Abstract
Coronary heart disease (CHD) is a complex disorder constituting a major health problem in Western societies. To assess the genetic background of CHD, we performed a genomewide linkage scan in two study samples from the genetically isolated population of Finland. An initial study sample consisted of family material from the northeastern part of Finland, settled by a small number of founders ∼300 years ago. A second study sample originated from the southwestern region of Finland, settled ∼2,000 years ago. Families were ascertained through probands exhibiting premature CHD, defined as >50% stenosis of at least two coronary arteries at a young age, as verified by coronary angiography. Both study samples and the pooled data set provided evidence for linkage in two chromosomal regions. A region on chromosome 2q21.1-22 yielded two-point LOD scores of 3.2, 1.9, and 3.7, in the affected sib-pair (ASP) analyses of the northeastern, southwestern, and pooled study samples. The corresponding multipoint maximum-likelihood scores (MLSs) for these three study samples were 2.4, 1.3, and 3.0. In addition, a region on chromosome Xq23-26 resulted in two-point LOD scores of 1.9, 3.5, and 2.9 and in multipoint MLSs of 3.4, 3.1, and 2.5, respectively. In conclusion, this study identifies two loci likely to contribute to premature CHD: one on chromosome 2q21.1-22 and another on chromosome Xq23-26.
Introduction
Coronary heart disease (CHD) is the leading cause of death in the Western world. For effective treatment and prevention, the complex molecular processes underlying CHD, as well as factors predisposing to atherosclerosis, should be identified. Clinical and epidemiological studies have documented that several types of risk factors—such as age, male sex, family history of myocardial infarction (MI), increased serum total and low-density lipoprotein cholesterol, decreased serum high-density lipoprotein cholesterol, smoking history, and presence of diabetes mellitus—predict the risk for atherogenesis (Rose 1964; Kannel et al. 1971; Pooling Project Research Group 1978; Keys 1980; Pyörälä et al. 1987; Grundy et al. 1990, 1998; Genest 1995; Wilson et al. 1998). Furthermore, both inflammation linked with disadvantageous plasma lipoprotein profile and chronic infections have also been proposed as risk factors for CHD (Saikku et al. 1988; Ridker et al. 1998; Noll 1998). Clearly, CHD is a multifactorial disease in which risk factors tend to cluster and interact in individuals and families to determine the level of risk. In addition, risk evaluation is further complicated by the unknown relationship among the underlying genetic and environmental risk factors.
The genetic background in common diseases merges from combinations of risk alleles that, together with environmental risk factors, lead to development of the disease phenotype (Risch and Merikangas 1996; Collins et al. 1997). Numerous studies have been carried out to identify the genes that have alleles that can predispose to CHD. Considerable evidence now indicates that variation in genes encoding the apolipoprotein E (APOE [MIM 107741]), apolipoprotein (a) (LPA [MIM 152200]), methyltetrahydrofolate reductase, and angiotensin-converting enzyme play a role in the development of CHD (Kraft et al. 1996; Cambien et al. 1992; Boushey et al. 1995; Wilson et al. 1996). However, the size and nature of the effects of these and other genes influencing the risk of CHD are still largely unknown.
Evidence for genetic involvement in development of CHD has been obtained from studies showing that family history of CHD increases the risk for CHD (Rose 1964; Sholtz et al. 1975; Jousilahti et al. 1996) and also from family studies estimating heritability of CHD to be 56%–63% (Nora et al. 1980). To increase the impact of genetic involvement and the possibilities of identifying contributing predisposing genes, a careful selection of the study sample is advised (Lander and Schork 1994; Wright et al. 1999; Terwilliger and Göring 2000a). Families with multiple affected individuals, extreme phenotypes, subjects with early-onset disease, and individuals originating from populations with restricted genetic variation may increase the power to identify genetic risk factors by decreasing etiological heterogeneity.
Genomewide linkage studies have been utilized to identify loci for complex disorders and to search for evidence of major gene effects. To date, however, no genome scans have been reported for premature CHD. We conducted a genomewide search for CHD loci in well-documented families affected by premature CHD that originate from the genetically isolated population of Finland. A two-stage strategy was used, in which the first stage of the scan, with an average marker interval of 10 cM, was followed by fine mapping of interesting regions. We also included two independent sample sets in both stages of the scan: one from the late-settlement region of Finland and the other from the early-settlement region. This strategy enabled us to identify two chromosomal regions, in both study samples, that may harbor loci with alleles that predispose to premature CHD: one on chromosome 2q21.1-22 and another on chromosome Xq23-26.
Subjects and Methods
Subjects
A total of 156 families with at least two individuals affected with premature CHD were collected. Of these, 89 “northeastern” families were collected as an initial study sample in the Kuopio University Hospital. For replication, an additional study sample of 67 “southwestern” families affected by premature CHD was collected in the Turku University Hospital. The probands for both study samples were selected from patients undergoing elective coronary angiography for clinically suspected coronary artery disease or from the angiography register. Inclusion criteria in both northeastern and southwestern study groups for the probands were ⩾50% stenosis in two or three coronary arteries confirmed by coronary angiography, age <55 for males and <65 for females. These age cutoffs take into account the fact that, in general, women fall sick with CHD ∼10 years later than men, and they thus reflect the well-known differences for CHD incidence between genders (Mosca et al. 1997).
Two diagnostic criteria were used in the analyses. In both of these criteria, the probands fulfilled the strict diagnostic criteria for CHD and age described above, but the criteria for the affected siblings varied. However, every additional family member coded as “affected” fulfilled the same age criteria as the proband (<55 years for males and <65 years for females). In criterion I, the CHD status of the affected sibling was determined as 2–3 vessel disease confirmed by coronary angiography or verified myocardial infarction, using the following criteria: (1) typical clinical symptoms; (2) definite electrocardiogram finding, according to the Minnesota coding (WHO criteria) (Rose et al. 1982); and (3) elevated levels of the creatine kinase (CK) enzyme and of its cardiac isoenzyme CK-MB, and age <55 for males and <65 for females. In criterion II, the CHD status of the affected sibling was determined as 1–3 vessel disease, confirmed by coronary angiography, or verified myocardial infarction. In addition, in the southwestern group, positive exercise-test information (i.e., at least a 2-mm ST segment depression) was available from six individuals, and this information was used in determination of the affection status of the affected sibling in diagnostic criterion II. Families in which probands were affected with familial hypercholesterolemia were excluded from the study samples. In addition, 150 first-degree relatives of affected individuals were genotyped to increase the phase information in the statistical analysis (see below). A total of 324 individuals were considered affected under criterion I, and 364 were considered affected under criterion II. The characteristics of these study samples are shown in table 1.
Table 1.
Characteristics of Northeastern-, Southwestern-, and Combined-Sample Families with CHD
| Criterion | SampleOrigina | No. ofAffectedSubjects | No. ofFamilies | AverageSibshipSize | FamilieswithTwoAffectedSibs | FamilieswithThreeAffectedSibs | FamilieswithFourAffectedSibs | No. ofMaleAffecteds | No. ofFemaleAffecteds | FamilieswithUnaffectedSibs | FamilieswithParentalData |
| I | NE | 197 | 89 | 2.2 | 73 | 13 | 3 | 140 | 57 | 64 | 5 |
| I | SW | 127 | 56 | 2.3 | 43 | 11 | 2 | 86 | 41 | 35 | 7 |
| I | All | 324 | 145 | 2.2 | 116 | 24 | 5 | 226 | 98 | 99 | 12 |
| II | NE | 202 | 89 | 2.3 | 71 | 12 | 6 | 142 | 60 | 61 | 5 |
| II | SW | 162 | 67 | 2.4 | 43 | 20 | 4 | 106 | 56 | 31 | 10 |
| II | All | 364 | 156 | 2.3 | 114 | 32 | 10 | 248 | 116 | 92 | 15 |
NE = northeastern; SW = southwestern.
The northeastern study was approved by the ethics committee of the Kuopio University Hospitals and the southwestern study by the ethics committee of the Turku University Hospital. All subjects gave informed consent. All the samples were taken according to the Helsinki declaration.
Genotyping
DNA was extracted from EDTA blood, according to a standard procedure. In stage I, a total of 303 microsatellite markers from a modified Weber screening set (version 6) (Sheffield et al. 1995) were genotyped in 345 individuals from northeastern and 181 individuals from southwestern CHD families. An additional 24 individuals from eight additional southwestern CHD families were genotyped in Stage II. In addition to genotyping affected individuals, 150/158 first-degree relatives were genotyped to increase phase information in stage I/II, but were coded as unknown phenotype in the statistical analysis. For the TDT and HRR analyses an additional 31 individuals were also genotyped from one southwestern and 11 northeastern singleton families in which the singleton had a 2–3-vessel disorder and fulfilled the age criteria.
For typing of markers in stage I of the project, fluorescently labeled PCR products were generated and then were pooled into panels before electrophoresis was done on ABI377 sequencers (PE Biosystems). Genotyping gels were analyzed in an automated system developed at the Whitehead Institute/MIT Center for Genome Research as previously described (Rioux et al. 1998). Microtiter-well PCR amplification of the markers was automated by a pipetting robot (Biomek 2000, Beckman).
In stage II, some areas resulting in maximum likelihood scores (MLSs) >1—either in the two-point or multipoint affected-sib-pair (ASP) analysis of stage 1 (or where marker informativeness was low)—were selected for denser marker mapping. An additional 82 markers were genotyped in stage II. The markers analyzed in stage II were selected and mapped on the basis of information derived from the genetic maps of Cooperative Human Linkage Center (Sheffield et al. 1995), Genethon (Dib et al. 1996) or GeneMap ’99.
For typing of markers in stage II of the project, microtiter-well PCR amplification of the markers was automated by a pipetting robot (Biomek 2000, Beckman). The fluorescently labeled PCR products were electrophoretically separated on an automated laser fluorescence DNA sequencer ABI 377 (PE Biosystems) with Genescan version 2.1 fragment-analysis software. The alleles were identified and verified by two persons independently for each marker, using the Genotyper program (version 2.0, PE Biosystems).
Statistical Analyses
Mendelian segregation was checked using the PedCheck program (O’Connel and Weeks 1998). Because the study material that we used consisted of small nuclear families with sparse phase information, we also searched for possible genotyping errors using the “error” option of the Mendel program, version 4.0 (kindly provided by K. Lange), in addition to the PedCheck program. The program MAPMAKER/SIBS (Kruglyak and Lander 1995) was used for two-point and multipoint maximum likelihood IBD calculations. For all analyses, the all independent pairs option was used when families with more than two affected siblings were analyzed. Two-point ASP analyses were also performed using the SIBPAIR program (Kuokkanen et al. 1996) of the ANALYZE package (Göring and Terwilliger 2000a, 2000c) to better address the problems associated with sibship size (Davis and Weeks 1997). For each marker, the allele frequencies were estimated from the total study sample of the particular study group—that is, the northeastern allele frequencies from the northeastern, the southwestern from the southwestern, and the combined allele frequencies from the combined study sample, respectively, using a gene-counting method (Smith 1957; Göring and Terwilliger 2000b).
To test for possible linkage disequilibrium (LD) on the linked regions on chromosomes 2 and X, family-based tests of association, the TDT (Terwilliger and Ott 1992) and/or haplotype-based haplotype relative risk (HRRR) (Terwilliger and Ott 1992), were conducted in the 156 families. The TDT option of the GENEHUNTER (Kruglyak et al. 1996) program and the HRRLAMB (Terwilliger and Ott 1992) program of the ANALYZE package (Göring and Terwilliger 2000a; Göring and Terwilliger 2000c) were performed on chromosome 2. For chromosome X, the TDT test is less useful, because this family-based association test by definition handles only data from heterozygous parents. Therefore, the possibility of LD on chromosome X was examined using only the HRRLAMB tool (Terwilliger and Ott 1992) of the ANALYZE package (Göring and Terwilliger 2000a; Göring and Terwilliger 2000c), in which homozygous parents are used as well. Further, this program takes only maternal nontransmitted alleles as controls, but both transmitted ones as case alleles. However, there was a large number of missing parental genotypes in these study samples. Therefore, to further test whether the allele frequencies differ between case and control subjects with one marker showing initial evidence of association in the HRRLAMB analysis, we genotyped 94 controls who were >55 years old and had no medical records of cardiovascular disorders. A total of 47 of the control subjects originated from northeastern Finland (30 males and 17 females) and 47 (29 males and 18 females) from southwestern Finland. The program DISLAMB (Terwilliger 1995) was then used to test differences between alleles of case and control subjects.
Potential genetic homogeneity between the northeastern and southwestern study samples was first tested by computing whether there are differences between the allele frequencies by the GENPOP program. Then, genetic distance between the two populations was computed on the basis of the allele-frequency information on common markers typed in unlinked regions in both study samples. This analysis was performed using the POPDIST program (Guldbrandtsen et al. 2000).
Results
The two study samples consisted of 89 families affected by carefully defined premature CHD from the late-settlement northeastern part of Finland and 67 families from the early-settlement southwestern part. Thus, altogether, 156 families affected by premature CHD were included in the study. The strategy was to test the initial findings from the northeastern study sample using an independent sample which fulfilled similar strict phenotypic criteria for CHD. The early-settlement sample provides an advantageous small founder size. In addition, the population of this region has an older genetic history than that of the late-settlement region. Since the late-settlement population originated from that of the early-settlement region, a random sampling of a selected set of CHD-predisposing genes could have taken place. In the late-settlement population, the effective population size is also smaller, and lifestyle and cultural risk factors are less variable, making this a potentially more powerful study population. Both samples, however, were genotyped in both stages of the genome scan to monitor possible regional differences in the genetic predisposition to CHD between the southwestern and northeastern parts of Finland and to allow for confounding effects of likely gene-environment interactions.
In the initial genome scan (stage I), the genotype data were analyzed using the strict diagnostic criterion I. However, to better utilize all familial information about CHD in fine mapping (stage II), we analyzed selected regions using criterion II as well. Affected individuals under criterion II fulfill the strict age limits, but include individuals with one-vessel disorder or positive ECG findings in the affected phenotype. Assuming that the same risk allele is segregating in one family, this should increase our power in fine mapping.
Stage I: Initial Genome Scan
Overall, we genotyped 303 markers in stage I. Multipoint ASP analyses of the northeastern sample showed maximum-likelihood scores (MLS) >1.0 at 9 positions on 7 chromosomes (2, 6, 10, 12, 15, 18, and X) (table 2). Multipoint MLSs >2.0 were observed at two locations on chromosome 2 (MLS 2.8), and on chromosome X (2.2). In the southwestern sample, seven positions on six chromosomes exhibited multipoint MLSs >1.0: chromosomes 2, 6, 9, 13, 21, and X (table 2), of which chromosomes 2 (MLS 2.2) and X (3.1) exhibited the MLSs >2. In the pooled analysis, only chromosome 2 (MLS 2.7) resulted in a multipoint MLS >2. These pooled results for all chromosomes using diagnostic criterion I are shown in table 2. In addition, the individual markers with the two-point MLSs >1.0 in the northeastern, southwestern, or combined analyses of stage I are indicated in table 3.
Table 2.
Multipoint MLS >1.0 Using the MAPMAKER/SIBS Program
|
Multipoint MLS (Nearest Marker) for Study Sample |
||||||
| Northeastern |
Combined |
Southwestern |
||||
| Chromosome | Stage I | Stage II | Stage I | Stage II | Stage I | Stage II |
| 1 | 1.04 (D1S1597) | |||||
| 2 | 1.10 (D2S1780), 2.81 (D2S125) | 2.40 (D2S2313) | 2.30 (D2S1326-D2S1353), 2.67 (D2S338-D2S125) | 2.99 (D2S2385-D2S2304) | 2.16 (D2S442) | 1.31 (D2S1776) |
| 6 | 1.24 (D6S1027) | 1.24 (D6S1027) | 1.32 (D6S503-D6S1027) | 1.32 (D6S503-D6S1027) | 1.02 (D6S1040) | 1.02 (D6S1040) |
| 9 | 1.41 (D9S1119-D9S910) | |||||
| 10 | 1.69 (D10S1412-GGAT1A4) | 1.01 (D10S1412-D10S585) | 1.29 (D10S1412) | |||
| 12 | 1.18 (D12S398) | 1.32 (D12S374-D12S391), 1.44 (D12S1090) | 1.36 (D12S374-D12S391) | |||
| 13 | 1.67 (D13S800-D13S317) | 1.20 (D13S317-D13S779) | ||||
| 14 | 1.12 (D14S588) | |||||
| 15 | 1.75 (D15S153) | 1.14 (D15S153) | ||||
| 18 | 1.68 (GATA26C03) | |||||
| 21 | 1.58 (D21S1432) | |||||
| X | 1.12 (DXS6800), 2.22 (DXS1047) | 3.44 (DXS8038-DXS8009) | 1.91 (DXS6799), 1.57 (DXS1047) | 2.46 (DXS8067-DXS1212) | 3.06 (DXS6810), 2.39 (DXS6796-DXS1047) | 3.09 (DXS1059) |
Table 3.
Two-Point MLSs for Regional Samples
|
Two-Point MLS for Sample |
||||
| Stage andChromosome | Marker | Northeastern | Combined | Southwestern |
| Stage I:a | ||||
| 1 | D1S1660 | 1.41 | ||
| 1 | D1S1665 | 1.04 | ||
| 2 | D2S125 | 2.00 | 1.17 | |
| 2 | D2S1326 | 1.23 | 1.17 | |
| 2 | D2S1328 | 1.72 | 1.52 | |
| 2 | D2S1353 | 1.52 | ||
| 2 | D2S1360 | 1.05 | 1.14 | |
| 2 | D2S1363 | 1.59 | 2.13 | |
| 2 | D2S1400 | 1.21 | ||
| 2 | D2S1776 | 1.74 | ||
| 2 | D2S1780 | 1.27 | ||
| 2 | D2S338 | 2.21 | 1.69 | |
| 2 | D2S434 | 1.26 | ||
| 3 | D3S2427 | 2.25 | 1.25 | |
| 4 | D4S408 | 1.44 | 1.84 | |
| 6 | D6S1027 | 1.30 | 1.18 | |
| 6 | D6S503 | 2.05 | 1.13 | |
| 8 | D8S1100 | 1.09 | 1.00 | |
| 9 | D9S1122 | 1.25 | ||
| 9 | D9S910 | 1.77 | ||
| 10 | D10S1412 | 1.30 | 1.27 | |
| 10 | D10S1426 | 1.21 | ||
| 10 | D10S169 | 1.13 | ||
| 11 | GATA67D03 | 1.35 | ||
| 12 | D12S1090 | 1.02 | 1.88 | 1.09 |
| 12 | D12S391 | 1.79 | 1.22 | |
| 14 | D14S588 | 1.11 | 1.52 | |
| 14 | D14S610 | 1.38 | 1.07 | |
| 15 | D15S153 | 2.19 | 1.07 | |
| 16 | D16S2619 | 1.01 | ||
| 18 | GATA26C03 | 2.12 | 1.38 | |
| 19 | D19S589 | 1.20 | 1.23 | |
| 20 | D20S478 | 1.11 | 1.82 | |
| 22 | D22S446 | 1.38 | 1.01 | |
| 22 | D22S685 | 1.53 | 1.23 | |
| X | DXS1047 | 2.46 | 1.67 | |
| X | DXS6789 | 1.03 | 1.80 | 1.03 |
| X | DXS6800 | 2.75 | ||
| X | DXS6810 | 3.31 | ||
| X | DXS8041 | 1.32 | ||
| Stage II:b | ||||
| 7 | D7S1808 | 1.01 | 1.30 | |
| 10 | D10S570 | 1.31 | 1.31 | |
| 10 | D10S585 | 1.28 | 1.28 | |
| 12 | D12S345 | 1.01 | ||
| 22 | D22S282 | 1.88 | 1.36 | |
Only markers resulting in two-point MLS >1 are shown.
Additional two-point MLS >1 observed with stage II markers (except for chromosomes 2 and X).
Table 3 further shows that the marker DXS6810 resulted in a two-point MLS of 3.3 in the southwestern study sample, whereas the MLS of this marker remained <1 in the northeastern and pooled analyses. To test if this difference is significant, we conducted a formal test of equality of linkage evidence between these populations. For the marker DXS6810, we calculated as follows: 4.6 × [MLSsouthwest + MLSnortheast− MLScombined], distributed as χ2 with 2 df, since each of the MLS statistics had two free parameters; thus, 4.6×[0+3.31-.19]=14.35, resulting in a P value <.001.
Thus, the MLS of 3.3 in the southwestern study sample is significantly different from the results in the northeastern sample. However, in stage 2 (see below), when more markers and individuals were genotyped in the southwestern study sample, the MLS with this marker decreased to 2.4 in the southwestern study sample using criteria II, and the MLSs of the nearest markers remained <1.0. The MLSs in the northeastern and the combined group also remained <1.0 in stage 2.
Stage II: Denser Map
In stage II, a total of 82 additional markers on chromosomes 1 (1 marker), 2 (29), 3 (2), 4 (1), 5 (7), 6 (1), 7 (5), 10 (6), 11 (2), 12 (2), 22 (3), and X (23) were genotyped in both sample sets. In the fine mapping, the two-point MLSs of the additional markers, as well as the multipoint MLSs, remained <2.0 in both sample sets, separately and combined, for all regions except for chromosomes 2 and X (tables 2 and 3).
Stage II: Chromosome 2
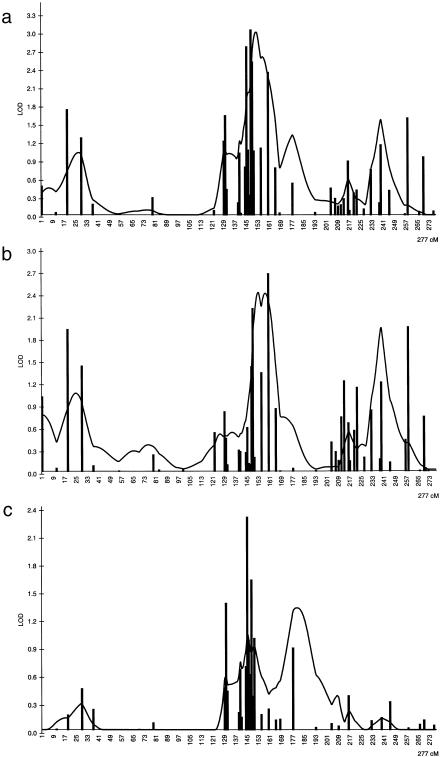
For chromosome 2, a total of 29 additional markers were genotyped in both study samples using diagnostic criteria I and II. The most significant results for chromosome 2 were obtained using diagnostic criterion II. The results with criterion I were similar to results obtained with criterion II but MLSs are slightly lower. The results of the two-point and multipoint ASP analyses using a high resolution marker map for chromosome 2 are shown in both study groups separately and combined in fig. 1.
Figure 1.
Results on chromosome 2 of two-point and multipoint ASP analysis using the MAPMAKER/SIBS program. Bars indicate the two-point results, and the solid line indicates the multipoint results. a, Results for the combined sample. b, Results for the northeastern study sample. c, Results for the southwestern study sample.
In the northeastern study sample, the highest two-point MLS was 2.7 with the marker D2S284 and the highest multipoint MLS was 2.4 (fig. 1b). In the replication material from southwestern Finland, the same region showed an MLS of 2.3 in two-point ASP analysis with the marker D2S2385 and an MLS of 1.3 in the multipoint analysis (fig. 1c). The pooled analysis resulted in MLSs of 3.0 with the marker D2S129 and of 3.0 in multipoint analyses (fig. 1a).
We searched for potential linkage equilibrium (LD) using TDT and HRRLAMB analyses in the combined sample set with all markers genotyped for the interesting region on chromosome 2. Marker D2S284 resulted in a P value of .02 in the TDT analysis, but the numbers of transmitted versus nontransmitted alleles (6 vs. 0) were too small to draw any conclusions. Further, the HRRLAMB analysis for this marker remained nonsignificant. In summary, strong evidence for LD was not observed with any of the loci tested on chromosome 2. The average marker interval was 2.5 cM in this analyzed 39.5-cM region (fig. 1).
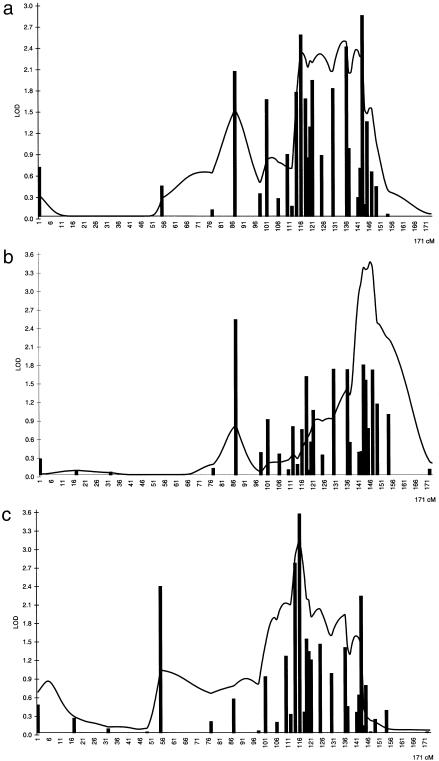
Stage II: Chromosome X
In stage II, 23 additional markers were genotyped in both sample sets for the region identified in stage I on chromosome X (fig. 2a–2c). As in the case of chromosome 2, the highest MLSs for chromosome X were obtained using diagnostic criterion II. Worth noting is that, for X-chromosomal markers, the MLS has only 1 df, because one can only evaluate identity by descent of chromosomes inherited from the mother’s side. So, an affected sib pair can share either 0 or 1 allele IBD. In the northeastern study sample, marker DXS6800 resulted in the highest MLS of 2.5 in two-point maximum-likelihood IBD analysis, whereas the maximum multipoint MLS was 3.4 when criterion II was used (fig. 2b). In the replication sample set from the southwestern Finland, the highest two-point MLS was 3.5, with the marker DXS1072, and the highest multipoint MLS was 3.1 in the ASP analysis (fig. 2c). However, the distance between the multipoint peaks in two regional materials was 23 cM (fig. 2b and 2c). In the combined analysis, the two-point analysis produced an MLS of 2.8 with the marker DXS1047. Furthermore, multipoint analysis of the combined data set resulted in a relatively wide peak of a 30-cM region with a maximum MLS of 2.5 (fig. 2a).
Figure 2.
Results on chromosome X of two-point and multipoint ASP analysis using the MAPMAKER/SIBS program. Bars indicate the two-point results, and the solid line indicates the multipoint results. a, Results for the combined sample. b, Results for the northeastern study sample. c, Results for the southwestern study sample.
The possibility of LD on chromosome X was tested using the HRRLAMB program which also uses information from homozygous parents. The TDT test is less informative when applied to genotype data from chromosome X, since it only considers data from heterozygous parents. A total of 22 markers, with an average marker interval of 2.4 cM, were analyzed in the potential region on chromosome X (fig. 2a–2c). One of the markers, DXS8053, showed initial hint of association using the HRRLAMB program (P<.01). To test whether the allele frequencies differ between cases and controls with this marker, we genotyped 94 unrelated controls, aged >55 years, who had no medical record of CHD: 47 individuals from northeastern (30 male and 17 female subjects) and 47 (29 male and 18 female subjects) from southwestern Finland. The distribution of the case and control alleles, with marker DX8053, were as follows: allele 1, 6 in case subjects versus 14 in control subjects; allele 2, 218 versus 113; allele 3, 18 versus 16; and allele 4, 76 versus 41, resulting in a P value of .007 (the 2×n table χ2 test) using the DISLAMB program. Separately, the southwestern families gave a P value of .04 with the southwestern control subjects and the northeastern families a P value of .2 with the northeastern control subjects, respectively.
Analysis of the Dense Mapping Regions Using the SIBPAIR Program
Since sibship size varies in the families with CHD included in the study (table 1), we also performed a two-point ASP analysis, using the SIBPAIR program of the ANALYZE package, to better address the problems associated with sibship size (Davis and Weeks 1997). On chromosome 2, the maximum two-point LOD scores with the SIBPAIR program were 3.7 (marker D2S2313) in the combined, 3.2 (D2S2313) in the northeastern, and 1.9 (D2S129) in the southwestern study groups. Marker D2S129 is the same marker at which the combined data set peaks in the MLS analysis, and the D2S1313 is its adjacent marker (1.6 cM). On chromosome X, the highest two-point LOD scores were 2.9 (DXS1072), 1.9 (DXS1212), and 3.5 (DXS1072). Marker DXS1072 is also the peak marker in the MLS analysis, and D2S1212 is located within the multipoint peak of the northeastern sample. Thus, these results support those produced by the MAPMAKER/SIBS program.
Testing for Genetic Heterogeneity between the Study Samples
We pursued the possible genetic heterogeneity between the southwestern (early settlement) and northeastern (late settlement) samples by comparison of allele frequencies and computation of the genetic distance between the two populations on the basis of the allele-frequency information on common markers typed in unlinked regions in both study samples. The GENEPOP analysis showed significant evidence of a difference between allele frequencies of multiple loci between the two populations. The analysis of genetic distance was performed using the POPDIST program (Guldbrandtsen et al. 2000). In this analysis, Nei’s genetic distance, Ds (Nei 1984), was .009 (standard error [SE] .001) between the study samples, providing some evidence for genetic heterogeneity between the southwestern and northeastern study samples.
Discussion
Premature CHD is a major cause of morbidity in the Western societies. In Finland, the occurrence of CHD is one of the highest in the world (Tunstall-Pedoe et al. 1994), with the highest incidence in eastern Finland (Salomaa et al. 1992; Jousilahti et al. 1998). In our genomewide scan for loci predisposing to premature CHD, we used two independent regional study samples, both originating from the genetically isolated Finnish population. The southwestern sample represents an early-settlement region of Finland (∼2,000 years old) which also has a less-heavy burden of environmental risk factors for cardiovascular disorders (Vartiainen et al. 1994). The northeastern study sample represents a late-settlement region, founded ∼300 years ago by founders mostly originating from the early-settlement region (Varilo et al. 2000). This northeastern region has a higher prevalence of CHD than other parts of Finland, probably because of genetic factors combined with a clustering of environmental risk factors for CHD (Vartiainen et al. 1994; Jousilahti et al. 1998). We also tested for possible genetic heterogeneity between the northeastern and southwestern study samples by computing the genetic distance between the two populations. Some evidence emerged for genetic heterogeneity between the northeastern and southwestern study samples, in accordance with the earlier study by Kittles et al. (1998), in which a genetic difference between the populations of these two regions was implicated. The population in the late-settlement region has gone through a bottleneck with restricted number of founders and this might still be visible in the allele frequencies of the late-settlement population. The same loci were, however, identified in both populations using angiographically confirmed severe-CHD index cases and affected family members who fulfilled strict diagnostic criteria for premature CHD in terms of the well-defined clinical diagnosis and their age.
Two chromosomal regions, one on chromosome 2q21.1-22 and another on chromosome Xq23-26, revealed evidence of linkage to premature CHD in both separate and pooled analyses. The chromosomal region on chromosome Xq23-26 is especially intriguing, because we also identified some evidence for LD with one marker in this region.
The multipoint ASP analysis of the X-chromosomal markers in the northeastern and southwestern study samples resulted in two peaks separated by 23 cM. Consequently, the peak produced in the pooled analysis remained wide (∼30 cM). This could indicate the existence of more than one X-chromosomal locus linked with premature CHD or could reflect well-known problems in multipoint analyses, such as genotyping and map errors. Other explanations are also possible. The small sample size of each study and, by definition, incorrect parameters in linkage analyses of complex traits may result in this kind of variation in the peak positions (Hovatta et al. 1998; Ekelund et al. 2000). Minor differences in the allele frequencies of markers and/or disease alleles could create differences in the informativeness of individual markers in separate study samples. Interestingly, despite the presence of two multipoint linkage peaks, only one marker (DXS8053) showed some hint of linkage disequilibrium. This marker is located in the middle of the 30-cM region, suggesting that a single locus may be responsible for the linkage to CHD in both study samples.
The full explanation for the higher frequency of premature CHD in men is still lacking. To some extent, a hormonal shelter helps protect women from premature CHD until menopause, since, after menopause, women’s likelihood of having CHD increases dramatically. This combined with higher load of cardiovascular risk factors is likely to predispose men to premature CHD (Jousilahti et al. 1999). Here, we found a region linked to premature CHD on chromosome X. Most of the linkage information emerges from men, because 65% of the affected individuals in the northeastern study material and 70% in the southwestern material were men. Since men are hemizygotes for disease alleles on chromosome X, they have higher probability to show the effects of recessively acting X-chromosomal susceptibility genes than do women. Although the background of CHD is complex, undoubtedly also including other genetic and environmental factors, this X-chromosomal finding may offer a new starting point for future studies on increased risk for premature CHD in men. Monitoring of intragenic single-nucleotide polymorphisms (SNP) in this chromosomal region could help pinpoint to the gene(s) associated with CHD.
On chromosome 2, a replication sample from the southwestern part of Finland also showed evidence of the potential linkage found in the northeastern sample set, resulting in two-point MLS of 2.3. The combined analysis produced an even higher MLS of 3.0 in two-point analysis, with the marker D2S129 supporting further the evidence for involvement of this chromosomal region in CHD. No evidence of LD, however, could be detected. This may well indicate that none of the genotyped markers is near enough to the causative gene. In fact, in complex, multifactorial diseases such as CHD, LD detected is likely to be restricted to much smaller regions around the causative genes than in monogenic diseases (Weiss 1995), even in genetic isolates (Kruglyak 1999; Lonjou et al. 1999; Weiss and Terwilliger, in press).
Earlier studies have shown that it is likely that genes encoding the apolipoprotein E, apolipoprotein (a), methyltetrahydrofolate reductase, and angiotensin-converting enzyme participate in the CHD pathogenesis (Kraft et al. 1996; Cambien et al. 1992; Boushey et al. 1995; Wilson et al. 1996). We therefore compared the locations of these genes to our CHD scan results in the northeastern, southwestern, and combined study groups in stage 1 and 2 of the genome scan. However, none of these genes were located in regions where we detected MLSs >1.0 in stage 1 or 2. Neither were they located on chromosomes 2 and X. These results suggest that additional genes are likely to have effects on the complex atherosclerotic process. We also looked for novel CHD candidate genes on chromosome 2q21.1-22 and Xq23-26. On chromosome Xq23-26, we found one potentially interesting candidate; the angiotensin receptor 2 (AGTR2) gene resides in that region. The function of the AGTR2 gene is largely unknown, but it is suggested to play a role in the cardiovascular and central nervous functions (Hein et al. 1995). In the identified region on chromosome 2, the mitochondrial glycerophosphate dehydrogenase (GPD2) gene is indirectly a potentially interesting candidate gene. Deficiency of the mitochondrial glycerophosphate dehydrogenase has been shown to contribute to impaired glucose-stimulated insulin release in animal models of noninsulin-dependent diabetes mellitus. However, no obvious CHD candidate genes are located in this chromosomal region, suggesting that a novel gene—or a gene the function of which in CHD is not clear yet—may underlie the linkage. Further studies are warranted involving genotyping of SNPs for these and other possible candidate genes in the regions to search for allelic association with CHD and, finally, to verify variants with functional significance.
In conclusion, CHD is a multifactorial disorder in which numerous factors determine an individual’s coronary risk. Our genome-scan data on severe premature cases of familial CHD, collected from a population with substantial environmental and genetic homogeneity provided evidence for two chromosomal regions, 2q21.1-22 and Xq23-26, which are candidates to harbor novel genes predisposing to CHD.
Acknowledgments
This work was supported by the Academy of Finland, Finnish Heart Foundation, Maud Kuistila Foundation, and the Duodecim Foundation. We thank M. Parkkonen for excellent laboratory technical assistance.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GeneMap ’99, http://www.ncbi.nlm.nih.gov/genemap
- GENPOP program, http://wbiomed.curtin.edu.au/genepop/history.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/omim (for APOE [MIM 107741], LPA [MIM 152200])
References
- Boushey CJ, Beresford SA, Omenn GS, Motulsky AG (1995) A quantitative assessment of plasma homocysteine as a risk factor for vascular disease: probable benefits of increasing folic acid intakes. JAMA 274:1049–1057 [DOI] [PubMed] [Google Scholar]
- Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, Luc G, Bard J-M, Bara L, Ricard S, Tiret L, Amouyel P, Alhenc-Gelas F, Soubrier F (1992) Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature 359:641–644 [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Chakravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278:1580–1581 [DOI] [PubMed] [Google Scholar]
- Davis S, Weeks DE (1997) Comparison of nonparametric statistics for detection of linkage in nuclear families: single-marker evaluation. Am J Hum Genet 61:1431–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, Juvonen H, Varilo T, Arajärvi R, Kokko-Sahin M-L, Lönnqvist J, Peltonen L (2000) Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet 9:1049–1057 [DOI] [PubMed] [Google Scholar]
- Genest J Jr, Cohn JS (1995) Clustering of cardiovascular risk factors: targeting high-risk individuals. Am J Cardiol Suppl 76:8A–20A [DOI] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000a) Gene mapping in the 20th and 21st centuries: statistical methods, data analysis, and experimental design. Hum Biol 72:63–132 [PubMed] [Google Scholar]
- ——— (2000b) Linkage analysis in the presence of errors III: marker loci and their map as nuisance parameters. Am J Hum Genet 66:1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2000c) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy SM, Wilhelmsen L, Rose G, Campbell RW, Assman G (1990) Coronary heart disease in high risk populations: lessons from Finland. Eur Heart J 11:462–471 [DOI] [PubMed] [Google Scholar]
- Grundy SM, Balady GJ, Criqui MH, Fletcher G, Greenland P, Hiratzka LF, Houston-Miller N, Kris-Etherton P, Krumholz HM, LaRosa J, Ockene IS, Pearson TA, Reed J, Washington R, Smith SC Jr (1998) Primary prevention of coronary heart disease: guidance from Framingham—a statement for healthcare professionals from the AHA Task Force on Risk Reduction. Circulation 97:1876–1887 [DOI] [PubMed] [Google Scholar]
- Guldbrandtsen B, Tokiuk J, Loeschcke V (2000) POPDIST, version 1.1.1: a program to calculate population genetic distance and identity measures. J Hered 91:178–179 [DOI] [PubMed] [Google Scholar]
- Hein L, Barsh GS, Pratt RE, Dzau VJ, Kobilka BK (1995) Behavioural and cardiovascular effects of disrupting the angiotensin II type-2 receptor in mice. Nature 377:744–747 [DOI] [PubMed] [Google Scholar]
- Hovatta I, Lichtermann D, Juvonen H, Suvisaari J, Terwilliger JD, Arajärvi R, Kokko-Sahin ML, Ekelund J, Lönnqvist J, Peltonen L (1998) Linkage analysis of putative schizophrenia gene candidate regions on chromosomes 3p, 5q, 6p, 8p, 20p and 22q in a population-based sampled Finnish family set. Mol Psychiatry 3:452–457 [DOI] [PubMed] [Google Scholar]
- Jousilahti P, Puska P, Vartiainen E, Pekkanen J, Tuomilehto J (1996) Parental history of premature coronary heart disease: an independent risk factor of myocardial infarction. J Clin Epidemiol 49:497–503 [DOI] [PubMed] [Google Scholar]
- Jousilahti P, Vartiainen E, Tuomilehto J, Pekkanen J, Puska, P (1998) Role of known risk factors in explaining the difference in the risk of coronary heart disease between eastern and southwestern Finland. Ann Med 30:481–487 [DOI] [PubMed] [Google Scholar]
- Jousilahti P, Vartiainen E, Tuomilehto J, Puska P (1999) Sex, age, cardiovascular risk factors, and coronary heart disease: a prospective follow-up study of 14,786 middle-aged men and women in Finland. Circulation 99:1165–1172 [DOI] [PubMed] [Google Scholar]
- Kannel WB, Castelli WP, Gordon T, McNamara, PM (1971) Serum cholesterol, lipoproteins, and the risk of coronary heart disease: the Framingham study. Ann Intern Med 74:1–12 [DOI] [PubMed] [Google Scholar]
- Keys A (1980) Seven countries: a multivariate analysis of death and coronary heart disease. Harvard University Press, Cambridge [Google Scholar]
- Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft HG, Lingenhel A, Kochl S, Hoppichler F, Kronenberg F, Abe A, Muhlberger V, Schonitzer D, Utermann G (1996) Apolipoprotein(a) kringle IV repeat number predicts risk for coronary heart disease. Arterioscler Thromb Vasc Biol 16:713–719 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S, Sundvall M, Terwilliger JD, Tienari PJ, Wikström J, Holmdahl R, Pettersson U, Peltonen L (1996) A putative vulnerability locus to multiple sclerosis maps to 5p14-p12 in a region syntenic to the murine locus Eae2. Nat Genet 13:477–480 [DOI] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048 [DOI] [PubMed] [Google Scholar]
- Lonjou C, Collins A, Morton NE (1999) Allelic association between marker loci. Proc Natl Acad Sci USA 96:1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca L, Manson JE, Sutherland SE, Langer RD, Manolio T, Barrett-Connor E (1997) Cardiovascular disease in women: a statement for healthcare professionals from the American Heart Association. Circulation 96:2468–2482 [DOI] [PubMed] [Google Scholar]
- Nei M (1984) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Noll G (1998) Pathogenesis of atherosclerosis: a possible relation to infection. Atherosclerosis Suppl 140:S3–S9 [DOI] [PubMed] [Google Scholar]
- Nora JJ, Lortscher RH, Spangler RD, Nora AH, Kimberling WJ (1980) Genetic-epidemiologic study of early-onset ischemic heart disease. Circulation 61:503–508 [DOI] [PubMed] [Google Scholar]
- O’Connel JR, Weeks DE (1998) Pedcheck: a program for identification of genotype imcompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooling Project Research Group, The (1978) Relationship of blood pressure, serum cholesterol, smoking habit, relative weight and ECG abnormalities to incidence of major coronary events. Monograph 60, American Heart Association, Dallas [DOI] [PubMed] [Google Scholar]
- Pyörälä K, Laakso M, Uusitupa M (1987) Diabetes and atherosclerosis: an epidemiologic view. Diabetes Metab Rev 3:463–542 [DOI] [PubMed] [Google Scholar]
- Ridker PM, Buring JE, Shih J, Matias M, Hennekens CH (1998) Prospective study of C-reactive protein and the risk of future cardiovascular events among apparently healthy women. Circulation 98:731–733 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Stone VA, Daly MJ, Cargill M, Green T, Nguyen H, Nutman T, Zimmerman PA, Tucker MA, Hudson T, Goldstein AM, Lander E, Lin AY (1998) Familial eosinophilia maps to the cytokine gene cluster on human chromosomal region 5q31-q33. Am J Hum Genet 63:1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Rose G (1964) Familial patterns in ischaemic heart disease. Br J Prev Soc Med 18:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Blackburn H, Gillum R (1982) Cardiovascular survey methods. 2nd ed. World Health Organization, Geneva [PubMed] [Google Scholar]
- Saikku P, Leinonen M, Mattila K, Ekman MR, Nieminen MS, Mäkelä PH, Huttunen JK, Valtonen V (1988) Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet 2:983–986 [DOI] [PubMed] [Google Scholar]
- Salomaa V, Arstila M, Kaarsalo E, Ketonen M, Kuulasmaa K, Lehto S, Miettinen H, Mustaniemi H, Niemela M, Palomäki P, Pyörälä K, Torppa J, Tuomilehto J, Vuorenmaa T (1992) Trends in the incidence of and mortality from coronary heart disease in Finland, 1983–1988. Am J Epidemiol 136:1303–1315 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, Pulido JC, Yandava C, Sunden SL, Mattes G, Businga T, McClain A, Beck J, Scherpier T, Gilliam J, Zhong J, Duyk GM (1995) A collection of tri- and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet 4:1837–1844 [DOI] [PubMed] [Google Scholar]
- Sholtz RI, Rosenma RH, Brand RJ (1975) The relationship of reported parental history to the incidence of coronary heart disease in Western Collaborative Group Study. Am J Epidemiol 102:350–356 [DOI] [PubMed] [Google Scholar]
- Smith CAB (1957) Counting methods in genetical statistics. Ann Hum Genet 21:254–276 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J (1992) A haplotype-based “haplotype relative risk” approach to detecting allelic associations. Hum Hered 42:337–346 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- Tunstall-Pedoe H, Kuulasmaa K, Amouyel P, Arveiler D, Rajakangas AM, Pajak A (1994) Myocardial infarction and coronary deaths in the World Health Organization MONICA Project Registration procedures, event rates, and case-fatality rates in 38 populations from 21 countries in four continents. Circulation 90:583–612 [DOI] [PubMed] [Google Scholar]
- Weiss KM (1995) Genetic variation and human disease: principles and evolutionary approaches. Cambridge University Press, Cambridge [Google Scholar]
- Weiss KM, Terwilliger JD (2000) How many diseases does it take to map a gene with SNPs? Nat Genet 26:151–157 [DOI] [PubMed] [Google Scholar]
- Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB (1998) Prediction of coronary heart disease using risk factor categories. Circulation 97:1837–1847 [DOI] [PubMed] [Google Scholar]
- Wilson PW, Schaefer EJ, Larson MG, Ordovas JM (1996) Apolipoprotein E alleles and risk of coronary disease A meta-analysis. Arterioscler Thromb Vasc Biol 16:1250–1255 [DOI] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 [DOI] [PubMed] [Google Scholar]
- Varilo T, Laan M, Hovatta I, Wiebe V, Terwilliger JD, Peltonen L (2000) Linkage disequilibrium in isolated populations: Finland and a young sub-population of Kuusamo. Eur J Hum Genet 8:604–612 [DOI] [PubMed] [Google Scholar]
- Vartiainen E, Puska P, Jousilahti P, Korhonen HJ, Tuomilehto J, Nissinen A (1994) Twenty-year trends in coronary risk factors in North Karelia and in other areas of Finland. Int J Epidemiol 23:495–504 [DOI] [PubMed] [Google Scholar]