Abstract
Three common protein isoforms of apolipoprotein E (apoE), encoded by the ε2, ε3, and ε4 alleles of the APOE gene, differ in their association with cardiovascular and Alzheimer's disease risk. To gain a better understanding of the genetic variation underlying this important polymorphism, we identified sequence haplotype variation in 5.5 kb of genomic DNA encompassing the whole of the APOE locus and adjoining flanking regions in 96 individuals from four populations: blacks from Jackson, MS (n=48 chromosomes), Mayans from Campeche, Mexico (n=48), Finns from North Karelia, Finland (n=48), and non-Hispanic whites from Rochester, MN (n=48). In the region sequenced, 23 sites varied (21 single nucleotide polymorphisms, or SNPs, 1 diallelic indel, and 1 multiallelic indel). The 22 diallelic sites defined 31 distinct haplotypes in the sample. The estimate of nucleotide diversity (site-specific heterozygosity) for the locus was 0.0005±0.0003. Sequence analysis of the chimpanzee APOE gene showed that it was most closely related to human ε4-type haplotypes, differing from the human consensus sequence at 67 synonymous (54 substitutions and 13 indels) and 9 nonsynonymous fixed positions. The evolutionary history of allelic divergence within humans was inferred from the pattern of haplotype relationships. This analysis suggests that haplotypes defining the ε3 and ε2 alleles are derived from the ancestral ε4s and that the ε3 group of haplotypes have increased in frequency, relative to ε4s, in the past 200,000 years. Substantial heterogeneity exists within all three classes of sequence haplotypes, and there are important interpopulation differences in the sequence variation underlying the protein isoforms that may be relevant to interpreting conflicting reports of phenotypic associations with variation in the common protein isoforms.
Introduction
Apolipoprotein E (apoE) (MIM 107741) is a plasma protein that plays a prominent role in lipid metabolism and cholesterol transport in human tissues (Davignon et al. 1988; Mahley and Huang 1999). In humans, apoE is polymorphic (Utermann et al. 1977). Three common isoforms of the protein are found in most populations: apoE2, apoE3, and apoE4, determined at the DNA level by the ε2, ε3, and ε4 alleles of the APOE gene on chromosome 19 (Weisgraber 1994). The alleles differ from one another by two nonsynonymous nucleotide polymorphisms causing amino acid substitutions at positions 112 and 158 of the protein (E2 is distinguished from E3 by cys→arg at 158, and E3 from E4 by cys→arg at 112) (Rall et al. 1982). The isoforms are metabolically distinct and differ in their affinity for lipoprotein particles, as well as in the extent to which they bind to apoE and low-density lipoprotein receptors (Hui et al. 1984).
The relative frequencies of the three common alleles have been investigated in a wide range of human populations, and no population studied to date has lacked polymorphism in APOE. The ε3 allele is the most common allele in all populations thus far investigated (de Knijff et al. 1994; Gerdes et al. 1996b; Corbo and Scacchi 1999), ranging in relative frequency from 0.536 in African Pygmies (n=70) (Zekraoui et al. 1997) to 0.911 in Mayans (n=135) (Kamboh 1995). The ε2 allele, on the other hand, is typically the least-common allele in most populations. The relative frequency of the ε4 allele ranges from 0.052 in Sardinians (n=280) (Corbo et al. 1995) to 0.407 in Pygmies (n=70) (Zekraoui et al. 1997) and is significantly negatively correlated with ε3 allele frequencies in African, Asian, and European populations (Corbo and Scacchi 1999). Statistically significant differences in allele frequencies, even among closely related white populations from Europe and the United States, have long been recognized (Davignon et al. 1988). Recently, a north-to-south decline in the relative frequency of the ε4 allele in Europe (and an associated increase in ε3 frequency) has been noted by several investigators (Corbo et al. 1995; Lucotte et al. 1997).
The large number of surveys of the APOE polymorphism reflect a long-standing interest in the relationship of allelic variation to interindividual differences in total serum cholesterol and LDL cholesterol levels (Sing and Davignon 1985), effects that make the gene a key candidate susceptibility locus for coronary artery disease (CAD) (Kaprio et al. 1991; Xhignesse et al. 1991). Numerous studies have confirmed that within a given population, individuals carrying at least one ε2 allele have higher levels of circulating apoE, lower levels of plasma total cholesterol, and higher triglyceride levels, whereas individuals with an ε4 allele have lower apoE levels, higher total cholesterol levels, and, accordingly, a higher risk of developing CAD (Davignon et al. 1988; de Knijff et al. 1994; Stengård et al. 1995). More recently, allelic variation at the APOE locus also has been associated with both familial and sporadic cases of Alzheimer disease (AD) (MIM 104300). It is now recognized that inheritance of ε4 alleles increases the risk of AD in a dose-dependent manner and predisposes carriers to an earlier age at onset than matched controls (Corder et al. 1993; Strittmatter et al. 1993). In relative terms, possession of an ε2 allele appears to protect carriers against the disease (Chartier-Harlin et al. 1994; Corder et al. 1994; Talbot et al. 1994). The ease with which such relationships may be described, however, belies the considerable context-dependence of the inferred associations. For example, the extent to which APOE variation predicts cholesterol level varies with genetic background, age, gender, and other environmental factors (diet, smoking, etc.) (Reilly et al. 1992; Davignon 1993). There are also ethnic differences in AD risk (see Tang et al. 1996, 1998; Sahota et al. 1997).
Somewhat surprisingly, the epidemiological focus has been almost exclusively on the classic isoforms; allelic heterogeneity in the APOE gene has been only incompletely characterized. The observation of enhanced levels of APOE mRNA in the brains and plasma of AD patients (Yamada et al. 1995; Lambert et al. 1997; Taddei et al. 1997) has led recently to the investigation of DNA-sequence variation in the APOE promoter region, resulting in the identification of five new polymorphic sites upstream of the gene (Artiga et al. 1998b; Lambert et al. 1998a). Single-site association studies focused on these variants, as well as on a site found at position +113 (Mui et al. 1996), have come to contradictory conclusions however, so that, to date, no clear consensus exists regarding the extent to which these sites may or may not be relevant to disease risk. Although the increasingly complex picture of APOE biological variation suggested by conflicting reports is sobering, it is important to recognize that even these many variants do not encompass the full extent of APOE variation present at the DNA-sequence level. In the first systematic resequencing study of the whole of the APOE locus and associated 5′ and 3′ flanking DNA, we recently identified 22 variable sites, 14 of which had not been previously reported (Nickerson et al., in press). The relationship of this newly identified sequence variation to the three common isoforms which have formed the basis of most epidemiological inquiry to date is the focus of this report.
Ultimately, the fundamental question dogging both the epidemiological and population-genetics literature is why different apoE isoforms exist at all, with generally similar frequencies around the world, particularly when nonhuman primate relatives show no such variation (Zannis et al. 1985). Several investigators have suggested that ε4 is the ancestral form of the gene (Hixson et al. 1988; Hanlon and Rubinsztein 1995; Seixas et al. 1999). If this is so, then the high relative frequency of the derived ε3 allele in all human populations clearly demands explanation. Has the ε3 allele drifted to its current frequency by chance, or is it selectively maintained? Have the amino acid differences that determine the ε3 isoform arisen once, or is there evidence that they have recurred multiple times? These questions can be addressed by consideration of the evolutionary history of APOE, recorded in the arrangement of polymorphic sites linked as DNA-sequence haplotypes. When variation is considered in this way, we can assess how and when the three common alleles arose and can determine what forces may have contributed to their current global distribution. We have undertaken such an analysis here. We show that there is considerable sequence heterogeneity among the haplotypes in each class of common allele (ε2, ε3, and ε4) and that the history of allelic divergence suggested by the observed haplotype variation is consistent with a global expansion of ε3 alleles, relative to the ancestral ε4 alleles. Detailed consideration of the heterogeneity underlying each of the common alleles also provides important new insights into the population-specific distribution of subclasses of alleles, which may go some way toward explaining reported discrepancies in the association between variation in risk of disease and variation in the gene marked by the common apoE isoforms.
Material and Methods
Populations Sampled
Individuals from four populations were sampled for sequence variation: blacks from Jackson, Mississippi (n=48 chromosomes), recruited as part of an ongoing study of hypertension in American blacks (see Family Blood Pressure Program Web site); whites from North Karelia, Finland (n=48), collected as part of the Finrisk Hemostasis Study (Salomaa et al. 1994); whites from Rochester, MN (n=48) from the Rochester Family Heart Study (Turner et al. 1989); and Mayans, with little external admixture (n=48), sampled by one of us (K.M.W.) in the state of Campeche on the Yucatan Peninsula of Mexico (Kidd et al. 1991). All subjects were selected for this survey without regard to their disease status or their levels of any risk-factor trait.
DNA Sequencing
DNA sequencing was done on templates prepared by PCR from diploid genomic DNA samples following procedures described in Nickerson et al. (in press). Extensive confirmatory resequencing was also done on a subset of the samples to aid haplotype determination, as described below.
Human and chimpanzee are sufficiently similar in DNA sequence at this locus for us to use the same array of PCR and sequencing primers for the chimpanzee-sequence analysis (exact details of the primers used are available on request). DNA from a single chimpanzee (Pan troglodytes) was sequenced as described above and was aligned to the human genomic consensus sequence, using the SIM sequence-alignment algorithm (Huang et al. 1990). The Genbank accession number for the chimpanzee APOE reference sequence is AF261280.
Haplotype Determination
Haplotypes were determined by a sequential procedure involving application of an algorithm that first identifies unambiguous haplotypes in homozygotes and single-site heterozygotes and then follows a series of inferential steps to arrive at a call of likely haplotypes (Clark 1990). To simplify inference, we began by ignoring six singleton polymorphisms (i.e., sites in which only a single copy of the less-common nucleotide variant was present in the combined sample—namely, 308, 545, 2907, 3106, 3673, and 3701), and a multiallelic indel polymorphism at site 5229B. Key pairs of sites were then identified for allele-specific PCR to assign phasing of a subset of multiply heterozygous individuals. This phase information then was incorporated into the inferential procedure, until all genotypes were resolved (see Clark et al. 1998). Allele-specific PCR was also done, to assign phase for every genotype that could have been composed of more than one pair of haplotypes that were unambiguously present in the sample. The phasings of the singleton variants were subsequently inferred either by context (e.g., the singletons at sites 545 and 3701 occurred in homozygous backgrounds and thus were unambiguously assigned) or by reference to haplotypes inferred in a larger survey of variation at the same locus (Nickerson et al., in press and unpublished data).
Population Genetic Analysis
Two measures of diversity were computed for each of the four population samples and the combined (pooled) sample: θ, an estimate of the expected per-site nucleotide heterozygosity, theoretically equal to the neutral mutation parameter 4Neμ (Watterson 1975) and π, the direct estimate of per-site heterozygosity derived from the average pairwise sequence difference (Nei 1987). The test statistic D (Tajima 1989) was used to compare these summary statistics. Under neutrality, the two estimates should be equal and D=0. Two test statistics—related to Tajima’s D, F, and DFL (Fu and Li 1993)—were used to compare the observed number of singleton polymorphisms in each sample with those expected under a neutral model (we used the form of the test statistics that explicitly incorporates outgroup information). Finally, a fourth test statistic, Fs (Fu 1997), was used to compare the observed number of sequence haplotypes in our samples to the number expected under the assumption of an infinite-sites (IS) model of mutation with no recombination. Again, under neutrality (and if no recombination is acting), the two estimates are expected to be equal. Significance values for each of the four test statistics were computed by comparison to a distribution of estimates, calculated for 1,000 random samples of the same size and level of polymorphism as the observed data and generated under a Wright-Fisher equilibrium model of the coalescent, with no recombination (Hudson 1990).
Heterogeneity in the D, F, and DFL test statistics across the length of the 5.5-kb sequenced region was examined by use of the sliding window feature of the program DnaSP v. 3.0 (Rozas and Rozas 1999). Statistics were calculated for overlapping windows of 750 bp, placed at 25-bp intervals along the sequence. Sequence divergence between the chimpanzee outgroup sequence and the human polymorphic sample was measured as K, the net sequence divergence (Nei 1987). Sliding-window estimation of π and K (using the same window and step size as in the test-statistic analyses) allowed consideration of cross-locus heterogeneity in the ratio of polymorphism to divergence. Pairwise linkage disequilibrium was measured with the linkage disequilibrium parameter D, calculated as D=Pij-pipj, where Pij is the frequency of the most common gametic type for a pair of sites, and pi and pj are the frequencies of the nucleotides in that haplotype (Hartl and Clark 1997). The genetic differentiation among population samples was measured with the statistic FST (Wright 1931), which measures the fraction of the total genetic variation that is between, rather than within, populations, estimated with the program Arlequin.
The mutational relationships among the inferred sequence haplotypes, which reflect the evolutionary history of genetic changes at the APOE locus, were visualized by means of the Reduced Median (RM) Network algorithm (Bandelt et al. 1995). The RM algorithm unambiguously links haplotypes that differ by a single variable site and then employs a frequency-based compatibility criterion to choose among equally likely mutational paths linking remaining haplotypes. In other words, the algorithm assumes that mutational events typically proceed from a more frequent haplotype to a less frequent one (a statistical assumption that is supported by coalescent theory). Although this assumption can be violated if selection is acting, for a data set such as the one described here—in which a large number of closely related haplotypes are compared—the assumption is rarely required; thus, it is unlikely this assumption biases the final result. In cases where apparent parallelisms cannot be resolved, the uncertainty is depicted as a reticulation, or loop, in the network. Haplotypes are represented in each network by open circles, and the area of each circle is proportional to the number of copies of that haplotype in the sample. RM networks were constructed for the combined total sample and each of the four population samples using the RM option of the program Network 2.0 by A. Röhl.
Maximum-likelihood (ML) estimates of the neutral mutation parameter θ were derived for the IS-compatible data only, following the approach of Griffiths and Tavaré (1997). Sites that exhibit homoplasies cannot be considered in this analysis, which also assumes stationary populations and selectively neutral variation. Estimates of diversity for the IS-compatible samples were in no case significantly different from those estimated for the original data, suggesting that no major biases were introduced by data truncation. The analysis was performed with the combined sample, as well as each of the individual population samples. The assumption of constant population size was tested explicitly by comparing likelihood estimates for models with and without population expansion. No significant improvement in likelihood was observed when exponential populational growth was included in the model, when either the combined sample or separate population samples were considered. Therefore, all calculations were performed assuming constant population size. Associated age estimates, for both the time to the most recent common ancestor (TMRCA) and individual mutations, were generated in units of 2Ne generations and converted to years using the estimate of effective population size derived from the ML estimate of θ (4Neμ) and an estimated generation time of 20 years. The latter conversion assumed a mutation rate, μ, of 1.29×10-4/locus/generation, calculated under the assumption of a human-chimp divergence of 5 million years before present (BP) (Horai et al. 1992), as described previously by Harris and Hey (1999). This estimate is calculated from the net number of sequence differences observed between human and chimpanzee (64.7), an estimate which ignores length variation/divergence and does not take functional constraint into account. The reported uncertainty in the age estimates also does not take uncertainty in the estimates of either Ne and μ into account. Although the simplifications preclude precision, the approach provides a general characterization of the evolutionary history of a gene. All calculations were carried out with the program Genetree (Griffiths and Tavaré 1997).
Results
APOE Diversity at the Nucleotide Level
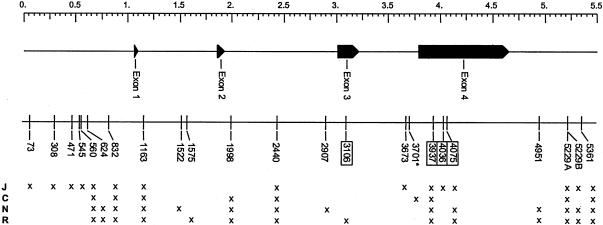
The nature, amount, and population distribution of nucleotide polymorphism identified at the APOE locus in three of the four populations investigated here is described in the report by Nickerson et al. (in press). In summary, 22 variable sites were identified in the 5,491-bp region surveyed: 21 diallelic single-nucleotide polymorphisms (SNPs) and one multiallelic insertion/deletion polymorphism (at 5229A, figure 1 and table 1). Five sites were singletons; that is, only a single copy of the rarer allele was observed in the sample of 144 chromosomes (i.e., sites 308, 545, 2907, 3106, and 3673; site 3106 is a nonsynonymous substitution), whereas another five “doubleton” sites had two copies of the rarer nucleotide (sites 73, 471, 1522, 1575, and 4036; site 4036 is a nonsynonymous substitution). In this study, APOE sequence variation in a fourth population, from the state of Campeche, Mexico, was also surveyed, bringing the total number of chromosomes investigated for sequence diversity to 192. One new, singleton insertion variant was identified in the Campeche sample, at position 3701 of the reference sequence (involving an insertion of CT dinucleotide) (fig. 1 and table 1). An additional eight polymorphic sites were present in the Campeche sample, all of which had been identified in the earlier investigation (560, 832, 1163, 1998, 2440, 3937, 4075, and 5229B). None of the previously identified singleton or doubleton variant alleles were observed in the new sample. The positions of all 23 variants, relative to the reference sequence reported by Nickerson et al. (in press), are shown in figure 1.
Figure 1.
Locations of polymorphic variants in and around the APOE gene. The genomic location of the 23 DNA variants, identified by sequencing 5.5 kb in 96 individuals, is shown, below the exon-intron structure of the APOE gene. An asterisk (*) marks the new variant identified in the Mayan sample from Campeche at position 3701, and x’s show the population distribution of the observed polymorphisms (J = Jackson, C = Campeche, N = North Karelia, and R = Rochester). Variants that result in amino acid substitutions are boxed.
Table 1.
Observed Haplotypes of APOE
| Siteb |
PopulationCountsc |
|||||||||||||||||||||||||||
| Haplotypea | 73 | 308 | 471 | 545 | 560 | 624 | 832 | 1163 | 1522 | 1575 | 1998 | 2440 | 2907 | 3106 | 3673 | 3701 | 3937* | 4036 | 4075* | 4951 | 5229A | 5229B | 5361 | J | C | N | R | P |
| Chimpd | C | C | A | C | A | T | G | G | G | C | G | G | T | T | C | — | C | C | C | A | nd | G | T | |||||
| ε2: | ||||||||||||||||||||||||||||
| 10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | T | . | 1 | 0 | 1 | 2 | 4 | |
| 16 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | T | . | T | . | T | . | 1 | 0 | 0 | 1 | 2 | |
| 22 | . | . | . | . | T | C | . | . | . | T | . | . | . | . | . | . | T | . | T | . | T | . | 0 | 0 | 0 | 1 | 1 | |
| 9 | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | T | . | T | . | T | . | 0 | 0 | 1 | 4 | 5 | |
| 24 | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | T | . | T | C | T | . | 0 | 0 | 0 | 1 | 1 | |
| ε3: | ||||||||||||||||||||||||||||
| 26 | . | . | . | . | T | C | T | C | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 0 | 0 | 0 | 1 | 1 | |
| 21 | . | . | . | . | . | C | T | C | . | T | . | . | . | . | . | . | T | . | . | . | . | . | 0 | 0 | 0 | 1 | 1 | |
| 4e | . | . | . | . | T | . | T | C | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 1 | 5 | 2 | 8 | 16 | |
| 29 | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | CT | T | . | . | . | . | . | 0 | 1 | 0 | 0 | 1 | |
| 1e | . | . | . | . | . | . | T | C | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 8 | 19 | 11 | 7 | 45 | |
| 30 | . | . | . | . | . | . | T | C | . | . | . | . | G | . | . | . | T | . | . | . | . | . | 0 | 0 | 1 | 0 | 1 | |
| 14 | . | . | . | . | . | . | T | C | A | . | . | . | . | . | . | . | T | . | . | . | . | C | 0 | 0 | 2 | 0 | 2 | |
| 19 | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 1 | 0 | 0 | 0 | 1 | |
| 6e | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 3 | 5 | 0 | 0 | 8 | |
| 27 | . | T | . | . | T | . | . | . | . | . | . | . | . | . | G | . | T | . | . | . | . | . | 1 | 0 | 0 | 0 | 1 | |
| 8e | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | 2 | 3 | 0 | 0 | 5 | |
| 3e | . | . | . | . | T | . | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | . | 8 | 3 | 3 | 1 | 15 | |
| 11e | . | . | . | . | . | C | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | . | 0 | 0 | 0 | 2 | 2 | |
| 2e | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | . | 15 | 6 | 11 | 11 | 43 | |
| 28 | . | . | . | T | . | . | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | . | 1 | 0 | 0 | 0 | 1 | |
| 7e | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | T | . | . | . | . | C | 1 | 1 | 4 | 2 | 8 | |
| 25 | . | . | . | . | . | . | . | . | . | . | A | A | . | . | . | . | T | . | . | . | . | C | 0 | 0 | 1 | 0 | 1 | |
| ε4: | ||||||||||||||||||||||||||||
| 17 | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | 2 | 0 | 0 | 0 | 2 | |
| 12 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 0 | 0 | 1 | 2 | |
| 13 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | 0 | 0 | 1 | 1 | 2 | |
| 20 | . | . | G | . | T | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 0 | 0 | 0 | 1 | |
| 23 | . | . | G | . | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 1 | 0 | 0 | 0 | 1 | |
| 15 | . | . | . | . | . | C | T | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | 0 | 0 | 0 | 2 | 2 | |
| 18 | . | . | . | . | . | . | T | . | . | . | A | . | . | . | . | . | . | . | . | C | . | C | 0 | 0 | 1 | 0 | 1 | |
| 5 | . | . | . | . | . | . | T | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | 0 | 5 | 9 | 1 | 15 | |
| 31 | . | . | . | . | . | . | T | . | . | . | A | . | . | C | . | . | . | . | . | . | . | . | 0 | 0 | 0 | 1 | 1 | |
Haplotypes are numbered, in descending order, according to their relative frequency in the pooled sample.
Location of the 23 sites, relative to the reference sequence reported in Nickerson et al. (in press). Asterisks (*) mark the nonsynonymous cSNPs that define the three common protein isoforms (as indicated in the left-most column). Bases identical to those in the chimp are marked with a dot. The phase of the multiallelic indel at site 5229A was not determined.
J = Jackson, MS; C = Campeche, Mexico; N = North Karelia, Finland; R = Rochester, MN; and P = pooled sample.
nd = ancestral state of the multiallelic indel at site 5229A could not be determined.
Haplotypes present as homozygotes or single-site heterozygotes.
Summary statistics describing the sequence diversity in the combined sample, and each of the four population samples, are presented in table 2. Overall, average per-nucleotide expected heterozygosity (θ) for the total sample, estimated from the observed number of polymorphic sites (Watterson 1975), was 0.0007±0.0002. An equivalent estimate, based on the average pairwise sequence difference π (Nei 1987), was slightly lower (0.0005±0.0003). These estimates are lower than, but not significantly different from, estimates of diversity reported for other autosomal (Li and Sadler 1991; Harding et al. 1997; Clark et al. 1998; Rana et al. 1999; Rieder et al. 1999; Hamblin and Di Rienzo 2000) and X-linked (Zietkiewicz et al. 1998; Harris and Hey 1999; Jaruzelska et al. 1999; Kaessmann et al. 1999) human genetic loci. Estimated values of Tajima’s (1989) D and Fu and Li’s (1993) DFL and F statistics did not differ significantly from 0. More than the expected number of haplotypes were observed in all four population samples, as well as the combined sample and this excess haplotype variation, as measured by the Fs statistic (Fu 1997), was highly significant in both the Jackson and combined samples. The observed excess haplotype diversity is likely to reflect the effects of recurrent mutation and/or interallelic recombination, as discussed below.
Table 2.
Diversity Estimates and Neutrality Tests for APOE
|
Results for Population Samples |
|||||
| Jackson | Campeche | North Karelia | Rochester | Pooled Sample | |
| na | 48 | 48 | 48 | 48 | 192 |
| Sb | 14 | 8 | 13 | 13 | 22 |
| sc | 4 | 2 | 2 | 1 | 6 |
| θ (× 10−4)d | 5.75 ± 2.17 | 3.28 ± 1.44 | 5.33 ± 2.05 | 5.33 ± 2.05 | 6.87 ± 2.05 |
| π (× 10−4)e | 4.36 ± 2.41 | 3.92 ± 2.21 | 5.62 ± 2.97 | 6.37 ± 3.30 | 5.31 ± 2.80 |
| D (Tajima 1989) | −.736 | .531 | .163 | .586 | −.619 |
| DFL(Fu and Li 1993) | −.438 | −.148 | .506 | 1.05 | −1.09 |
| F (Fu and Li 1993) | −.650 | −.090 | .464 | 1.07 | −1.09 |
| No. of haplotypes | 16 | 9 | 13 | 18 | 31 |
| Expected no. of haplotypesf | 7.8 ± 2.2 | 7.3 ± 2.1 | 9.1 ± 2.4 | 9.8 ± 2.5 | 12.9 ± 3.1 |
| Fs (Fu 1997) | −7.080*** | −.963 | −2.378 | −6.203* | −15.055*** |
Number of chromosomes surveyed.
Number of polymorphic sites, excluding site 5229A indel.
Number of singleton sites.
Expected heterozygosity per nucleotide, SEs derived from variance estimate assuming no recombination (Watterson 1975).
Average pairwise sequence difference, SEs derived from stochastic and sampling variance, assuming no recombination (Nei 1987).
Mean ± SD of estimates derived from distribution of 1,000 random samples (see Material and Methods).
P<.05.
P<.001.
Haplotype Variation Underlying the Common APOE Alleles
The 22 variants observed in the total sample of 192 chromosomes were found to segregate as 31 distinct sequence haplotypes (the phase of the multiallelic indel at site 5229A was not determined). These haplotypes, numbered in order of descending relative frequency in the combined sample, aligned with respect to overall sequence similarity, and arranged according to the classical protein-allele that their variation at positions 3937 and 4075 defines (i.e., ε2, ε3, ε4), are shown in table 1. We observed no examples of the fourth configuration of isoform-determining cSNPs in our sample (i.e., the haplotype combination C-T at sites 3937 and 4075, as opposed to T-T [ε2], T-C [ε3], or C-C [ε4]). The failure to observe the C-T haplotype in our sample is consistent with previous surveys of apoE polymorphism, which have never observed that combination of amino acid substitutions (de Knijff et al. 1994).
In the combined sample, 13 (0.068) of the observed sequence haplotypes comprise ε2 alleles, 152 (0.792) are ε3 alleles, and 27 (0.140) are ε4 alleles (table 1). The relative proportions of the three alleles vary among population samples, however, with the greatest relative frequency differences associated with ε2-type sequence haplotypes (table 1). This latter class of sequence haplotype was not observed at all in the Campeche sample, for example (relative frequencies of ε2, ε3, and ε4 were 0, 0.896, and 0.104, respectively), and is much more common in the Rochester sample (relative frequencies 0.188, 0.687, and 0.125) than in either of the other two population samples (relative frequencies in the Jackson sample were 0.042, 0.854, and 0.104, and those in the North Karelia sample were 0.042, 0.729, and 0.229). Marked interpopulation differences in the relative frequency of the ε2 allele (χ2=8.29, P=.004) are consistent with previous observations (Corbo and Scacchi 1999).
Full sequence analysis and haplotype determination reveal substantial heterogeneity within each of the three protein allelic classes (table 1). The rarest allelic class, ε2, is comprised of five distinct sequence haplotypes in the total sample, with variation at four nucleotide sites (table 1). Similarly, the most common allelic class, ε3, comprises 17 distinct sequence haplotypes, which vary at 14 sites. Finally, nine different ε4-type haplotypes were observed, which vary at 10 positions. One half of the polymorphic sites found in either the ε2 or the ε3 group of haplotypes occur as singletons in those groups, and five of the six singletons found in the combined sample are linked to ε3-type sequence haplotypes. In contrast, only 2 of the 10 sites segregating within the ε4 group occur as singletons. The large number of variable sites found among the comparatively less common ε4 class of haplotypes results in an estimate of θ for this group of alleles that is the same as that observed for the ε3s (0.0005±0.0002 in both cases). ε2-type haplotypes are less internally polymorphic by the same measure (θ=0.0002±0.0001).
Comparison of the homologous genomic region in chimpanzee (Pan troglodytes) identified 76 fixed sequence differences between the human consensus (Genbank accession AF261279) and chimpanzee (GB AF261280) sequences, including 63 nucleotide substitutions and 13 length differences ranging from 1 to 36 bp in length. The associated estimate of net sequence divergence between human and chimpanzee, which takes into account human polymorphic variation (Nei 1987), was 64.7 (calculation of this estimate ignored length variation/divergence). For every diallelic nucleotide polymorphism observed in humans, one of the two alleles was present at the homologous position in chimpanzee (table 1). In all but one case (site 3937), the more common human variant corresponded to the presumed ancestral allele. Although the chimpanzee haplotype sequenced here cannot be considered as representative of all APOE variation in that species, the nucleotide observed at this position in chimpanzee (C) is also present in other nonhuman primate species (Hanlon and Rubinsztein 1995), suggesting that it is, in fact, the true ancestral state at this position. Haplotype 12, one of the ε4 haplotypes, has the same configuration of nucleotides at the 22 variable positions as is observed at homologous sequence positions in chimpanzee, but this haplotype still differs from that of the chimpanzee at 75 additional positions. This observation is consistent with previous analyses, which have suggested that the ε4 allele is the ancestral APOE allele on the basis either of sequence comparisons with nonhuman primates (Hixson et al. 1988; Hanlon and Rubinsztein 1995) or of patterns of linkage disequilibrium between APOE and the downstream locus APOCI (Seixas et al. 1999).
Evolutionary History of APOE Allelic Divergence
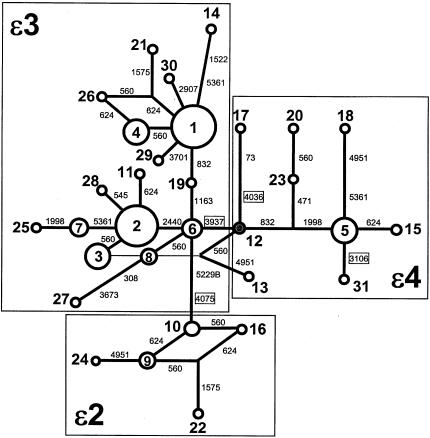
The likely genealogical history of the observed variation was inferred from RM networks (Bandelt et al. 1995) constructed for haplotypes in the combined sample (fig. 2) and each of the separate population samples (fig. 3). In a RM network, circles correspond to individually distinct haplotypes (as presented in table 1), with the size of the circles scaled to reflect the relative frequency of each haplotype in the sample. The haplotypes are connected to one another by branches, along which mutational differences are indicated. For example, in figure 2, haplotypes 5 and 18 differ from one another by mutational changes at sites 4951 and 5361, whereas haplotypes 5 and 15 differ by a change at site 624 only. When the connections among the haplotypes are traced, the history of mutational changes at the locus can be inferred, particularly if outgroup information allows identification of the “root” haplotype in the network (haplotype 12, as noted above).
Figure 2.
RM network of APOE sequence haplotypes in the pooled total sample. Mutational relationships are indicated by lines linking the 31 unique haplotypes, indicated in the network as circles (lighter lines indicate inferred mutational relationships that are less likely if homoplasy caused by recombination or recurrent mutation involving site 560 is assumed). The size of each circle is proportional to the relative frequency of the haplotype in the total sample. Mutational differences between haplotypes are indicated on the branches of the network (variants and haplotypes are numbered as in table 1; mutations that result in amino acid substitutions are boxed). Haplotype 12 (shaded) is the root haplotype in the network. The three large boxes indicate the groups of haplotypes that define the ε2, ε3, and ε4 APOE alleles.
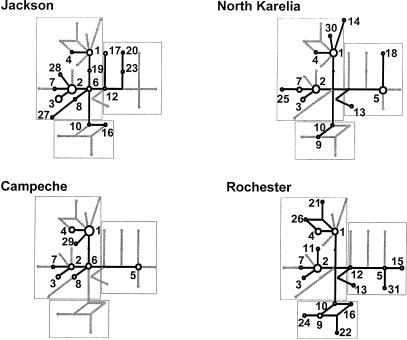
Figure 3.
RM networks of APOE haplotypes in the four population samples. Networks describing haplotype relationships, as they are found in each individual sample, are shown in black. Other branches, which are present in the total network but missing in a particular sample, are indicated in light gray. Numbers label haplotypes only (mutations are not indicated). Boxes indicate groups of haplotypes that define the ε2, ε3, and ε4 alleles, as presented in figure 2.
As shown in the RM network for the total sample (fig. 2), the 31 APOE haplotypes fall into four major lineages, or clades, which correspond coherently to the protein allelic classes defined by variation at sites 3937 and 4075, consistent with the arrangement depicted in table 1. The ε2 and ε4 groups of haplotypes each comprise phylogenetically distinct lineages. The ε3 group of haplotypes, however, is comprised of two discrete, equally frequent clades, separated by three mutational steps (involving changes at sites 832, 1163, and 2440) and two transitional haplotypes (6 and 19), suggesting a significant bifurcation in the lineage undetected by previous surveys of either protein-level or DNA-level variation. A third minor subclade, leading to haplotypes 8 and 27, also extends from the core haplotype (6) in this group.
The root haplotype in the network is haplotype 12, which encodes an apoE4 isoform. Only two copies of this haplotype are present in the total sample, one copy in Jackson and one in Rochester (table 1). The presence of the root in the sample from Jackson is consistent with other studies of human genetic variation, which have found root haplotypes in black African populations (e.g., Harding et al. 1997; Harris and Hey 1999). The RM network shows clearly that the predominant ε3 allelic class is derived from the ε4 group by a single-step change from the root haplotype, involving site 3937, the cSNP that causes the cys→arg substitution at residue 112 of the apoE protein. ε2-type haplotypes are derived from the core haplotype of the ε3 group by two changes, one at site 5229B and one at the previously recognized cSNP at site 4075. Apart from the core ε3 haplotype (haplotype 6), all other haplotypes in the combined sample are two mutational steps from the root haplotype.
Homoplasy of Upstream Regulatory-Region Variants
A prominent feature of the data illustrated in the RM network is the homoplasy (the inferred occurrence of multiple independent evolutionary events giving rise to the same allelic state at a variable site) that affects sites in and around the APOE locus. In all, 7 of the 22 variants in the phased haplotypes show some evidence of homoplasy, and two sites in particular, 560 and 624 (corresponding, respectively, to the regulatory region variants at −491 and −427 reported previously by Artiga et al. 1998b) occur repeatedly in the RM network. Two unresolved loops and a double loop broken by an assumption of recurrent mutation at site 560, all suggest multiple mutational and/or short recombinational events at those sites. Site 560 (−491) is particularly homoplasic, occurring eight separate times in the network of the combined sample (between haplotypes 1 and 4, 1 and 26, 12 and 13, 6 and 8, 2 and 3, 20 and 23, 10 and 16, and 9 and 22 in fig. 2). In fact, homoplasy at site 560 was so pronounced that confirmatory allele-specific amplifications on >100 individuals were done to ensure that the site had been correctly phased. Site 624 (−427) occurs six times in the network. Other homoplasic sites include 4951 and 5361, in the 3′ flanking region (with three occurrences each), and sites 832 (−219), 1575, and 1998 (with two occurrences each) (table 3). The inferred homoplasy is not restricted to any particular class of allele: occurrences of the same mutational state at different places in the network are associated with ε2-, ε3-, and ε4-type haplotypes. This observation is consistent with previous reports, which have suggested that the association of variation at these sites with AD risk is often independent of ε4 allelic status (Artiga et al. 1998a; Bullido et al. 1998).
Table 3.
APOE Site Heterozygosity and Observed Homoplasy versus Potential Mutability
| Sitea | Heterozygosity | Homoplasyb | Repeat Motif | Mutability Typec | Surrounding Sequenced |
| 73 | .02 | 1 | MIR | 3 | AGGATTCACGcCCTGGCAATT |
| 308 | .01 | 1 | 0 | CCACCCCTCCcATCCCACTTC | |
| 471 | .02 | 1 | AluSq | 0 | CCAAGTAGCTaGGATTACAGG |
| 545 | .01 | 1 | AluSq | 0 | ACCATGTTGGcCAGGCTGGTC |
| 560 | .35 | 8 | AluSq | 0 | CTGGTCTCAAaCTCCTGACCT |
| 624 | .13 | 6 | AluSq | 0 | AGGCGTGAGCtACCGCCCCCA |
| 832 | .50 | 2 | 0 | AGGGTGTCTGgATTACTGGGC | |
| 1163 | .46 | 1 | 0 | ACCCTGGGAAgCCCTGGCCTC | |
| 1522 | .02 | 1 | 3 | TGAGGTTGGAgCTTAGAATGT | |
| 1575 | .02 | 2 | 1 | GAGATGGAACcGGCGGTGGGG | |
| 1998 | .19 | 2 | 0 | CCCCATTCAGgCAGACCCTGG | |
| 2440 | .46 | 1 | 0 | CTGGCTGGGAgTTAGAGGTTT | |
| 2907 | .01 | 1 | 3 | CTGCCCACCAtGGCTCCAAAG | |
| 3106 | .01 | 1 | 0 | CGCTGGGAACtGGCACTGGGT | |
| 3673 | .01 | 1 | 0 | GCCTCTGCCCcGTTCCTTCTC | |
| 3701 | .01 | 1 | 0 | TGGTCTCTCT^GGCTCATCCC | |
| 3937 | .24 | 1 | 1 | GGAGGACGTGcGCGGCCGCCT | |
| 4036 | .02 | 1 | 1 | GCGCAAGCTGcGTAAGCGGCT | |
| 4075 | .13 | 1 | 1 | CCTGCAGAAGcGCCTGGCAGT | |
| 4951 | .04 | 3 | AluSg | 0 | AGTAGAGACGaGCTTTCACCA |
| 5229Ae | .63 | … | AluJo | 2 | TGGGGGGGGG^GTGGTGTGTG |
| 5229B | .13 | 1 | AluJo | 2 | TGGGGGGGGGgTGGTGTGTGT |
| 5361 | .12 | 3 | 2 | CCCAGCTTTTtATTATTATTT |
Location of polymorphic site relative to reference sequence, as presented in Nickerson et al. (in press).
Homoplasy = homoplasy count, i.e. number of independent mutations that must be invoked to explain observed haplotype diversity in the combined sample.
Type 0 = no known mutational motif; type 1 = CpG; type 2 = mononucleotide run; type 3 = alpha polymerase arrest site (Templeton et al. 2000).
^ = location of polymorphic indel.
Multiallelic indel (haplotype phase undetermined), all other sites are diallelic SNPs or indels.
The observed homoplasy, which is consistent with the excess haplotype diversity suggested by the Fs test statistic, can be explained as arising either from recurrent mutation, or as a consequence of interhaplotype recombination (either reciprocal exchange or gene conversion). Reciprocal recombination would be expected to lead, at least some of the time, to exchanges of multiple adjacent sites, like those described in a recent investigation of variation at the LPL locus (Templeton et al. 2000), but these are not observed. Also, unlike in LPL (Clark et al. 1998), only a small proportion of site pairs show all four gametic types (38, or 16.5%, of 231 comparisons in the combined sample) (table 4), suggesting that the data are dominated more by extreme homoplasy in a few sites than extensive recombination across the whole of the sequenced region. That homoplasy affects sites located at the 5′ and 3′ ends of the sequenced region is consistent with the inferred lack of reciprocal exchange events, and yet, as noted earlier, no instances of the T-C cSNP haplotype involving sites 3937 and 4075 were observed.
Table 4.
Significant Pairwise Linkage Disequilibria (D) and Four-Gamete Site Pair Counts
| Jackson | Campeche | North Karelia | Rochester | Pooled Sample | |
|
Da for Sample |
|||||
| Site 1/Site 2: | |||||
| 73/4036 | .040*** | .010*** | |||
| 624/4075 | .078** | .032*** | |||
| 624/5229B | .078** | .032*** | |||
| 832/1163 | .140*** | .195*** | −.153*** | .199*** | .187*** |
| 832/1998 | −.039 | −.084** | .047* | .051*** | |
| 832/2440 | .119*** | .130*** | .214*** | −.146*** | −.167*** |
| 832/3937 | .018 | −.039 | −.084** | .029 | .045*** |
| 832/4075 | −.010 | .023 | −.082** | −.031*** | |
| 832/5229B | −.010 | .023 | −.082** | −.031*** | |
| 1163/1998 | .054* | −.076** | −.030 | −.037*** | |
| 1163/2440 | .109*** | .109*** | −.132*** | −.118*** | −.129*** |
| 1163/3937 | −.022 | .054* | −.076** | −.044 | −.050*** |
| 1998/3937 | .093*** | .156*** | .073*** | .084*** | |
| 2440/3937 | .054* | −.022 | −.091** | −.042 | −.051*** |
| 4075/5229B | .040*** |
.040*** |
.152*** |
.063*** |
|
| Data Summary for Sample |
|||||
| Total no. of pairwise LD comparisons | 45 | 15 | 55 | 66 | 120 |
| No. significant at P< .001 | 5 | 4 | 5 | 5 | 15 |
| Proportion | 11.1% | 26.7% | 9.1% | 7.6% | 12.5% |
| Total no. of pairwise four-gamete comparisons | 91 | 28 | 78 | 78 | 231 |
| No. with four gametes | 9 | 3 | 17 | 25 | 38 |
| Proportion | 9.9% | 10.7% | 21.8% | 32.0% | 16.5% |
Disequilibrium statistics are reported as D=Pij-pipj, where Pij is the frequency of the most common gametic type for a pair of sites, pi and pj are the frequencies of the nucleotides in that haplotype. Tail probability is reported for Fisher's Exact Test.
.01<P<.05.
.001<P<.01.
P<.001.
Although nearly half of the variable positions identified at APOE arise at known mutational hotspot motifs, only two of the seven homoplasic sites (sites 1575 and 5361) occur in such regions (table 3). This would argue against recurrent mutation as generating the inferred homoplasy. The two most homoplasic sites, sites 560 and 624, fall in the midst of an Alu repeat sequence, as does the moderately homoplasic site 4951 (table 3). The level of sequence polymorphism associated with such repetitive sequences in the 5.5 kb surveyed is not higher than that observed in nonrepeat regions (Nickerson et al., in press) so the significance, if any, of these associations is unclear. In the absence of direct empirical evidence, it is impossible to decide whether gene conversion or recurrent mutation is a more plausible explanation of the observed patterns. Interestingly, although the two sites that define the ε2-ε3-ε4 polymorphism (3937 and 4075) both fall in CpG dinucleotides (that are known to be methylated in APOE [Larsen et al. 1993] and thus, in principle, are likely to be hotspots of mutation [Cooper and Krawczak 1989]), the RM network does not suggest homoplasy at these sites.
The high degree of site homoplasy, combined with the large number of low frequency variants, explains the comparatively low level of linkage disequilibrium (LD) observed among site pairs at the APOE locus within each of the sampled populations. Five or fewer site pairs in each population sample were found with a significant level of LD (Fisher's exact test P<.001) and only 15 pairs (12.5% of all pairwise comparisons) were significant at this level in the pooled sample (table 4). Site 832 was in significant LD with the largest number of sites in the combined sample (1163, 1998, 2440, 3937, 4075, and 5229B). However, only associations between site 832 and sites 1163 and 2440 were consistently observed across separate population samples. It is also important to note that, whereas 7 of the 15 significant site pairs in the combined sample involve the commonly-typed cSNPs at sites 3937 and 4075, only two pairs involving these sites (1998-3937 and 4075-5229B) show consistent associations in the separate population samples (table 4). The pattern of LD observed in this small sample is generally similar to LD estimated from a much larger sample of unphased genotype data for the same sites (Nickerson et al., in press and unpublished data). More precise understanding of the LD pattern in APOE awaits detailed haplotype-based analysis of that larger sample.
Population Differences in APOE Diversity
Sequence haplotype diversity, as it is found in the separate samples, suggests small but significant differences in the genetic make-up of the four populations surveyed. The overall estimate of FST (Weir 1996) for the combined sample was calculated as 0.060 (P<.001, by haplotype permutation). Pairwise estimates of FST (table 5) ranged from a high of 0.127 between samples from Jackson and Campeche (P<.001) to a low of 0.034 between Rochester and North Karelia (P<.05). These estimates are comparable to those calculated from an average of site-specific estimates, where an overall FST value of 0.045 was observed (Nickerson et al., in press). The higher degree of differentiation of the populations at the haplotype level is consistent with the population-specific distribution of haplotypes: 18 (0.581) of the 31 haplotypes identified occur in single population samples (table 1), whereas 11 (0.478) of the 23 variable sites are restricted to one sample only (Nickerson et al., in press) (table 1). The greatest proportion of unique haplotypes (7 of 18, or 0.389) was found in the Rochester sample. Six haplotypes were shared by two samples only, two haplotypes by three samples, and five haplotypes (1, 2, 3, 4, and 7) by all four population samples (table 1). Haplotype 5 (which belongs to the ε4 group of sequence haplotypes) is missing from the Jackson sample, despite the fact that it is found at a higher overall relative frequency than haplotype 7 in the combined sample.
Table 5.
Pairwise Estimates of Population Subdivision (FST)
| Jackson | Campeche | North Karelia | Rochester | |
| Jackson | … | |||
| Campeche | .127*** | … | ||
| North Karelia | .068** | .035* | … | |
| Rochester | .052*** | .054*** | .034* | … |
P<.05 (by haplotype-based permutation).
P<.01 (by haplotype-based permutation).
P<.001 (by haplotype-based permutation).
Differences in the population-distribution of the observed haplotypes are clearly illustrated by the sample-specific RM networks presented in figure 3. Most apparent is the absence of the ε2 clade of sequence haplotypes in the Mayan sample from Campeche, an observation consistent with the low overall level of sequence diversity in that sample (table 2), as well as previous reports of apoE polymorphism, which have noted the absence of ε2 alleles in other New World population samples (Crews et al. 1993; Kamboh 1995; Scacchi et al. 1997). The extent of ε2 haplotype variation found in the other three samples also differs: ε2s are represented by pairs of singleton haplotypes in the Jackson and North Karelia samples, whereas the full repertoire of ε2 variation is present in the sample from Rochester. Population differences in the distribution of ε3 haplotypes are less striking. The two most common types of ε3s (haplotypes 1 and 2) are observed in all four samples, as are the closely associated derivative haplotypes, 3, 4, and 7. The core ε3 haplotype (haplotype 6) is, however, absent in the European samples (North Karelia and Rochester) and the rare haplotype 19, which links haplotype 6 to haplotype 1, is observed only in the black sample from Jackson. The remaining interpopulation differences in the distribution of ε3 haplotypes largely involve rare variants, which differ from the common ε3 haplotypes by one or, at most, two sites. Homoplasy at sites 560 and 624, which results in the unresolved loops in the ε2 and ε3 clades of the total RM network (fig. 2), is observed in the Rochester sample only (fig. 3).
The most important difference among the population sample networks involves the composition of the ε4 clade in the four samples. The two subbranches of the ε4 group observed in the sample from Jackson (leading to haplotypes 17 and 20) are completely absent in the other three population samples, whereas the portion of the ε4 clade observed in the Mayan and European-derived population samples (dominated by haplotype 5, as noted above) is missing in the sample from Jackson. The degree to which these branches are present in all populations—but at low frequency and, hence, not captured in samples of 48 chromosomes from each population—remains to be seen from other and/or larger samples. In any case, the observed sample differences suggest an ethnic and/or geographic difference in the relative frequency of ε4 subtypes, which clearly merits further investigation in the context of conflicting risk associations in the literature. The nucleotide polymorphism that distinguishes haplotype 5 from the Jackson-specific ε4s is site 1998, a previously unrecognized variant that falls at the 5′ end of intron 2 (fig. 1).
Time Depth of APOE Variation
RM networks summarize the mutational relationships among sequence haplotypes but provide no indication of the time period over which the sequence differences have arisen. Determination of the time depth of the ε2-ε3-ε4 APOE polymorphism is relevant both to understanding the current geographic distribution of the protein-level polymorphism and to assessing the relative impact of demographic and selective processes on the observed variation. The most straightforward way to estimate the time depth of polymorphism is to compare the observed level of intraspecific diversity within humans to the number of fixed interspecific differences observed between humans and chimpanzees. As noted above, the net sequence divergence between human and chimp APOE genes was estimated as 64.7; the average pairwise sequence difference among human haplotypes in the same genomic region was 2.93. If a constant molecular clock is assumed and the humans-chimpanzee time of divergence is taken as 5 million years BP (Horai et al. 1992), the time required to generate the observed variation would be ∼226,000 years.
To get a fuller picture of the timing of APOE allelic divergence, we also analyzed the variation using an ML method based on a coalescent model of neutral sequence evolution (Griffiths and Tavaré 1997), which uses all of the information in a haplotype network to infer the TMRCA of the observed variation, as well as ages of individual mutations (for indicative previous analyses using the same methodology, see Harding et al. [1997] and Harris and Hey [1999]). Estimates of the total time depth of APOE variation and specific mutational ages were calculated for the combined sample and each of the separate population samples (table 6). One gene tree, depicting the evolutionary relationships among haplotypes in the combined sample and summarizing age estimates for mutations in that tree, is shown in figure 4. The main features of this tree are found in each of the trees generated for the separate population samples, although the overall time depth of the variation (summarized by the TMRCA estimates in table 6) varies, due to differences in the haplotype composition of the respective samples.
Table 6.
Estimates of Population Genetic Parameters from Genetree
| Sample | na | Sb | θWc | θMLd | Nee | TMRCAf (×105 years) | Agef of Site 3937(×105 years) | Average Agefof Other Sites(×105 years) |
| Jackson | 48 | 12 | 2.7 | 3.23 | 6,260 | 2.92 (1.59–5.40) | 2.09 (1.08–4.13) | .54 |
| Campeche | 48 | 6 | 1.35 | 1.32 | 2,558 | 1.99 (.95–4.03) | 1.44 (.68–3.10) | .46 |
| North Karelia | 46 | 10 | 2.28 | 2.44 | 4,729 | 2.73 (1.49–5.03) | 2.0 (1.04–3.92) | .63 |
| Rochester | 46 | 10 | 2.28 | 2.5 | 4,845 | 2.7 (1.45–5.04) | 2.06 (1.04–4.03) | .56 |
| Pooled | 183 | 18 | 3.11 | 3.66 | 7,093 | 3.11 (1.76–5.79) | 2.2 (1.22–4.40) | .45 |
Sample size after removal of non–IS-compatible data (details of sites and haplotypes excluded available on request).
Number of IS-compatible polymorphic sites.
Watterson (1975) estimator of θ per locus, based on S.
ML estimate of θ, per locus, from Genetree program (see Material and Methods).
Effective population size, derived from θML and assuming μ=1.29×10-4/locus/generation.
Median age estimates, derived assuming the given Ne and a generation time of 20 years. In parentheses, 2.5% and 97.5% points of the age distribution are given.
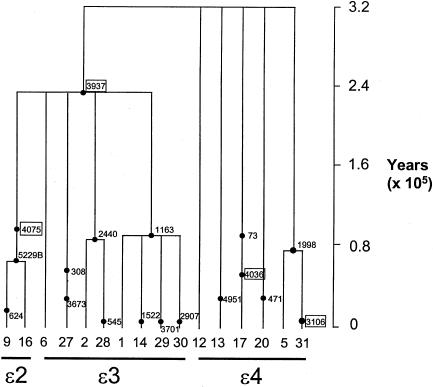
Figure 4.
Scaled coalescent gene tree of IS-compatible APOE variation in the total (pooled) sample. The tree shows the inferred genealogical history of the 16 haplotypes compatible with the IS model (indicated by the numbered labels marking each terminal branch at the bottom of the tree; haplotype numbering as in table 1). The mean estimated ages of variants in the tree, indicated as numbered points within the genealogy, were derived by the program Genetree. For branches with multiple mutations, the order of mutations in time is arbitrary. Horizontal lines are drawn underneath the groups of haplotypes that define the ε2, ε3, and ε4 APOE alleles, respectively. Mutations that result in amino acid substitutions are boxed.
There are three main features of interest. First, the estimate of the TMRCA (i.e., the total time-depth of the tree) is 311,000 years BP (with a 95% credibility interval ranging from 176,000 to 579,000 years BP). Estimates are of the order of 200,000–300,000 years BP, when variation in the separate population samples is considered (table 6). This time depth is lower than, but not significantly different from, an equivalently derived estimate of 770,000±420,000 years for the TMRCA of human β-globin diversity, the time depth of which is roughly that expected of a neutral autosomal locus (Harding et al. 1997). Second, site 3937, the cSNP that distinguishes ε2 and ε3 alleles from ε4 alleles, is the oldest mutation in the tree and is approximately twice as ancient as any of the other mutations in the IS-compatible data set (fig. 4). The age of site 3937 suggested by the ML analysis ranges from 150,000–220,000 years BP (table 6). Finally, all of the other mutations in the tree are estimated to have arisen within the past 90,000 years, and several within the past 10,000 years. The average age of all sites other than site 3937 is ∼50,000 years in each of the population samples examined (table 6). Even if we could have incorporated all homoplasic sites in this analysis (as described in the Material and Methods section, these were excluded in order to conform to the requirement for an IS-compatible data set), this striking difference in the relative age of site 3937, which reflects the high relative frequency of the derived allele at this site in our sample, would have remained.
A key assumption underlying these analyses is that the variation is selectively neutral. If this assumption is met, then these analyses suggest that the divergence between ε4 and the ε2 and ε3 clades (dated by the mutation at site 3937) began ∼200,000 years ago and that most of the subsequent intraallelic divergence has occurred only comparatively recently in human history, within the past 60,000 years. If, instead, selection has acted to increase the relative frequency of the mutation at site 3937 (as discussed below), then the inferred ages may overestimate the antiquity of the ε2-ε3-ε4 polymorphism, because sites in the sample would then be at a higher relative frequency than expected, given the influence of random genetic drift alone. In either case, the recent origin of the APOE protein polymorphism suggested by these age estimates is not consistent with the long-standing maintenance of the ε2, ε3, and ε4 alleles in human populations as a balanced polymorphism.
Evidence for Departure from Selective Neutrality
Of the four test statistics estimated from patterns of variation found in the whole 5.5-kb sequenced region surveyed, three (D, DFL, and F) suggested no statistically significant deviation from selective neutrality (table 2). Although the fourth, Fs, did suggest departures from neutral expectation for the Jackson, Rochester, and combined samples, as discussed earlier, the extensive site homoplasy in these samples suggests that this deviation is more likely explained by the effects of interhaplotype recombination than genetic hitchhiking. In addition, HKA test (Hudson et al. 1987) comparisons of levels of APOE polymorphism and divergence to that observed at two other equivalently sampled human autosomal genes, LPL (Clark et al. 1998; Nickerson et al. 1998) and β-globin (Harding et al. 1997), suggested no significant differences (P>.05, data not shown). By all of these standard general measures, then, APOE sequence variation indicates a remarkably good fit to neutral expectation.
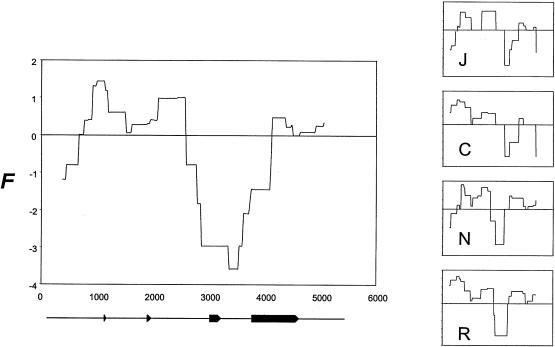
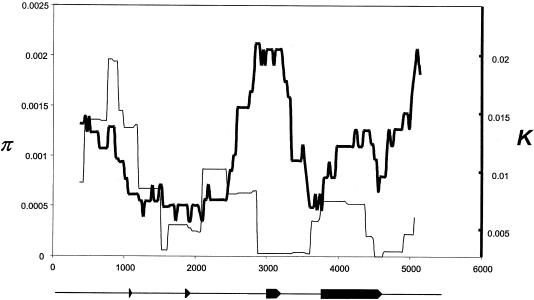
Nevertheless, several features of the data appear broadly consistent with the selective expansion of the ε3-type sequence haplotypes defined by site 3937. As noted earlier, five of the six singleton variants in the combined sample are linked to ε3-type sequence haplotypes. Low frequency variation is expected to accompany the rise in frequency of an advantageous mutant, as preexisting variation in a population is replaced in the “sweep” to fixation of the haplotype containing the advantageous site (Fu and Li 1993). The size of the genomic region affected depends on the strength of selection and the recombination rate; if the recombination rate is high, the region of sequence affected by the sweep will be small, and only sites in the immediate genomic vicinity of the selected site will show the expected frequency disturbance (Kaplan et al. 1989). In this context, it is relevant that the singleton variants in our sample all fall immediately upstream of the fourth exon, where the mutation at site 3937 is located (fig. 1). Sliding window estimates of the F test statistic (which measures the extent to which polymorphism is characterized by excess singleton variation), suggests a consistent excess of low frequency variation in the center of the sequenced region, encompassing bases 3000–4500 (fig. 5). Moreover, although the 1 kb of sequence that lies upstream of exon 4 is characterized by a low level of polymorphism (measured as average pairwise sequence difference π), interspecific sequence divergence (measured by net sequence divergence, K) in this central segment is higher than in adjoining regions, reflecting a marked discordance between patterns of polymorphism and divergence in this portion of the sequenced region (fig. 6). This difference is consistent with the recent loss of variation in a region with a normal-to-high underlying rate of neutral mutation.
Figure 5.
Sliding-window analysis of the F test statistic (Fu and Li 1993). Estimates of F were calculated for overlapping 750-bp-wide windows placed at 25-bp steps along the 5.5-kb sequenced region. The value for each window was plotted at the midpoint of the window and these points were linked by a continuous line for ease of inspection. Sliding window plots for the pooled sample and each of the separate population samples are shown (J = Jackson, C = Campeche, N = North Karelia, R = Rochester). The location of the APOE exons within the sequenced region are shown directly beneath the plot for the combined sample.
Figure 6.
Sliding-window analysis of APOE polymorphism (π) and interspecific divergence (K). Estimates of average pairwise sequence difference, π, among the human haplotypes and net sequence divergence, K, between the haplotypes and the chimpanzee APOE sequence, were calculated for overlapping 750-bp-wide windows placed at 25-bp steps along the 5.5-kb sequenced region. π is plotted as a light line (and with respect to the scale on the left) and K is plotted in bold (and with respect to the scale on the right). The location of the APOE exons within the sequenced region are shown beneath the plot.
Discussion
The global ubiquity of the three common apoE isoforms and, in particular, the persistently high relative frequency of the ε3 allele in all human populations, has puzzled investigators and prompted much speculation regarding the evolutionary forces responsible for the observed polymorphism (e.g., Hanlon and Rubinsztein 1995; Corbo and Scacchi 1999; Finch and Sapolsky 1999). As has been recognized in the population-genetics literature for many years, however, it is nearly impossible to distinguish among competing explanations when variation at the amino acid level alone is considered (Lewontin 1985). It is only when the nucleotide-level diversity underlying protein polymorphism is considered that relevant inferences can be made (see, e.g., Kreitman 1983).
When we look beneath the surface of apoE protein polymorphism, we discover something unexpected. Although considerable, previously unrecognized variation underlies the three common isoforms, the observed heterogeneity is far less than would be expected if the three alleles had persisted for a long time in human populations as a balanced polymorphism. Despite the widespread major polymorphism at the amino acid level, the APOE locus remains one of the less variable genes that has been examined by in-depth resequencing. With an average nucleotide diversity (π) of 0.0005, APOE is less variable than LPL (π=0.002) (Clark et al. 1998; Nickerson et al. 1998), ACE (π=0.0009) (Rieder et al. 1999), β-globin (π=0.002) (Harding et al. 1997) or MC1R (π=0.002) (Rana et al. 1999; Harding et al. 2000). The low level of variation is not a function of a low neutral mutation rate: the mutation rate estimated here, 1.29×10-4/locus/generation, is of the same order of magnitude as estimates for many other human nuclear loci (Harding et al. 1997; Clark et al. 1998; Zietkiewicz et al. 1998; Harris and Hey 1999; Jaruzelska et al. 1999). Nor is the low level of sequence diversity consistent with expected effects of balancing selection, whose influence would serve to allow greater variation to accumulate in the regions flanking the target of selection as a consequence of genetic hitchhiking. Instead, the observed polymorphism is more consistent with a reduction in variation associated with the rise in frequency of an advantageous mutation (i.e., the amino acid–altering T allele at 3937), which, in the process of increasing in frequency toward fixation, has “swept” away linked variation.
However, none of the test statistics applied to the observed variation suggest anything other than a good fit to the expectations of selective neutrality. The failure to reject neutrality with these tests is likely to reflect the narrow window of the sequence affected by the suggested perturbations, the relatively low level of polymorphism in our sample, and the fact that the null distributions of the statistics are generated under the assumption of a conservative model in which no recombination is acting (Wall 1999). Even given the peculiarities of the data set analyzed here, these test statistics are notorious for their lack of power (see, e.g., Simonsen et al. 1995), and at least one author has argued that this lack of power may stem directly from inappropriateness of the equilibrium neutral model normally assumed (Gillespie 2000). What is clear is that these (and related) test statistics have failed to indicate departures from neutrality for at least two other human loci for which there is compelling, independent evidence of selective effects, namely β-globin (Harding et al. 1997) and the Duffy blood group locus (Hamblin and Di Rienzo 2000). Therefore, although we did not find statistically significant evidence of the effects of natural selection on APOE variation, it is not unreasonable to infer that selection has acted. The value of examining this possibility closely is heightened by the variety of phenotypes with which the protein isoforms have been associated.
Precisely when the new variant at site 3937, and its associated haplotypes, began to expand in frequency relative to the ancestral allele is uncertain and is unlikely to be resolved using analytical methods that assume selective neutrality (such as those employed here). The presence of both ε3 sublineages in each of the four populations investigated, in the absence of evidence for recurrent mutation at the sites involved, is consistent with site 3937 having arisen prior to the major population expansions that accompanied the spread of anatomically modern humans <100,000 years ago. Alternatively, strong selective pressure may have facilitated the global distribution of the variant much more recently. Indirect evidence in support of the latter hypothesis is the fact that the basal haplotype of the ε3 clade (haplotype 6) is absent in the European samples from Rochester and North Karelia (hence, if present, likely to be at low frequency in those populations), suggesting that the rise in frequency of the ε3s in Europe may have occurred after the differentiation of the clade into two primary lineages. The low overall level of population heterogeneity (FST = 0.06) is also consistent with a recent rapid expansion of ε3. In either case, the lower relative frequency and patchy geographical distribution of the ε2 haplotypes, as well as their clear derivation from the ε3 clade, suggest that the variant defining these latter alleles (at site 4075) arose subsequently.
A relevant question is whether known phenotypic effects associated with the observed variation contribute to current differences in reproductive fitness. There is strong evidence that the inheritance of an ε4 allele places carriers at a higher risk of succumbing to CAD or AD, at least in European and Asian populations (Davignon et al. 1988; Roses 1996). The deleteriousness of ε4, relative to that of ε3, is consistent with the history of long-term genetic change at the APOE locus, but CAD and AD are both diseases of late adulthood or old age, and increased susceptibility to these conditions would not be expected to result in important differences in reproductive success (for an opposing argument, see Finch and Sapolsky [1999]). The widely expressed protein does play many different roles in the body, including facilitation of lipid absorption, neural growth and regeneration, and immune function (Mahley and Huang 1999), one or more of which could have a direct effect on fertility or could contribute to differential survival and reproductive success. One study has suggested that men carrying at least one ε3 allele have, on average, more children than men with other APOE genotypes (Gerdes et al. 1996a). Alternatively, APOE variation may reflect an adaptation to changing diets (Hanlon and Rubinsztein 1995), such as those which accompanied the transition from subsistence to agricultural economies (Corbo and Scacchi 1999), may play a key role in neurological response to head injury (Friedman et al. 1999), or may mediate susceptibility to lipophylic pathogens (Martin 1999).
Population-level differences in the distribution of APOE variation are relevant to a comprehensive prediction of risk and understanding of disease etiology. Despite the low overall level of polymorphism at the APOE gene, considerable heterogeneity characterizes each of the three common alleles at the sequence level, heterogeneity which helps explain previously perplexing association results. For example, in a 5-year prospective epidemiological study of AD incidence among different ethnic groups, Tang et al. (1998) found that, compared to ε3/ε3 homozygotes, the relative risk (RR) of AD associated with one or more copies of the ε4 allele was significantly increased among whites (RR 2.5) but not among blacks (RR 1.0) or Hispanics (RR 1.1). These results confirmed previous reports suggesting that the association of ε4 with AD is weaker or nonexistent among blacks living in New York City (Tang et al. 1996) and Indiana (Sahota et al. 1997), as well as among black Nigerians (Osuntokun et al. 1995) and East Africans (Sayi et al. 1997). Similar variation is found regarding lipids (Xu et al. 1999). These observations take on new significance in the light of our finding that geographic and/or ethnic differences exist in the distribution of haplotypes within the ε4 class (fig. 3). If, as our results suggest, different types of ε4 alleles are found at different relative frequencies in different geographic regions, this heterogeneity (which may be related to nonneutral forces acting on the locus) can be—indeed, must be—accounted for.
Similarly, our data provide an invaluable evolutionary context in which to interpret more circumscribed analyses. There has, for instance, recently been much interest in characterizing APOE promoter-region polymorphism and examining the association of such variants with AD and CAD risk. These analyses have met with only limited success. Either the observed variants have turned out not to explain any greater proportion of the observed variance in phenotype than explained by the common allelic variants or positive associations in one population have failed to be replicated in subsequent studies. The most problematic discrepancies in this regard have centered on the role of the −491 variant (Artiga et al. 1998b). Some workers have suggested that variation at this site is a strong determinant of AD risk (Artiga et al 1998a; Bullido et al 1998), whereas other researchers have either questioned the importance of the polymorphism relative to other regulatory region variants (Lambert et al. 1998b; Town et al. 1998), or failed to replicate the association altogether (Roks et al. 1998; Song et al. 1998). Our analysis suggests a possible explanation: −491 (site 560 here) appears to be particularly susceptible to recurrent mutation and/or gene conversion, placing it in association with different allelic backgrounds, with different functional effects, in different populations. In this context, it is unsurprising that results have conflicted. On the other hand, the prominent placement of site 832 (−219) in our inferred haplotype network, as a site defining major subtypes of both ε3 and ε4 haplotypes in multiple populations, appears consistent with the association of this site with differences in both AD risk (Lambert et al. 1998a, 1998b) and myocardial infarction (Lambert et al. 2000). The relationship of other variants with unique positions in the APOE gene tree (particularly sites 1163 and 2440 among ε3s and site 1998 among ε4s) clearly merit detailed investigation. When large samples are typed at these variable sites, and relevant phenotypes are scored, it will be possible to test directly the independence of effects of the variable sites on lipid phenotypes.
Our survey of DNA-sequence variation at the human apolipoprotein E locus has been simultaneously exhaustive and wholly preliminary. We have not characterized the full extent of sequence variation as it arises at the gene on a global scale, nor have we attempted to identify polymorphisms in linked regions that might contribute in important ways to gene expression and disease etiology. Nevertheless, our investigation has done what previous studies had only approximated: a systematic investigation of sequence haplotype variation, as it occurs in the whole of the APOE gene and its nearby flanking regions, in samples drawn from nonclinical populations. The analysis reveals a young polymorphism with its genesis in recent human history and yet, despite this recency, encompassing an intricate and largely unforeseen heterogeneity with important implications for our understanding of the biology that variation encodes. Because it has been so extensively studied, APOE is an excellent model gene to illustrate the importance of considering genetic variation thoroughly (e.g., Martin et al. 2000), preferably without making prior assumptions about causation. It is clear from that study as well as from our own work that there is no consistent or specifically predictable relationship among sites within a gene. In particular, convenient or even simplistic assumptions that only coding variation is important, or that the effect of each variable site in a gene acts independently of the others, can be tested. The recent discovery of regulatory sites with apparent effects on lipid and AD risk factors demonstrates this clearly, and this gene has more surprises of the same type in store (C. F. Sing, unpublished data).
Acknowledgments
We thank B. P. Lazzaro and E. T. Dermitzakis for comments on the manuscript. We are also grateful to two anonymous reviewers for their suggestions. We acknowledge support from National Heart, Lung, and Blood Institute grants HL39107, HL58238, HL58239, and HL58240.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Arlequin, http://lgb.unige.ch/arlequin (for Arlequin program)
- DnaSP, http://www.bio.ub.es/~julio/DnaSP.html (for DnaSP software)
- Family Blood Pressure Program Web site, http://www.hypertensiongenetics.org
- Fluxus Engineering, http://www.fluxus-engineering.com/sharenet.htm (for Network 2.0 software)
- Genbank, http://www.ncbi.nlm.nih.gov/entrez/ (for chimpanzee APOE reference sequence [AF261280])
- Genetree, http://www.maths.monash.edu.au/~mbahlo/mpg/gtree.html (for Genetree software)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for apoE [MIM 107741] and AD [MIM 104300])
References
- Artiga MJ, Bullido MJ, Frank A, Sastre I, Recuero M, Garcia MA, Lendon CL, Han SW, Morris JC, Vazquez J, Goate A, Valdivieso F (1998a) Risk for Alzheimer's disease correlates with transcriptional activity of the APOE gene. Hum Mol Genet 7:1887–1892 [DOI] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Garcia MA, Aldudo J, Vazquez J, Valdivieso F (1998b) Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. Fed Europ Bioch Soc Letters 421:105–108 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, Lendon C, Han SW, Morris JC, Frank A, Vazquez J, Goate A, Valdivieso F (1998) A polymorphism in the regulatory region of APOE associated with risk for Alzheimer's dementia. Nat Genet 18:69–71 [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Parfitt M, Legrain S, Perez-Tur J, Brousseau T, Evans A, Berr C, Vidal O, Roques P, Gourlet V, Fruchart JC, Delacourte A, Rossor M, Amouyel P (1994) Apolipoprotein E, epsilon 4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer's disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet 3:569–574 [DOI] [PubMed] [Google Scholar]
- Clark AG (1990) Inference of haplotypes from PCR-amplified samples of diploid populations. Mol Biol Evol 7:111–122 [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengård J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M (1989) Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 83:181–188 [DOI] [PubMed] [Google Scholar]
- Corbo RM, Scacchi R (1999) Apolipoprotein E (APOE) allele distribution in the world: is APOE*4 a “thrifty” allele? Ann Hum Genet 63:301–310 [DOI] [PubMed] [Google Scholar]
- Corbo RM, Scacchi R, Mureddu L, Mulas G, Alfano G (1995) Apolipoprotein E polymorphism in Italy investigated in native plasma by a simple polyacrylamide gel isoelectric focusing technique. Comparison with frequency data of other European populations. Ann Hum Genet 59:197–209 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC Jr, Rimmler JB, Locke PA, Coneally PM, Schmader KE, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1994) Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet 7:180–184 [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (1993) Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261:921–923 [DOI] [PubMed] [Google Scholar]
- Crews DE, Kamboh MI, Mancilha-Carvalho JJ, Kottke B (1993) Population genetics of apolipoprotein A-4, E, and H polymorphisms in Yanomami Indians of northwestern Brazil: associations with lipids, lipoproteins, and carbohydrate metabolism. Hum Biol 65:211–24 [PubMed] [Google Scholar]
- Davignon J (1993) Apolipoprotein polymorphism and atherosclerosis. In: Born GVR, Schwartz CJ (eds) New horizons in coronary heart disease. Current Science, London, pp 5.1–5.21 [Google Scholar]
- Davignon J, Gregg RE, Sing CF (1988) Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis 8:1–21 [DOI] [PubMed] [Google Scholar]
- de Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM (1994) Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels. Hum Mutat 4:178–194 [DOI] [PubMed] [Google Scholar]
- Finch CE, Sapolsky RM (1999) The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol Aging 20:407–428 [DOI] [PubMed] [Google Scholar]
- Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda B, Groswasser Z (1999) Apolipoprotein E-ε4 genotype predicts a poor outcome in survivors of traumatic brain injury. Neurology 52:244–248 [DOI] [PubMed] [Google Scholar]
- Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking, and background selection. Genetics 147:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H (1993) New statistical tests of neutrality for DNA samples from a population. Genetics 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Gerdes C, Hansen PS, Klausen IC, Faergeman O (1996a) Are men carrying the apolipoprotein epsilon 4- or epsilon 2 allele less fertile than epsilon 3/epsilon 3 genotypes? Hum Genet 98:239–242 [DOI] [PubMed] [Google Scholar]
- Gerdes LU, Gerdes C, Hansen PS, Klausen IC, Faergeman O, Dyerberg J (1996b) The apolipoprotein E polymorphism in Greenland Inuit in its global perspective. Hum Genet 98:546–550 [DOI] [PubMed] [Google Scholar]
- Gillespie JH (2000) Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155:909–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RC, Tavaré S (1997) Computational methods for the coalescent. In: Progress in population genetics and human evolution. Donnelly P, Tavaré S (eds) Springer-Verlag, New York, pp 165–182 [Google Scholar]
- Hamblin MT, Di Rienzo A (2000) Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am J Hum Genet 66:1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon CS, Rubinsztein DC (1995) Arginine residues at codons 112 and 158 in the apolipoprotein E gene correspond to the ancestral state in humans. Atherosclerosis 112:85–90 [DOI] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB (1997) Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 60:772–789 [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, Rees JL (2000) Evidence for variable selective pressures at MC1R. Am J Hum Genet 66:1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EE, Hey J (1999) X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA 96:3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG (1997) Principles of population genetics. 3d ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- Hixson JE, Cox LA, Borenstein S (1988) The baboon apolipoprotein E gene: structure, expression, and linkage with the gene for apolipoprotein C-1. Genomics 2:315–323 [DOI] [PubMed] [Google Scholar]
- Horai S, Satta Y, Hayasaka K, Kondo R, Inoue T, Ishida T, Hayashi S, Takahata N (1992) Man’s place in Hominoidea revealed by mitochondrial DNA genealogy. J Mol Evol 35: 32–43 [DOI] [PubMed] [Google Scholar]
- Huang XQ, Hardison RC, Miller W (1990) A space-efficient algorithm for local similarities. Comput Appl Biosci 6:373–381 [DOI] [PubMed] [Google Scholar]
- Hui DY, Innerarity TL, Mahley RW (1984) Defective hepatic lipoprotein receptor binding of B-very low density lipoporetins from type III hyperlipoproteinemic patients. J Biol Chem 259:860–869 [PubMed] [Google Scholar]
- Hudson RR (1990) Gene genealogies and the coalescent process. Oxf Surv Evol Biol 7:1–44 [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruzelska J, Zietkiewicz E, Batzer M, Cole DE, Moisan JP, Scozzari R, Tavaré S, Labuda D (1999) Spatial and temporal distribution of the neutral polymorphisms in the last ZFX intron: analysis of the haplotype structure and genealogy. Genetics 152:1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non- coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 [DOI] [PubMed] [Google Scholar]
- Kamboh MI (1995) Apolipoprotein E polymorphism and susceptibility to Alzheimer's disease. Hum Biol 67:195–215 [PubMed] [Google Scholar]
- Kaplan NL, Hudson RR, Langley CH (1989) The “hitchhiking effect” revisited. Genetics 123:887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J, Ferrell RE, Kottke BA, Kamboh MI, Sing CF (1991)Effects of polymorphisms in apolipoproteins E, A-IV, and H on quantitative traits related to risk for cardiovascular disease. Arterioscler Thromb 11:1330–1348 [DOI] [PubMed] [Google Scholar]
- Kidd JR, Black FL, Weiss KM, Balazs I, Kidd KK (1991) Studies of three Amerindian populations using nuclear DNA polymorphisms. Hum Biol 63:775–794 [PubMed] [Google Scholar]
- Kreitman M (1983) Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304:412–417 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Perez-Tur J, Dupire M-J, Galasko D, Mann D, Amouyel P, Hardy J, Delacourte A, Chartier-Harlin MC (1997) Distortion of allelic expression of apolipoprotein E in Alzheimer's disease. Hum Mol Genet 6:2151–2154 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Berr C, Pasquier F, Delacourte A, Frigard B, Cottel D, Perez-Tur J, Mouroux V, Mohr M, Cecyre D, Galasko D, Lendon C, Poirier J, Hardy J, Mann D, Amouyel P, Chartier-Harlin MC (1998b) Pronounced impact of Th1/E47cs mutation compared with - 491 AT mutation on neural APOE gene expression and risk of developing Alzheimer's Disease. Hum Mol Genet 7:1511–1516 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Brousseau T, Defosse V, Evans A, Arveiler D, Ruidavets J-B, Haas B, Cambou JP, Luc G, Ducimetiere P, Cambien F, Chartier-Harlin MC, Amouyel P (2000) Independent association of an APOE gene promoter polymorphism with increased risk of myocardial infarction and decreased APOE plasma concentrations—the ECTIM study. Hum Mol Genet 9:57–61 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Pasquier F, Cottel D, Frigard B, Amouyel P, Chartier-Harlin M-C (1998a) A new polymorphism in the APOE promoter associated with risk of developing Alzheimer's Disease. Hum Mol Genet 7:533–540 [DOI] [PubMed] [Google Scholar]
- Larsen F, Solheim J, Prydz H (1993) A methylated CpG island 3′ in the apolipoprotein-E gene does not repress its transcription. Hum Mol Genet 2:775–780 [DOI] [PubMed] [Google Scholar]
- Li WH, Sadler LA (1991) Low nucleotide diversity in man. Genetics 129:513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC (1985) Population genetics. Annu Rev Genet 19:81–102 [DOI] [PubMed] [Google Scholar]
- Lucotte G, Loirat F, Hazout S (1997) Pattern of gradient of apolipoprotein E allele *4 frequencies in western Europe. Hum Biol 69:253–262 [PubMed] [Google Scholar]
- Mahley RW, Huang Y (1999) Apolipoprotein E: from atherosclerosis to Alzheimer's disease and beyond. Curr Opin Lipidol 10:207–217 [DOI] [PubMed] [Google Scholar]
- Martin ER, Lai EH, Gilbert JR, Rogala AR, Afshari AJ, Riley J, Finch KL, Stevens JF, Livak KJ, Slotterbeck BD, Slifer SH, Warren LL, Conneally PM, Schmechel DE, Purvis I, Pericak-Vance MA, Roses AD, Vance JM (2000) SNPing away at complex diseases: analysis of single-nucleotide polymorphisms around APOE in Alzheimer disease. Am J Hum Genet 67:383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM (1999) APOE alleles and lipophylic pathogens. Neurobiol Aging 20:441–443 [DOI] [PubMed] [Google Scholar]
- Mui S, Briggs M, Chung H, Wallace RB, Gomez-Isla T, Rebeck GW, Hyman BT (1996) A newly identified polymorphism in the apolipoprotein E enhancer gene region is associated with Alzheimer's disease and strongly with the ε4 allele. Neurology 47:196–201 [DOI] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Nickerson DA, Taylor SL, Weiss KM, Clark AG, Hutchinson TG, Stengård J, Salomaa V, Vartiainen E, Boerwinkle E, Sing CF (1998) DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene.. Nat Genet 19:233–240 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, Stengård J, Boerwinkle E, Sing CF. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuntokun BO, Sahota A, Ogunniyi AO, Gureje O, Baiyewu O, Adeyinka A, Oluwole SO, et al (1995) Lack of an association between apolipoprotein E epsilon 4 and Alzheimer's disease in elderly Nigerians. Ann Neurol 38:463–465 [DOI] [PubMed] [Google Scholar]
- Rall SC Jr, Weisgraber KH, Mahley RW (1982) Human apolipoprotein E: the complete amino acid sequence. J Biol Chem 257:4171–4178 [PubMed] [Google Scholar]
- Rana BK, Hewett-Emmett D, Jin L, Chang BH, Sambuughin N, Lin M, Watkins S, Bamshad M, Jorde LB, Ramsay M, Jenkins T, Li WH (1999) High polymorphism at the human melanocortin 1 receptor locus. Genetics 151:1547–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SL, Ferrell RE, Kottke BA, Sing CF (1992) The gender-specific apolipoprotein E genotype influence on the distribution of plasma lipids and apolipoproteins in the population of Rochester, Minnesota. II. Regression relationships with concomitants. Am J Hum Genet 51:1311–1324 [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nat Genet 22:59–62 [DOI] [PubMed] [Google Scholar]
- Roks G, Cruts M, Bullido MJ, Backhovens H, Artiga MJ, Hofman A, Valdivieso F, Van Broeckhoven C, Van Duijn CM (1998) The −491 A/T polymorphism in the regulatory region of the apolipoprotein E gene and early-onset Alzheimer's disease. Neurosci Lett 258:65–68 [DOI] [PubMed] [Google Scholar]
- Roses AD (1996) Apolipoprotein E alleles as risk factors in Alzheimer's disease. Annu Rev Med 47:387–400 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- Sahota A, Yang M, Gao S, Hui SL, Baiyewu O, Gureje O, Oluwole S, Ogunniyi A, Hall KS, Hendrie HC (1997) Apolipoprotein E-associated risk for Alzheimer's disease in the African-American population is genotype dependent. Ann Neurol 42:659–661 [DOI] [PubMed] [Google Scholar]
- Salomaa VV, Rasi VP, Vahtera EM, Pekkanen J, Pursiainen M, Jauhiainen M, Vartiainen E, Ehnholm CP, Myllyla G (1994) Haemostatic factors and lipoprotein (a) in three geographical areas in Finland: the Finrisk Haemostasis Study. J Cardiovasc Risk 1:241–248 [DOI] [PubMed] [Google Scholar]
- Sayi JG, Patel NB, Premkumar DR, Adem A, Winblad B, Matuja WB, Mtui EP, et al (1997) Apolipoprotein E polymorphism in elderly east Africans. East Afr Med J 74:668–70 [PubMed] [Google Scholar]
- Scacchi R, Corbo RM, Rickards O, Mantuano E, Guevara A, De Stefano GF (1997) Apolipoprotein B and E genetic polymorphisms in the Cayapa Indians of Ecuador. Hum Biol 69:375–382 [PubMed] [Google Scholar]
- Seixas S, Trovoada MJ, Rocha J (1999) Haplotype analysis of the apolipoprotein E and apolipoprotein C1 loci in Portugal and Sao Tome e Principe (Gulf of Guinea): linkage disequilibrium evidence that APOE*4 is the ancestral APOE allele. Hum Biol 71:1001–1008 [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF (1995) Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141:413–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing CF, Davignon J (1985) Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet 37:268–285 [PMC free article] [PubMed] [Google Scholar]
- Song Y-Q, Rogaeva E, Premkumar S, Brindle N, Kawarai T, Orlacchio A, Yu G, Levesque G, Nishimura M, Ikeda M, Pei Y, O'Toole C, Duara R, Barker W, Sorbi S, Freedman M, Farrer L, St George-Hyslop P (1998) Absence of association between Alzheimer's disease and the −491 regulatory region polymorphism of APOE. Neurosci Lett 250:189–192 [DOI] [PubMed] [Google Scholar]
- Stengård JH, Zerba KE, Pekkanen J, Ehnholm C, Nissinen A, Sing CF (1995) Apolipoprotein E polymorphism predicts death from coronary heart disease in a longitudinal study of elderly Finnish men. Circulation 91:265–269 [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (1993) Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA 90:1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei K, Clarnette R, Gandy SE, Martins RN (1997) Increased plasma apolipoprotein E (apoE) levels in Alzheimer's disease. Neurosci Lett 223:29–32 [DOI] [PubMed] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphisms. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot C, Lendon C, Craddock N, Shears S, Morris JC, Goate A (1994) Protection against Alzheimer's disease with apoE epsilon 2. Lancet 343:1432–1433 [DOI] [PubMed] [Google Scholar]
- Tang MX, Maestre G, Tsai WY, Liu XH, Feng L, Chung WY, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R (1996) Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet 58:574–584 [PMC free article] [PubMed] [Google Scholar]
- Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, Andrews H, Feng L, Tycko B, Mayeux R (1998) The APOE-ε4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA 279:751–755 [DOI] [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet 66:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town T, Paris D, Fallin D, Duara R, Barker W, Gold M, Crawford F, Mullan M (1998) The −491 A/T apolipoprotein E promoter polymorphism association with Alzheimer's disease: independent risk and linkage disequilibrium with the known APOE polymorphism. Neurosci Lett 252:95–98 [DOI] [PubMed] [Google Scholar]
- Turner ST, Weidman WH, Michels VV, Reed TJ, Ormson CL, Fuller T, Sing CF (1989) Distribution of sodium-lithium countertransport and blood pressure in Caucasians five to eighty-nine years of age. Hypertension 13:378–391 [DOI] [PubMed] [Google Scholar]
- Utermann G, Hees M, Steinmetz A (1977) Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinemia in man. Nature 269:604–607 [DOI] [PubMed] [Google Scholar]
- Wall JD (1999) Recombination and the power of statistical tests of neutrality. Genet Res 74:65–79 [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Pop Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis II. Sinauer Associates, Sunderland, MA [Google Scholar]
- Weisgraber KH (1994) Apolipoprotein E: structure-function relationships. Adv Protein Chem 45:249–302 [DOI] [PubMed] [Google Scholar]
- Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhignesse M, Lussier-Cacan S, Sing CF, Kessling AM, Davignon J (1991) Influences of common variants of apolipoprotein E on measures of lipid metabolism in a sample selected for health. Arterioscler Thromb 11:1100–1110 [DOI] [PubMed] [Google Scholar]
- Xu Y, Berglund L, Ramakrishnan R, Mayeux R, Ngai C, Holleran S, Tycko B, Leff T, Schacter N (1999) A common HpaI RFLP of apolipoprotein C-I increases gene transcription and exhibits an ethnically distinct pattern of linkage disequilibrium with the alleles of apolipoprotein E. J Lipid Res 40:50–58 [PubMed] [Google Scholar]
- Yamada T, Kondo A, Takamatsu J, Tateishi J, Goto I (1995) Apolipoprotein E mRNA in the brains of patients with Alzheimer's disease. J Neurol Sci 129:56–61 [DOI] [PubMed] [Google Scholar]
- Zannis VI, Nicolosi RJ, Jensen E, Breslow JL, Hayes KC (1985) Plasma and hepatic apoE isoproteins of nonhuman primates: differences in apoE among humans, apes, and New and Old World monkeys. J Lipid Res 26:1421–30 [PubMed] [Google Scholar]
- Zekraoui L, Lagarde JP, Raisonnier A, Gerard N, Aouizerate A, Lucotte G (1997) High frequency of the apolipoprotein E *4 allele in African pygmies and most of the African populations in sub-Saharan Africa. Hum Biol 69:575–581 [PubMed] [Google Scholar]
- Zietkiewicz E, Yotova V, Jarnik M, Korab-Laskowska M, Kidd KK, Modiano D, Scozzari R, Stoneking M, Tishkoff S, Batzer M, Labuda D (1998) Genetic structure of the ancestral population of modern humans. J Mol Evol 47:146–155 [DOI] [PubMed] [Google Scholar]