Abstract
The Toll/NF-κB pathway is a common, evolutionarily conserved innate immune pathway that modulates the responses of animal cells to microbe-associated molecular patterns (MAMPs). Because MAMPs have been implicated as critical elements in the signaling of symbiont-induced development, an expressed sequence tag library from the juvenile light organ of Euprymna scolopes was used to identify members of the Toll/NF-κB pathway. Full-length transcripts were identified by using 5′ and 3′ RACE PCR. Seven transcripts critical for MAMP-induced triggering of the Toll/NF-κB phosphorylation cascade have been identified, including receptors, signal transducers, and a transcription factor. Further investigations should elucidate the role of the Toll/NF-κB pathway in the initiation of the beneficial symbiosis between E. scolopes and Vibrio fischeri.
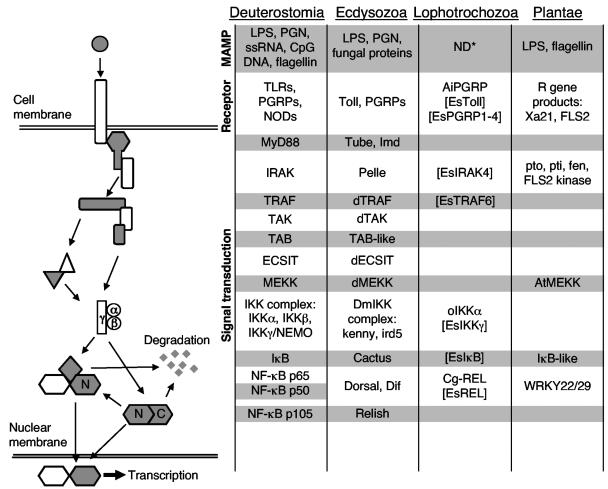
Microbe-associated molecular patterns (MAMPs), such as lipopolysaccharide (LPS), peptidoglycan (PGN), and CpG DNA, are active in initiating innate immune responses in both animals and plants (32, 39, 70). In such responses, the binding of MAMPs to receptors often triggers signal transduction pathways, including the Toll/NF-κB pathway, that have striking similarities among these phylogenetically diverse organisms (Fig. 1) (5, 30, 51). The highly conserved Toll/NF-κB signaling pathway has been studied extensively for its role in microbial pathogenesis in two of the three major branches of the animal kingdom, the vertebrates (Deuterostomia) and the insects (Ecdysozoa) (1, 21, 64). The NF-κB family proteins exist in unstimulated cells of these animals as homo- or heterodimers that are sequestered in the cytoplasm by a member of the Inhibitor of NF-κB (IκB) proteins. Through interaction with a receptor, MAMPs elicit an intracellular phosphorylation cascade. This phosphorylation cascade converges on, and activates, a complex of serine kinases termed IκB kinases (IKKs) (35). The IKK complex phosphorylates serine residues of IκB, leading to its ubiquitination and degradation by the proteosome (25). Degradation of IκB allows the NF-κB dimer to translocate into the nucleus and activate transcription of target genes possessing the κB promoter site. Although evidence is available for the presence of the NF-κB pathway in a mollusk, the Pacific oyster (20, 28, 48), the signaling pathways activating the immune responses in mollusks and other Lophotrochozoa, the members of which form the third branch of the animal kingdom (2, 29, 59), are largely unknown (Fig. 1).
FIG. 1.
The signaling pathways activating the innate immune responses of plants and animals are similar. NF-κB p105 and Relish have a REL homology domain located at their N terminus (N) and an IκB-like domain at their C terminus. The C terminus domain can be proteolytically cleaved and degraded. Brackets enclose pathway members identified in the present study. The molecules listed in the figure represent full-length sequences. Ai, Argopecten irradians; At, Arabidopsis thaliana; C, C terminus; Cg, Crassostrea gigas; CpG DNA, unmethylated CG dinucleotides prevalent in bacterial DNA; d, drosophila; ECSIT, evolutionarily conserved signaling intermediate in Toll pathways; Es, Euprymna scolopes; IκB, inhibitor of NF-κB; IKK, IκB kinase; Imd, immune deficiency; IRAK, interleukin-1-receptor-associated kinase; ird, immune response deficient; LPS, lipopolysaccharide; MEKK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase; MyD88, myeloid differentiation primary response gene 88; N, N terminus; ND*, not determined that MAMPs activate this pathway in this superphylum; NEMO, NF-κB essential modulator; NF-κB, nuclear factor κB; NIK, NF-κB inducing kinase; NOD, nucleotide-binding oligomerization domain protein; o, oyster; PGN, peptidoglycan; PGRP, PGN recognition proteins; R gene, plant resistance genes; REL, proteins containing regions of homology to the rel oncogene, the definition of the NF-κB family of genes; ssRNA, single-stranded RNA; TAB, TAK-binding protein; TAK, transforming growth factor β-activated kinase 1; TLR, Toll-like receptor; Tollip, Toll interacting protein; TRAF, tumor necrosis factor receptor-associated factor.
Euprymna scolopes is a cephalopod mollusk that has been used as a model for the study of the establishment and maintenance of beneficial animal-bacterial symbioses (53). This Hawaiian squid species harbors the marine luminous bacterium Vibrio fischeri in a complex light organ in the center of its body cavity. Several studies of this association have implicated MAMPs as the molecules that mediate the early symbiont-induced development of the juvenile light organ. Specifically, the PGN fragments released by environmental bacteria induce the newly hatched, uncolonized juvenile E. scolopes to secrete mucus from a complex field of epithelial cells on the surface of the organ (52, 54). After V. fischeri gathers in this mucus outside the light organ, it enters pores and travels down ducts to epithelium-lined crypt spaces, where it induces mucus shedding by the crypt epithelial cells (52). Once inside the crypts, the symbiont also triggers the cessation of mucus shedding by the superficial field of ciliated epithelial cells (52) and loss of this field over the first 4 days of the symbiosis by apoptosis (24) and regression (18, 49). Experimental analysis of these developmental events has demonstrated that the synergistic interactions of derivatives of symbiont LPS and PGN are capable of inducing the exact pattern of V. fischeri-induced light organ morphogenesis (23, 39).
Both biochemical and cellular evidence have suggested that, during the complex program of early host light organ development, V. fischeri MAMPs signal and activate host cells through pathways either analogous or homologous to the Toll/NF-κB signaling pathway. For example, mucus secretion, as a response to interactions with bacteria, is controlled by the Toll/NF-κB pathway in other systems (34, 40, 63). In addition, the activities of halide peroxidase (58, 65, 71) and nitric oxide synthase (16) have been implicated in the dynamics of the squid-vibrio system, and the transcription of the genes that encode these proteins is either directly or indirectly activated by NF-κB in other hosts (4, 60). At the cellular level, the recruitment of macrophage-like hemocytes into the light organ (39) and apoptosis of the cells of the superficial ciliated epithelium (24) are types of cellular behaviors modulated by activity of the NF-κB pathway in many host-pathogen interactions (13, 33, 36, 41, 55).
In the present study, we used juvenile light organ expressed sequence tag (EST) libraries to search for transcripts encoding proteins of the Toll/NF-κB pathway and rapid amplification of cDNA ends (RACE) PCR to obtain full-length sequences. Our data provide evidence that the pathway is present in E. scolopes, a finding that paves the way for the study of its regulation in beneficial animal-bacterial interactions using the squid-vibrio model.
MATERIALS AND METHODS
General procedures.
Adult E. scolopes organisms were collected from shallow sand flats of Oahu, Hawaii, and breeding colonies were maintained as described previously (18, 23). Upon hatching, the juvenile E. scolopes organisms were rinsed and placed individually in 5 ml of either offshore seawater, which does not contain sufficient quantities of V. fischeri to result in colonization, or offshore seawater containing 5 × 103 cells ml−1 of wild-type V. fischeri strain ES114 (10). Colonization of the light organ was monitored by using a photometer (TD-20/20 luminometer; Turner Designs, Sunnydale, CA) to measure bacterial luminescence emitted by each animal. Unless otherwise stated, reagents used were purchased from Sigma-Aldrich (St. Louis, MO).
Molecular analysis.
Candidate sequences were identified by analyses of a light organ EST database that had been constructed by using juvenile E. scolopes light organs (EST sequences available through The Wellcome Trust Sanger Institute and European Bioinformatics Institute [http://trace.ensembl.org/]). The sequences from the EST library were translated and compared to protein sequences already in GenBank by using the BlastX algorithm of the Blastcl3 netclient software (NCBI [ftp://ftp.ncbi.nih.gov/BLAST/blastcl3]) (3). For use in cloning and RACE (rapid amplification of cDNA ends) PCR techniques, total RNA was extracted from 100 juvenile light organs by using the MasterPure complete DNA and RNA purification kit (Epicentre, Madison, WI) according to the manufacturer's protocol. 5′- and 3′-RACE PCR was performed by using the SMART RACE cDNA amplification kit including the Advantage II PCR kit (BD Biosciences, Palo Alto, CA) and the FirstChoice RLM-RACE kit (Ambion, Austin, TX) according to the manufacturers' instructions. 5′- and 3′-RACE-ready cDNA was produced by using 1 μg of symbiotic juvenile light organ mRNA. RACE primers were constructed from the EST sequences identified to have similarity to a transcript of interest. The resulting amplification products were gel purified by using a Geneclean kit (Bio 101, Carlsbad, CA) or the QIAquick gel extraction kit (QIAGEN, Valencia, CA). Purified RACE products were ligated into pGEM-T Easy Vector (Promega, Madison, WI). Calcium-competent cells of Escherichia coli DH5α were transformed with the resultant plasmids. Recombinant colonies were screened for the insert of the predicted length, and plasmids were prepared for sequencing by using QIAprep spin miniprep kit columns (QIAGEN). PCR products were sequenced from the recombinant plasmid by using T7 and SP6 primers (Promega, Madison, WI).
Sequence analysis.
EST library clones that contained transcripts of interest were fully sequenced. In cases where the EST library clone did not contain the full-length transcript, 5′ or 3′ RACE PCR was used to obtain the complete transcript. For each RACE amplification product, a consensus contiguous sequence was generated by aligning all sequenced clones using MacVector and AssemblyLIGN or the DSgene software package (Accelrys, San Diego, CA), ensuring a three times independent PCR coverage per base. The full sequence of the transcript was determined by (i) the inability of 5′ RACE PCR to amplify any further sequence, coupled with the presence of in-frame stop codons upstream of the prospective methionine start site at the 5′ end, and (ii) the presence of a poly(A) tail following the prospective stop codon at the 3′ end of the transcript. The resulting sequence was analyzed by BLAST searches of GenBank using the default settings (3). Potential open reading frames (ORFs) were predicted by alignment with sequences in GenBank and codon preference analysis (GCG Sequence Analysis; Genetics Computer Group, Inc.). Predicted protein sequences were generated by MacVector or DSgene. The predicted proteins were compared to protein sequences in GenBank by using conserved domain searches within BLAST (44) and SMART (61), secondary structure prediction (Jpred) (14), and alignment. Bootstrapped (1,000 iterations) neighbor-joining trees were constructed based on the Kimura two-parameter correction by using CLUSTAL X (69) with gapped positions included. Maximum-likelihood analyses were also performed on these data using the default parameters of the program proML with global rearrangement in the PHYLIP (version 3.63) suite of programs (22). Trees constructed by the neighbor-joining and maximum-likelihood methods were compared using the Kishino-Hasegawa-Templeton test in proML (38).
RESULTS
Analysis of the EST library showed that the transcriptome of the light organ of E. scolopes contains several clusters of transcripts that have high similarity to molecules of the Toll/NF-κB pathway (Table 1 and Fig. 1), including molecules of the receptor complex, signaling molecules, and proteins with high similarity to members of the NF-κB family. The transcripts of genes that are regulated by the Toll/NF-κB pathway in other systems are also present (Table 1 and Fig. 1). With the exception of peptidoglycan recognition protein 3 (PGRP3), the 3′ ESTs are similar to the 3′ region of the most similar molecule in the database, adding to the confidence of their identity. The PGRP3 3′ EST was not found to have sequence similarity with sequences in GenBank because it included only a small portion of the ORF. This EST was identified as a candidate PGRP by a BLASTn comparison of E. scolopes PGRP1, -2, and -4 against the entire squid 3′ EST database.
TABLE 1.
E. scolopes 3′ ESTs most similar to the molecules of the Toll/NF-κB pathwaye
| Most similar complete sequence in the database
|
E. scolopes 3′ EST
|
||||
|---|---|---|---|---|---|
| Protein | Organisma | Protein length (aa) | Length (bp) | Region of hitb (aa) | E value |
| Receptors | |||||
| Toll-like receptor | Apis mellifera | 1,248 | 691 | 721-947 | 2e−29 |
| PGRP1 | Apis mellifera | 172 | 785 | 140-169 | 1e−2 |
| PGRP2 | Xenopus laevis | 182 | 802 | 28-180 | 3e−41 |
| PGRP3 | No hitc | 760 | |||
| PGRP4 | Argopecten irradians | 189 | 727 | 15-188 | 6e−42 |
| LBP | Crassostrea gigas | 477 | 785 | 267-473 | 1e−8 |
| Signaling molecules | |||||
| Tollip | Mus musculus | 274 | 794 | 38-273 | 1e−66 |
| IRAK4 | Homo sapiens | 460 | 748 | 261-458 | 1e−36 |
| TRAF6 | Rattus norvegicus | 738 | 716 | 522-715 | 6e−35 |
| ECSIT | Mus musculus | 435 | 678 | 214-393 | 8e−22 |
| MEKK | Dictyostelium discoideum | 942 | 634 | 287-429 | 2e−38 |
| IKKγ/NEMO | Homo sapiens | 419 | 697 | 235-416 | 4e−15 |
| IκBα | Gallus gallus | 318 | 799 | 118-275 | 5e−28 |
| NF-κB molecules | |||||
| RelA/p65 | Crassostrea gigas Rel1 | 615 | 618 | 313-475 | 2e−15 |
| RELISH/p105 | Mus musculus | 971 | 736 | 732-894 | 2e−11 |
| Regulated genes | |||||
| MMPd | Homo sapiens | 1,037 | 658 | 839-1,034 | 9e−36 |
| Mucin | Homo sapiens | 2,448 | 697 | 2,253-2,390 | 2e−3 |
| Nitric oxide synthase | Aplysia californica | 1,387 | 505 | 1,223-1,348 | 6e−19 |
Apis mellifera, honeybee; Aplysia california, California sea hare; Argopecten irradians, bay scallop; Bombyx mori, silk moth; Crassostrea gigas, Pacific oyster; Dictyostelium discoideum, slime mold; Gallus gallus, chicken; Homo sapiens, human; Mus musculus, house mouse; Rattus norvegicus, brown rat; Xenopus laevis, African clawed frog.
Region of most similar complete sequence in the database that matches the E. scolopes EST.
Identified by BLASTn comparison of E. scolopes PGRP1, -2, and -4 against the entire squid 3′ EST database.
Matrix metalloproteinase.
Of the 3′ ESTs, ten molecules that are key components of the Toll/NF-κB pathway were investigated further, namely, a Toll-like receptor (TLR), the four PGRPs, interleukin-1 receptor-associated kinase 4 (IRAK4), TNF-α receptor associated factor 6 (TRAF6), inhibitor of κB kinase γ (IKKγ), inhibitor of κB α (IκBα), and REL (Table 2). In all cases, sequencing of the entire coding region improved the similarity to a sequence in the database. However, in the case of Toll, the PGRPs, IRAK4, TRAF6, IκBα, and REL, the best hit to the entire transcript was to the same protein but from a different organism than that of the 3′ EST (Table 2). Trees constructed using the neighbor-joining and maximum likelihood with global rearrangement methods were not found to be statistically different.
TABLE 2.
Full-length transcripts of E. scolopes most similar to key molecules of the Toll/NF-κB pathwayc
| Most similar complete sequence in the database
|
E. scolopes complete sequence
|
||||
|---|---|---|---|---|---|
| Protein | Organisma | Protein length (aa) | Coding sequence length (aa) | Region of hitb (aa) | E value |
| Receptors | |||||
| Toll-like receptor | Drosophila melanogaster | 1,346 | 1,191 | 55-1212 | 1e−122 |
| PGRP1 | Argopecten irradians | 189 | 207 | 15-183 | 9e−42 |
| PGRP2 | Argopecten irradians | 189 | 201 | 15-183 | 4e−45 |
| PGRP3 | Anopheles gambiae | 188 | 243 | 25-187 | 7e−42 |
| PGRP4 | Argopecten irradians | 189 | 270 | 15-188 | 6e−42 |
| Signaling molecules | |||||
| IRAK4 | Rattus norvegicus | 520 | 573 | 1-516 | 2e−59 |
| TRAF6 | Danio rerio | 542 | 555 | 53-533 | 3e−66 |
| IKKγ/NEMO | Homo sapiens | 419 | 287 | 154-416 | 5e−20 |
| IκBα | Rattus norvegicus p105 | 1,719 | 339 | 1,283-1,473 | 2e−37 |
| NF-κB molecules | |||||
| RelA/p65 | Crassostrea gigas Rel2 | 614 | 466 | 85-475 | 4e−96 |
Anopheles gambiae, mosquito; Argopecten irradians, bay scallop; Crassostrea gigas, Pacific oyster; Danio rerio, zebrafish; Drosophila melanogaster, fruit fly; Homo sapiens, human; Rattus norvegicus, brown rat.
Region of most similar complete sequence in the database that matches the E. scolopes complete sequence.
TLR.
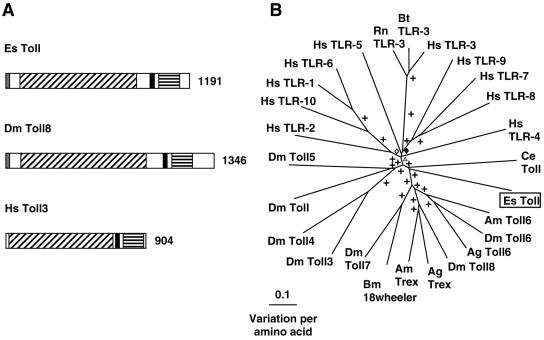
5′ and 3′ RACE revealed a 4,186-bp transcript that contains an ORF encoding a 1,191-amino-acid (aa) protein (Table 2 and Fig. 2). The conserved regions of the E. scolopes Toll-like protein share 26% identity and 46% positive substitutions with Toll-8 from D. melanogaster. The predicted Toll-like protein contains a signal peptide, N-terminal leucine-rich repeat (LRR) domains and a C-terminal Toll/IL-1 receptor (TIR) domain (conserved domain search [CDS] E value of 4e−13) separated by a transmembrane domain. This domain architecture is consistent with the known architecture of previously described Toll-like proteins from both mammals and insects (Fig. 2A and Fig. S1 in the supplemental material). Therefore, the E. scolopes Toll-like protein is predicted to be an integral membrane protein with the N terminus on the extracellular membrane face. Because of the inherent variability of the MAMP-recognizing LRR domains in TLRs (8, 51), phylogenetic comparison of Toll-like sequences results in a tree exhibiting long branch lengths. Consequently, the Toll sequences of only relatively recently diverged organisms group together (for example, the TLR3s from vertebrates; Fig. 2B). Therefore, phylogenetic analysis revealed that the E. scolopes sequence was not closely associated with any other Toll-like sequence in the database.
FIG. 2.
(A) Comparison of Toll-like receptor proteins from E. scolopes, fruit fly, and human. Gray regions indicate the signal peptide, black diagonal shading indicates the LRR region, black regions indicate a transmembrane domain, and black horizontal shading indicates the TIR domain. The number indicates the size of the derived amino acid sequence. Accession numbers are AY956809, NP_524757, and NP_003256, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between Toll-like protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. Symbols on the branches represent percentage bootstrap values: +, 90 to 100%; ⋄, 80 to 89%; ⧫, 70 to 79%; ▵, 60 to 69%. Accession numbers clockwise from E. scolopes are as follows: AY956809, XP_393712, AAF86226, AAL37902, NP_524757, AAL37904, XP_393713, BAB85498, AAF86225, NP_649719, NP_523519, NP_733166, AAF86227, NP_003255, NP_112218, NP_003254, NP_006059, NP_003259, NP_942086, NP_001008664, NP_003256, NP_059138, AAF78035, NP_057694, NP_612564, and AAK37544. Abbreviations: Ag, Anopheles gambiae (mosquito); Am, Apis mellifera (honeybee); Bm, Bombyx mori (silk moth); Bt, Bos taurus (cow); Ce, Caenorhabditis elegans (nematode); Dm, Drosophila melanogaster (fruit fly); Es, Euprymna scolopes (Hawaiian bobtail squid); Hs, Homo sapiens (human); Rn, Rattus norvegicus (rat).
PGRPs.
Four PGRP transcripts—EsPGRP1, EsPGRP2, EsPGRP3, and EsPGRP4—were identified among the 3′ EST sequences.
(i) EsPGRP1.
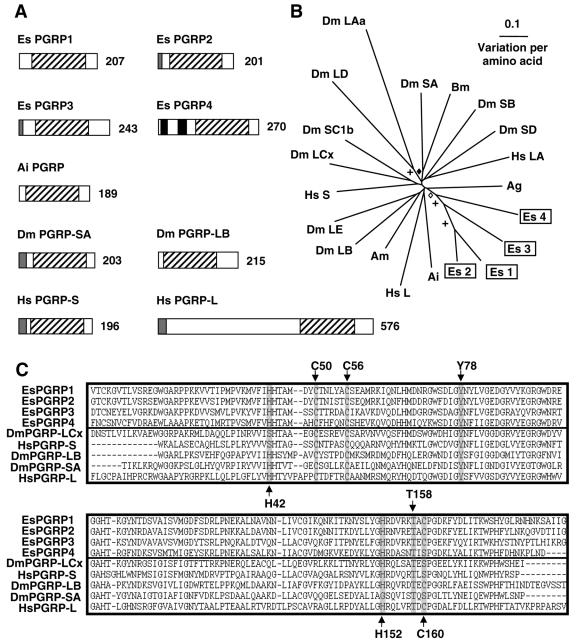
5′ and 3′ RACE revealed a 1,362-bp transcript that is predicted to encode a 207-aa protein (Table 2 and Fig. 3A). The putative protein sequence for EsPGRP1 contains a conserved PGRP domain (CDS E value of 1e−43). Because no other protein domains, such as a signal peptide, were strongly suggested by in silico analysis of the protein structure, EsPGRP1 is predicted to have a cytosolic cellular localization.
FIG. 3.
(A) Comparison of PGRP proteins from E. scolopes, bay scallop, fruit fly, and human sources. Gray regions indicate the signal peptide, black regions indicate a transmembrane domain, and black diagonal shading indicate the PGRP domain. The number indicates the size of the derived amino acid sequence. Reading from left to right and down from the top, accession numbers are AY956811, AY956812, AY956813, AY956814, AAR92030, NP_572727, NP_731576, NP_005082, and AAL05629, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between PGRP protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. Symbols on the branches represent percentage bootstrap values: +, 90 to 100%; ⋄, 80 to 89%; ⧫, 70 to 79%. Values below 70% are not shown. Accession numbers clockwise from E. scolopes PGRP1 are as follows: AY956811, AY956812, AAR92030, AAL05629, XP_395941, NP_731576, AAG32064, NP_005082, NP_729468, AAG23736, NP_788467, AAK00295, NP_572727, BAA77210, CAD89136, CAD89193, NP_443123, EAA10910, AY956814, and AY956813. (C) Alignment of the PGRP domains of the EsPGRPs and those of other organisms. Residues critical to PGRP function are highlighted. Accession numbers from top to bottom are as follows: AY956811, AY956812, AY956813, AY956814, NP_729468, NP_005082, NP_731576, NP_572727, and AAL05629. Abbreviations: Ag, Anopheles gambiae (mosquito); Ai, Argopecten irradians (bay scallop); Am, Apis mellifera (honeybee); Bm, Bombyx mori (silk moth); Dm, Drosophila melanogaster (fruit fly); Es, Euprymna scolopes (Hawaiian bobtail squid); Hs, Homo sapiens (human).
(ii) EsPGRP2.
5′ and 3′ RACE results yielded a 949-bp transcript that contains an ORF encoding a 201-aa protein (Table 2 and Fig. 3A). The sequence of EsPGRP2 is very similar to that of EsPGRP1, sharing 82% identity when their coding nucleotide sequences are aligned. At the protein level, EsPGRP1 and EsPGRP2 share 74% identity. The EsPGRP2 protein contains a PGRP domain (CDS E value of 4e−48). An N-terminal signal peptide sequence is predicted, suggesting that EsPGRP2 is secreted into the extracellular environment, similar to PGRP-SA from Drosophila melanogaster (46).
(iii) EsPGRP3.
5′ RACE PCR completed a 2,156-bp EsPGRP3 transcript encoding a 243-aa protein (Table 2 and Fig. 3A). At the nucleotide level EsPGRP3 does not share significant similarity to any of the other EsPGRPs, but the putative EsPGRP3 protein is strongly similar to that of EsPGRP1 and EsPGRP2 (60% identity) with a somewhat weaker similarity to EsPGRP4 (48% identity). EsPGRP3 protein is predicted to have a signal peptide and a PGRP domain (CDS E value of 6.4e−68). It is also strongly predicted to have a glycosylphosphatidylinositol (GPI)-anchor, which would bind the PGRP to the outer surface of the cell membrane, with a possible GPI-anchor cleavage site at position 216. Therefore, we predict that EsPGRP3 is an extracellular extrinsic membrane protein that may be released into the extracellular environment through cleavage of the GPI-anchor.
(iv) EsPGRP4.
The 5′ and 3′ RACE PCR product sequence indicated that the EsPGRP4 transcript is 1,527 bp encoding a 270-aa protein (Table 2 and Fig. 3A). At the nucleotide level, the coding sequence of EsPGRP4 does not share significant similarity to either EsPGRP1 or EsPGRP2 (using the BLASTn algorithm). The EsPGRP4 protein sequence is about 45 and 52% identical to EsPGRP1 and EsPGRP2, respectively. EsPGRP4 contains a putative PGRP domain (CDS E value of 1e−42). The prediction of two N-terminal transmembrane domains suggests that EsPGRP4 is an integral membrane protein.
When the E. scolopes PGRPs (EsPGRPs) are compared to PGRP molecules from other organisms (Fig. 3B), the E. scolopes molecules group together with strong bootstrap support (>70%). Apart from this cluster and the cluster containing the PGRP-LD and PGRP-LAa from D. melanogaster, there is no other strong bootstrap support for the PGRPs of a single organism grouping together. EsPGRPs also did not group with a PGRP from another mollusk, Argopecten irradians (the bay scallop). These data suggest that the PGRPs identified from the E. scolopes light organ were derived from a single ancestral EsPGRP. Such findings do not preclude the possibility of other EsPGRPs that would cluster outside of those already found, and are derived from a different PGRP.
Some PGRPs have T7 lysozyme-like catalytic activity, including PGRP-LB from D. melanogaster (37, 45), which coordinates a zinc ion required for N-acetylmuramic acid l->alanine amidase (NAMLAA) activity with a conserved tetrad of two histidines, a critical cysteine, and tyrosine residue (H42, Y78, H152, and C160, PGRP-LB numbering). EsPGRP1, EsPGRP2, and EsPGRP3 contain all four zinc coordinating residues in the PGRP-binding domain, suggesting zinc-dependent NAMLAA activity in these three proteins (Fig. 3C). At the homologous position of the critical cysteine 160, EsPGRP4 contains a serine substitution similar to D. melanogaster PGRP-SA, which does not coordinate a zinc ion in the binding pocket (56). It is likely that EsPGRP4 does not act as a catalytic PGRP and is limited to PGN recognition and/or signal transduction functions. All four EsPGRPs contain a pair of cysteines C50 and C56 (using PGRP-LB numbering) at synonymous positions with those essential for the structure of the PGN binding pocket and a highly conserved threonine, T158 (using PGRP-LB numbering) that is required in the binding of PGN by PGRP-LB from D. melanogaster (37).
IRAK4.
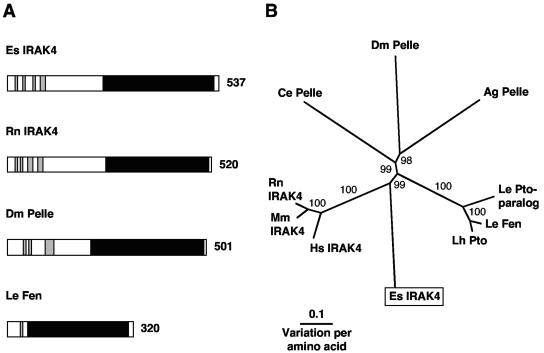
5′ and 3′ RACE PCR of juvenile light organ RNA resulted in a 1,791-bp transcript that contains an ORF encoding a 537-aa protein (Table 2 and Fig. 4). The translated transcript exhibited a C-terminal serine/threonine protein kinase domain (CDS E value of 5e−34) and at least four N-terminal α-helices of a possible death domain (31), characteristic of IRAK proteins (Fig. 4A and Fig. S2 in the supplemental material). When the E. scolopes IRAK4 is compared to IRAK4-like molecules from other organisms, the E. scolopes molecule clusters with the vertebrate IRAK4 protein sequences with strong bootstrap support (Fig. 4B). However, the branch lengths within this cluster indicate that the E. scolopes sequence is separate from the vertebrate IRAK4 cluster.
FIG. 4.
(A) Comparison of IRAK4-like proteins from E. scolopes, rat, fruit fly, and tomato. Black regions indicate the serine/threonine protein kinase domain, and gray regions indicate the alpha helices of a possible death domain. Number indicates size of the derived amino acid sequence. The accession numbers are AY956810, XP_217026, NP_476971, and T07416, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between IRAK4 protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. The numbers on the branches are percent bootstrap values. Accession numbers clockwise from E. scolopes are as follows: AY956810, NP_057207, NP_084202, XP_217026, NP_502587, NP_476971, XP_311931, AAF76311, T07416, and AAK11568. Abbreviations: Ag, Anopheles gambiae (mosquito); Ce, Caenorhabditis elegans (nematode); Dm, Drosophila melanogaster (fuit fly); Es, Euprymna scolopes (Hawaiian bobtail squid); Hs, Homo sapiens (human); Le, Lycopersicon esculentum (tomato); Lh, Lycopersicon hirsutum (tomato); Mm, Mus musculus (mouse); Rn, Rattus norvegicus (rat).
TRAF6.
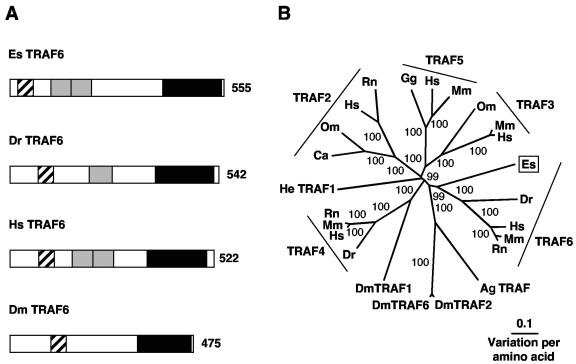
5′ and 3′ RACE resulted in a 2,019-bp transcript that contains an ORF encoding a 555-aa protein (Table 2 and Fig. 5). The derived amino acid sequence of the E. scolopes transcript has a number of characteristic TRAF domains (64) (Fig. 5A and Fig. S3 in the supplemental material). All TRAF proteins share a common C-terminal TRAF domain (CDS E value of 7e−42) required for the interaction with upstream factors such as IRAK4 (12). The putative protein also contains two TRAF-type zinc-finger domains (CDS E value of 2e−7) N-terminal to the TRAF domain and a RING finger motif (CDS E value of e−7) at its extreme N terminus. The N-terminal region of TRAF6 and especially the RING finger motif are required for efficient IKK and NF-κB activation (7, 12, 15). Comparison of the E. scolopes sequence with TRAF sequences from other organisms supports its classification as TRAF6 (Fig. 5B). E. scolopes TRAF6 clusters with the vertebrate TRAF6 sequences with strong bootstrap support. E. scolopes TRAF6 is 33% similar to human TRAF6, whereas only sharing similarities of 26, 28, 26, and 29% to TRAF2, TRAF3, TRAF4, and TRAF5 of humans, respectively. TRAF-like proteins from D. melanogaster and A. gambiae branch one node away from TRAF6 of E. scolopes and vertebrates, and this grouping is also well supported. The branch lengths indicate that E. scolopes, vertebrates, and insects are separate from each other within the TRAF6-like sequence cluster.
FIG. 5.
(A) Comparison of TRAF6 proteins from E. scolopes, zebrafish, human, and fruit fly with characteristic domains indicated. Black hatched shading indicates the RING finger domain, gray regions indicate the TRAF-type zinc finger domains, and black regions indicate the TRAF homology domain. The number indicates size of the derived amino acid sequence. The accession numbers are AY956816, NP_956115, NP_004611, and AAD47895, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between TRAF protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. Numbers on the branches are percentage bootstrap values. Accession numbers clockwise from E. scolopes are as follows: AY956816, NP_956115, NP_004611, P70196, XP_230377, XP_311879, AAD34345, AAD47895, AAD34346, NP_991325, NP_004286, NP_033449, XP_220640, CAE88928, CAC82653, CAD69021, NP_066961, XP_231032, NP_989550, NP_004610, NP_035763, CAD57164, NP_035762, and Q13114. Abbreviations: Ag, Anopheles gambiae (mosquito); Ca, Carassius auratus (goldfish); Dm, Drosophila melanogaster (fruit fly); Dr, Danio rerio (zebrafish); Es, Euprymna scolopes (Hawaiian bobtail squid); Gg, Gallus gallus (chicken); He, Hydractinia echinata (hydrozoa); Hs, Homo sapiens (human); Mm, Mus musculus (mouse); Om, Oncorhynchus mykiss (rainbow trout); Rn, Rattus norvegicus (rat).
IKKγ.
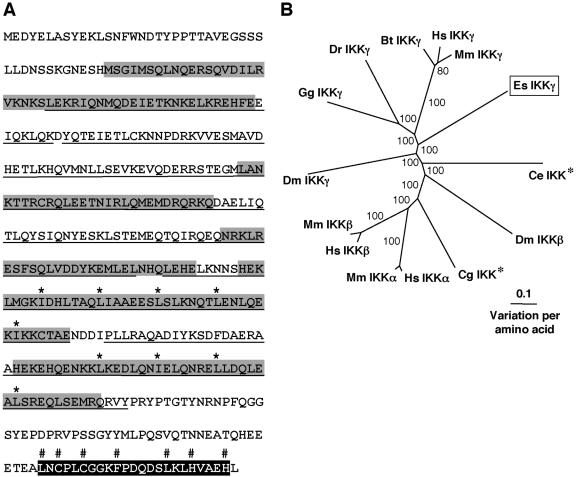
5′ and 3′ RACE PCR amplified a 2,323-bp transcript that contains an ORF encoding a 431-aa protein (Table 2 and Fig. 6). Identification of IKKγ proteins requires secondary structure analysis, as opposed to conserved domain searches, because IKKγ proteins do not contain unique IKKγ domains. IKKγ proteins are predominantly helical with large stretches of coiled-coil structure, including a leucine zipper motif and a zinc finger motif at the C terminus (35, 57). These motifs are likely to be involved in protein-protein interactions. Secondary structure analysis of the derived amino acid sequence (Fig. 6A and Fig. S4 in the supplemental material) indicates that the E. scolopes protein is a canonical IKKγ (35, 57). Similarly, the E. scolopes sequence clusters with IKKγ proteins of vertebrates with strong bootstrap support, although the branch lengths indicate the derived amino acid sequence of E. scolopes is separate from the vertebrate sequences (Fig. 6B). IKKγ from D. melanogaster branches one node away and is separated by a long branch from IKKγ of E. scolopes and vertebrates. The insect, vertebrate, and mollusk proteins form a well-supported IKKγ grouping that is separate from IKKα and IKKβ sequences.
FIG. 6.
(A) The characteristic domains of IKKγ exhibited by the derived amino acid sequence from E. scolopes. The underlined region represents a helical structure containing coiled coil regions (gray shading) and a leucine zipper motif with specific leucines (and hydrophobic isoleucine) marked with an asterisk. The C-terminal zinc finger motif is shaded in black with characteristic amino acids labeled with a “#” symbol. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between IKK protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. Numbers on the branches are percentage bootstrap values. *, The original description does not indicate assignment to α, β, or γ classes of IKK. Accession numbers clockwise from E. scolopes are as follows: AY956817, NP_499831, AAG02485, AAC05683, NP_001269, AAH18243, NP_001547, NP_034676, AAG02486, NP_989567, AAH63986, NP_776779, NP_003630, and AAM54725. Abbreviations: Bt, Bos taurus (cow); Ce, Caernorhabditis elegans (nematode); Cg, Crassostrea gigas (Pacific oyster); Dm, Drosophila melanogaster (fruit fly); Dr, Danio rerio (zebrafish); Es, Euprymna scolopes (Hawaiian bobtail squid); Gg, Gallus gallus (chicken); Hs, Homo sapiens (human); Mm, Mus musculus (mouse).
IκB.
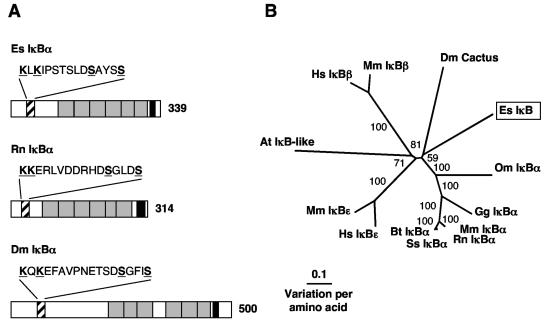
5′ and 3′ RACE resulted in a 1,463-bp transcript that contains an ORF encoding a 339-aa protein (Table 2 and Fig. 7). Similar to other IκB family members, the derived amino acid sequence exhibited six ankyrin repeat domains (CDS E value of 9e−23), an N-terminal regulatory region, and a C-terminal region rich in Pro, Glu/Asp, Ser, and Thr residues (PEST-rich region) (Fig. 7A and Fig. S5 in the supplemental material). Proteins of the IκB family contain either six or seven ankyrin repeats that mediate binding to the REL homology domain and mask the nuclear localization signal of REL family proteins (35). The N-terminal regulatory region is required for stimulus-induced degradation of the protein, the key step in NF-κB activation. Only the “professional” IκBs (74), namely, IκBα, IκBβ, IκBɛ, and Cactus, contain this short peptide stretch centered around two serine residues that can be inducibly phosphorylated (35). Mutations at either serine residue of this region results in the stabilization of the protein (6, 50, 74). The presence of this N-terminal regulatory region, coupled with the inability of 5′ RACE PCR to amplify any further sequence and in-frame stop codons upstream of the prospective start site, provides evidence that the sequence is complete and is not the C terminus of a p105-like or RELISH-like transcript that also has IκB-like characteristics (27, 41, 64). However, the sequence of the regulatory region in the E. scolopes protein has a conserved substitution in the third position, i.e., instead of the DSGXXS characteristic of the vertebrate IκBs and the drosophila homolog Cactus, the sequence of this region of the E. scolopes IκB is DSAXXS. The ubiquitin conjugation site, a lysine residue located 9 to 12 aa N-terminal to the recognition site of professional IκB is, however, shared by the E. scolopes sequence (74). The C-terminal contains a PEST-rich region. PEST sequences are often found in proteins with a rapid turnover rate and may be necessary for the degradation of IκB. For example, in mouse cell lines that were transfected with human IκBα missing the final 41 aa, including the PEST sequence, the mutant IκBα was phosphorylated but not degraded (11). The E. scolopes sequence clustered with IκBα-like proteins (Fig. 7B). Within this IκBα cluster, the putative protein sequence from E. scolopes did not group with either D. melanogaster Cactus or vertebrate IκBα proteins with strong bootstrap support, and branch lengths indicated that the vertebrates, D. melanogaster, and E. scolopes sequences were distinct from each other (Fig. 7B).
FIG. 7.
(A) Comparison of IκB family molecules from E. scolopes, rat, and fruit fly. Black diagonal shading represents the N-terminal regulatory region, with the serines of the phosphorylation site and the lysines of the possible ubiquitin conjugation sites underlined and highlighted in boldface. The ankyrin repeat domains are shaded in gray, and the PEST-rich region is shaded in black. The number indicates the size of the derived amino-acid sequence. The accession numbers are AY956818, XP_343066, and NP_476942, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between IκB protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. The numbers on the branches are percent bootstrap values. The accession numbers clockwise from E. scolopes are as follows: AY956818, CAC85086, Q91974, NP_035037, XP_343066, Q08353, CAD12786, O00221, NP_032716, AAB58262, NP_002494, NP_035038, and NP_476942. Abbreviations: At, Arabidopsis thaliana (thale cress); Bt, Bos taurus (cow); Dm, Drosophila melanogaster (fruit fly); Es, Euprymna scolopes (Hawaiian bobtail squid); Gg, Gallus gallus (chicken); Hs, Homo sapiens (human); Mm, Mus musculus (mouse); Om, Oncorhynchus mykiss (rainbow trout); Rn, Rattus norvegicus (rat); Ss, Sus scrofa (pig).
REL.
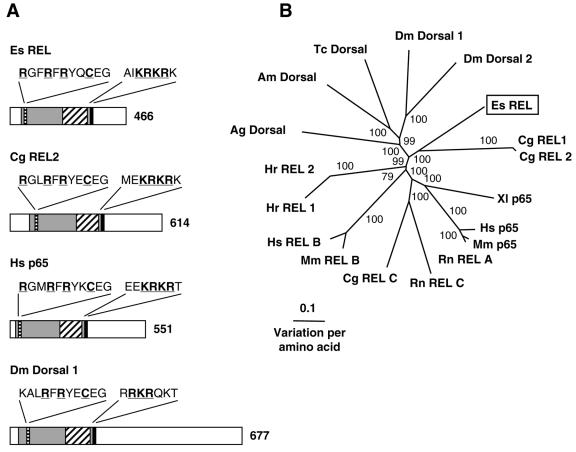
5′ and 3′ RACE of light organ RNA resulted in a 1,557-bp transcript that contains an ORF encoding a 466-aa protein (Table 2 and Fig. 8). The derived amino acid sequence of the transcript exhibited a REL homology domain (RHD), containing an immunoglobulin-like plexin domain (CDS E value of 3e−35) and a conserved nuclear localization motif (Fig. 8A and Fig. S6 in the supplemental material). The RHD (CDS E value of 5e−62) is characteristic of REL family transcription factors and is responsible for dimerization, DNA-binding via the conserved motif RXXRXRXXC and interaction with IκB (35). The transactivation domain, C-terminal of the RHD, is less well-conserved and is thought to be involved in the activation of transcription (41). No regions that are critical for transcriptional activation have been identified within the transactivation domain, apart from the presence of the transactivation domain itself (26, 27, 41, 47). The REL family of proteins can be separated into two distinct classes: class I contains the precursor REL proteins with C-terminal domains that have IκB-like characteristics, such as vertebrate and insect p100, p105, and Relish; class II contains the NF-κB subunit REL proteins, including p65/RelA, RelB, c-REL, v-Rel, and Dorsal (27). Compared to the other class II REL proteins, the E. scolopes REL sequence is relatively short (Fig. 8A). The difference appears to be in the length of the transactivation domain, since the size of the RHD of both proteins is similar. Since there is a 3′ stop codon and 3′ RACE PCR failed to amplify any sequence further 3′ of the poly(A) tail and, since the transactivation domain is known to be less conserved, we conclude that the sequence is complete. Compared to class II REL proteins from other organisms, the E. scolopes sequence clusters with the Crassostrea gigas REL sequences with strong bootstrap support (Fig. 8B). Since the insects and the vertebrate class II REL proteins, respectively, also form groupings that are strongly supported and the branch lengths separating the sequences are long, it is concluded that the mollusk sequences are distinct from the vertebrate and insect class II REL proteins.
FIG. 8.
(A) Comparison of REL family proteins from E. scolopes, oyster, human, and fruit fly. The RHD is indicated by gray shading and contains the following: the conserved residues of the DNA-binding motif (black horizontal shading) underlined and highlighted in boldface; the immunoglobulin-like plexin domain, found in all REL family molecules, indicated by black diagonal shading; and the nuclear localization signal indicated by solid black shading, with conserved residues underlined and highlighted in boldface. The unshaded region that is C-terminal to the RHD represents the transactivation domain. The number indicates size of the derived amino acid sequence. Accession numbers are AY956819, AAK72691, NP_068810, and NP_724052, respectively. (B) Unrooted phylogenetic tree produced by the neighbor-joining method, using Kimura two-parameter distances between REL protein sequences of the organisms named on the tree. Gapped positions were included in the analysis. Numbers on the branches are percentage bootstrap values. Accession numbers clockwise from E. scolopes are as follows: AY956819, AAK72690, AAK72691, Q04865, NP_068810, NP_033071, XP_238994, XP_223688, P16236, NP_033072, Q01201, BAB47172, BAB47173, XP_310177, AAP23055, AAG22858, NP_724052, and NP_724054. Abbreviations: Ag, Anopheles gambiae (mosquito); Am, Apis mellifera (honeybee); Cg, Crassostrea gigas (Pacific oyster); Dm, Drosophila melanogaster (fruit fly); Es, Euprymna scolopes (Hawaiian bobtail squid); Hr, Halocynthia roretzi (ascidian); Hs, Homo sapiens (human); Mm, Mus musculus (mouse); Rn, Rattus norvegicus (rat); Tc, Tribolium castaneum (beetle); XI, Xenopus laevis (African clawed frog).
DISCUSSION
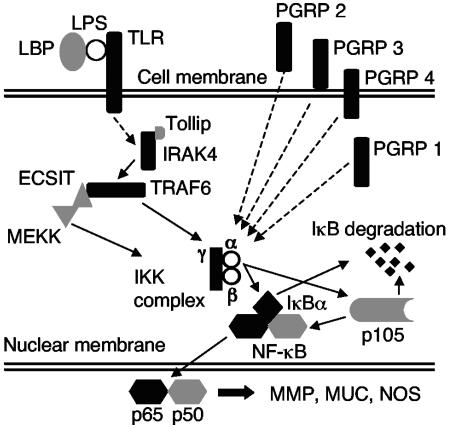
The results of our analyses provide evidence for the presence of several transcripts encoding components of the Toll/NF-κB pathway in E. scolopes, including receptors, signal transducers, and targets of the pathway (Fig. 9). Although this pathway appears to be present, its characteristics suggest certain unique features. This finding is not surprising, since E. scolopes is phylogenetically distant from the other animals for which this pathway has been described in detail.
FIG. 9.
Putative signaling pathway of the evolutionarily conserved Toll/NF-κB pathway utilizing the E. scolopes transcripts identified in the present study. Black shading indicates identification of full-length transcripts, and gray shading indicates identification of partial length transcripts (see Tables 1 and 2). Dashed lines indicate a pathway in which not all of the signaling intermediates have been found in E. scolopes.
Beginning with the receptors, only one TLR was identified in the EST library. This finding was surprising since multiple Tolls and TLRs have been identified in the insects and vertebrates, respectively (9, 68). The light organ may only express a single TLR gene that may be a global, rather than a specific, microbe receptor, although it is possible that other TLR candidates were not recognized from the 3′ EST library. The large genome size of E. scolopes (3.7Gb; T. R. Gregory, Animal Genome Size Database [http://www.genomesize.com]) and the inherent difficulties with working with cephalopod DNA (62, 66) have thus far precluded us from performing reliable Southern blots and genomic DNA amplification to identify any other TLRs. In contrast, several PGRPs were identified, but analyses of the E. scolopes PGRPs revealed that they do not readily fall into the classes of PGRPs identified in insect and mammals (19, 42, 72). Most notably, unlike the PGRPs of other animals, all of the EsPGRPs that we have identified thus far occur in a well-supported group, suggesting that this cohort of proteins was derived from a single ancestral PGRP. Further, the sequence data and structure predictions of the EsPGRPs do not support the classification of these proteins by their biochemical characteristics in the same ways that have been applied to other PGRPs. For example, the characteristics of molecular mass, as well as the patterns of occurrence of signal sequences and transmembrane domains, do not group the EsPGRPs into distinct classes as they do in the insects (72). Because each of the EsPGRPs is predicted to have unique biochemical characteristics, it is likely that they have distinct functions. The finding of four EsPGRPs in the light organ does not preclude the possibility that others exist there or in other parts of the body.
Within the pathway, several elements critical for MAMP-induced triggering of the NF-κB pathway were identified, including IRAK4, TRAF6, IKKγ, IκB-like, and REL-like transcripts (17, 25, 35, 43, 57, 64, 67, 73). In another mollusk, Crassostrea gigas (the Pacific oyster), full-length transcripts of an IKK (20), which appears to be an IKKβ, and a REL-like protein (48) were identified, lending further support for the presence of the pathway in the mollusks. An element not identified in the E. scolopes EST database was a MyD88-like transcript, which in other systems encodes a critical protein that links the Toll-like receptor to the IRAK protein. However, a partial sequence of a molluscan MyD88-like transcript has been identified in C. gigas (28), suggesting that E. scolopes may have this gene, although it may not be expressed at high levels in the light organ. Attempts to identify this transcript in other E. scolopes tissues by reverse transcription-PCR amplification were not successful.
The transcripts described in the present study can be used to propose a putative Toll/NF-κB signaling pathway in E. scolopes (Fig. 9). Taken together, the data for E. scolopes and the distantly related C. gigas provide strong evidence that this pathway is likely to occur throughout the Phylum Mollusca, as well as across the entire superphylum containing the mollusks, i.e., the Lophotrochozoa. The phylogenetic trees for the E. scolopes proteins do not clarify whether the E. scolopes sequences group closer to the vertebrates or the insects, although most of the closest BLAST hits are to vertebrate sequences. The relationship among the superphyla of the animals is controversial, with most phylogenies aligning the Lophotrochozoa with the Ecdysozoa, rather than the Deuterostomia (59). Further analysis of the E. scolopes EST library and those of other Lophotrochozoa will be necessary to address this issue rigorously.
The complex patterns of bacteria-induced development in the squid light organ, along with the different functions of MAMPs in the process, suggest that the NF-κB pathway may be modulated differently over time and in location through the early development of the light organ. The high levels of nitric oxide synthase (NOS) (16) and the mucus shedding (52, 54) in response to environmental PGN would be consistent with activity of the pathway in the superficial field of ciliated cells during the initial phases of colonization. However, nitric oxide/NOS levels are attenuated (16), mucus shedding ceases (52), and apoptosis (24) is induced in this field of cells by the subsequent interactions of the host and symbiont within the crypt space, which is several cell layers away. These observations suggest that remote signaling by the symbionts may turn down the activity of the pathway in the superficial field of ciliated cells. In contrast, the MAMPs responsible for development, i.e., derivatives of PGN and LPS, are presented directly to the crypt cells. These receptor-ligand interactions may increase the activity of the pathway in the cells with which the bacteria directly interact. In support of this possibility is the observation that mucus shedding by the crypt cells results from colonization of the crypts with symbionts (52).
The data presented here demonstrate the presence of the NF-κB pathway in the E. scolopes light organ, and the patterns of development and the involvement of MAMPs provide circumstantial evidence for its activity during the onset of the symbiosis. A full understanding of the connection among these findings awaits an in-depth analysis of the activity of the pathway in the various tissues of the light organ.
Supplementary Material
Acknowledgments
We thank W. Crookes-Goodson and A. Wier for helpful discussion and comments on the manuscript.
This research was supported by a grant from the W. M. Keck Foundation (to E. P. Greenberg and M.M.-N.), by NSF grant IBN 0451102 (to M.M.-N. and E. G. Ruby), by NIH grant AI 50611 (to M.M.-N.), by NIH grant RR12294 (to E. G. Ruby and M.M.-N.), and by a Career Development Award from Research to Prevent Blindness (to T.E.S).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Aguinaldo, A. M., J. M. Turbeville, L. S. Linford, M. C. Rivera, J. R. Garey, R. A. Raff, and J. A. Lake. 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 387:489-493. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Anton, P. M., V. Theodorou, S. Roy, J. Fioramonti, and L. Bueno. 2002. Pathways involved in mild gastrointestinal inflammation induced by a low level exposure to a food contaminant. Dig. Dis. Sci. 47:1308-1315. [DOI] [PubMed] [Google Scholar]
- 5.Asai, T., G. Tena, J. Plotnikova, M. R. Willmann, W. L. Chiu, L. Gomez-Gomez, T. Boller, F. M. Ausubel, and J. Sheen. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977-983. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin, A. S., Jr. 1996. The NF-κB and IκB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649-683. [DOI] [PubMed] [Google Scholar]
- 7.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, J. K., G. E. Mullen, C. A. Leifer, A. Mazzoni, D. R. Davies, and D. M. Segal. 2003. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24:528-533. [DOI] [PubMed] [Google Scholar]
- 9.Beutler, B. 2004. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430:257-263. [DOI] [PubMed] [Google Scholar]
- 10.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science 267:1485-1488. [DOI] [PubMed] [Google Scholar]
- 12.Cao, Z., W. J. Henzel, and X. Gao. 1996. IRAK: a kinase associated with the interleukin-1 receptor. Science 271:1128-1131. [DOI] [PubMed] [Google Scholar]
- 13.Clifton, D. R., R. A. Goss, S. K. Sahni, D. van Antwerp, R. B. Baggs, V. J. Marder, D. J. Silverman, and L. A. Sporn. 1998. NF-κB-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc. Natl. Acad. Sci. USA 95:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 15.Dadgostar, H., and G. Cheng. 1998. An intact zinc ring finger is required for tumor necrosis factor receptor-associated factor-mediated nuclear factor-κB activation but is dispensable for c-Jun N-terminal kinase signaling. J. Biol. Chem. 273:24775-24780. [DOI] [PubMed] [Google Scholar]
- 16.Davidson, S. K., T. A. Koropatnick, R. Kossmehl, L. Sycuro, and M. J. McFall-Ngai. 2004. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 6:1139-1151. [DOI] [PubMed] [Google Scholar]
- 17.Devin, A., Y. Lin, S. Yamaoka, Z. Li, M. Karin, and Z. Liu. 2001. The α and β subunits of IκB kinase (IKK) mediate TRAF2-dependent IKK recruitment to tumor necrosis factor (TNF) receptor 1 in response to TNF. Mol. Cell. Biol. 21:3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doino, J. A., and M. J. McFall-Ngai. 1995. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol. Bull. 189:347-355. [DOI] [PubMed] [Google Scholar]
- 19.Dziarski, R. 2004. Peptidoglycan recognition proteins (PGRPs). Mol. Immunol. 40:877-886. [DOI] [PubMed] [Google Scholar]
- 20.Escoubas, J. M., L. Briant, C. Montagnani, S. Hez, C. Devaux, and P. Roch. 1999. Oyster IKK-like protein shares structural and functional properties with its mammalian homologues. FEBS Lett. 453:293-298. [DOI] [PubMed] [Google Scholar]
- 21.Fallon, P. G., R. L. Allen, and T. Rich. 2001. Primitive Toll signalling: bugs, flies, worms and man. Trends Immunol. 22:63-66. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein, J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 23.Foster, J. S., M. A. Apicella, and M. J. McFall-Ngai. 2000. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226:242-254. [DOI] [PubMed] [Google Scholar]
- 24.Foster, J. S., and M. J. McFall-Ngai. 1998. Induction of apoptosis by cooperative bacteria in the morphogenesis of host epithelial tissues. Dev. Genes Evol. 208:295-303. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 26.Gilmore, T. D. 1990. NF-κB, KBF1, dorsal, and related matters. Cell 62:841-843. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore, T. D. 1999. The Rel/NF-κB signal transduction pathway: introduction. Oncogene 18:6842-6844. [DOI] [PubMed] [Google Scholar]
- 28.Gueguen, Y., J. P. Cadoret, D. Flament, C. Barreau-Roumiguiere, A. L. Girardot, J. Garnier, A. Hoareau, E. Bachere, and J. M. Escoubas. 2003. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene 303:139-145. [DOI] [PubMed] [Google Scholar]
- 29.Halanych, K. M., J. D. Bacheller, A. M. Aguinaldo, S. M. Liva, D. M. Hillis, and J. A. Lake. 1995. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science 267:1641-1643. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, Jr., and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 31.Huang, B., M. Eberstadt, E. T. Olejniczak, R. P. Meadows, and S. W. Fesik. 1996. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 384:638-641. [DOI] [PubMed] [Google Scholar]
- 32.Janeway, C. A., Jr. 1989. Approaching an asymptote? Evolution and revolution in immunology. Cold Spring Harbor Symp. Quant. Biol. 54:1-13. [DOI] [PubMed] [Google Scholar]
- 33.Jobin, C., and R. B. Sartor. 2000. The IκB/NF-κB system: a key determinant of mucosal inflammation and protection. Am. J. Physiol. Cell Physiol. 278:C451-C462. [DOI] [PubMed] [Google Scholar]
- 34.Jono, H., T. Shuto, H. Xu, H. Kai, D. J. Lim, J. R. Gum, Jr., Y. S. Kim, S. Yamaoka, X. H. Feng, and J. D. Li. 2002. Transforming growth factor-α-Smad signaling pathway cooperates with NF-κB to mediate nontypeable Haemophilus influenzae-induced MUC2 mucin transcription. J. Biol. Chem. 277:45547-45557. [DOI] [PubMed] [Google Scholar]
- 35.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 36.Karin, M., and A. Lin. 2002. NF-κB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 37.Kim, M. S., M. Byun, and B. H. Oh. 2003. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 4:787-793. [DOI] [PubMed] [Google Scholar]
- 38.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 39.Koropatnick, T. A., J. T. Engle, M. A. Apicella, E. V. Stabb, W. E. Goldman, and M. J. McFall-Ngai. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186-1188. [DOI] [PubMed] [Google Scholar]
- 40.Li, J. D., W. Feng, M. Gallup, J. H. Kim, J. Gum, Y. Kim, and C. Basbaum. 1998. Activation of NF-κB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for Pseudomonas aeruginosa-induced mucin overproduction in epithelial cells. Proc. Natl. Acad. Sci. USA 95:5718-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li, Q., and I. M. Verma. 2002. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2:725-734. [DOI] [PubMed] [Google Scholar]
- 42.Liu, C., Z. Xu, D. Gupta, and R. Dziarski. 2001. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276:34686-34694. [DOI] [PubMed] [Google Scholar]
- 43.Lomaga, M. A., W. C. Yeh, I. Sarosi, G. S. Duncan, C. Furlonger, A. Ho, S. Morony, C. Capparelli, G. Van, S. Kaufman, A. van der Heiden, A. Itie, A. Wakeham, W. Khoo, T. Sasaki, Z. Cao, J. M. Penninger, C. J. Paige, D. L. Lacey, C. R. Dunstan, W. J. Boyle, D. V. Goeddel, and T. W. Mak. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellroth, P., J. Karlsson, and H. Steiner. 2003. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem. 278:7059-7064. [DOI] [PubMed] [Google Scholar]
- 46.Michel, T., J. M. Reichhart, J. A. Hoffmann, and J. Royet. 2001. Drosophila Toll is activated by gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756-759. [DOI] [PubMed] [Google Scholar]
- 47.Miyamoto, S., and I. M. Verma. 1995. Rel/NF-κB/IκB story. Adv. Cancer Res. 66:255-292. [PubMed] [Google Scholar]
- 48.Montagnani, C., C. Kappler, J. M. Reichhart, and J. M. Escoubas. 2004. Cg-Rel, the first Rel/NF-κB homolog characterized in a mollusk, the Pacific oyster Crassostrea gigas. FEBS Lett. 561:75-82. [DOI] [PubMed] [Google Scholar]
- 49.Montgomery, M. K., and M. McFall-Ngai. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719-1729. [DOI] [PubMed] [Google Scholar]
- 50.Morin, P. J., A. B. Sparks, V. Korinek, N. Barker, H. Clevers, B. Vogelstein, and K. W. Kinzler. 1997. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 275:1787-1790. [DOI] [PubMed] [Google Scholar]
- 51.Nurnberger, T., and F. Brunner. 2002. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5:318-324. [DOI] [PubMed] [Google Scholar]
- 52.Nyholm, S. V., B. Deplancke, H. R. Gaskins, M. A. Apicella, and M. J. McFall-Ngai. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyholm, S. V., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2:632-642. [DOI] [PubMed] [Google Scholar]
- 54.Nyholm, S. V., E. V. Stabb, E. G. Ruby, and M. J. McFall-Ngai. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 97:10231-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pikarsky, E., R. M. Porat, I. Stein, R. Abramovitch, S. Amit, S. Kasem, E. Gutkovich-Pyest, S. Urieli-Shoval, E. Galun, and Y. Ben-Neriah. 2004. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature 431:461-466. [DOI] [PubMed] [Google Scholar]
- 56.Reiser, J. B., L. Teyton, and I. A. Wilson. 2004. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 Å resolution. J. Mol. Biol. 340:909-917. [DOI] [PubMed] [Google Scholar]
- 57.Rothwarf, D. M., E. Zandi, G. Natoli, and M. Karin. 1998. IKK-γ is an essential regulatory subunit of the IκB kinase complex. Nature 395:297-300. [DOI] [PubMed] [Google Scholar]
- 58.Ruby, E. G., and M. J. McFall-Ngai. 1999. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 7:414-420. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz-Trillo, I., J. Paps, M. Loukota, C. Ribera, U. Jondelius, J. Baguna, and M. Riutort. 2002. A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proc. Natl. Acad. Sci. USA 99:11246-11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzman, A. L., T. Eaves-Pyles, S. C. Linn, A. G. Denenberg, and C. Szabo. 1998. Bacterial induction of inducible nitric oxide synthase in cultured human intestinal epithelial cells. Gastroenterology 114:93-102. [DOI] [PubMed] [Google Scholar]
- 61.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw, P. W. 1997. Polymorphic microsatellite markers in a cephalopod: the veined squid Loligo forbesi. Mol. Ecol. 6:297-298. [Google Scholar]
- 63.Shekels, L. L., and S. B. Ho. 2003. Characterization of the mouse Muc3 membrane bound intestinal mucin 5′ coding and promoter regions: regulation by inflammatory cytokines. Biochim. Biophys. Acta 1627:90-100. [DOI] [PubMed] [Google Scholar]
- 64.Silverman, N., and T. Maniatis. 2001. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15:2321-2342. [DOI] [PubMed] [Google Scholar]
- 65.Small, A. L., and M. J. McFall-Ngai. 1999. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J. Cell. Biochem. 72:445-457. [PubMed] [Google Scholar]
- 66.Sokolov, E. P. 2000. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Moll. Stud. 66:573-575. [Google Scholar]
- 67.Suzuki, N., S. Suzuki, G. S. Duncan, D. G. Millar, T. Wada, C. Mirtsos, H. Takada, A. Wakeham, A. Itie, S. Li, J. M. Penninger, H. Wesche, P. S. Ohashi, T. W. Mak, and W. C. Yeh. 2002. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416:750-756. [DOI] [PubMed] [Google Scholar]
- 68.Tauszig, S., E. Jouanguy, J. A. Hoffmann, and J. L. Imler. 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl. Acad. Sci. USA 97:10520-10525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tlaskalova-Hogenova, H., R. Stepankova, T. Hudcovic, L. Tuckova, B. Cukrowska, R. Lodinova-Zadnikova, H. Kozakova, P. Rossmann, J. Bartova, D. Sokol, D. P. Funda, D. Borovska, Z. Rehakova, J. Sinkora, J. Hofman, P. Drastich, and A. Kokesova. 2004. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol. Lett. 93:97-108. [DOI] [PubMed] [Google Scholar]
- 71.Weis, V. M., A. L. Small, and M. J. McFall-Ngai. 1996. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc. Natl. Acad. Sci. USA 93:13683-13688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Werner, T., G. Liu, D. Kang, S. Ekengren, H. Steiner, and D. Hultmark. 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97:13772-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamaoka, S., G. Courtois, C. Bessia, S. T. Whiteside, R. Weil, F. Agou, H. E. Kirk, R. J. Kay, and A. Israel. 1998. Complementation cloning of NEMO, a component of the IκB kinase complex essential for NF-κB activation. Cell 93:1231-1240. [DOI] [PubMed] [Google Scholar]
- 74.Yaron, A., H. Gonen, I. Alkalay, A. Hatzubai, S. Jung, S. Beyth, F. Mercurio, A. M. Manning, A. Ciechanover, and Y. Ben-Neriah. 1997. Inhibition of NF-κB cellular function via specific targeting of the IκB-ubiquitin ligase. EMBO J. 16:6486-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.