − | + | + | − | − | − |
| porA100F-porA1101R | + | + | − | + | + | − | + | − |
| porA100F-porB230R | + and S | + and S | − | + and S | + and S | + and S | + and S | − |
| porA658F-porB230R | + | + | + and S | + | + | − | + | − |
| oorA67F-oorA177R | + | + | + | + | + and S | + | + | − |
| oorA67F-oorA345R | + | + | + | + | − | + | − | − |
| oorA67F-oorB68R | + and S | + and S | + and S | + and S | − | + and S | + and S | − |
| Calvin-Benson cycle | ||||||||
| cbbL379F-cbbL634R | − | − | − | − | − | − | − | + and S |
| cbbM591F-cbbM918R | − | − | − | − | − | − | − | + and S |
| Soluble hydrogenase | ||||||||
| hynS330F-hynL410R | + and Sb | − | + | + and S | − | + | − | − |
| hynS330F-hynL630R | + and Sb | − | + and S | − | − | + and S | − | − |
| hynL110F-hynL410R | + and Sb | − | + | + | − | + | + and S | − |
| hynL110F-hynL630R | + and Sb | − | + | − | − | + | − | − |
+, PCR amplification was successful; −, PCR amplification was unsuccessful; S, fragments that were sequenced.
These DNA fragments were cloned, and 25 clones of the library constructed by each primer set were sequenced.
PCR amplification and sequencing of genes for soluble hydrogenase.
For this study, we designed degenerate primers to amplify genes encoding HydAB (HynSL) (6, 62). Primer design was based on amino acid alignments for group I hydrogenases from a variety of phylogroups of Bacteria (6, 62). The primers used in this study include hynS330F (5′-GARCCWAAYTTYTGGGAYAC-3′), hynL110F (5′-GTSCAYTTYTAYCASTTACA-3′), hynL410R (5′-GGSGGYTTGATCCASGWRTAYTT-3′), and hynL630R (5′-CASGCGAWGCASGGRTCRAA-3′).
Reaction mixtures were prepared in which the concentration of each oligonucleotide primer was 0.4 μM and that of the DNA template was 100 ng μl−1. Thermal cycling was performed using the GeneAmp 9700 (Applied Biosystems, Foster City, CA) under the following conditions: denaturation at 96°C for 25 s, annealing at 45°C for 45 s, and extension at 72°C for 120 s for a total of 25 cycles. Not all strains amplified with all primer sets, as outlined in Table 2. Those amplicons with the expected sizes were excised from 1% agarose gels and purified by means of a Gel Spin DNA purification kit (MoBio, Carlsbad, CA). The amplification products were directly sequenced by the dideoxynucleotide chain termination method using a dRhodamine sequencing kit and an ABI Genetic Analyzer 3100 (both from Applied Biosystems).
Phylogenetic analysis.
Nucleic acid sequences were translated and aligned to the related sequences obtained from the GenBank/EMBL/DDBJ database in the DNASTAR sequence analysis program using Clustal W. Neighbor-joining distance trees as well as bootstrapped protein parsimony trees (based on 100 replicates) were generated with the PHYLIP package in GDE as described previously (8). Similarity matrices were generated in PHYLIP using the amino acid similarity matrix algorithm. Trees were generally based on alignments of somewhat less than the full-length amplified region. Insertions and deletions were not included in the calculations. Trees were viewed in Treeview X, version 0.4.1 (http://darwin.zoology.gla.ac.uk/∼rpage/treeviewx/).
Nucleotide sequence accession numbers.
All sequences have been deposited in the GenBank/EMBL/DDBJ database with the following accession numbers: AY929800 to AY929823 (rTCA cycle genes), AY958067 to AY958069 (cbbM and cbbL), and AB206664 to 206668 (hynS and hynL).
RESULTS
Enzymatic activities of carbon metabolisms.
The key enzymatic activities of potential CO2 assimilation pathways were examined in cell extracts prepared from chemolithoautotrophically grown deep-sea Epsilon- and Gammaproteobacteria (Table 3). Activities of ACL, the key enzyme of the rTCA cycle, were detected in all the deep-sea Epsilonproteobacteria but not in the gammaproteobacterium Thiomicrospira thermophila (Table 3). The primary CO2 assimilation enzymes of the rTCA cycle, OGOR and POR, were also found among all the deep-sea Epsilonproteobacteria but not in T. thermophila (Table 3). Since the OGOR and POR reactions are reversible, the occurrence of these enzyme activities does not always indicate the operation of the rTCA cycle. However, the concurrence of ACL, OGOR, and POR activities strongly suggests an operative rTCA cycle in these deep-sea Epsilonproteobacteria. In contrast, all the deep-sea Epsilonproteobacteria tested lacked RubisCO activity, the key enzyme of the Calvin-Benson cycle, while T. thermophila had RubisCO activity (Table 3). Other CO2 assimilation enzymes associated with the rTCA cycle, such as ICDH, PEPC, and PC, were also measured. Most of the deep-sea Epsilonproteobacteria had relatively lower but evident ICDH activity for the reductive reaction, whereas all of them lacked the replenishing CO2-assimilating PEPC and PC activities (Table 3). Compared to the specific activities of rTCA cycle enzymes in Thiomicrospira denitrificans and “Arcobacter sulfidicus” reported by Hügler et al. (21), all the deep-sea Epsilonproteobacteria represented similar specific activities of ACL but much higher specific activities of OGOR and POR (Table 3). The higher specific activities of OGOR and POR might have been obtained due to the anaerobic preparation of cell extracts in this study. Indeed, exposure of cell extracts to air reduced the specific activities of OGOR and POR, although no quantitative data were retrieved. In contrast, the ICDH specific activities of the deep-sea Epsilonproteobacteria were lower than those in T. denitrificans and “Arcobacter sulfidicus” (Table 3). Since Hügler et al. reported that epsilonproteobacterial ICDH was NADP or NADPH dependent (21), the absence or the low level of ICDH specific activity could be caused by miscoupling with NADH as a cofactor in this study.
TABLE 3.
Summary of the characterization of the activities of the key enzymes involved in the potential carbon and energy metabolisms of deep-sea Epsilon- and Gammaproteobacteria
| Enzyme (substrate)a | Activity of the indicated enzyme (nmol of product min−1 mg−1) for the following organism (primary energy source):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Hydrogenimonas thermophila (hydrogen) | Sulfurimonas autotrophica (reduced sulfur compounds) | “Sulfurimonas paralvinella” (hydrogen) | Lebetimonas acidiphila (hydrogen) | Sulfurovum lithotrophicum (reduced sulfur compounds) | Nitratifractor salsuginis (hydrogen) | Thioreductor micantisoli (hydrogen) | Thiomicrospira thermophila (reduced sulfur compounds) | |
| rTCA cycle | ||||||||
| ACL (citrate) | 386 ± 40 | 222 ± 45 | 14.5 ± 1.8 | 375 ± 30 | 20.8 ± 3.2 | 73.0 ± 8.2 | 147 ± 18 | ND |
| OGOR (oxoglutarate) | 7,800 ± 530 | 12,600 ± 900 | 5,520 ± 300 | 5,430 ± 240 | 1,770 ± 100 | 12,500 ± 800 | 15,800 ± 1,100 | ND |
| POR (pyruvate) | 2,660 ± 320 | 2,840 ± 600 | 257 ± 32 | 12,730 ± 520 | 2,190 ± 420 | 1,380 ± 120 | 16,900 ± 1,800 | ND |
| ICDH (isocitrate) | 5.33 ± 0.6 | 87.5 ± 12.2 | 115 ± 25 | ND | 33.9 ± 2.5 | ND | ND | 11.4 ± 2.2 |
| Replenishing pathway | ||||||||
| PEPC (PEP) | NDb | ND | ND | ND | ND | ND | ND | ND |
| PC (pyruvate) | ND | ND | ND | ND | ND | ND | ND | 3.4 ± 0.5 |
| Calvin-Benson cycle, RubisCO (RBP) | ND | ND | ND | ND | ND | ND | ND | 222 ± 40c |
| Hydrogen oxidation soluble hydrogenase:MV (H2) | 32,400 ± 740 | ND | 2,180 ± 220 | 25,500 ± 1,200 | ND | 1,700 ± 250 | 97,400 ± 8,000 | ND |
| Sulfur oxidation | ||||||||
| SOR:ferricyanide (sulfite) | ND | 4,120 ± 590 | 90 ± 65 | ND | 2,320 ± 400 | 460 ± 25 | ND | ND |
| APSR:ferricyanide (sulfite) | ND | ND | ND | ND | ND | ND | ND | 623 ± 120 |
| TSO:ferricyanide (thiosulfate) | ND | ND | ND | ND | ND | ND | ND | 1,820 ± 120 |
PEP, phosphoenolpyruvate; RBP, ribulose 1,5-bisphosphate.
ND, not detected.
HPLC.
In the comparison of CO2 assimilation enzyme activities among the deep-sea Epsilonproteobacteria, different patterns were found. H. thermophila, Sulfurimonas autotrophica, “S. paralvinella,” and N. salsuginis had much higher specific activities of OGOR than of POR, while L. acidiphila and Thioreductor micantisoli had specific activities of POR higher than or similar to those of OGOR (Table 3). Although it is still unclear how the different modes of two major CO2 assimilation enzyme activities are involved in the cellular physiological differences of the deep-sea Epsilonproteobacteria, POR might be a key enzyme controlling the fate of acetyl-CoA, directing it to either gluconeogenesis or fatty acid synthesis. This is also the decisive step for the stable carbon isotopic disproportionation between the cellular carbon and fatty acid carbon components, as previously demonstrated (51).
Radioisotope labeling experiment.
The cell extracts of H. thermophila and L. acidiphila were incubated with radioisotope-labeled sodium bicarbonate (NaH14CO3) with succinyl-CoA, Mg ATP, and NADH under H2 gas (100%, 1 atm), and the reaction products were analyzed by HPTLC. Citrate, pyruvate, and acetyl-CoA were identified as the major products with radioactivity in both deep-sea Epsilonproteobacteria (data not shown). These results support the operation of the rTCA cycle by these deep-sea Epsilonproteobacteria via the reductive reaction of succinyl-CoA coupled with carbon assimilation.
Enzymatic activities of energy metabolisms.
The key enzymatic activities of potential energy metabolisms were examined in cell extracts of Epsilonproteobacteria (Table 3). It is assumed that soluble hydrogen uptake hydrogenase is derived from cytoplasmic [NiFe] hydrogenase (group III) and/or from membrane-anchored, periplasmically oriented [NiFe] hydrogenase (group I) (62). The group I [NiFe] hydrogenase is usually membrane anchored, but its two hydrophilic subunits (HynS and HynL [previously HydA and HydB]) are the catalytic subunits and might be incorporated into the soluble fraction. Since the activity of the group III [NiFe] hydrogenase is coupled with an acceptor such as NAD or NADP (62), soluble hydrogenase activity was examined by using MV or NAD as an acceptor. The acceptor:MV hydrogenase activity was detected in many of the deep-sea Epsilonproteobacteria tested (Table 3). The occurrence of soluble hydrogenase activity was clearly restricted to the hydrogen-oxidizing Epsilonproteobacteria (Table 3).
The energy metabolisms of reduced sulfur compounds are completely unknown within the Epsilonproteobacteria. Some deep-sea Epsilonproteobacteria obtain energy solely by the oxidation of reduced sulfur compounds, in most cases via complete oxidation to sulfate (23, 24). In this study, we focused on TSO and sulfite oxidation pathways. TSO include enzymes catalyzing the oxidation of thiosulfate to tetrathionate (thiosulfate:acceptor oxidoreductase) or to sulfite (rhodanese). While the involvement of rhodanese in respiratory sulfur-oxidizing metabolism has been questioned, thiosulfate:acceptor oxidoreductase might play an important role in the sulfur oxidation of various sulfur-oxidizing chemolithoautotrophs (14, 15, 28). In addition, the complete oxidation of the reduced sulfur compounds to sulfate could accompany the enzymatic oxidation of sulfite. For the oxidation of sulfite, direct and indirect pathways are known, and SOR and APSR catalyze direct and indirect sulfite oxidation, respectively (27).
Among the deep-sea Epsilonproteobacteria tested, SOR activity was detected in many but neither APSR nor TSO activities were found (Table 3). In contrast, T. thermophila had both APSR and TSO activities but no SOR activity (Table 3). As observed with soluble hydrogenase activity, SOR activity was detected only in deep-sea Epsilonproteobacteria that were able to oxidize reduced sulfur compounds concomitantly with growth.
Zymogram of soluble hydrogenases.
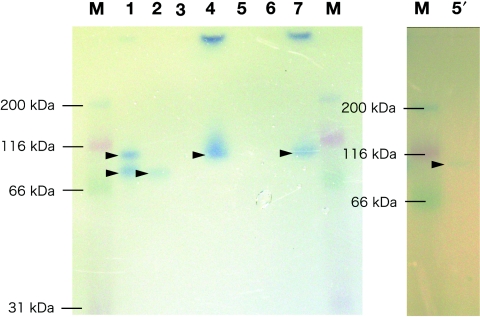
In order to check the isoform of soluble hydrogenase in deep-sea Epsilonproteobacteria, zymograms of soluble hydrogenase in native PAGE were conducted (Fig. 2). The hydrogen-oxidizing deep-sea Epsilonproteobacteria other than H. thermophila had single soluble hydrogenases (Fig. 2). H. thermophila had a high-molecular-weight and a low-molecular-weight hydrogenase (Fig. 2). Although the predicted molecular weight in the native PAGE was not precise, it is possible that the molecular weights of these two types of hydrogenase correspond to the molecular weights of two hydrophilic subunits of previously identified HydAB (HynSL) from the Epsilonproteobacteria. Alternatively, the two types of hydrogenase shown in the native PAGE may represent the presence or absence of an additional small subunit such as cytochrome b (6). The zymogram experiment indicated that at least one soluble hydrogenase was commonly distributed among the hydrogen-oxidizing deep-sea Epsilonproteobacteria.
FIG. 2.
Zymogram of soluble acceptor:MV hydrogenases among the deep-sea Epsilonproteobacteria. Each cell extract (100 μg protein for lanes 1 to 7) was run in a 4 to 20% (wt/vol) gradient native polyacrylamide gel. Lanes: 1, H. thermophila; 2, “S. paralvinella”; 3, S. autotrophica; 4, L. acidiphila; 5, N. salsuginis; 6, S. lithotrophicum; 7, T. micantisoli; 5′, N. salsuginis (500 μg protein); M, marker (Kaleidoscope prestained standards). Arrowheads indicate the visualized hydrogenase bands.
Genetic characterization of carbon metabolisms.
PCR amplification of the genes for ACL (aclB), POR (porAB), OGOR (oorAB), and RubisCO (cbbL and cbbM) was performed on these deep-sea Epsilon- and Gammaproteobacteria (Table 2). Although not all primer sets amplified all Epsilonproteobacteria, gene fragments of aclB, porAB, and oorAB were successfully recovered from the deep-sea Epsilonproteobacteria (Table 2). No gene fragments of cbbL or cbbM were amplified from the deep-sea Epsilonproteobacteria. In contrast, no gene fragments of aclB, porAB, or oorAB were recovered from the genomic DNA of T. thermophila, but gene fragments of both cbbL and cbbM were recovered (Table 2). The amplification pattern of gene fragments completely matched the activity pattern of the enzymes among all the deep-sea Epsilon- and Gammaproteobacteria. This provides clear evidence of carbon assimilation via the rTCA cycle in the deep-sea Epsilonproteobacteria on both the enzymatic and genetic levels.
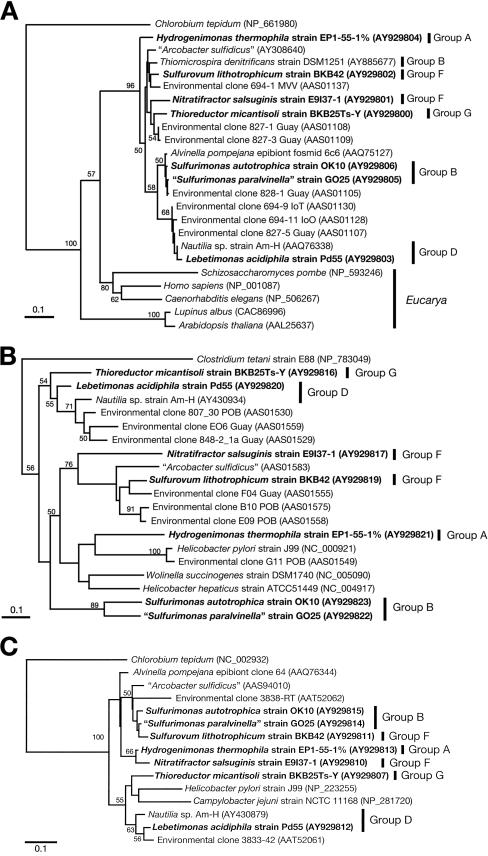
Phylogenetic analyses of the gene sequences of ACL, POR, and OGOR demonstrated topologies which differed from that inferred from the 16S rRNA gene sequences (Fig. 3). As indicated in Fig. 1, the phylogenetic tree based on the 16S rRNA gene sequences revealed distinctive affiliations for subgroups of the Epsilonproteobacteria. In the phylogenetic tree of aclB, however, group B (T. denitrificans), group F (Sulfurovum lithotrophicum and N. salsuginis), and group G (T. micantisoli) were clustered together (Fig. 3A), and in the porA tree, phylogenetic affiliations of group D and group G, group F and “Arcobacter sulfidicus,” and group A and Helicobacter and Wolinella species were demonstrated (Fig. 3B). Although the phylogenetic tree of oorA was constructed based on very small protein fragments (35 amino acids) and was not statistically reliable, the tree topology was also different from that of the 16S rRNA genes (Fig. 3C). The phylogenetic analyses of aclB and porA provided important insights into the phylogenetic affiliation of the environmental clones with the cultured deep-sea Epsilonproteobacteria. Most of the environmental clones previously recovered from Alvinella pompejana tubes and chimneys were phylogenetically associated with groups B, D, F, and G in the aclB tree and with groups D and F in the porA tree (Fig. 3). In culture-independent molecular phylogenetic surveys using 16S rRNA genes, a diversity of environmental rRNA gene clones were, for the most part, affiliated with groups B, D, and F (10, 20, 31, 36, 42, 43, 54, 55). The sequences of the rTCA cycle genes from the deep-sea Epsilonproteobacteria would be a good phylogenetic index for identifying the environmental clones of the genes.
FIG. 3.
Phylogenetic distance trees of representative bacterial strains inferred from the deduced amino acid sequences of the aclB (A), porA (B), and oorA (C) genes by the neighbor-joining method using 96 (aclB), 60 (porA), or 35 (oorA) homologous residues for each organism. The deep-sea epsilonproteobacterial strains examined in this study are boldfaced. Bootstrap values of >50% for the branches are shown (based on 100 replicates). Each bar represents the indicated number of changes per amino acid position. Database accession numbers are given in parentheses.
Genetic characterization of hydrogenase.
Based on previous genetic and physiological studies of the hydrogen respiration of the epsilonproteobacterium Wolinella succinogenes, it was demonstrated that a [NiFe] hydrogenase with two hydrophilic subunits (HydA and HydB) and one hydrophobic subunit (HydC) was the only hydrogenase in W. succinogenes (18, 19). In addition, the results of genome-sequencing analyses of four Epsilonproteobacteria, Helicobacter pylori strain 26695 (60), H. pylori strain J99 (2), Campylobacter jejuni strain NCTC11168 (41), and W. succinogenes strain DSMZ1740 (4), showed that the hydABCD operon (presently proposed as the hyn operon) (62) is the only gene cluster encoding hydrogen uptake within the Epsilonproteobacteria. Since most of the deep-sea Epsilonproteobacteria tested in this study contained at least one soluble hydrogenase, as suggested from the results of the zymogram assay (Fig. 2), we assumed that the soluble hydrogenases were potential hyn operon-encoded group I hydrogenases and examined the occurrence of hynS and hynL genes in the deep-sea Epsilonproteobacteria tested (Table 2).
Primer sets designed to bridge hynS and hynL genes were used to successfully amplify the gene fragments of hynS and hynL from the genomic DNAs of H. thermophila, “S. paralvinella,” and N. salsuginis (Table 2). The hynL fragments of L. acidiphila and T. micantisoli were also recovered by using hynL-targeting primers (Table 2). Gene fragments of hynS and hynL were not amplified from the deep-sea Epsilon- and Gammaproteobacteria lacking soluble hydrogenase activity and cellular hydrogen oxidation ability (Table 2). Since H. thermophila showed two soluble hydrogenases in the native PAGE (Fig. 2), the gene fragments amplified by all primer sets for H. thermophila were cloned and 25 clones of each primer set were sequenced. All the DNA sequences matched the DNA sequence of the longest fragment (data not shown). Hence, the hyn operon might be present singly in the genomic DNA of H. thermophila. The PCR amplification patterns of the hynS and hynL genes provided evidence that the hydrogen-dependent energy metabolism of the deep-sea Epsilonproteobacteria may be genetically attributed to the occurrence of the hyn operon.
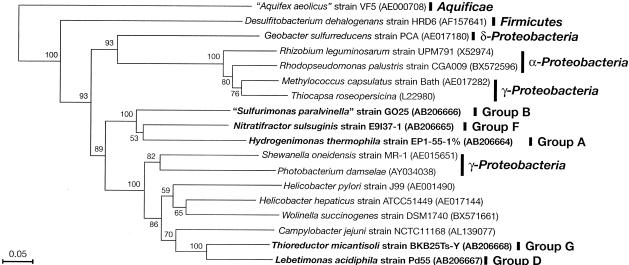
Phylogenetic analysis of the hynL gene sequences indicated a novel phylogenetic trait of the group I hydrogenase: hynL sequences from the gammaproteobacteria Photobacterium damselae and Shewanella oneidensis were grouped within the epsilonproteobacterial hynL sequences (Fig. 4). Since other gammaproteobacterial hynL sequences, such as those from Methylococcus capsulatus and Thiocapsa roseopersicina, were not clustered with the epsilonproteobacterial hynL, these gammaproteobacterial hynL sequences were most likely derived from the epsilonproteobacterial hynL by means of lateral gene transfer. The deepest lineage of epsilonproteobacterial hynL consisted of group A (H. thermophila), group B (“S. paralvinella”), and group F (N. salsuginis), and the other lineage included group D (L. acidiphila) and group G (T. micantisoli). As with the phylogenetic trees of the aclB, porA, and oorA genes, the phylogenetic affiliation of the deep-sea epsilonproteobacterial hynL was considerably different from that of the 16S rRNA genes (Fig. 4).
FIG. 4.
Phylogenetic tree of representative bacterial strains inferred from the deduced amino acid sequences of the hynL genes by the neighbor-joining method using 216 homologous residues for each organism. The deep-sea epsilonproteobacterial strains examined in this study are boldfaced. Bootstrap values of >50% for the branches are shown (based on 100 replicates). Bar, 5 substitutions per 100 residues. Database accession numbers are given in parentheses.
DISCUSSION
Comprehensive analyses of carbon and energy metabolisms were performed for a variety of phylogenetic subgroups of chemolithoautotrophic Epsilonproteobacteria isolated from deep-sea hydrothermal environments. Cellular physiological characterizations of these recently isolated deep-sea Epsilonproteobacteria have provided important insights into carbon and energy utilization profiles (1, 23, 24, 34, 35, 37-39, 53, 56, 58). Most of the deep-sea Epsilonproteobacteria are strict chemolithoautotrophs and use molecular hydrogen and/or reduced sulfur compounds as energy sources. However, the enzymatic and genetic aspects of the carbon and energy metabolisms of these deep-sea Epsilonproteobacteria had not been characterized until this study. Based on the absence of RubisCO activity and the carbon isotopic fractionation between the cellular carbon and the inorganic carbon source in “Arcobacter sulfidicus,” Wirsen et al. (63) first pointed out the possibility of carbon assimilation via the rTCA cycle in the Epsilonproteobacteria. Subsequently, Campbell et al. (8) and Campbell and Cary (9) demonstrated that genes for key enzymes of the rTCA cycle (aclB, porA, and oorA) of deep-sea Epsilonproteobacteria were widely and abundantly distributed in the episymbiotic and epilithic deep-sea hydrothermal vent habitats. Very recently, Hügler et al. (21) demonstrated the first enzymatic and genetic evidence of operation of the rTCA cycle as autotrophic CO2 fixation in the marine chemolithoautotrophic epsilonproteobacterial strains Thiomicrospira denitrificans and “Arcobacter sulfidicus.” Based on these results, it has been strongly suggested that the operative rTCA cycle might serve as a carbon assimilation pathway of the deep-sea Epsilonproteobacteria. In this study, the activities of the key enzymes of the rTCA cycle were determined for representative isolates of deep-sea Epsilonproteobacteria. Gene fragments of aclB, porAB, and oorAB were also recovered from each of the deep-sea Epsilonproteobacteria. The results provide strong enzymatic and genetic evidence of the operation of the rTCA cycle for primary production in the Epsilonproteobacteria tested and suggest that it occurs among all deep-sea chemolithoautotrophic Epsilonproteobacteria. In addition, Thiomicrospira thermophila, a member of the Gammaproteobacteria that has been historically cultivated as a predominant sulfur-oxidizing chemolithoautotroph in deep-sea hydrothermal systems (26, 44, 58), was examined in parallel. This gammaproteobacterium had RubisCO activity and the cbbL and cbbM genes but lacked both rTCA enzymes and genes. Since these deep-sea Epsilon- and Gammaproteobacteria likely inhabit similar niches in relatively low temperature mixing zones, key genetic or proteomic tools described in this study could be used to differentiate carbon assimilation pathways in order to estimate the ecophysiological functions of the deep-sea Epsilon- and Gammaproteobacteria, as partly demonstrated by Campbell and Cary (9). Furthermore, as new rTCA cycle gene sequences from a variety of phylogenetically distinct cultured deep-sea Epsilonproteobacteria become available, molecular ecological surveys of deep-sea hydrothermal habitats will be more efficiently performed via phylogenetic comparisons to closely related isolates whose cellular physiological properties are known.
The enzymatic and genetic aspects of energy metabolisms of deep-sea Epsilonproteobacteria have been much less characterized than those of carbon metabolisms. The hydrogen-dependent energy metabolism of the hydrogen-oxidizing, anaerobically respiring epsilonproteobacterium Wolinella succinogenes has been determined (29, 48). In W. succinogenes, it was demonstrated that a membrane-anchored, periplasm-oriented [NiFe] hydrogenase with two hydrophilic subunits (HydA and HydB) and one hydrophobic subunit (HydC) was the only hydrogenase (18). In addition, throughout the genome sequences of four Epsilonproteobacteria, Helicobacter pylori strain 26695 (60), H. pylori strain J99 (2), Campylobacter jejuni strain NCTC11168 (41), and W. succinogenes strain DSMZ1740 (4), the hyn operon (62) was the only gene found encoding group I [NiFe] hydrogenases. In this study, the soluble acceptor:MV hydrogenase activity was detected only in the hydrogen-oxidizing deep-sea Epsilonproteobacteria (Table 3) and was identified as a single isozyme in all the isolates other than H. thermophila (Fig. 2). The genetic incidence of the hyn operon completely matched the enzymatic incidence of the soluble hydrogenase, and both hydrogenase activities and genes were found only in the deep-sea Epsilonproteobacteria capable of growing by hydrogen oxidation (Tables 2 and 3). Based on the culture-dependent and -independent characterizations of the distribution, diversity, and metabolic versatility of deep-sea Epsilonproteobacteria, Nakagawa et al. (39) have demonstrated the lack of substantial correlation between the utilization of electron donors/acceptors and 16S rRNA phylogeny, particularly with strains of groups A, B, and F. In addition, the importance of molecular hydrogen as the principal energy source for deep-sea Epsilonproteobacteria has been demonstrated (39). Thus, the hyn operon has great potential for genetic elucidation of hydrogen-dependent energy metabolisms within the complex in situ communities of deep-sea Epsilonproteobacteria.
The enzymatic characterization in the present study also provides new insights into energy metabolisms of reduced sulfur compounds in the deep-sea Epsilonproteobacteria. Recently, increasing knowledge of sulfur oxidation mechanisms has been accumulated, mostly by genome-sequencing analyses with Paracoccus and Rhodopseudomonas species within the Alphaproteobacteria and Allochromatium and Acidithiobacillus species within the Gammaproteobacteria (14, 15, 27). Several potential pathways for the oxidation of reduced sulfur compounds, linked with the Sox system, the oxidative dissimilatory sulfite reductase (DSR) reaction, and sulfite oxidation systems such as SOR, APSR, and ATP sulfurylase, have been proposed for these sulfur-oxidizing bacteria (14, 15, 27). However, even after the genome-sequencing analyses of four Epsilonproteobacteria (2, 4, 41, 60), no potential homologs for the Sox families, DSR, or APSR have been identified. In this study, we focused on the TSO and the sulfite oxidation pathways, because some of the sulfur-oxidizing isolates of deep-sea Epsilonproteobacteria obtain energy preferentially by the oxidation of thiosulfate to sulfate (23, 24). The sulfur-oxidizing deep-sea Epsilonproteobacteria had SOR activity for the direct sulfite oxidation pathway but no APSR activity for the indirect pathway. The incidence of SOR activity was restricted to the deep-sea Epsilonproteobacteria capable of energy metabolism with reduced sulfur compounds. However, they lacked TSO activity. In contrast, T. thermophila had both APSR and TSO activities but no SOR activity. As in the case of carbon dioxide and molecular hydrogen metabolisms, the incidence of the sulfur-metabolizing enzyme activities can probably be attributed to the incidence of the genes coding for each of the enzymes in deep-sea Epsilon- and Gammaproteobacteria.
The present study significantly strengthens the enzymatic and genetic aspects of deep-sea epsilonproteobacterial carbon and energy metabolisms, although genetic characterization of the sulfur oxidation mechanisms must still be conducted. The metabolisms of the deep-sea Epsilonproteobacteria were significantly different from that of the chemolithoautotrophic gammaproteobacterium, even though they occupy similar niches within deep-sea hydrothermal environments. This study demonstrates the utility of enzymatic and genetic testing to specifically characterize the ecophysiological functions of these chemolithoautotrophic epsilon- and gammaproteobacterial components, which most likely sustain not only unique microbial communities but also chemosynthetic faunal communities in various physical and geochemical niches in deep-sea hydrothermal environments. As demonstrated currently by Suzuki et al. (51) and recently by Campbell et al. (8), the deep-sea Epsilonproteobacteria could also play an important role in endo- and episymbiotic primary production in certain chemosynthetic animals endemic in hydrothermal vents. Our work provides insights important for the elucidation of ecophysiological functions of deep-sea Epsilonproteobacteria in complex biogeochemical processes that occur in deep-sea hydrothermal environments.
Acknowledgments
This work was partially supported by NSF grants to B.J.C. and S.C.C. (OCE-0120648 and EF-0333203).
REFERENCES
- 1.Alain, K., J. Querellou, F. Lesongeur, P. Pignet, P. Crassous, G. Raguenes, V. Cueff, and M.-A. Cambon-Bonavita. 2002. Caminibacter hydrogeniphilus gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing bacterium isolated from an East Pacific Rise hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52:1317-1323. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L.-S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Angert, E. R., D. E. Northup, A.-L. Reysenbach, A. S. Peek, B. M. Goebel, and N. R. Pace. 1998. Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, Kentucky. Am. Mineral. 83:1583-1592. [Google Scholar]
- 4.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugna-Guiral, M., P. Tron, W. Nitschke, K. O. Stetter, B. Burlat, B. Guigliarelli, M. Bruschi, and M. T. Giudici-Orticoni. 2003. [NiFe] hydrogenases from the hyperthermophilic bacterium Aquifex aeolicus: properties, function, and phylogenetics. Extremophiles 7:145-157. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, B. J., C. Jeanthon, J. E. Kostka, G. W. Luther III, and S. C. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, B. J., J. L. Stein, and S. C. Cary. 2003. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 69:5070-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, B. J., and S. C. Cary. 2004. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl. Environ. Microbiol. 70:6282-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corre, E., A.-L. Reysenbach, and D. Prieur. 2001. Epsilon-proteobacterial diversity from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. FEMS Microbiol. Lett. 205:329-335. [DOI] [PubMed] [Google Scholar]
- 11.de Jong, G. A. H., J. A. Tang, P. Bos, S. de Vries, and G. J. Kuenen. 2000. Purification and characterization of a sulfite:cytochrome c oxidoreductase from Thiobacillus acidophilus. J. Mol. Catal. B 8:61-67. [Google Scholar]
- 12.Engberg, J., S. L. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezaki, S., N. Maeda, T. Kishimoto, H. Atomi, and T. Imanaka. 1999. Presence of a structurally novel type ribulose-bisphosphate carboxylase/oxygenase in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J. Biol. Chem. 274:5078-5082. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fritz, G., T. Buchert, and P. M. H. Kroneck. 2002. The function of the [4Fe-4S] clusters and FAD in bacterial and archaeal adenylsulfate reductases. J. Biol. Chem. 277:26066-26073. [DOI] [PubMed] [Google Scholar]
- 17.Gevertz, D., A. J. Telang, G. Voordouw, and G. E. Jenneman. 2000. Isolation and characterization of strains CVO and FWKOB, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66:2491-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, R., J. Simon, F. Theis, and A. Kroger. 1998. Two membrane anchors of Wolinella succinogenes hydrogenase and their function in fumarate and polysulfide respiration. Arch. Microbiol. 170:50-58. [DOI] [PubMed] [Google Scholar]
- 19.Gross, R., and J. Simon. 2003. The hydE gene is essential for the formation of Wolinella succinogenes NiFe-hydrogenase. FEMS Microbiol. Lett. 227:197-202. [DOI] [PubMed] [Google Scholar]
- 20.Haddad, A., F. Camacho, P. Durand, and S. C. Cary. 1995. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl. Environ. Microbiol. 61:1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hügler, M., C. O. Wirsen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ɛ subdivision of Proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inagaki, F., Y. Sakihama, A. Inoue, C. Kato, and K. Horikoshi. 2002. Molecular phylogenetic analyses of reverse-transcribed bacterial rRNA obtained from deep-sea cold seep sediments. Environ. Microbiol. 4:277-286. [DOI] [PubMed] [Google Scholar]
- 23.Inagaki, F., K. Takai, H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing ɛ-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 53:1801-1805. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki, F., K. Takai, K. H. Nealson, and K. Horikoshi. 2004. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ɛ-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54:1477-1482. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, M., S. Takishita, T. Iwasaki, Y. Peerapornpisal, J. Yoshino, T. Kodama, and Y. Igarashi. 2000. Purification and characterization of membrane-bound hydrogenase from Hydrogenobacter thermophilus strain TK-6, an obligately autotrophic, thermophilic, hydrogen-oxidizing bacterium. Biosci. Biotechnol. Biochem. 64:492-502. [DOI] [PubMed] [Google Scholar]
- 26.Jannasch, H. W., C. O. Wirsen, D. C. Nelson, and L. A. Robertson. 1985. Thiomicrospira crunogena sp. nov., a colorless sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Bacteriol. 35:422-424. [Google Scholar]
- 27.Kappler, U., and C. Dahl. 2001. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203:1-9. [DOI] [PubMed] [Google Scholar]
- 28.Kletzin, A., T. Urich, F. Muller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36:77-91. [DOI] [PubMed] [Google Scholar]
- 29.Kroger, A., S. Biel, J. Simon, R. Gross, G. Unden, and C. R. D. Lancaster. 2002. Fumarate respiration of Wolinella succinogenes: enzymology, energetics, and coupling mechanism. Biochim. Biophys. Acta 1553:23-38. [DOI] [PubMed] [Google Scholar]
- 30.Li, L., C. Kato, and K. Horikoshi. 1998. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 31.Lopez-Garcia, P., S. Duperron, P. Philippot, J. Foriel, J. Susini, and D. Moreira. 2003. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5:961-976. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, N., T. Kanai, H. Atomi, and T. Imanaka. 2002. The unique pentagonal structure of an archaeal Rubisco is essential for its high thermostability. J. Biol. Chem. 277:31654-31662. [DOI] [PubMed] [Google Scholar]
- 33.Marmur, J., and P. Doty. 1962. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 5:109-118. [DOI] [PubMed] [Google Scholar]
- 34.Miroshnichenko, M. L., N. A. Kostrikina, S. L'Haridon, C. Jeanthon, H. Hippe, E. Stackebrandt, and E. A. Bonch-Osmolovskaya. 2002. Nautilia lithotrophica gen. nov., sp. nov., a thermophilic sulfur-reducing ɛ-proteobacterium isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 52:1299-1304. [DOI] [PubMed] [Google Scholar]
- 35.Miroshnichenko, M. L., S. L'Haridon, P. Schumann, S. Spring, E. A. Bonch-Osmolovskaya, C. Jeanthon, and E. Stackebrandt. 2004. Caminibacter profundus sp. nov., a novel thermophile of Nautiliales ord. nov. within the class ‘Epsilonproteobacteria’, isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 54:41-45. [DOI] [PubMed] [Google Scholar]
- 36.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1995. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 61:1555-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa, S., F. Inagaki, K. Takai, K. Horikoshi, and Y. Sako. 2005. Thioreductor micantisoli gen. nov., sp. nov., a novel mesophilic, sulfur-reducing chemolithoautotroph within the ɛ-Proteobacteria isolated from the hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55:599-605. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa, S., K. Takai, F. Inagaki, K. Horikoshi, and Y. Sako. 2005. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ɛ-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55:925-933. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa, S., K. Takai, F. Inagaki, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 40.On, S. L. W. 2001. Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: current status, future prospects and immediate concerns. J. Appl. Microbiol. 90:1S-15S. [DOI] [PubMed] [Google Scholar]
- 41.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev., S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 42.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reysenbach, A.-L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruby, E. G., C. O. Wirsen, and H. W. Jannasch. 1981. Chemolithoautotrophic sulfur-oxidizing bacteria from the Galapagos Rift hydrothermal vents. Appl. Environ. Microbiol. 42:317-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudolph, C., G. Wanner, and R. Huber. 2001. Natural communities of novel archaea and bacteria growing in cold sulfurous springs with a string-of-pearls-like morphology. Appl. Environ. Microbiol. 67:2336-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1982. The deficient carbohydrate metabolic pathways and the incomplete tricarboxylic acid cycle in an obligately autotrophic hydrogen-oxidizing bacterium. Agric. Biol. Chem. 46:2341-2345. [Google Scholar]
- 47.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 48.Simon, J. 2002. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 49.Singleton, D. R., and D. W. Smith. 1988. Improved assay for rhodanese in Thiobacillus spp. Appl. Environ. Microbiol. 54:2866-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snaider, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Scheifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki, Y., T. Sasaki, M. Suzuki, Y. Nogi, T. Miwa, K. Takai, K. H. Nealson, and K. Horikoshi. 2005. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 71:5440-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takai, K., Y. Sako, A. Uchida, and Y. Ishida. 1997. Extremely thermostable phosphoenolpyruvate carboxylase from an extreme thermophile, Rhodothermus obamensis. J. Biochem. 122:32-40. [DOI] [PubMed] [Google Scholar]
- 53.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 54.Takai, K., H. Oida, Y. Suzuki, H. Hirayama, S. Nakagawa, T. Nunoura, F. Inagaki, K. H. Nealson, and K. Horikoshi. 2004. Spatial distribution of marine crenarchaeota group I in the vicinity of deep-sea hydrothermal systems. Appl. Environ. Microbiol. 70:2404-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takai, K., T. Gamo, U. Tsunogai, N. Nakayama, H. Hirayama, K. H. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 56.Takai, K., K. H. Nealson, and K. Horikoshi. 2004. Hydrogenimonas thermophila gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing chemolithoautotroph within the ɛ-Proteobacteria, isolated from a black smoker in a Central Indian Ridge hydrothermal field. Int. J. Syst. Evol. Microbiol. 54:25-32. [DOI] [PubMed] [Google Scholar]
- 57.Takai, K., H. Hirayama, T. Nakagawa, Y. Suzuki, K. H. Nealson, and K. Horikoshi. 2004. Thiomicrospira thermophila sp. nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int. J. Syst. Evol. Microbiol. 54:2325-2333. [DOI] [PubMed] [Google Scholar]
- 58.Takai, K., H. Hirayama, T. Nakagawa, Y. Suzuki, K. H. Nealson, and K. Horikoshi. 2005. Lebetimonas acidiphila gen. nov., sp. nov., a novel thermophilic, acidophilic, hydrogen-oxidizing chemolithoautotroph within the ‘Epsilonproteobacteria’, isolated from a deep-sea hydrothermal fumarole in the Mariana Arc. Int. J. Syst. Evol. Microbiol. 55:183-189. [DOI] [PubMed] [Google Scholar]
- 59.Takeda, Y., F. Suzuki, and H. Inoue. 1969. ATP citrate lyase (citrate-cleavage enzyme). Methods Enzymol. 13:153-160. [Google Scholar]
- 60.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H.-P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 61.Tuttle, J. H., J. H. Schwartz, and G. M. Whited. 1983. Some properties of thiosulfate-oxidizing enzyme from marine heterotroph 16B. Appl. Environ. Microbiol. 46:438-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 63.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]