Abstract
Whole-genome association studies will be a powerful tool to identify genes responsible for common human diseases. A crucial task for association-mapping studies is the evaluation of the relationship between linkage disequilibrium (LD) and physical distance for the genomic region under study. Since it is known that the extent of LD is nonuniformly distributed throughout the human genome, the required marker density has to be determined specifically for the region under study. These regions may be related to isochores and chromosomal bands, as indicated by earlier cytogenetic findings concerning chiasma distribution in meiosis. Therefore we analyzed the neurofibromatosis type 1 (NF1) gene region on chromosome 17q11.2, which is characterized by a nonuniform LD pattern and an L1-to-H2 isochore transition. Long-range LD within the NF1 gene was found to extend over 200 kb (D′ = 0.937) in the L1 isochore, whereas, in the neighboring H2 isochore, no LD is apparent between markers spaced by 26 kb (D′ = 0.144). Recombination frequencies derived from the LD are at .00019 (high LD) and .01659 (low LD) per megabase, the latter identical to the average value from segregation analysis. The boundary between these regions coincides precisely with a transition in the GC content of the sequences, with low values (37.2%) in the region with long-range LD and high values (51%) in the other. Our results suggest a correlation between the LD pattern and the isochores, at least in the NF1 region. If this correlation can be generalized, the marker densities required for association studies have to be adjusted to the regional GC content and may be chosen according to the isochores.
Introduction
The localization and identification of genes that are responsible for common human diseases is one of the major goals of human genetics in the near future. Whole-genome association studies between markers and traits are considered to be a possible approach for the identification of genes underlying complex genetic disorders. This approach depends on the detection of linkage disequilibrium (LD) between a marker and a risk locus. Simulation studies (Kruglyak 1999; Zöllner and von Haeseler 2000) showed that the extent of LD depends on the history of the population from which the probands are selected but covers only a few kilobases in most cases. On the basis of these results, it has been estimated that ∼500,000 markers have to be employed in whole-genome association studies (Kruglyak 1999), which is not yet feasible. However, the extent of LD is not uniform within the human genome. Data from the human lipoprotein lipase gene (Clark et al. 1998; Nickerson et al. 1998) fit fairly well into the simulation model of Zöllner and von Haeseler (2000). On the other hand, LD can be observed between markers that are several megabases away from each other in the human MHC locus. However, this long-distance LD may be atypical and represents a special situation at the MHC region (Carrington 1999; Sanchez-Mazas et al. 2000). For the neurofibromatosis type 1 (NF1 [MIM 162200]) gene region on chromosome 17q11.2, it has been demonstrated that markers at a distance of up to 350 kb from each other are in strong linkage disequilibrium in different populations (Jorde et al. 1993; Messiaen et al. 1993; Purandare et al. 1996). A marker located 68 kb downstream of the NF1 gene shows only weak LD with intragenic markers, indicating that, at the NF1 gene locus, two genomic regions face each other with differing degrees of LD. In this case, a regional difference in recombination frequency (θ) would most plausibly explain the observed differences between the extent of LD. This renders the NF1 locus a good model to study the variation in the extent of LD between different genomic regions.
Why different genomic regions show different values of θ is an open question. From cytogenetic analysis, it is known that meiotic chiasmata, the visible correlates of meiotic recombination, are more often seen in R bands and T bands than in G bands (reviewed by Holmquist [1992]), suggesting that θ is higher in R- and T-band sequences than in G-band sequences. This observation relates θ to compositional genomic patterns. The human genome, like the genomes of all warm-blooded vertebrates, is composed of long stretches of DNA homogeneous in GC content, which were called isochores. According to their buoyant density, five classes of isochores were defined: two light ones, L1 and L2, and three heavy ones, H1, H2, and H3, with GC contents of ∼39%, 41%, 45%, 49%, and 53%, respectively (Bernardi 2000). The topographic distribution of isochores is clearly related to chromosomal banding patterns. G bands contain almost exclusively DNA sequences belonging to the light isochore classes. R bands are more heterogeneous and contain both heavy and light isochores (Saccone et al. 1993). H3 isochores can be found only in a subset of R bands called T bands (Saccone et al. 1996, 1999). However, a direct correlation between θ and isochore classes has not been demonstrated, up to now. The data on the stability of multiple-marker haplotypes in the NF1 gene region and the availability of long-range DNA sequences from this region prompted us to analyze the extent of LD between markers from the region in greater detail and to examine its relationship to the isochore structure at the NF1 locus.
Material and Methods
Subjects and DNA Methods
Ninety-three individuals, unrelated and selected at random from the southern German population, were genotyped in this study. DNA was isolated from white blood cells, using a simple salting-out procedure (Miller et al. 1988).
All polymorphisms except one are based on single nucleotide exchanges, and typing of individuals was performed by PCR amplification from genomic DNA, digestion of PCR products with the appropriate restriction enzyme, and standard agarose gel electrophoresis of restriction fragments. The other polymorphism (Pin28) is a 21-bp duplication in intron 28 (Eisenbarth et al. 2000). A typical PCR amplification reaction was performed with the following components: ∼100 ng genomic DNA, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.25 mM of forward and reverse primer, and 2.5 U Taq polymerase (Amersham Pharmacia Biotech), for 35 cycles of 1 min at 92°C, 1 min at 56°C, and 1 min at 72°C, after an initial predenaturation step for 1 min at 93°C.
Primers and PCR conditions for the published polymorphisms were used as described elsewhere (Rsa in Hoffmeyer and Assum 1994; Pin28 in Eisenbarth et al. 2000). In case of the Taq RFLP (designated as “10647” by Purandare et al. [1996]), we used the primer pair 87763-AAAAATACTGATATTTCCATAATCG and 87964-CCTCAAAAGTCAGAAGTTCTTTA. For Bgl, described as RFLP detected with probe f7G4GL.2 by Jorde et al. (1993), the primers were 92462-TTTGAGTCTGGCCCAAAGTTG and 92731-TGGTGCTCAAATTGAGTGACAA (numbering according to the Genbank sequence L05367.1).
For amplification of the polymorphic sites first described in this article, the primers are SNP1: 207828-CACAGCCTTATCCATACCTGT and 208189-CTCTAGGAAGAAACAAGTTTGAT, SNP2: 230036-GGAATGCCAGGCTCAAACCT and 230423-GGGACTTAGCTGGGATCTCT, SNP2.2: 265290-CTTCTGTCCCTTCTGGCATTT and 265802-GGACAATGTTTTCTCCAATTCTT, and SNP3: 292210-GGGAAATGACAAGACCTGCC and 292591-CCACCTAACTCTTCCCGATTC (numbering according to Genbank sequence AC004526). PCR products were digested with the appropriate restriction enzymes (HaeIII for SNP1, MboI for SNP2, HpaII for SNP2.2, and MboI for SNP3) and typed after agarose gel electrophoresis.
Statistical Methods
Haplotype frequencies were calculated with the EH program of Terwilliger and Ott (1994), available at the Human Genome Mapping Project Web site. EH was also used to test the significance of allelic association with the χ2 test. For LD, D′ and d2 were calculated as described elsewhere (Devlin et al. 1995). θ was deduced from d2 with the formula θ=1-d2(1/2N), where N is the assumed age of the variant in generations. The θ values were then normalized to the physical distance of marker pairs and presented as θ/1 Mb.
Results
LD in the NF1 Gene Region
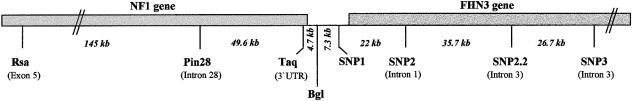
Three known intragenic NF1 polymorphisms (Rsa, Pin28, and Taq) and the flanking marker Bgl were tested to corroborate the strong LD in our population sample of unrelated, randomly chosen individuals originating from the southern part of Germany (n=93). Four new polymorphic markers mapping distal of the NF1 gene (SNP1, SNP2, SNP2.2, SNP3) were isolated to extend the region into the neighboring part, where the LD diminishes. The precise location of markers in relation to the genes mapping in this genomic interval is depicted in figure 1. The Rsa polymorphism is located in exon 5, Pin28 is located in intron 28, and Taq is a 3′ UTR marker of the NF1 gene. Two of the markers (Bgl and SNP1) map to a short 12-kb intergenic region, whereas the other three markers (SNP2, SNP2.2, and SNP3) are located in introns of the next gene, FHN3 (H. Kehrer-Sawatzki, personal communication). The allele frequencies and rates of heterozygosity of the polymorphisms observed in our population sample are given in table 1. All markers show allele frequencies of 27%–43% for the rarer allele and therefore can be assumed to represent relatively ancient polymorphisms. The mean rate of heterozygosity is 0.44. The Rsa polymorphism from exon 5 of the NF1 gene was used as an anchor marker, and d2/D′ values were calculated for the distal flanking markers, to move—step by step—into the genomic region supposed to exhibit recombination. As shown in table 2, the marker pairs Rsa/Pin28, Rsa/Taq and Rsa/Bgl are in absolute linkage disequilibrium, as reflected by a D′ value of 1. A strong linkage disequilibrium is still present between Rsa and SNP1, which are separated by a genomic distance of 200 kb (D′ = 0.937). The disequilibrium decreases with Rsa/SNP2 (0.7168), and a similar D′ value is obtained for the SNP1/SNP2 pair (0.6244), indicating that recombination begins to occur in the interval between SNP1 and SNP2 (22 kb). No allelic association is apparent between the following markers, SNP2/SNP2.2 and SNP2.2/SNP3, which are separated by 35.7 kb and 26.7 kb, respectively (D′ = 0.1712 and 0.1436). These data demonstrate that a strong LD is maintained over a region of >200 kb and disappears within the next 30 kb. The high degree of LD in the proximal region is also apparent from the haplotype data: only three of the four possible haplotypes have been found with the proximal markers, whereas all four haplotypes have been identified with the distal marker pairs of the region without allelic association.
Figure 1.
Polymorphic markers used for LD analyses and their position in the NF1 gene, in the short intergenic region and in the flanking FHN3 gene.
Table 1.
Allele Frequencies and Heterozygosity Rates of Polymorphic Markers in the NF1 Gene Region
| PolymorphicMarker | + | − | Heterozygosity |
| Rsa | .32 | .68 | .42 |
| Pin28 | .41 | .59 | .45 |
| Taq | .32 | .68 | .41 |
| Bgl | .57 | .43 | .45 |
| SNP1 | .60 | .40 | .44 |
| SNP2 | .53 | .47 | .46 |
| SNP2.2 | .63 | .37 | .52 |
| SNP3 | .73 | .27 | .38 |
Table 2.
Observed Haplotype Frequencies and Calculated D′ and d2 Values for Marker Pairs from the NF1 Gene Region
|
Marker Pair |
||||||||
| Haplotypeor LDMeasure | Rsa/Pin28 | Rsa/Taq | Rsa/Bgl | Rsa/SNP1 | Rsa/SNP2 | SNP1/SNP2 | SNP2/SNP2.2 | SNP2.2/SNP3 |
| ++ | .317 | .317 | 0 | .012 | .275 | .199 | .308 | .486 |
| +− | 0 | 0 | .317 | .305 | .042 | .397 | .224 | .149 |
| −+ | .091 | .005 | .570 | .585 | .257 | .333 | .326 | .24 |
| −− | .592 | .678 | .113 | .098 | .426 | .071 | .142 | .125 |
| d2 | .7509 | .9854 | .6966 | .6709 | .2425 | .2420 | .0138 | .0119 |
| D′ | 1 | 1 | 1 | .937 | .7168 | .6244 | .1712 | .1436 |
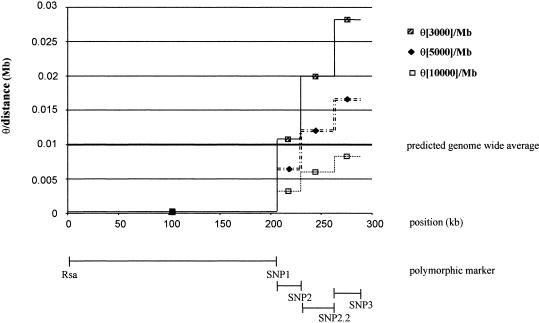
To obtain more insight into the allelic association of makers located in the genomic region with strong LD and the flanking region without LD, we calculated the θ values for representative marker pairs, assuming different ages of the variants (n=3,000, n=5,000, and n=10,000). After correction for the physical distance (in Mb) between the marker pairs, we obtained the results shown in table 3. θ/Mb values range between 0.00032194 (n=3,000) and 0.00009658 (n=10,000) for Rsa/SNP1 located in the proximal region. θ is ∼50 times lower in this region than the genomewide average (1 cM/Mb) and increases independently of the assumed age in the flanking region (SNP1 to SNP3) to values that are close to the generally accepted genomic average (0.003224–0.027657/Mb). The dramatic change in LD (and θ) is most obvious in a graphic representation (fig. 2). Independent of the assumed number of generations there is nearly no recombination between Rsa and SNP1. The θ/Mb values rise for all flanking markers, depending on the assumed age to values that are within the range of 0.01/Mb. These differences can not be explained by features like coding or noncoding region, since SNP2, SNP2.2, and SNP3 are located in introns of the FHN3 gene just like the markers with high LD in the NF1 gene.
Table 3.
θ, Calculated for Different Ages of the Variants (N), in Relation to the Physical Distance of the Markers
| θ for N = |
θ/Mb for N = |
|||||||
| MarkerPair | d2 | 3,000 | 5,000 | 10,000 | PhysicalDistance(Mb) | 3,000 | 5,000 | 10,000 |
| Rsa/SNP1 | .6709 | 6.651×10−5 | 3.990×10−5 | 1.995×10−5 | .2066 | 3.219×10−4 | 1.931×10−4 | 9.658×10−5 |
| SNP1/SNP2 | .2420 | 2.364×10−4 | 1.418×10−4 | 7.093×10−5 | .022 | .010746 | .006448 | .003224 |
| SNP2/SNP2.2 | .0138 | 7.131×10−4 | 4.279×10−4 | 2.139×10−4 | .0357 | .019975 | .011989 | .005994 |
| SNP2.2/SNP3 | .0118 | 7.384×10−4 | 4.431×10−4 | 2.215×10−4 | .0267 | .027657 | .016599 | .008299 |
Figure 2.
Values of θ/Mb between representative marker pairs, under the assumption of three different ages of the variants (N=3,000, 5,000, or 10,000 generations).
To test our initial hypothesis that there may be a correlation between the compositional pattern of the sequence and the recombinational behavior of the region, we determined the long-range GC composition of the genomic interval under investigation.
Long-Range GC% Structures in the Human NF1 Locus
A continuously sequenced region of 476 kb is publicly available with three Genbank fragments (accession numbers AC004222, AC004526, AC003101) that cover most of the genomic region of the NF1 locus and extends in telomeric direction. The only part of the NF1 gene that is still unknown comprises parts of the large intron 1 which is thought to span ∼105 kb. We used these Genbank fragments to determine the GC content of the sequences and to define the isochore class to which the genomic interval belongs. The proportion of GC nucleotides was calculated with the Window program of the GCG program package (Wisconsin Package Version 10.0, Genetics Computer Group). The analysis was done using 5-kb fragments with steps of 5 kb to gradually move along the region (fig. 3). The NF1 gene itself is relatively GC-poor, with an average content of 37.2%, and represents an L1 isochore. Immediately after the 3′ end of the gene, there is a transition into a GC-rich H2 isochore with an average GC content of 51%. At the L1-H2 transition, between nucleotide position numbers 240,000 and 260,000, we observed the first GC value that is above the range of variation of the light isochore at position 242,500 ± 2,500 bp (44.8%). The next value, 5 kb downstream (position 247,500 ± 2,500 bp), is already 56.4%, which is a typical score for heavy isochores. These data indicate that the transition is very sharp and takes place within an interval of 10 kb.
Figure 3.
Long-range GC composition of the genomic interval using the indicated Genbank sequences. The NF1 gene in the proximal part is GC-poor, with an average content of 37.2% (L1 isochore); after the 3′ end of the gene, the GC content rises to 51% (H2 isochore).
To estimate the length of the isochores, the GC content of two further Genbank fragments (AC004666 and AC004523) were also analyzed flanking in proximal and distal directions, respectively (data not shown). Neither sequence overlaps the sequences from the NF1 region, leaving a gap in either direction. AC004666, the sequence fragment that follows in the centromeric direction, has an average GC content of 45% (H1 isochore) and the neighboring fragment in telomeric direction AC004523 shows an average score of 43%, which is intermediate between L2 and H1 (data not shown). The fact that the AC004666 sequence was mapped ∼250 kb away from NF1 (Dorschner et al. 2000) limits the length of the L1 isochore between 250 and 600 kb (250 kb of the sequenced part of the NF1 gene plus an unknown part of the sequences located between NF1 and AC004666). The length of the H2 isochore flanking NF1 in telomeric direction can not be estimated, since the distance between NF1 and the L2/H1 typical sequence AC004523 is not known.
Extent of Linkage Disequilibrium in Relation to the Pattern of Sequence Composition
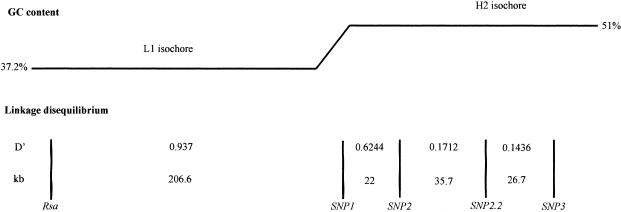
We refined the extent of linkage disequilibrium in the NF1 gene region in order to relate the nonuniform disequilibrium pattern to properties of the underlying DNA. Figure 4 summarizes the results: the region with the low GC content is characterized by a large extent of LD. In the small transition region, between the light and the heavy isochores, we also observed high D′ values involving markers Bgl and SNP1 (fig. 4 and data not shown). Immediately distal of the transition region, at the beginning of the heavy isochore (∼20 kb apart) the D′ values decrease for the markers around SNP2, and there is no allelic association between any marker pair in the heavy isochore.
Figure 4.
Relation between the compositional pattern of the DNA sequence from the NF1 gene region and the extent of linkage disequilibrium (D′) observed between representative marker pairs (Rsa, SNP1, SNP2, SNP2.2, and SNP3). LD was found to range over the whole L1 isochore, whereas, in the H2 isochore, no allelic association is apparent with the chosen marker spacing.
Discussion
At the NF1 gene locus, two genomic regions face each other that are characterized by strongly different extents of LD between biallelic markers. The markers within the NF1 gene are in complete LD over a distance of 195 kb. In contrast, no LD was observed between markers with pairwise distances of 36 kb and 27 kb in the region distal of the NF1 gene. The differences in the extent of LD observed in the two regions can not be explained by differences in population history as the observations were made within the same sample of probands, but most probably reflect regional genomic differences in θ values. Within the NF1 gene region, the θ value calculated from the d2 value observed between markers Rsa and SNP1 is severely reduced, compared to the genomic average of 1 cM/Mb. It is unlikely that this reduction is due to the overall reduced θ of the pericentromeric regions of human chromosomes, since marker UT172, located 250 kb centromeric to NF1, is in equilibrium with the intragenic NF1 markers (data not shown).
The d2 values found between markers located distal from NF1 fit well within the values obtained by Kruglyak (1999) by theoretical simulations for a population expanding for the last 5,000 generations and θ 1cM/Mb. The θ values per Mb calculated here from d2 are ∼.01/Mb, for a population founded 5,000–10,000 generations ago, as can be assumed for the general human population (Harpending et al. 1998). The boundary between the genomic regions with different values of θ is sharp, with marker SNP1 clearly belonging to the region where long range LD can be observed, followed by a 22-kb region, between markers SNP1 and SNP2, showing half of the θ/Mb as can be found further distal. The boundary precisely coincides with a transition in the long range GC content from 37.2% GC to 51% GC, as demonstrated by the analysis of 470 kb of contiguous DNA sequences. If this correlation between isochore structure and θ turns out to be a general phenomenon of the human genome, it has a strong impact on the marker densities that have to be used for successful association studies. In GC-rich genomic regions, marker densities of one marker every 6 kb may indeed be necessary, as suggested by Kruglyak (1999). But the marker density, and thereby the typing effort, possibly can be highly reduced for loci located in GC-poor genomic regions, which account for 65% of the human genome and show a low density of coding genes.
In this context, information about the extent of the long-range LD proximal to marker Rsa and the length of the L1 isochore would be of interest. The long range LD extends over a region between 340 and 590 kb in length. Strong LD has been demonstrated between marker Bgl (named “f7G4GL.2” by Jorde et al.) and marker fHB5-E3, which is located at the 5′ end of the NF1 gene, 340 kb away from Bgl (Jorde et al. 1993), whereas marker UT172, which is located 250 kb further proximal, shows no LD with intragenic NF1 markers in our population sample (data not shown). Therefore, the proximal boundary of the region with long-range LD ends within the 250 kb between the 5′ end of NF1 and marker UT172. The proximal boundary of the L1 isochore is located within the same region, because marker UT172 maps to the DNA sequence AC004666, which is characterized by a GC content of 45%, a typical score for H1 isochores (data not shown). Because of the lack of sequence information, more precise location of both boundaries is not possible at this time.
Another genomic region where the isochore structure has been analyzed in detail is the human MHC gene cluster. In this case, two isochore transitions were found within a 1.5-Mb region, one L2/H2 transition within the class II gene region, and another L2/H3 transition between the class II and class III genes (Stephens et al. 1999). For the latter region, a precise switching of replication timing at the isochore boundary has been described (Tenzen et al. 1997). This boundary is further characterized by the presence of a pseudoautosomal boundary–like sequence (PABL), a cluster of L1 sequences on the L2 side, and a cluster of Alu sequences on the H3 side (Fukagawa et al. 1995, 1996). Within the sequences from the NF1 gene locus, neither a PABL sequence nor clustering of repetitive elements was found (data not shown). Therefore, the presence of these features does not seem to be a general characteristic of isochore boundaries.
When all the evidence is taken together, a sharp boundary exists at the NF1 gene locus between two genomic regions with different values of θ, and this boundary colocates precisely with the L1-H2 isochore boundary. Our results mediate a molecular basis for the cytogenetic observations that different values of θ can be assigned to different chromosomal bands. We describe for the first time a genomic region where the extent of LD changes with the GC content of the underlying DNA sequence. Of course, more studies should be carried out at various genomic loci before this observation can be generalized to the entire human genome.
Acknowledgments
We would like to thank Margot Brugger, Bärbel Dieske, Britta Pietsch, and Sigrid Wieland-Lange for excellent technical assistance.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Human Genome Mapping Project (HGMP), http://www.hgmp.mrc.ac.uk
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for NF1 [MIM 162200] [PubMed]
References
- Bernardi G (2000) Isochores and the evolutionary genomics of vertebrates. Gene 241:3–17 [DOI] [PubMed] [Google Scholar]
- Carrington M (1999) Recombination within the human MHC. Immunol Rev 167:245–256 [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Risch N (1995) A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29:311–322 [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephans K (2000) NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 9:35–46 [DOI] [PubMed] [Google Scholar]
- Eisenbarth I, Beyer K, Krone W, Assum G (2000) Toward a survey of somatic mutation of the NF1 gene in benign neurofibromas of patients with neurofibromatosis type 1. Am J Hum Genet 66:393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Sugaya K, Matsumoto K, Okumura K, Ando A, Inoko H, Ikemura T (1995) A boundary of long-range G+C% mosaic domains in the human MHC locus: pseudoautosomal boundary–like sequence exists near the boundary. Genomics 25:184–191 [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nakamura Y, Okumura K, Nogami M, Ando A, Inoko H, Saitou N, Ikemura T (1996) Human pseudoautosomal boundary–like sequences: expression and involvement in evolutionary formation of the present-day pseudoautosomal boundary of sex chromosomes. Hum Mol Genet 5:23–32 [DOI] [PubMed] [Google Scholar]
- Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1961–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer S, Assum G (1994) An RsaI polymorphism in the transcribed region of the neurofibromatosis (NF1)-gene. Hum Genet 93:481–482 [DOI] [PubMed] [Google Scholar]
- Holmquist GP (1992) Chromosome bands, their chromatin flavors, and their functional features. Am J Hum Genet 51:17–37 [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Viskochil D, O`Connell P, Ward K (1993) Linkage disequilibrium in the neurofibromatosis 1 (NF1) region: implications for gene mapping. Am J Hum Genet 53: 1038–1050 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Messiaen L, De Bie S, Moens T, Van den Enden A, Leroy J (1993) Lack of independence between five DNA polymorphisms in the NF1 gene. Hum Mol Genet 2:485 [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson TG, Stengard J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF (1998) DNA sequence diversity in a 9.7 kb region of the human lipoprotein lipase gene. Nat Genet 19:233–240 [DOI] [PubMed] [Google Scholar]
- Purandare SM, Cawthon R, Nelson LM, Sawada S, Watkins WS, Ward D, Jorde LB, Viskochil DH (1996) Genotyping of PCR-based polymorphisms and linkage-disequilibrium analysis at the NF1 locus. Am J Hum Genet 59:159–166 [PMC free article] [PubMed] [Google Scholar]
- Saccone S, De Sario A, Wiegant J, Raap AK, Della Valle G, Bernardi G (1993) Correlations between isochores and chromosomal bands in the human genome. Proc Natl Acad Sci USA 90: 11929–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone S, Caccio S, Kusuda J, Andreozzi L, Bernardi G (1996) Identification of the gene-richest bands in human chromosomes. Gene 174:85–94 [DOI] [PubMed] [Google Scholar]
- Saccone S, Federico C, Solovei I, Croquette M-F, Della Valle G, Bernardi G (1999) Identification of the gene-richest bands in human prometaphase chromosomes. Chromosome Res 7:379–386 [DOI] [PubMed] [Google Scholar]
- Sanchez-Mazas A, Djoulah S, Busson M, Le Monnier de Gouville I, Poirier JC, Dehay C, Charron D, Excoffier L, Schneider S, Langaney A, Dausset J, Hors J (2000) A linkage disequilibrium map of the MHC region based on the analysis of 14 loci haplotypes in 50 French families. Eur J Hum Genet 8:33–41 [DOI] [PubMed] [Google Scholar]
- Stephens R, Horton R, Humphray S, Rowen L, Trowsdale J, Beck S (1999) Gene organisation, sequence variation and isochore structure at the centromeric boundary of the human MHC. J Mol Biol 291:789–799 [DOI] [PubMed] [Google Scholar]
- Tenzen T, Yamagata T, Fukagawa T, Sugaya K, Ando A, Inoko H, Gojobori T, Fujiyama A, Okumura K, Ikemura T (1997) Precise switching of DNA replication timing in the GC content transition area in the human major histocompatibility complex. Mol Cell Biol 17:4043–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. John Hopkins University Press, Baltimore [Google Scholar]
- Zöllner S, von Haeseler A (2000) A coalescent approach to study linkage disequilibrium between single-nucleotide polymorphisms. Am J Hum Genet 66: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]