Abstract
Understanding the complexities of DNA has been a hallmark of science for over a half century, and one of the important topics in DNA research is recognizing the occurrence of mutations in the base-stack. In this article, we present a study of SNPs by direct-contact electrical measurements to a single DNA duplex. We have used short, 11- and 12-bp dsDNA to investigate the change in conductance that occurs if a single base pair, a single base, or two separate bases in the stack are modified. All measurements are carried out in aqueous solution with the DNA chemically bound to the electrodes. These measurements demonstrate that the presence of a single base pair mismatch can be identified by the conductance of the molecule and can cause a change in the conductance of dsDNA by as much as an order of magnitude, depending on the specific details of the double helix and the single nucleotide polymorphism.
Keywords: molecular electronics, scanning tunneling microscope break junction
The measurement and understanding of the physical properties of DNA are crucial to understanding the biological functions of DNA and developing DNA-based technologies. For example, thermal melting of DNA has become an indispensable parameter in the design of PCR primers and DNA probes and also in the analysis of mutations. In this regard, the understanding and detection of an SNP in a dsDNA molecule has become a topic of significant value in attempts to understand the causes of cancer, the inherent properties of DNA, and its interactions with various proteins. A variety of methods for detecting and understanding SNPs in DNA have been described (1–3), yet a simple, reliable and unambiguous method for recognizing these mutations would be an important advance. The focus of the present work is to address the questions as to what effect the alteration of a single base or base pair in the stack has on the molecule's conductivity, if the change is large enough to effectively measure this important biological alteration, and how such a study improves our current understanding of charge transport in DNA.
Charge transport in DNA has been studied by photochemical (4–7), biochemical (8–11), electrochemical (12–14), and direct electrical measurements (15–20). The direct measurement of DNA conductivity requires wiring the molecule to two probing electrodes (21). Depending on the experimental method, insulating, semiconducting, and conducting behaviors have been reported for DNA. This large disparity is partially due to the difficulty of forming good molecule–electrode contacts and variations in the experimental conditions (22). However, most measurements on relatively short DNA duplexes have demonstrated that the molecule is capable of charge transport, although the exact mechanism is still a source of debate.
To investigate the conductance differences that occur due to an SNP and to better elucidate the inherent charge transport mechanisms available in the stack, six dsDNA structures were studied in this work. We show that the alteration of a single base in the stack can either increase or decrease the conductivity of the dsDNA helix, depending on the type of the mismatched base(s), and demonstrate the possibility of detecting SNPs by means of direct conductance measurement.
Methods
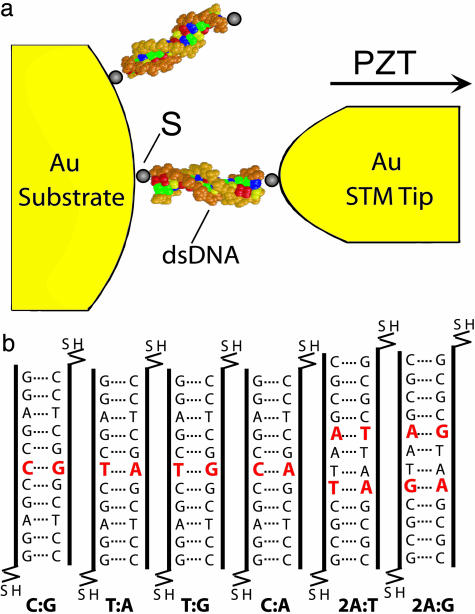
In this work, the conductivities of individual dsDNA helices chemically bound to two gold electrodes were measured in aqueous solution by using the scanning tunneling microscope (STM) break-junction approach (4–7, 23). The details of the approach have been described in ref. 23. Briefly, it works by repeatedly bringing a gold STM tip into and out of contact with a gold substrate covered with a monolayer of thiolated dsDNA molecules to create Au-S-DNA-S-Au structures. As the tip is pulled away from the substrate, the current is monitored, and steps in the current decay indicate the presence of an attached molecule. By repeating these measurements thousands of times, a histogram can be constructed from the current decay curves that exhibit steps. The histogram reveals peaks located near integer multiples of a constant value, which we attribute to the attachment of one, two, or three molecules between the tip and the surface, provided they are multiples of the smallest visible peak.
The six molecules that were chosen are shown in Fig. 1; the four 11-bp-length molecules will be referred to by the center base pair: T:A, C:G, T:G, and C:A. The other two DNA molecules will be referred to by the fact that they demonstrate a 2-bp polymorphism, as either 2A:T or 2A:G. All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA), which were HPLC purified and terminated with 3′ thiol linkers in the form of C3H6S2C3H6OH. The DNA molecules were left in this protected form before use. Upon receipt of the oligonucleotides, 10-μM ssDNA solutions were created in 10 mM phosphate buffer solution (PBS) at pH 7.4 and 100 mM NaCl. The concentrations were determined by using the Biospec-Mini UV-Visible spectrophotometer. Tris(2-carboxyethyl)phosphine (TCEP) (10 mM) was added to the solution to reduce the disulfide bond and remove the mercaptopropanol-protecting group (24). The solution was allowed to incubate for 3 h at room temperature to allow for the complete deprotection of the linker units. The oligonucleotides were then separated from the cleaved mercaptopropanol groups with the use of a spin column (Quick Spin Columns G-25, Roche Applied Science). At this point, the appropriate oligomers were combined as needed to achieve the desired dsDNA helix. The combined solutions were then heated to 70°C in a bath of 1 liter of water and allowed to return to room temperature over several hours to anneal the dsDNA. Yields of dsDNA were determined by UV absorbance at 260 nm and were typically in the range of 2–4 μM dsDNA in solution.
Fig. 1.
Depiction of the experimental method and molecules. (a) Schematic diagram of experimental setup. The STM tip is driven into contact with the DNA monolayer and then retracted. Chemically bound molecules give a conductance signature during the pulling process that can be determined by means of the histogram. (b) This graphic shows six dsDNA molecules tested during this series of experiments. The first four molecules represent the SNP series, and the last two represent the change of a 2-bp mismatch in the stack. The name given below each of the molecules is used to refer to that specific molecule throughout the paper.
Once the dsDNA was acquired, it was added to 100 mM NaClO4 and placed on a freshly annealed gold substrate for measurements. Electrical measurements were carried out by using a Nanoscope IIIA controller from Digital Instruments (Santa Barbara, CA) and a Molecular Imaging (Tempe, AZ) STM scanner. The repeated creation of the metal–molecule–metal junctions was controlled with labview software designed in-house. The STM tips were prepared by cutting 0.25-mm gold wires (99.998%), which were then coated with Apiezon wax to limit the ionic and polarization currents to the order of the noise in the amplifier, which is in the pA range. The substrates were prepared by thermally evaporating 130 nm of gold on mica in a ultra high vacuum (UHV) chamber. Before each experiment, the substrate was briefly annealed in a hydrogen flame.
It is also important to ascertain whether the DNA is in biological, or B-form, during the measurements. For this purpose, melting temperature (Tm) experiments were carried out on a Varian Cary 300 Bio UV spectrophotometer with Peltier thermal controller to demonstrate appropriate melting temperatures. Melting temperatures were determined by taking the peak of the first derivative of the melting curves for the respective dsDNA. Similarly, temperature sweeps were repeated a number of times for each sample to ensure repeatability. The temperature sweeps were carried out at a rate of 0.2°C/min, with the absorbance at 260 nm recorded every 30 s.
Although melting temperatures ensure that a stable helix is formed at room temperature, CD is needed to validate the formation of a B-form helix. As such, CD measurements were carried out on a Jasco (Easton, MD) J-1710 Spectropolarimeter to demonstrate that the DNA was in B-form. For both sets of photometric measurements, positive controls were carried out for each of the four strands used in the SNP series to demonstrate that the linkers themselves have no effect on the melting temperature or the conformation of the DNA. The data shown were acquired with a wavelength sweep rate of 100 nm/min and smoothed with a Savitzky-Golay second-order smoothing algorithm with a 10-point window.
Results and Discussion
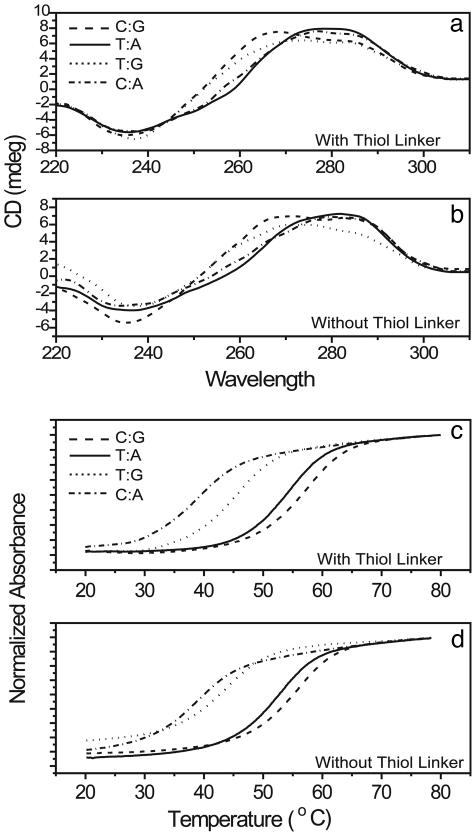
To ensure that the DNA under investigation is in the desired conformation, the photometric measurements were carried out as described above. As can be seen in Fig. 2, the melting temperature of each of the strands in the SNP series was measured with and without the thiol linker. In all cases Tm is well above room temperature, with the minimum value at 38.22°C for the C:A mismatch, as expected and shown in Table 1. Furthermore, the Tm of each of the linker-modified dsDNA molecules is within 3°C of the Tm of the unmodified DNA. These data demonstrate that the linker has little to no effect on the free energy of the helix. Secondly, the melting temperatures predicted by HyTherm are consistently lower than the measured values, meaning that the helices are actually slightly more stable than predicted.
Fig. 2.
Assays of DNA helical structure with and without thiol linkers present. (a) Melting temperature curves for each of the molecules in the SNP series without a thiol linker attached to the molecule. (b) Melting curves for each of the molecules with thiol linkers attached to each of the 3′ ends. (c) CD measurements on the samples without linkers. (d) The CD curves from the molecules with linkers. Data show that the thiol linkers have no effect upon melting or structure, and that the DNA is in a stable B-form at experimental temperature. —, T:A; - - -, C:G; · · · ·, T:G; - · - · -, C:A.
Table 1. The melting temperature and conductance values measured in the reported experiments.
| DNA sequence | Tm, °C | Measured conductance, G/Go |
|---|---|---|
| GGAG CCC GAGG | 56.17 (54.1) | 1.3 × 10–4 |
| CCTC GGG CTCC | ||
| GGAG CTC GAGG | 53.17 (49.6) | 5 × 10–5 |
| CCTC GAG CTCC | ||
| GGAG CTC GAGG | 44.22 (41.4) | 7.5 × 10–5 |
| CCTC GGG CTCC | ||
| GGAG CCC GAGG | 38.22 (32.6) | 1 × 10–5 |
| CCTC GAG CTCC | ||
| CGCG AATT CGCG | – (56.0) | 3.6 × 10–6 |
| GCGC TTAA GCGC | ||
| CGCG AATG CGCG | – (38.2) | 1.7 × 10–6 |
| GCGC GTAA GCGC | ||
| GGAG CCC GAGG* | 57.97 (54.1) | – |
| CCTC GGG CTCC | ||
| GGAG CTC GAGG* | 53.97 (49.6) | – |
| CCTC GAG CTCC | ||
| GGAG CTC GAGG* | 46.98 (41.4) | – |
| CCTC GGG CTCC | ||
| GGAG CCC GAGG* | 38.97 (32.6) | – |
| CCTC GAG CTCC |
Values in parentheses predicted by HyTherm. Bold letters indicate points where mutations were placed in the DNA stack. Dashes indicate points where data were not/could not be acquired.
These molecules measured without thiol linkers
The CD measurements were also carried out on the SNP series both with and without the thiol linkers, and, as can be seen in Fig. 2, the linker has no effect on the conformation of the DNA. Nevertheless, there are some differences between the various molecules, demonstrating that there may be some structural differences between them based on sequence, as is expected for short dsDNA helices (25). It is also worth noting that the C:G and T:G molecules seem to be more similar to one another than the two perfectly matched sequences are to each other. However, despite this slight change in structure, the CD measurement clearly demonstrates that each of the samples in the SNP series is in B-form in solution phase.
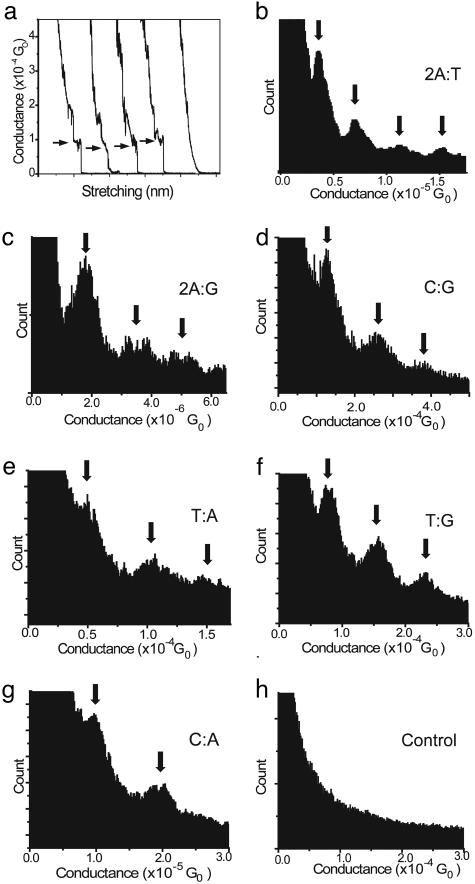
Knowing that the dsDNA molecules have the proper conformation in solution and the required stability for measurements, one can begin to investigate the charge transport properties of the dsDNA and the effects of mutation on the conductance of the molecule. Fig. 3a shows several conductance curves recorded during the separation of the STM tip from contacting with the substrate. About 20% of the curves show stepwise decay in the conductance, which is due to the breakdown of individual molecular junctions as discussed earlier. It also shows an example of the exponential decay. Such exponential decay is expected for simple electron tunneling between the STM tip and the substrate, and this decay corresponds to the incidents where no DNA molecules bridge the two electrodes. The curves in Fig. 3a are from T:G at 100-mV bias. It is important to note that each of the dsDNA helices investigated was analyzed at several different biases between 50 and 400 mV, and, for each specific sequence, the conductance value obtained was the same regardless of bias, thus suggesting a linear current response with bias change for this small potential window.
Fig. 3.
DNA conductance data. (a) Several representative curves that occur during the stretching of the STM break junction. These curves are sampled and added together to form the conductance histograms. (b) Conductance histogram for 2A:T showing a conductance of 3.6 × 10-6 Go.(c) Histogram for 2A:G showing a conductance of 1.7 × 10-6 Go. (d) C:G histogram showing a conductance of 1.3 × 10-5 Go. (e) Histogram for T:A showing a conductance of 5 × 10-5 Go.(f) T:G histogram showing a conductance of 7.5 × 10-5 Go. (g) C:A histogram showing a conductance of 1 × 10-5 Go. (h) Histogram for a control experiment without DNA present in 10 mM TCEP, 10 mM PBS at pH 7.4, and 50 mM NaClO4, demonstrating no conductance peaks.
The histograms were obtained from curves such as the ones shown in Fig. 3a by using the procedures described in Methods. A representative histogram of each of the six molecules studied is shown in Fig. 3. In conjunction with discussing the conductance values of the various DNA molecules, it is important to analyze the overall sequence of the molecules used. The sequence 5′-CGCGAATTCGCG-3′-C3H6S is matched first with its complement (molecule 2A:T) and shows a conductance value of 3.6 × 10-6 Go, in accordance with the prior results (23), where Go is the fundamental conductance quantum of 77.5 μS, as is shown in Fig. 3b. However, upon replacement of two of the Ts by Gs, two mismatched A:G base-pairs were created in the helix. The corresponding conductance of the mutated DNA drops to a value of 1.7 × 10-6 Go (Fig. 3c). Thus, these data suggest that the presence of mismatches in the DNA duplexes can cause a measurable decrease in the conductance of the molecule, presumably due to decreased coupling between bases or structural instabilities.
The SNP series has a sequence of the form 5′-GGAGCXCGAGG-3′-C3H6S, where X is either a C or a T, and represents the point of mutation of the SNP. Before discussing the SNP results, it is important to analyze the differences in sequence between the molecules chosen for the SNP study and those described above. In general, the sequence for the SNP study is symmetric about the mutation point. More importantly, rather than having an alternating GC or AT sequence, as was analyzed previously, this sequence often has similar bases adjacent to one another. Thus, this sequence will allow for better coupling between Gs than the alternating structure used previously if the G-hopping mechanism is the dominant mode in these molecules (26). Furthermore, this structure includes the presence of two separated A:T barriers in the C:G molecule and three A:T barriers in the T:A molecule. These differences in sequence and the potential roles that they play in charge transport and nearest-neighbor coupling must be kept in mind when analyzing the data from the SNP series of molecules.
The first molecule to be discussed in the SNP series is the C:G molecule. This molecule is 11 bp in length and has a measured conductance of 1.3 × 10-4 Go as is shown in Fig. 3d. Alternatively, if the center C:G is replaced by a T:A base pair, the conductance drops by a factor of 2.6 to 5 × 10-5 Go (Fig. 3e). These data clearly demonstrate that the change of a single base pair in the DNA stack is capable of significantly changing the charge transport properties of the system. Furthermore, this result is in good agreement with the idea that G has a relatively low HOMO (highest occupied molecular orbital), which favors charge transport, and short A:T sequences create a tunneling barrier for charge hopping through Gs in a DNA stack (23, 27).
In the presence of an SNP, one would expect that both the electronic states of each base and the structural instability due to a mutated base play a role in any conductance change in the molecule. In the case of molecule T:G, as is shown in Fig. 3f, the conductance of this molecule is actually between the values of the C:G and T:A molecules at 7.5 × 10-5 Go. However, when the alternate mismatch is explored, the total conductance of the molecule drops significantly to 1 × 10-5 Go, as is shown in Fig. 3g. As such, the conductance of this molecule is an order of magnitude lower than that of C:G, just by changing a single base from a G to an A. Therefore, it is possible to recognize differences in conductance due to simple, single base pair point mutations in the stack.
It is widely believed that charge transport in DNA is highly dependent on the coupling of adjacent and alternating bases, and therefore also on structure (26–29). So, one expects that the conductance of dsDNA depends critically upon the sequence of the specific molecule because the sequence determines the base coupling and local structure. This point is made by the observation that, although the C:G and T:G molecules would have the same pathway in a guanine-hopping scheme, they actually have quantifiable conductivity differences. As such, these data contribute specifically to the idea that the coupling of an individual base to its neighbors and the stability of the molecule are extremely important in the charge transfer dynamics of DNA. Furthermore, this idea of coupling can be extrapolated to point out some differences between the values obtained in this work and the previous results of Xu et al. (23), where G:C bases play a dominant role in the conductivity of the helix. In the case of molecule C:G, even though this is a molecule that is longer than the 10-bp molecule (5′-CGCGATCGCG-3′) that also has two A:T base pairs in it that was reported by Xu et al. (23), the C:G molecule actually has a higher conductivity. This observation can be attributed to the fact that the two A:T base pairs are separated in the C:G molecule, whereas they are adjacent in the molecule of Xu et al., and the fact that quite often in the C:G molecule similar bases are adjacent to one another, potentially leading to better coupling and charge transfer.
The results from the SNP series also seem to strengthen the case for incoherent guanine-hopping along the stack, as is predicted in some theoretical models (26), because the molecule actually increases in conductivity with the change to a G:T mismatch. However, this case cannot be generalized because it is obvious, from the case of the 12-bp molecules studied in this paper, that the introduction of two G:A mismatches in the stack still caused a decrease in the conductivity. This decrease of a factor of ≈2.1 is much smaller than that caused by the C:A mismatch, and it is known that G:As typically stack well with the helix, whereas C:A mismatches do not (6). However, the point that there is still a decrease is interesting where there was an increase in the T:G molecule. A possible explanation is that the conductance gain by the extra guanine bases in 2A:G is offset by the structural instability introduced by the two G:A mismatches. This result implies that the structure of the DNA itself and the coupling with nearest neighbors must play a crucial role in DNA charge transfer.
Control measurements must be carried out to verify that any of the component pieces of the system cannot contribute to any of the measured conductance values. To this end, control experiments were done with an ssDNA connected to a 3′ thiol linker in the same buffer solution. Measurements were also done in PBS at pH 7.4, in a 10 mM PBS with 10 mM TCEP and 50 mM NaClO4 present to directly mimic the experimental conditions without the DNA present, in an NaClO4-only solution, with unmodified dsDNA, with mercaptopropanol in PBS, which is the group left when the DNA is deprotected, and in a 10-mM TCEP solution. These control experiments all yielded histograms similar to that of Fig. 3h. Specifically, Fig. 3h displays a histogram constructed as described above from a control experiment done in a solution of 10 mM PBS at pH 7.4, 10 mM TCEP, and 50 mM NaClO4 without any DNA present. Furthermore, all control experiments demonstrate clearly that low current can be observed with any of these items in solution, that ionic current plays little to no role in the measured current, and that all steps in the current curves, and therefore, the peaks in the conductance histograms, are due to the molecule of interest for that specific experiment.
Summary
The detection of SNPs in a DNA double helix is of significant value in the quest for understanding and preventing genetic diseases. This research demonstrates that a small mutation in dsDNA can cause a significant change in the conductance of the molecule, which may be used for SNP analysis. It also shows that a full understanding of the charge transport properties of DNA will require the exploration of the effects of nearest neighbors and the structural distortions that occur simply because of the sequence (25), as well as the effects of the proximity to, and coupling of, the electrodes to the DNA (30).
Acknowledgments
This work was supported by a grant from the National Science Foundation (to J.H.) and by Department of Energy Grant DE-FG02-01ER45943 (to B.X.).
Author contributions: J.H., B.X., P.Z., and N.T. designed research; J.H. and B.X. performed research; J.H. and B.X. analyzed data; P.Z. contributed new reagents/analytic tools; and J.H. and N.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: STM, scanning tunneling microscope; TCEP, Tris(2-carboxyethyl)phoshine; PBS, phosphate buffer solution.
References
- 1.Jackson, B. A. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 12986-12987. [Google Scholar]
- 2.Wei, F., Chen, C., Zhai, L., Zhang, N. & Zhao, X. S. (2005) J. Am. Chem. Soc. 127, 5306-5307. [DOI] [PubMed] [Google Scholar]
- 3.Weizmann, Y., Patolsky, F. & Willner, I. (2001) Analyst 126, 1502-1504. [DOI] [PubMed] [Google Scholar]
- 4.Murphy, C. J., Arkin, M. R., Jenkins, Y., Ghatlia, N. D., Bossmann, S. H., Turro, N. J. & Barton, J. K. (1993) Science 262, 1025-1029. [DOI] [PubMed] [Google Scholar]
- 5.Lewis, F., Wu, T., Zhang, Y., Letsinger, R., Greenfield, S. & Wasielewski, M. (1997) Science 277, 673-676. [DOI] [PubMed] [Google Scholar]
- 6.Kelley, S. O., Holmlin, R. E., Stemp, E. D. A. & Barton, J. K. (1997) J. Am. Chem. Soc. 119, 9861-9870. [Google Scholar]
- 7.Meade, T. J. & Kayyem, J. F. (1995) Angew. Chem. Int. Ed. Engl. 34, 352-354. [Google Scholar]
- 8.Hall, D. B., Holmlin, R. E. & Barton, J. K. (1996) Nature 382, 731-735. [DOI] [PubMed] [Google Scholar]
- 9.Ly, D., Sanii, L. & Schuster, G. B. (1999) J. Am. Chem. Soc. 121, 9400-9410. [Google Scholar]
- 10.Meggers, E., Kusch, D., Spichty, M., Wille, U. & Giese, B. (1998) Angew. Chem. Int. Ed. 37, 460-462. [DOI] [PubMed] [Google Scholar]
- 11.Saito, I., Nakamura, T., Nakatani, K., Yoshioka, Y., Yamaguchi, K. & Sugiyama, H. (1998) J. Am. Chem. Soc. 120, 12686-12687. [Google Scholar]
- 12.Kelley, S. O., Barton, J. K., Jackson, N. M. & Hill, M. G. (1997) Bioconjugate Chem. 8, 31-37. [DOI] [PubMed] [Google Scholar]
- 13.Kelley, S. O., Jackson, N. M., Hill, M. G. & Barton, J. K. (1999) Angew. Chem. Int. Ed. 38, 941-945. [DOI] [PubMed] [Google Scholar]
- 14.Hartwich, G., Caruana, D. J., de lumley-Woodyear, T., Wu, Y., Campbell, C. N. & Heller, A. (1999) J. Am. Chem. Soc. 121, 10803-10812. [Google Scholar]
- 15.Okahata, Y., Kobayashi, T., Tanaka, K. & Shimomura, M. (1998) J. Am. Chem. Soc. 120, 6165-6166. [Google Scholar]
- 16.Braun, E., Eichen, Y., Sivan, U. & Ben-Yoseph, G. (1998) Nature 391, 775-778. [DOI] [PubMed] [Google Scholar]
- 17.Porath, D., Bezryadin, A., de Vries, S. & Dekker, C. (2000) Nature 403, 635-638. [DOI] [PubMed] [Google Scholar]
- 18.Fink, H.-W. & Schonenberger, C. (1999) Nature 398, 407-410. [DOI] [PubMed] [Google Scholar]
- 19.Kasumov, A. Y., Kociak, M., Gueron, S., Reulet, B., Volkov, V. T., Klinov, D. V. & Bouchiat, H. (2001) Science 291, 280-282. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Y., Austin, R. H., Kraeft, J., Cox, E. C. & Ong, N. P. (2002) Phys. Rev. Lett. 89, 198102. [DOI] [PubMed] [Google Scholar]
- 21.Dekker, C. & Ratner, M. A. (2001) Physics World 14, 29-33. [Google Scholar]
- 22.Hipps, K. W. (2001) Science 294, 536-537. [DOI] [PubMed] [Google Scholar]
- 23.Xu, B., Zhang, P., Li, X. & Tao, N. (2004) Nano Lett. 4, 1105-1108. [Google Scholar]
- 24.Aqua, T., Naaman, R. & Daube, S. S. (2003) Langmuir 19, 10573-10580. [Google Scholar]
- 25.Hays, F. A., Teegarden, A., Jones, Z. J. R., Harms, M., Raup, D., Watson, J., Cavaliere, E. & Ho, P. S. (2005) Proc. Natl. Acad. Sci. USA 102, 7157-7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jortner, J., Bixon, M., Langenbacher, T. & Michel-Beyerle, M. E. (1998) Proc. Natl. Acad. Sci. USA 95, 12759-12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bixon, M., Giese, B., Wessely, S., Langenbacher, T., Michel-Beyerle, M. E. & Jortner, J. (1999) Proc. Natl. Acad. Sci. USA 96, 11713-11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voityuk, A. A., Rosch, N., Bixon, M. & Jortner, J. (2000) J. Phys. Chem. B 104, 9740-9745. [Google Scholar]
- 29.Conwell, E. M. & Rakhmanova, S. V. (2000) Proc. Natl. Acad. Sci. USA 97, 4556-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia, E., Triozon, F. & Roche, S. (2005) Phys. Rev. B Condens. Matter 71, 113106. [Google Scholar]