Abstract
A mecA-containing Staphylococcus aureus strain was grown in the presence of high concentrations of d-serine, d-threonine, and d-phenylalanine. These growth conditions resulted in the replacement of the carboxyl-terminal (fifth) d-alanine residue of peptidoglycan stem peptides with the d-amino acid present in the growth medium and a reduced ability to grow in the presence of methicillin. The most dramatic effect was seen with d-serine. With 32 mM d-serine, strains that had been able to grow in the presence of 800 μg of methicillin per ml were only able to grow in the presence of less than 50 μg/ml. The results also suggest that in S. aureus vancomycin resistance mediated through the incorporation of precursors not terminating in d-alanyl-d-alanine would be mutually exclusive with expression of mecA-mediated methicillin resistance.
Expression of methicillin resistance in Staphylococcus aureus is mediated by the mecA gene product, penicillin-binding protein (PBP) 2A (PBP 2A) (15). PBP 2A is assumed to act as a peptidoglycan-synthesizing enzyme (8, 12), and because it has much lower affinities for β-lactam antibiotics compared to those of the set of regular staphylococcal peptidoglycan-synthesizing enzymes (4), PBP 2A could allow continued peptidoglycan biosynthesis in the presence of concentrations of β-lactam antibiotics in the medium that inhibit the other peptidoglycan-synthesizing enzymes. Therefore, mecA-containing S. aureus strains are able to grow in the presence of high concentrations of β-lactam antibiotics. Although mecA expression is required, mecA expression itself is not sufficient for expression of methicillin resistance in certain backgrounds. Several studies have identified other important genes (2, 11). Some of those genes, called fem, have been recognized as determinants in the biosynthesis of peptidoglycan precursors (6, 7, 14, 18, 19, 20).
Environmental conditions that cause changes in peptidoglycan precursor biosynthesis may also alter expression of methicillin resistance in S. aureus. Growth of a mecA-containing S. aureus strain in the presence of high concentrations of glycine produced peptidoglycan in which the carboxyl-terminal d-alanine residue of the stem peptides was replaced with glycine, and this substitution was paralleled by heterogeneous expression and a reduction in the level of methicillin resistance (9). The replacement of the carboxyl-terminal residue of peptidoglycan stem peptides apparently resulted from the synthesis of peptidoglycan precursors terminating in glycine rather than d-alanine. This situation is analogous to that in the fem mutants, in which altered precursors also led to a reduced susceptibility to methicillin (6, 7, 14, 18, 19, 20).
In order to determine whether modifications in peptidoglycan precursors are always accompanied by a change in the level of methicillin resistance, a mecA-containing S. aureus strain was grown in the presence of high concentrations of a variety of d-amino acids. Such conditions are known to lead in many bacteria to a peptidoglycan in which the carboxyl-terminal d-alanine of the stem peptides is replaced by the d-amino acid present in the growth medium (17, 21). Provided that this also occurs in S. aureus, these growth conditions would be expected to render a mecA-containing strain more susceptible to methicillin.
MATERIALS AND METHODS
Growth conditions and growth in the presence of methicillin.
A total of 107 cells of an overnight culture of the mecA-containing S. aureus strain COL (11) grown in tryptic soy broth (TSB; Difco) was transferred to 2 ml of TSB supplemented with 0.25 M glycine, l-serine, d-serine, d-phenylalanine, and d-threonine (Sigma) and twofold increasing concentrations of methicillin. Growth was measured after 24 and 48 h of incubation at 37°C. In another set of experiments, the concentration of methicillin was kept constant at 5 or 50 μg/ml and the amount of d-amino acid was varied.
Peptidoglycan and muropeptide analysis.
Cells were grown in TSB with the addition of d-amino acids. Peptidoglycan was isolated from exponentially grown cells, and its muropeptide composition was analyzed by reversed-phased high-performance liquid chromatography (HPLC) as described before (5). The structures of the muropeptides were determined by a combination of amino acid analysis and mass spectrometric (MS) techniques (23).
RESULTS AND DISCUSSION
Peptidoglycan composition and muropeptide structures.
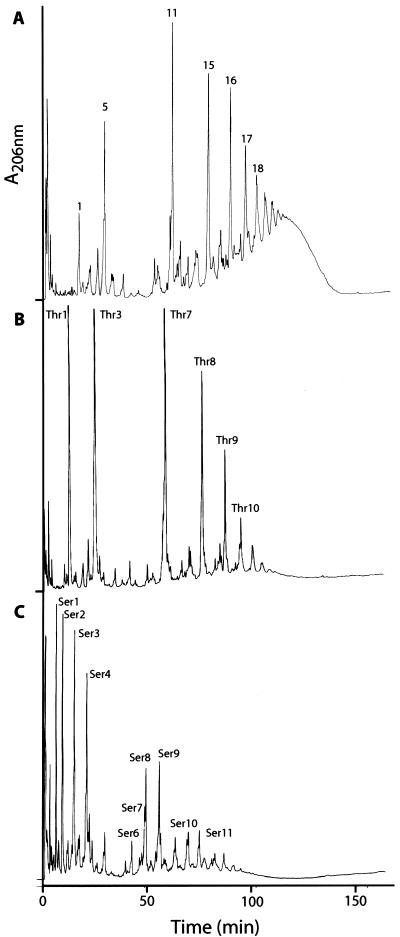
Growth in the presence of 0.25 M d-serine and d-threonine altered the peptidoglycan composition of S. aureus dramatically (Fig. 1). The d-alanine-terminating pentapeptide-containing muropeptides (unsubstituted monomeric species and pentaglycine-substituted mono-, di-, tri-, tetra-, penta-, and hexameric species [peaks 1, 5, 11, 15, 16, 17, and 18, respectively]) present in peptidoglycan in the absence of the d-amino acids (Fig. 1A) virtually disappeared and were replaced by peaks with different retention times (Thr1, Thr3, Thr7 to Thr10 [Fig. 1B], and Ser1 to Ser11 [Fig. 1C]). Amino acid analysis showed that when d-threonine was present in the medium, muropeptides of peptidoglycan contained a threonine residue, and when cells were grown in the presence of 0.25 M d-serine, at least one serine residue was incorporated into the peptidoglycan (Table 1). MS-MS analysis revealed the amino acid sequence and showed threonine and serine to be at the carboxyl-terminus position of the muropeptide stem peptides (data not shown).
FIG. 1.
HPLC elution profiles of muropeptides isolated from peptidoglycan of S. aureus COL grown in TSB (A), TSB supplemented with 0.25 M d-threonine (B), and TSB supplemented with 0.25 M d-serine (C). For structures of peaks, see Table 1 and reference 5.
TABLE 1.
Amino acid compositions, molecular masses and proposed structures of muropeptides present in peptidoglycan of S. aureus strain COL grown in the presence of d-serine or d-threonine
| Peaka | Amino acid compositionb
|
Molecular mass [M-H]− | Proposed structurec | |||||
|---|---|---|---|---|---|---|---|---|
| GLX | LYS | ALA | GLY | SER | THR | |||
| Thr1 | 1 | 1.3 | 2.4 | —d | 0.3 | 1.1 | 996.8 | G-M-Ala-Gln-Lys-Ala-Thr |
| Thr3 | 1 | 1.2 | 2.3 | 5.4 | 0.3 | 1.0 | 1,282.1 | G-M-Ala-Gln-Lys (Gly)5-Ala-Thr |
| Thr7 | 1 | 1.2 | 2.3 | 5.6 | 0.3 | 0.5 | 2,446.6 | G-M-Ala-Gln-Lys (Gly)5-Ala-Thr |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Thr8 | NDe | ND | ND | ND | ND | ND | 3,609.8 | G-M-Ala-Gln-Lys (Gly)5-Ala-Thr |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Thr9 | ND | ND | ND | ND | ND | ND | 4,774.9 | G-M-Ala-Gln-Lys (Gly)5-Ala-Thr |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Thr10 | ND | ND | ND | ND | ND | ND | 5,939.3 | G-M-Ala-Gln-Lys (Gly)5-Ala-Thr |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Ser1 | 1 | 1.1 | 1.1 | — | 1.9 | — | ND | G-M-Ala-Gln-Lys-Ser-Ser |
| Ser2 | 1 | 1.2 | 2.3 | — | 1.3 | — | 983.1 | G-M-Ala-Gln-Lys-Ala-Ser |
| Ser3 | 1 | 1.2 | 1.1 | 5.4 | 2.2 | — | 1,284.3 | G-M-Ala-Gln-Lys (Gly)5-Ser-Ser |
| Ser4 | 1 | 1.2 | 2.4 | 5.6 | 1.3 | — | 1,268.3 | G-M-Ala-Gln-Lys (Gly)5-Ala-Ser |
| Ser6 | 1 | 1.0 | 1.2 | 5.2 | 1.5 | — | 2,464.7 | G-M-Ala-Gln-Lys (Gly)5-Ser-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ser | ||||||||
| Ser7 | 1 | 1.1 | 1.7 | 5.6 | 1.3 | — | 2,448.4 | G-M-Ala-Gln-Lys (Gly)5-Ala-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ser | ||||||||
| Ser8 | 1 | 1.1 | 1.7 | 5.5 | 1.3 | — | 2,448.4 | G-M-Ala-Gln-Lys (Gly)5-Ser-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Ser9 | 1 | 1.1 | 2.3 | 5.7 | 0.8 | — | 2,432.2 | G-M-Ala-Gln-Lys (Gly)5-Ala-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Ser10 | ND | ND | ND | ND | ND | ND | 3,612.7 | G-M-Ala-Gln-Lys (Gly)5-Ser-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| Ser11 | ND | ND | ND | ND | ND | ND | 3,595.3 | G-M-Ala-Gln-Lys (Gly)5-Ala-Ser |
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
| | | ||||||||
| G-M-Ala-Gln-Lys (Gly)5-Ala | ||||||||
See Fig. 1.
Values are normalized to that for (glutamine/glutamate) (GLX). LYS, lysine; ALA, alanine; GLY, glycine; THR, threonine; SER, serine.
G, N-acetylglucosamine; M, N-acetyl muramic acid.
—, less than 0.1.
ND, not determined.
The elution times and mass spectrometric analyses of Thr3, Thr7, Thr8, Thr9, and Thr10 indicated that these peaks are pentaglycine-substituted threonine-terminating mono-, di-, tri-, tetra-, and pentameric species, respectively (Table 1). Similarly, in the presence of d-serine, monomeric (Ser1 to Ser4), dimeric (Ser6 to Ser9), and trimeric (Ser10 and Ser11) serine-terminating pentapeptide species could be detected (Table 1). The proposed structures of the peaks were confirmed by MS-MS analysis (data not shown).
Carboxyl-terminal d-threonine and d-serine in peptidoglycan stem peptides have been reported before in other bacteria grown under similar conditions (3, 21), as has the presence of two serine residues in stem peptides of peptidoglycan (22).
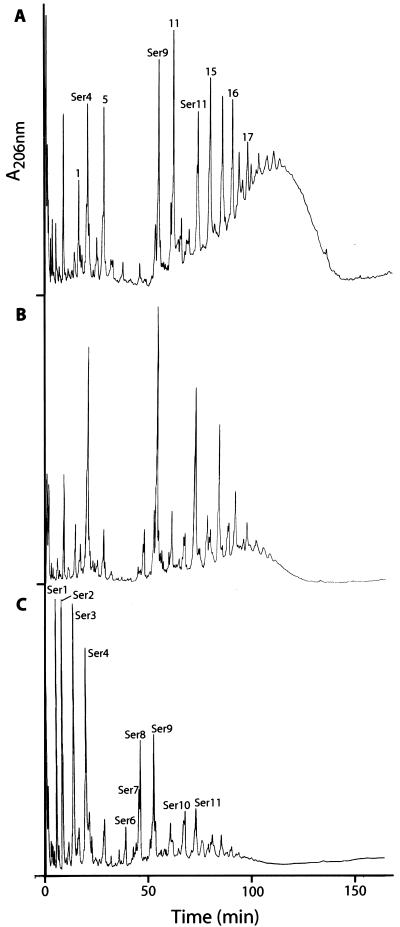
The relative abundance of the peaks depended on the concentration of d-amino acids in the medium, as shown in Fig. 2 for d-serine. The presence of d-amino acids other than d-alanine in the peptidoglycan must be the result of the incorporation of d-alanyl--d-amino acid-terminating peptidoglycan precursors synthesized in the cytoplasm. Such d-alanyl-d-amino acid-terminating precursors are likely the result of the addition of the d-alanyl--d-amino acid rather than d-alanyl--d-alanine to the UDP-linked tripeptide at the cytoplasmic stage of peptidoglycan precursor synthesis (21). Information on the sequence of the S. aureus genome did not reveal the presence of more than one ligase in this organism (13), and therefore, synthesis of the altered dipeptide is likely to occur by the d-alanyl--d-alanine ligase. Such proposed broad substrate specificities for the d-alanyl--d-alanine ligases have been demonstrated for the enzymes of Escherichia coli (24).
FIG. 2.
HPLC elution profiles of muropeptides isolated from peptidoglycan of S. aureus COL grown in TSB supplemented with 0.01 M d-serine (A), TSB supplemented with 0.1 M d-serine (B), and TSB supplemented with 0.25 M d-serine (C). For structures of peaks see Table 1 and reference 5.
Equimolar concentrations of l-serine in the growth medium did not affect the muropeptide profile of peptidoglycan (data not shown).
d-Amino acids in growth medium and ability to grow in the presence of methicillin.
Growth in the presence of d-amino acids also had a profound effect on the ability to grow in the presence of methicillin. Addition of 0.25 M d-serine, d-threonine, or d-phenylalanine to the growth medium allowed growth in the presence of methicillin only at concentrations below 10 μg/ml, whereas growth occurred in the presence of methicillin at concentrations of 800 μg/ml when the medium was not supplemented with d-amino acids (Table 2). The degree of growth reduction in the presence of methicillin differed for each d-amino acid. Addition of 0.25 M l-serine to the growth medium did not affect the ability to grow in the presence of methicillin (Table 2).
TABLE 2.
Growth of S. aureus strain COL in the presence of 0.25 M amino and methicillin
| Amino acid and time (h) of incubation | Methicillin concn (μg/ml)a |
|---|---|
| None | |
| 24 | >800 |
| 48 | >800 |
| Glycine | |
| 24 | 12.5 |
| 48 | >800 |
| d-Serine | |
| 24 | 0.75 |
| 48 | 1.6 |
| l-Serine | |
| 24 | 800 |
| 48 | >800 |
| d-Threonine | |
| 24 | 3.1 |
| 48 | 12.5 |
| d-Phenylalanine | |
| 24 | 0.75 |
| 48 | 0.75 |
Methicllin concentration at which growth was inhibited.
The extent of growth reduction in the presence of methicillin depended on the concentration of the d-amino acid in the medium. Addition of 0.125 M d-threonine allowed cell growth in the presence of methicillin at concentrations above 50 μg/ml, whereas 0.25 M d-threonine allowed growth in the presence of methicillin only at concentrations below 5 μg/ml (Table 3). d-Serine was more effective in reducing cell growth in the presence of methicillin, and it was effective at even lower concentrations of the amino acid than those of d-threonine. Addition of 0.125 M serine did not allow cell growth in the presence of methicillin at concentrations above 5 μg/ml in the medium, and addition of 0.032 M d-serine halted cell growth in the presence of between 5 and 50 μg of methicillin per ml in the growth medium (Table 3). It was observed that growth did occur upon prolonged incubation (Tables 2 and 3), and this may have been caused by a heterogeneous expression of resistance (16). Population analysis profiles showed that this was indeed the case (data not shown).
TABLE 3.
Growth of S. aureus strain COL in the presence of 5 and 50 μg of methicillin per ml and d-serine or d-threonine
| Amino acid, methicillin concn, and incubation time (h) | Amino acid concn (mM)a |
|---|---|
| d-Serine | |
| 5 μg/ml | |
| 24 | 32 |
| 48 | 125 |
| 50 μg/ml | |
| 24 | 16 |
| 48 | 32 |
| d-Threonine | |
| 5 μg/ml | |
| 24 | 250 |
| 48 | 250 |
| 50 μg/ml | |
| 24 | 250 |
| 48 | 250 |
Concentration of amino acid (mM) at which growth was inhibited.
The presence of these high concentrations of d-amino acids had little effect on cell growth in the presence of vancomycin (data not shown).
Cell growth in the presence of methicillin and peptidoglycan composition.
This study extends the observation that there is a correlation between the presence of a d-amino acid other than d-alanine at the fifth (terminal) position of peptidoglycan stem peptides and the expression of methicillin resistance in S. aureus (9). The presence of d-amino acids other than d-alanine at the carboxyl terminus of these stem peptides must have resulted from the incorporation of d-alanyl-d-amino acid-terminating peptidoglycan precursors synthesized in the cytoplasm. In the absence of methicillin, synthesis and incorporation of these precursors had little effect on cell growth, indicating that these precursors are suitable substrates for the enzymes that usually process d-alanyl-d-alanine-terminating species, i.e., the staphylococcal PBPs. It should be noted, though, that the peptidoglycan of these cells contained a reduced amount of oligopeptides, as evidenced by the disappearance of the hump at the end of the chromatogram (Fig. 1B and C). The reduction in cross-linking itself could make the cells more susceptible to methicillin (and β-lactams in general) and thus explain the correlation between the reduced methicillin susceptibility and the altered peptidoglycan composition. Alternatively, it is possible that d-alanyl--d-serine and d-alanyl--d-threonine are poor substrates for PBP 2A, and biosynthesis of these precursors would therefore fail to support growth under conditions in which PBP 2A is the only active PBP, i.e., when methicillin is present in the growth medium and all the staphylococcal PBPs with the exception of the low-affinity PBP 2A are inactivated (4, 10).
This study presents another example of how alterations in peptidoglycan precursor biosynthesis in S. aureus affect susceptibility to methicillin (6, 7, 9, 10, 14, 18, 19). It demonstrated that substitution of the carboxyl-terminal d-alanine residue resulted in decreased methicillin susceptibility. It therefore seems most likely that expression of vancomycin resistance through incorporation of d-alanyl--d-lactate-terminating peptidoglycan precursors, as seen in enterococci (1), is mutually exclusive with the expression of methicillin resistance mediated through PBP 2A.
Acknowledgments
We thank A. Tomasz and A. Severin for helpful and stimulating discussions.
Amino acid determination was provided by The Rockefeller University Protein Sequence Facility, which is supported in part by NIH shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment. MS analysis was performed at the MSU MS facility, which is in part supported by NIH grant RR00480 (to D.G.).
REFERENCES
- 1.Arthur, M., and P. Courvalin. 1993. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37:1563-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger-Bachi, B., A. Strassle, J. E. Gustafson, and F. H. Kayser. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 36:1367-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caparros, M., A. G. Pisabarro, and M. A. de Pedro. 1992. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol. 174:5549-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers, H. F., and M. Sachdeva. 1990. Binding of beta-lactam antibiotics to penicillin-binding protein in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 161:1170-1176. [DOI] [PubMed] [Google Scholar]
- 5.De Jonge, B., Y.-S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain: the role of penicillin-binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 6.De Jonge, B., Y.-S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J. Biol. Chem. 267:11255-11259. [PubMed] [Google Scholar]
- 7.De Jonge, B. L. M., T. Sidow, Y.-S. Chang, H. Labischinski, B. Berger-Bachi, D. Gage, and A. Tomasz. 1993. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J. Bacteriol. 175:2779-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jonge, B. L. M., and A. Tomasz. 1993. Abnormal peptidoglycan produced in a methicillin-resistant strain of Staphylococcus aureus grown in the presence of methicillin: functional role for penicillin-binding protein 2A in cell wall synthesis. Antimicrob. Agents Chemother. 37:342-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.deJonge, B. L. M., Y.-S. Chang, N. Xu, and D. Gage. 1996. Effect of exogenous glycine on peptidoglycan composition and resistance in a methicillin-resistant Staphylococcus aureus strain. Antimicrob. Agents Chemother. 40:1490-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lencastre, H., B. L. M. de Jonge, P. R. Matthews, and A. Tomasz. 1994. Molecular aspects of methicillin resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 33:7-24. [DOI] [PubMed] [Google Scholar]
- 11.de Lencastre, H., and A. Tomasz. 1994. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 38:2590-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaisford, W. C., and E. Reynolds. 1989. Methicillin resistance in Staphylococcus epidermidis: relationship between the additional penicillin-binding protein and an attachment transpeptidase. Eur. J. Biochem. 185:211-218. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Maidhof, H., B. Reinicke, P. Blumel, B. Berger-Bachi, and H. Labischinski. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuhashi, M., M. D. Song, F. Ishino, M. Wachi, M. Doi, M. Inoue, K. Ubukata, N. Yamashita, and M Konno. 1986. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to β-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 167:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews, P. R., and P. R. Stewart. 1984. Resistance heterogeneity in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 22:161-166. [Google Scholar]
- 17.Neuhaus, F. C., and W. P. Hammes. 1981. Inhibition of cell wall biosynthesis by analogues of alanine. Pharmacol. Ther. 14:265-319. [DOI] [PubMed] [Google Scholar]
- 18.Ornelas-Soares, O., H. de Lencastre, B. L. M. de Jonge, D. Gage, Y.-S. Chang, and A. Tomasz. 1993. The peptidoglycan composition of a Staphylococcus aureus mutant selected for reduced methicillin resistance. J. Biol. Chem. 268:26268-26272. [PubMed] [Google Scholar]
- 19.Ornelas-Soares, O., H. de Lencastre, B. L. M. de Jonge, and A. Tomasz. 1994. Reduced methicillin resistance in a new Staphylococcus aureus transposon mutant that incorporates muramyl dipeptides into cell wall peptidoglycan. J. Biol. Chem. 269:27246-27250. [PubMed] [Google Scholar]
- 20.Rohrer, S., K. Ehlert, M. Tschierske, H. Labischinski, and B. Berger-Bachi. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc. Natl. Acad. Sci. USA 96:9351-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleifer, K. H., W. Hammes, and O. Kandler. 1976. Effects of exogenous and endogenous factors on the primary structure of bacterial peptidoglycan. Adv. Microbiol. Physiol. 13:245-288. [DOI] [PubMed] [Google Scholar]
- 22.Trippen, B., W. P. Hammes, K.-H. Schleifer, and O. Kandler. 1976. Die Wirkung von d-Aminosauren auf die Struktur und Biosynthese des Peptidoglycans. Arch. Microbiol 109:247-261. [DOI] [PubMed] [Google Scholar]
- 23.Xu, N., Z.-H. Huang, B. L. M. de Jonge, and D. A. Gage. 1997. Structural characterization of peptidoglcycan muropeptides by matrix-assisted laser desorption ionization mass spectrometry and postsource decay analysis. Anal. Biochem. 248:7-14. [DOI] [PubMed] [Google Scholar]
- 24.Zawadzke, L. E., T. D. Bugg, and C. T. Walsh. 1991. Existence of two d-alanine:d-alanine ligases in Escherichia coli: cloning and sequencing of the ddlA gene and purification and characterization of the DdlA and DdlB enzymes. Biochemistry 30:1673-1682. [DOI] [PubMed] [Google Scholar]