Abstract
Background
Ciliates employ massive chromatid breakage and de novo telomere formation during generation of the somatic macronucleus. Positions flanking the 81-MAC locus are reproducibly cut. But those flanking the Common Region are proposed to often escape cutting, generating three nested macronuclear chromosomes, two retaining "arms" still appended to the Common Region. Arm-distal positions must differ (in cis) from the Common Region flanks.
Results
The Common-Region-flanking positions also differ from the arm-distal positions in that they are "multi-TAS" regions: anchored PCR shows heterogeneous patterns of telomere addition sites, but arm-distal sites do not. The multi-TAS patterns are reproducible, but are sensitive to the sequence of the allele being processed. Thus, random degradation following chromatid cutting does not create this heterogeneity; these telomere addition sites also must be dictated by cis-acting sequences.
Conclusions
Most ciliates show such micro-heterogeneity in the precise positions of telomere addition sites. Telomerase is believed to be tightly associated with, and act in concert with, the chromatid-cutting nuclease: heterogeneity must be the result of intervening erosion activity. Our "weak-sites" hypothesis explains the correlation between alternative chromatid cutting at the Common Region boundaries and their multi-TAS character: when the chromatid-breakage machine encounters either a weak binding site or a weak cut site at these regions, then telomerase dissociates prematurely, leaving the new end subject to erosion by an exonuclease, which pauses at cis-acting sequences; telomerase eventually heals these resected termini. Finally, we observe TAS positioning influenced by trans-allelic interactions, reminiscent of transvection.
Background
Telomeres cap the tips of eukaryotic chromosomes and protect them from involvement in the double-strand break repair mechanism and in classic breakage-fusion-bridge catastrophes described by McClintock [1]. Telomeres are maintained at full length, against their shortening at replication, by restoration of 3' terminal repeats by telomerase. However, telomerase is not expressed in most mammalian somatic cells, and once somatic lineages expose their chromosome ends, crisis ensues and normally results in lineage senescence. Oncogenic transformation requires the re-expression of telomerase [2]. We study telomeres in ciliated protozoa, because telomeres comprise an unprecedented mass of the ciliate somatic genome, and because telomeres are developmentally created de novo during its development.
Telomeres consist of tandem repeats of a short dG-rich sequence, polymerized by the reverse transcriptase telomerase, templated by an RNA component, and primed by a 3'OH end that pairs with the template (review: [3]). The dG-rich strand protrudes beyond the underhanging 5' strand. For example, Oxytricha macronuclear telomeres consist of the 20 bp, dGGGGTTTTGGGGTTTTGGGG, with a 3' protrusion of the 16 nts TTTTGGGGTTTTGGGG [4]. These telomeric repeats comprise a biochemically significant fraction (~2%) of Oxytricha macronuclear DNA; they also tip the telomeres of the mitotic chromosomes of the Oxytricha micronucleus [5].
Raw, broken ends are occasionally "healed" by telomerase, but healing is a normal event in the massive somatic genome re-organizations seen in ascarid worms [6] and in ciliated protozoa. Molecular biology of telomeres began in ciliates [7,8], rich sources of telomeres and the proteins that create and service them, including telomerase (reviews: [3,9]). Ciliates provide the opportunity to study the molecular genetics of de novo telomere formation during the programmed development of the somatic macronucleus (MAC) from a mitotic copy of the germline micronucleus (MIC). Telomere components and telomerase are homologous across the range of crown eukaryotes – with a few notable exceptions (review: [3]) – from which ciliates diverged maybe one billion years ago [10]. Thus, characters shared with mammals high-light critical features of telomere metabolism.
Additionally, ancestors of modern ciliate classes have been diverging for a large fraction of these billion years [10], and thus the study of telomere biology in various ciliates also provides a rich diversity for comparison. Molecular biology of telomeres has been studied primarily in two ciliate clades, the Spirotrichs and the Oligohymenophorans. In the Spirotrichs (previously Hypotrichs), the Stichotrichs – Oxytricha and Stylonychia species – and the early-diverged Euplotids have been intensely studied for their dramatic extents of developmental chromosome fragmentation and elimination of MIC-limited sequences. Of the Spirotrich telomerases, Euplotes' has been studied most intensely, although likely it is very similar to Oxytricha's [11-13]. The Oligohymenophorans Tetrahymena and Paramecium, while not displaying such dramatic extents of chromosome fragmentation, have played a unique role in telomere biology [3].
Ciliate nuclear dimorphism
Following pair-wise mating (conjugation), each exconjugant replaces its old parental MAC with a new MAC, developed from a mitotic daughter of its genotypically new zygotic nucleus (review: [14]). The mitotic sister of the MAC precursor (anlage) is set aside as the new diploid germline MIC, which is then heterochromatically silenced; all known gene expression is from the MAC or MAC anlage [15-17]. Once the anlage begins differentiation, the old MAC is apoptotically destroyed [18]. The mature exconjugant resumes feeding and establishes a vegetative clone (a karyonide) propagated by binary fission, in which the MIC replicates as a typical diploid mitotic nucleus and the MAC replicates amitotically.
Macronuclear development
Ciliates employ a strategy of irreversible genome alterations to generate the MAC, a specialized gene-expression organelle. Analogous developmental strategies have evolved several times outside the ciliates, scattered across phyla [15,19]; thus, mammalian immunity depends on irreversible somatic genome alterations [20]. Initially the MAC anlage's chromosomes are endo-reduplicated, evidenced by visible polyteny in a variety of ciliates. The ploidy can reach ~64 in some Spirotrichs [21,22]. Thus, each sequence that survives subsequent elimination (MAC-destined) is represented by numerous polytene chromatids; in principle, each could be processed differently. Near the end of endo-replication in Spirotrichs many transposons and transposon-like sequences are eliminated by precise excision (Internal Eliminated Sequences or IESs [23]). Excision directly precedes massive chromatid fragmentation and de novo telomere formation ("chromosome healing" [9]) and the degradation of IESs and of "spacer" DNAs that lie between blocks of sequences destined for different MAC chromosomes. The surviving sequences (as few as 5% of the germline sequences) reside on many small MAC chromosomes, which are then highly amplified to the level of the mature MAC.
The Oxytricha MAC genome is typical of those of other Spirotrichs. It consists ~1000 copies of ~20,000 different tiny, acentric linear chromosomes. They range in length from ~0.5 kbp to >15 kbp (number average length, ~2.4 kbp; J. Garrett, K.R.W., D. Witherspoon and G.H., unpublished), comprised almost exclusively of transcription units. Many carry only one gene (review: [24]), but some carry two or three genes each ([25]; N. Sanders, B. Fausett, T.G.D., K.R.W. and G.H., unpublished).
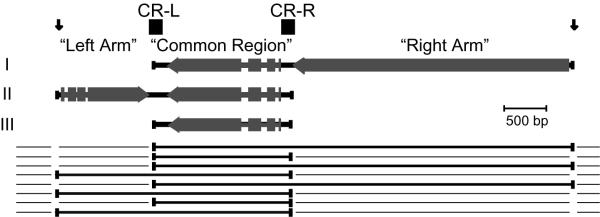
Not all identical polytene chromatids of the anlage are processed identically, as evidenced by the generation of alternative processing families of MAC chromosomes. We have studied alternative processing of the 81 MAC locus, which generates a family of three overlapping chromosomes ([26,27]; see Fig. 1, below). Analogous phenomena have been described in other ciliates [28-31].
Figure 1.
Alternative processing of the 81-MAC locus. In the center are maps of the three MAC chromosomes generated from the 81-MAC locus (after [25]), with their genes' exons indicated as blocks separated by gaps representing introns; the 3' of each gene is indicated by an arrowhead. Above, indicated by arrows and blocks, are the four Telomere Addition Site regions of the locus. Below is a diagram illustrating the "partial-digest" model, in which only some chromatids are cut at the CR-L and CR-R regions, and telomeres (signified by terminal blocks) are added to each chromosome carrying a Common Region (signified by a thick line; see text).
Chromatid breakage and de novo telomere formation
Telomeres are not encoded in situ at the ends of MAC-destined blocks [32,33], but are polymerized onto new ends de novo by telomerase. These processes have been best studied in Tetrahymena thermophila and in various Spirotrichs, including Euplotes crassus. Breakage occurs at ~200 "cut" sites in Tetrahymena, and at ~40,000 sites in Spirotrichs. A short 15 bp cis-acting sequence, cbs, is necessary and sufficient to direct breakage at or near it in Tetrahymena, presumably being the binding site of the breakage enzyme – the "cutter" [34-36]. The cbs sits in eliminated sequence, a short, variable distance from the positions that receive telomeres. An analogous sequence, E-cbs, has been characterized in Euplotes by sequence comparisons and analyses of DNA intermediates and products of MAC development [37-39]. At a fixed distance from itself, E-cbs directs a 6-nucleotide staggered cut, with a protruding 3'OH to which telomerase adds telomere repeats. The E-cbs can sit within the MAC-destined sequence, or in the adjacent "spacer" DNA; in some cases both generated ends receive telomere repeats, so that the two resultant telomere-tailed ends overlap by 6 bp. A cbs has been tentatively identified in Stylonychia lemnae [40]; no cbs is known in Oxytrichs, although we assume it exists (see Discussion).
The in vitro properties of telomerase are consistent with its known role in creation of MAC telomeres in Tetrahymena [41], as well as, presumably, in all other ciliates [9]. Telomerases from exconjugants of Tetrahymena and Euplotes have been extensively studied in vitro. The E. crassus enzyme can only act on oligomers that pair with telomerase template RNA at their 3' terminus, unless assisted by a dissociable factor (chromosome healing factor), which contributes extra mass to the enzyme in exconjugants [42]. Telomerase, assisted by this factor, still requires a block of dGs or telomere repeats internal to the 3' primer end [43]. In contrast, the Tetrahymena exconjugant enzyme can add to a primer completely devoid of telomere repeats in special reaction conditions, and shows no evidence of increased mass beyond that of the vegetative enzyme [44].
Although chromatid breakage requires an endonuclease activity, the cutter, directed by a Cbs-binding activity, no Cbs-dependent "binder/cutter" activity has been detected in vitro. The cutter could be an activity of telomerase itself [39,43]. Telomerase of both Tetrahymena and Euplotes has a single-strand endonuclease activity [45,43]. It can trim potential primers back, so that the 3' terminus pairs with template RNA [46]. Thus, while it may serve a 3' editing function, the existence of a nuclease activity in potential proximity to the site of chromatid breakage is provocative, suggesting that telomerase provides the double-strand cutting activity, and thereby explaining coupled action between chromatid cutting and telomere addition (see below). However, Niu et al. [47], in a study of a similar endonuclease activity of the yeast telomerase, argue that since yeast perform no developmental fragmentation, this activity plays some role other than in fragmentation.
It is generally believed that the developmental action of telomerase is tightly coupled to cleavage. The Oxytricha multi-TAS regions we describe here seem to cause an uncoupling of these steps; our models invoke the ability of telomerase to act independently of the binder/cutter. Tetrahymena telomerase and the binder/cutter appear to act concertedly in vivo, as if in a physical complex [48,49]. However, this contrasts with in vitro properties of the enzyme from exconjugants: 1.) it acts on non-telomeric primers [44,50], and 2.) it apparently is no larger than the vegetative enzyme [44]. Furthermore, telomerase can act in the vegetative MAC of Paramecium, uncoupled from chromatid breakage [51]. It remains to be seen if a complex between telomerase and binder/cutter activity does exist. We consider models that assume the two activities exist separately, in a loose complex. This assumption is supported by our results – as well as those of Möllenbeck and Klobutcher [52] – which indicate that exonucleolytic erosion of new ends must sometimes occur before telomerase caps them. It is possible that the nuclease activity of telomerase causes the erosion.
Telomere addition sites (TASs)
The exact nucleotide positions where telomere repeats are added have been examined in several ciliates (reviewed by [23,53]). Cloned chromosome ends from 81 MAC family indicated heterogeneity of TAS placement, which we attributed to exonuclease erosion of the broken ends prior to telomerase action [27]. Analogous multi-TAS regions have been described in Tetrahymena ([49]; E. Hamilton and E. Orias, pers. comm.) and in Paramecium (A. Le Mouël and E. Meyer, pers. comm.). In stark contrast, no heterogeneity of TAS positions has been reported for E. crassus MAC chromosomes, although recently Möllenbeck and Klobutcher [52] report multiple TASs at the end of a MIC-limited "spacer" segment. Thus, in Tetrahymena and Paramecium telomeres are added at a heterogeneous collection of clustered sites, which we here refer to as "multi-TAS regions," whereas in E. crassus only one TAS is used for each MAC-destined end ("single TAS regions"). Here we describe both kinds of regions in Oxytricha, both involved in the generation of the 81-MAC family of MAC chromosomes.
Alternative processing of the 81-MAC locus
A family of three nested MAC chromosomes was discovered when the MAC chromosome cloned in pMA81 was hybridized to Southern-blotted MAC DNA [26]. The chromosomes consist of three contiguous segments of germline sequence, referred to as the "common region" (CR) and the left and right "arms" (Figure 1). Each segment bears a protein-coding gene [54,25]. The smallest chromosome, MAC III, consists solely of the common region with telomeres. Arm-alone chromosomes have never been observed in vegetative MAC DNA, and are proposed to be replication-incompetent [27]. The two large chromosomes each have a common region: MAC I carries a right arm, and MAC II, a left arm. Therefore, the latter two chromosomes each carry two genes [25], a novel finding in Spirotrichs (see [24]). The identities of the genes have not suggested any selective advantage to such couplings, but the alternative processing of this locus has been conserved within the Oxytrichs, and with variation, in some Stylonychids (A. Seegmiller and K.R.W., unpublished).
We proposed that this nested set is generated by alternative breakage patterns of polytene chromatids of the 81 locus DNA ([27]; bottom, Fig. 1). While the arm-distal positions must be efficiently cut on each chromatid, alternative breakage at the regions bounding the CR was proposed. Thus, not all CR-arm junctions are cut on each chromatid (analogous to an incomplete DNA restriction digest), leaving arm DNA appended to CR DNA in some cases (MACs I and II), and not others (MAC III). Given the apparent functioning of exonucleolytic erosion following chromatid cutting, note that another way to explain the generation of arm-less chromosomes is to assume that chromatids are only cut at arm-distal sites, and that erosion alternatively stops at the arm-distal TAS position, or proceeds completely through the arm to stop in the CR-boundary multi-TAS region. By this model arm-alone chromosomes never exist-even transiently; use of anchored PCR (see below) is intended, to search for such species and to distinguish between this massive-erosion model and the incomplete-cutting model. The latter model seems more parsimonious to us, and we assume it in discussing the results of this report.
Incomplete cutting at the CR boundaries might be caused by poor binding sites or poor cut-sites for the binder/cutter. In the work presented here we employed an anchored PCR procedure to precisely map telomere addition sites (TASs). We show that the two CR boundaries, CR-Right and CR-Left, are multiple-TAS regions, but the two arm-distal regions have single TASs. Models are proposed to explain why the two putatively incompletely-cut CR boundary regions also are multiple-TAS regions.
Results
From previous work we were aware that multiple sites are used for telomere addition at the ends of the 81-MAC common region [27]. To study the full range of positions, a ligation-mediated PCR (LM-PCR [55]) strategy was employed to display products that represent the different extents of the 5' termini of a region. LM-PCR relies on phosphorylation of these 5' termini, and the 5' termini of Oxytrichid macronuclear DNAs are phosphorylated (R.D. Wesley and G.A.H., unpublished; L. Klobutcher, pers. comm.; see [4,56].
LM-PCR employs a locus-specific, nested set of oligomers (see Fig. 2A), each directed toward the termini to be mapped. In the first step, the 5'-termini are converted to precisely blunt double-strand ends, by denaturing the MAC DNA, annealing a distal primer, "Extend," and extending it with Vent polymerase to create a blunt end. Other MAC DNA chains remain single-stranded. To the resulting 5'P/3'OH blunt end a double-strand blunt linker is ligated, thereby fastening a PCR "anchor" to the 5'P terminus. A round of PCR is employed to impose specificity, using the nested primer "P1." The P1-anchor products are then analyzed.
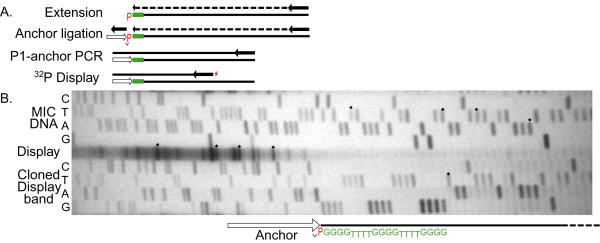
Figure 2.
Telomere Addition Site mapping by LM-PCR. 2A. LM-PCR strategy, described in the Materials and Methods, and in Results. The native 5'P at the end of the MAC DNA telomere is indicated in red, and the telomere G4T4 repeats are indicated in green. Short, thick arrows indicate primers. Ligation to the 3'OH of the anchor linker is indicated by an inverted caret; anchor primer 7A is indicated by hollow arrows. The asterisk at the end of the display primer indicates its 5' 32P label. 2B. Autoradiogram of electrophoresis gel displaying end-labeled display products derived from a multi-TAS region. TAS positions are determined as described in the text, by running the display chains (central lane) next to the lanes of sequencing reactions derived from the MIC DNA that is processed. Four display bands and corresponding TAS positions on the MIC DNA sequence lanes are marked with black diamonds. This early CR-R result has not been further analyzed: analysed results are shown in Figures 3 and 4. Sequencing lanes derived from a cloned P1-anchor PCR product are run on the other side of the central display lane. This clone carries a product with telomere repeats added to its dot-marked dT, represented by the second-slowest, marked display band. Along side these lanes are diagrammed the extent of the 7A anchor primer, ligated to the 5'P of the telomere G4T4 repeats.
First, the product can be directly sequenced with the P1 primer. If heterogeneity of the product is indicated (not shown), then the products can be cloned and individually sequenced, or a display of the heterogeneity is generated by denaturation and annealing of a final primer, "Display," to the denatured P1-anchor products, and extending it to their ends, with Taq polymerase. "Display" is 5'-32P end-labeled so that the extension products can be autoradiographically visualized on a high-resolution polyacrylamide gel.
The central lane of Figure 2B shows the products of such a display, representing the TASs of the right end of the 81-MAC common region. Many different bands are seen, many more than inferred from previous analyses of cloned chromosomes [27]. Run in parallel are lanes carrying dideoxy-terminated reactions that show the sequence across the region of micronuclear DNA from which these ends are derived. The sites of telomere-repeat addition can be directly mapped onto this sequence, after taking into account the length of the PCR anchor oligomer, the 5'-terminal 20 nucleotides of telomere repeats, and the untemplated dT added by Taq polymerase to the display chains [57]. As an example, sequencing chains from a cloned display band are run in parallel, verifying the calculated mapping of the TAS it represents. Note that telomeres often are added to dT ends. We will return to the results of mapping TASs after first examining the genetic basis of the TAS heterogeneity.
It has been assumed that TAS heterogeneity is the result of random exonuclease resection of termini generated by site-specific chromatid breakage. However, the heterogeneous pattern seen here is not random, but is highly reproducible each time a new MAC is generated in an exconjugant, and it is dependent of the allelic sequence of the substrate, leading to the conclusion that the TAS positions are genetically determined, dictated by (linked) cis-acting sequences. These results will be presented in turn.
Karyonidal reproducibility
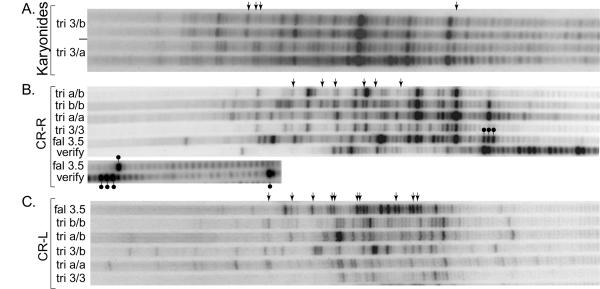
A karyonide is a clone of cells established by an emerging exconjugant – that is, a clone of cells that have the same ancestral, developed new MAC [58]. If random erosion is responsible for the heterogeneity we see, then it should be different in each different karyonide. Figure 3A shows displays of the CR-R TASs from two sets of karyonides with different alleles of the 81-MAC locus. The displays for the different genotypes differ at many positions, but inter-karyonidal variation is very slight, by comparison. In a separate experiment (not shown), similarly-identical display patterns of CR-R were generated from three independent a/b heterozygote karyonides (displays from both P1-anchor and P2-anchor PCR products were identical, demonstrating that P2-anchor PCR does not generate artifacts). While the allele composition of the 81-locus is identical in each pair of karyonides, the karyonides are not otherwise genetically identical (unlike sister karyonides in many ciliates; e.g., see [59]), but are independently derived, so that the backgrounds differ between karyonides, which might explain these slight variations. Perhaps more interestingly, these inter-karyonidal variations could be attributed to the limited number of chromatids available to be partitioned into the various TAS classes represented by the display bands; however, the over-riding reproducibility implies that the positions of these TASs are dictated genetically by cis-acting sequences.
Figure 3.
LM-PCR evidence that cis-acting sequences determine TAS positions. 3A. Karyonidal reproducibility of CR-Right TAS display patterns. Two display lanes represent CR-R TASs in two karyonides with identical heterozygous 81-locus genotypes, 310/510b. Also shown are two lanes representing two karyonides with 310/510a genotypes. Arrows indicate the positions of distinct allele-specific bands. Specific oligomers used: Extend = ST2; P1 = PEB; Display = VHO. 3B. Allele-specificity of TAS positions, and "verification" of one set of display bands. DNAs from Oxytricha strains with various 81-locus allele compositions were used as templates for LM-PCR and display (see text). Arrows indicate the positions of distinct allele-specific bands. Specific oligomers used: Extend = ST2; P1 = PEB; Display = VHO. A verification PCR reaction was performed with the P1-7A PCR product from O. fallax 3.5, and display lanes are run along side (see text for explanation of this verification procedure). Because short chains ran especially rapidly, the bottoms of the "fal 3.5" lanes are presented to the side of the main panel. Four corresponding bands in the P1-anchor PCR display and the verification display are marked. 3C. Allele-specificity of TAS display pattern for the multi-TAS region bounding the Left end of Common Region. DNAs from strains with various 81 locus genotypes were used as templates for LM-PCR and display (see text). Arrows indicate the positions of distinct allele-specific bands. Specific oligomers used: Extend = PEB-prime; P1 = oRG; P2 primer = T1target; Display = 180L-.
Allelic specificity
Note that if TAS positions are dictated solely by cis-acting sequences, then the pattern of the a/b heterozygote should be equivalent to the sum of the two homozygote patterns (a/a + b/b), but this is not the case (Fig. 3B). Indeed, we've observed analogous non-additivity at CR-R for each of the three heterozygotes, a/b, a/3 and 3/b, and at CR-L for a/b and 3/b (data not shown). Since independent isolates of a given genotype have essentially the same pattern, this is unlikely to be caused by different genetic backgrounds. Rather, it is possible that this phenomenon indicates allelic-interactions, particularly given that the alleles are closely associated in a polytene chromosome at the time they are processed (homologs are paired in the polytene anlage of Stichotrichs [60,61]). For instance, transvection is a phenomenon dependent on homolog pairing; transvection normally is evidenced by rescued gene expression in heterozygotes of two alleles defective in different cis-acting sequence features, which interact (in trans) to essentially complement one another (review: [62]).
Whatever the explanation of this complexity, it does not negate the conclusion that the patterns are genotype-sensitive and that cis-acting sequences must be operating in the process of TAS determination. Thus, even if alleles interact to determine the TAS pattern, nonetheless the alleles are determining the pattern – that is, the patterns are determined by cis-acting components of the allele sequences. The results suggest that allelic sequences predominantly determine TAS positions autonomously (additively), but the non-additivity results indicate that the allele sequences must also interact in trans.
These two multi-TAS regions roughly lie between their neighboring genes' coding regions, and no TAS site interrupts the common region transcription unit (Fig. 4, below; CR-L data not shown).
Figure 4.
Map positions of TASs of the Right End of the 81-MAC Common Region. Sequences of three O. trifallax alleles and three O. fallax 3.5 alleles across the multi-TAS region right of the Common Region are aligned. TAS positions, defined as the nucleotide 5' of the telomere repeats block, dGGGGTTTTGGGGTTTTGGGG, were inferred from mobilities of display bands (see Fig. 2), and are shaded in this figure. Three mapped O. fallax TASs are known from sequences of cloned chromosomes [27]; they are indicated by white letters with black backgrounds. The two start sites of transcription of the CR gene, mapped by Williams and Herrick [54], are indicated below the beginning of the alignment, in a region of nearly perfect conservation, excepting a bold-faced T in the O. fallax vA sequence. It is tightly associated with a dA TAS in this allele, which is indicated with a "/" below it. The stop codon of the Right Arm gene [25] is underlined. Asterisks above the alignment mark positions with a TAS unique to one allele; daggers mark positions where some, but not all alleles have a TAS. In 5 cases, the inferred position is followed directly by a dG. Because telomerase can complete GGGG runs, it is possible (discussed by [38]) that in each of these cases, the TAS was not at the inferred dT or dA, but instead could have been at the following dG; to indicate these ambiguous cases, the T or A and its 3' G are shown in lower case.
The CR-R multi-TAS region of O. fallax 3.5 is larger than that of O. trifallax (Fig. 3B). To display the fastest bands, representing TASs most proximal to the CR gene, the images of the "bottom" of the 3.5 lanes are shown along side the images of their respective lanes. Furthermore, these clone 3.5 patterns are displayed in two parallel lanes which illustrate the band-confirmation technique of "telomere step-in PCR." In this procedure, PCR is performed with the P1-anchor PCR product as template, using P1 in combination with an oligomer of 3' telomere repeats (see "Anchor" and "G4T4G4T4G4" marked in Fig. 2B), generating a set of products shortened by removal of the anchor from beyond the telomere repeats, and once again display chains are generated. Bands representing bone fide telomere-bearing 5'P termini "shift" down by a predicted size decrement, thereby verifying each responsible TAS. Note essentially all the bands in the "3.5" display shift down in concert, including the extremely slow band, a marked short-chain triplet, and the marked shortest-chain band. The significance of the extreme TAS position represented by the latter band will be considered below.
Figure 4 shows the positions of detected TASs mapped onto the sequences of six alleles of the CR-R multi-TAS region. Homologous nucleotides are aligned. Forty six positions are used as TASs. As the displays indicate (Fig. 3), uses of many positions are influenced by the allele sequence. Illustrating that allele-specific cis-acting sequences must dictate TAS-position use, alleles differ at most TAS positions: only 3 are used by all alleles; 24 positions (†-marked) are used by more than one, but not all alleles, and 19 positions (*-marked) are used by only one allele.
For instance, note the most CR-proximal dA TAS, only mapped to O. fallax 3.5 vA; it resides in a segment of highly conserved sequences, with the exception of a vA-specific dT 5 nts 5' of the TAS. Might this single mutation have caused the use of this unique TAS position? Given the interactions between alleles we observe in the O. trifallax data (Fig. 3), it may be that the vA TAS might not be dictated by a vA sequence feature acting in cis, but could have been induced on vA by the vB and/or vC alleles also present in the anlage that established strain 3.5 macronuclei. This vA segment is immediately 5' of the Common Region gene's transcription start sites (Fig. 4; [54]), and presumably is conserved by selection focused on expression of that gene [27,54,63].
The position of this most-proximal TAS in vA is surprising, indicating that some vA MAC I or II chromosomes have as few as 3–5 base pairs between the telomere DNA and the transcription start site of the Common Region gene (Fig. 4; [54]). The two kinds of possibilities are that these chromosomes cannot express their Common Region gene for lack of promoter elements, or that they do, noting that the use of this verified TAS is genetically dictated, and hence is tested for fitness by natural selection. Thus, if the CR genes are silenced on these close-trimmed chromosomes, the implication is that generation of these chromosomes with inoperative genes is somehow useful to Oxytricha, perhaps adjusting the stoichiometry of expressed 81-locus genes.
Alternatively these chromosomes may be able express their Common Region gene, despite the extreme lack of 5' sequences. Assuming that some promoter element is required and is not provided by the 3–5 bp 5' of the transcription start, perhaps the telomere repeats serve this purpose (see [64]). It is also possible that a promoter element resides internal to this region: we have identified a highly conserved sequence block in the gene's first intron and suggested that it might serve as a promoter element [65].
Perhaps the most trivial interpretation of the existence of these short chromosomes is that the responsible TAS is positioned due to a recent mutation that arose in the vA allele, subsequent to its relatively recent divergence from vC [65]; thus, the mutation is relatively new, and despite its potential impairment of the CR gene on these short chromosomes, the mutation may not yet have been expunged by selection.
Single TAS regions, distal to the left and right arms of the 81-MAC locus
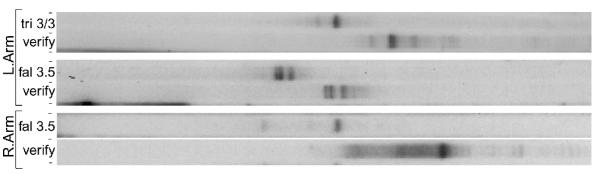
In contrast to the multi-TAS regions bounding the Common Region, telomeres are added to single positions at the extreme, arm-distal boundaries of the 81-MAC locus. Figure 5 presents LM-PCR displays in which both the left-arm- and right-arm-distal TAS regions show single bands, "step-in" verified. An exception is the triple band for left-arm-distal region of O. fallax 3.5. A more complete study of single TAS regions will be presented elsewhere (K.R.W., T.G.D., D.J. Witherspoon, G.A.H., manuscript in preparation). Clones of these three bands have been sequenced (K.R.W., unpubl. results), showing that the three bands represent the three versions/alleles of this locus, each of which has a different TAS. Sequence differences that might be responsible for the different TAS locations are not obvious.
Figure 5.
Single TAS regions bounding the 81-MAC locus. Three pairs of display lanes are shown. Display reactions were performed from 7A-anchored products and from verification, tel-anchored templates. Left Arm-distal TAS region: displays and verifications are shown representing the O. trifallax 310 homozygote and O. fallax 3.5. The latter products show three bands, representing the strain's three alleles vA, vB, and vC (not shown, see text). Specific oligomers used: Extend = -123-; P1 = -387-; P2 primer = -903-; Display = LA4. Right Arm-distal TAS region of O. fallax 3.5, specific oligomers used: Extend = RA1.1; P1 = 3239; P2 primer = -894; Display = 247+.
Discussion
We show that two distinct processes are determined by cis-acting sequences. First, cis-acting sequences must determine that the arm-distal TAS regions only generate single TASs, while the common-region boundaries each generate multiple TASs. Below we propose that the responsible sequences are the sites for binding or cutting by the chromatid binder/cutter. Second, we show that the heterogeneity of TAS positions within the CR-bounding multi-TAS regions are genetically determined: the pattern is not random but reproducible from karyonide to karyonide, and it is allele-specific, indicating that each TAS in these regions is dictated by its own cis-acting sequence, which we model below as a pause site for an exonuclease that resects the new MAC end before telomerase can act.
By trying to explain why alternatively- or incompletely-cut TAS regions are also multi-TAS regions, while completely-cut positions have a single TAS, we have attempted to infer properties of the trans-acting machinery that works at these sites. Toward that we now present three models involving a set of cis-acting sequences and trans-acting components.
A series of assumptions has been made, some of which will be examined after the models are presented, as we consider alternative explanations. First, we assume the previous model for alternative processing, that the CR boundaries are not efficiently broken on each chromatid, sometimes leaving the common region linked to a left or right arm [27]. We assume three trans-acting components and three kinds of cis-acting sites. The trans actors assumed are a loose, non-covalent complex between the binder/cutter enzyme and telomerase, and a resecting exonuclease that pauses. The assumed cis-acting sites are the binding, cutting and exonuclease-pause sites. Finally, we assume that enzyme can act either coupled to the binder/cutter action or on its own at ends exposed by the exonuclease.
The first model is what we take to be the accepted model in the field for chromatid breakage and telomere formation at a single TAS, and what we propose also to be the mechanism operating at the arm-distal single-TAS regions. In this case all the components function with full efficiency: first, a complex of telomerase and binder/cutter bind to the binding site, then cutting of the cut-site ensues with immediate, coupled telomerase capping of the MAC-destined end, with no interposed erosion.
Two further, alternative, mechanisms are offered to generate the spectrum of TASs at the CR boundaries. These models share several features. Each is a kinetic, chain-of-events scenario. Each invokes a sub-maximal, or weak cis-acting sequence (compared to that at a single-TAS region) as the cause of the ensuing events, and in each enzyme is portrayed anthropomorphically for heuristic and didactic reasons.
In the "hesitant cutter" model, the complex binds, but the endonuclease is slow to cut, because the cut-site is weak (recalcitrant to cutting). Meanwhile the "impatient" telomerase dissociates from the complex, and while it is away from the site, the cutter finally cuts, and the raw end is not capped and is exposed to the exonuclease, which progresses in fits and starts, stalling at each TAS. When telomerase eventually acts, it usually encounters the eroder paused at a pause site and adds telomeric repeats to those resected chromatid ends, thus generating the reproducible spectrum of TASs observed.
In the "frustrated telomerase" model, the cut-site is normal, but, instead, the binding site is weak, so that the complex binds, but with a high off-rate. In this chain of events, the complex binds transiently and cuts, and dissociates from the binding site, removing telomerase before it can cap the new end. As in the previous scenario, exonuclease erodes the end and pauses, telomerase returns and caps the ends it finds, and the observed spectrum of TASs is generated. In principle, the binding- and cut-sites might be coincident or overlap, as for Type II restriction enzymes [66]; in that case, cutting would destroy the binding-site and this frustrated-telomerase situation would apply. This predicts that multiple TASs should result, even at a TAS region with an optimal binding and cut-site, but this is contrary to the behavior of the arm-distal TAS regions. Further, Euplotes binding- and cut-sites do not overlap [38].
A central and potentially contentious assumption in these two weak-sites models is that telomerase is capable of acting independently of the binder/cutter, that is, outside the widely inferred complex. However, the ciliate literature is unclear about whether Tetrahymena telomerase might be able to act uncoupled from the action of the binder/cutter (see Introduction). Furthermore, the in vitro exconjugant enzyme of Euplotes crassus can cap non-telomeric DNA, aided by Chromosome Healing Factor [42]. That Euplotes is more closely related to Oxytricha than to Tetrahymena tentatively supports our assumption that telomerase can heal ends well after the binder/cutter has acted. In support of this, we note that the level of telomerase activity must be much higher in the Oxytricha anlage than in the Tetrahymena anlage, in that the density of broken ends would be, all else being equal, ~100× higher (0.5 ends/kbp in Oxytricha MIC DNA vs. 0.005 ends/kbp in Tetrahymena).
Might telomerase nuclease activity be the cutter [39,43]? If so, obviously the cutter and telomerase cannot physically dissociate. The weak-sites models invoke dissociation, specifically to explain the correlation between the two properties of the boundaries of the CR: their partial cutting, and their broad distribution of TASs. Thus, we argue that the cutter (and binder) activities must reside in some entity other than telomerase. Also arguing against both activities being in one inseparable entity is the apparent erosion interposed between cutting and telomerase action. Specifically, erosion requires that nuclease activities have access to the ends of the break.
Might the telomerase endonuclease activity erode the 3' ends? This nuclease clips 3' tails that protrude beyond internal segments that pair with the RNA template AAAACCCCAAAA segment [9]. While many (76%) of the TASs in the CR-R multi-TAS region have 3'dT's, which could pair with template As, several cannot, specifically the 25 dAs and 5 dCs (Fig. 4). In any case, telomerase in vitro requires that the 3' primer end protrudes on a single-strand tail (review: [9]). Thus another activity is required to resect the 5' underhanging strand (see [67]), and this enzyme too would need access to the break. A variety of 5'-3' exoncleases are known in eukaryotes (review: [68]), so it is reasonable to assume such an activity operates during macronuclear development.
Conclusions
We have shown that the two CR boundaries share the property of being multi-TAS regions, in contrast to the two arm-distal regions. These properties correlate with the previously-inferred incompleteness of chromatid cutting that occurs in each of these regions. Cis-acting sequences must dictate these two contrasting behaviors of the chromatid-breakage and telomerase activities. Reproducibility and sensitivity to allele sequence show that the positions within multi-TAS regions also are dictated by cis-acting sequences. The proposed weak-sites models make several predictions. We predict that a loose complex will be found in ciliates between telomerase and a cbs-directed binder-cutter activity. In Oxytricha, other alternatively processed loci like the 81 locus should have multi-TAS regions that define the boundaries of segments that are shared between different-sized chromosomes; we are searching for such loci in a growing data set of MAC chromosome end-sequences being generated in collaboration with R. Weiss and D. Dunn of the Utah Genome Center. We have begun to examine this data-set for TAS-adjacent sequence motifs that potentially define the binding-sites and/or cut-sites of single-TAS MAC ends, as Klobutcher et al. [38] have done for E. crassus. We can then test the prediction that multi-TAS regions bear either weak binding- or cut-sites. Finally, we are expanding our data set of allelic and orthologous variants of the 81-MAC multi-TAS regions, to be able to infer the identity of their cis-acting pause site sequences. As the details of multi-TAS region processing unravel, we should be able to illuminate the processes of chromatid breakage, and healing by telomerase.
Methods
DNAs
Oxytricha was cultured and whole cell DNAs were harvested as previously described [69]. Genomic DNAs are stored in "DNAb/50" (3 mM NaCl, 3 mM Tris-HCl, pH 8.1, 0.3 mM Na2EDTA). Oligomers are stored in deionized, distilled water. Their names and sequences (5' to 3') are: -123-, ACCCTCTCCAAAATCCCATAA; 1370, AATCTTTCATAGTTCATGCGTT; 180L-, CTCACCCTTCAGGAAAAGAAAATCTCGC; 247+, GGTGGAGAGTACGAAAGATTA; 3239, AATTTATATGGAATGCTACGATT; 387-, AATAACCTCCAAATTGACCTG; 7A, GGACCGTGGCTAGCATTAGT; 7B, ACTAATGCTAG; 894, CTTCAGTTTTGCGTCTTATTTC; 903-, TTCTCACCTTAATGGAGCTATAC; LA4, TTCCTGCAAATGATCCACATA; oRG, AGAGGTGCTGGTGCCAAC; PEB prime, CACTCGATATGGTTAGAATCAG; PEB, AGATCTGATTCTAACCATATCGAGTGG; RA1.1, TCTTCGATTGTGATGATTGTAA; ST2, AGGACCAAAGAAGAAGTAGGA; T1target, ACTTTGGATCATTCTATGCAG; Tel, CCCCAAAACCCCAAAACCC; VHO, GGCATCTTTCTTGATTGC.
LM-PCR directed at MAC telomeres
The technique of Mueller and Wold [55] was followed, diagrammed in Figure 2A. Significant modifications and details are described here, or in the appropriate figure legend.
Extension step
200–400 ng of whole-cell DNA was denatured and annealed with an "Extend" primer using Vent (exo-) polymerase (New England BioLabs).
Anchor ligation
A blunt-end linker oligomer was designed according to Mueller and Wold [55], and was formed by annealing a mixture of oligomers 7A and 7B, 20 μm each, in DNAb/50, by first heating in a 100° temperature block for 1 minute, and then allowing the block to cool to room temperature, sitting on the bench top. The linker was ligated to the Vent-extended preparation in 37.5 µl, at 17°C for 16–20 hrs. Ligates were made 0.3 M sodium acetate, precipitated with 0.75× volume of isopropanol, washed with 80% v/v ethanol, and dissolved in 30 μl DNAb/50.
Anchored PCR
PCR between a specific "P1" primer and 7A was performed with 10 μl of dissolved ligate. Cycle conditions were: 95° 20 sec, 58° 1 min, 72° 90 sec., 35 cycles. For further specificity, a second PCR reaction is sometimes run on the P1-7A product, using a "step-in" or nested primer, "P2." "Step-in" verification PCR is performed on 0.1 μl of P1+7A product, again with P1 but Tel instead of 7A.
Display
an appropriate nested primer is 5'-32P-labeled as previously described [54]; 5 μl of the P1+7A reaction or the step-in verification reaction is denatured and annealed with 1 pmol of the end-labeled display primer and 0.5 units of Taq polymerase (Qiagen) in 10 μl final volume of 1× "Taq salts" (Qiagen). The mixture is subjected to 1–5 cycles (depending on amount of template product examined by ethidium bromide-staining of an agarose electrophoresis gel) of PCR, 95° 20 sec, 58° 1 min, 72° 2 min. Verify reactions are generated when P1-PCR is performed with Tel primer instead of the 7A anchor primer. See Figure 2 legend for a further description. Display chains are displayed on a 6% polyacrylamide gel, which is fixed in 10% methanol, 10% acetic acid, and vacuum-dried under heat. Bands are visualized by autoradiography.
DNA cloning
P1-7A PCR products were cloned, using Invitrogen's TopoTA cloning kit, according to the supplied directions. This step is used to verify – by sequencing with the Thermo Sequenase Radiolabeled Termintor Cycle Sequencing Kit (USB) – the composition of a particular band, which can be cut out, eluted and reamplified (not shown), or simply cloned from the P1-7A reaction (see Fig. 2B).
Abbreviations used
LM-PCR, ligation-mediated polymerase chain reaction; MAC, macronucleus; MIC, micronucleus; TASs, telomere addition sites; CR-R, CR-L, Common Region-Right, -Left.
Authors' contributions
K.R.W. adapted LM-PCR to this application, and generated the data. T.G.D. participated in data analysis, and writing of the manuscript. G.H. procured funding, directed the research, and wrote the manuscript. All authors contributed to interpretation of the results and design of experiments, and have read and approved the final manuscript.
Authors' note
A highly-useful review will soon appear [70].
Acknowledgments
Acknowledgements
This work was supported by funding from NIH grant GM-25203 and NSF grant MCB-0110952. We thank Larry Klobutcher and Dorothy Shippen for many insightful discussions.
Contributor Information
Kevin R Williams, Email: kevin.williams@path.utah.edu.
Thomas G Doak, Email: tom.doak@path.utah.edu.
Glenn Herrick, Email: glenn.herrick@path.utah.edu.
References
- B McClintock. The fusion of broken ends of sister half-chromatids following chromatid breakage at meiotic anaphases. Missouri Agricultural Experiment Station Research Bulletin. 1938;290:1–48. [Google Scholar]
- J Shay, W Wright. When Do Telomeres Matter? Science. 2001;291:839–840. doi: 10.1126/science.1058546. [DOI] [PubMed] [Google Scholar]
- E Blackburn. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/S0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- A Pluta, B Kaine, B Spear. The terminal organization of macronuclear DNA in Oxytricha fallax. Nucleic Acids Res. 1982;10:8145–8154. doi: 10.1093/nar/10.24.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Dawson, G Herrick. Telomeric properties of C4A4-homologous long sequence blocks in micronuclear DNA of Oxytricha fallax. Cell. 1984;36:171–177. doi: 10.1016/0092-8674(84)90086-2. [DOI] [PubMed] [Google Scholar]
- Müller F, H Tobler. Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int J Parasitol. 2000;30:391–399. doi: 10.1016/S0020-7519(99)00199-X. [DOI] [PubMed] [Google Scholar]
- R Wesley. Inverted repetitious sequences in the macronuclear DNA of hypotrichous ciliates. Proc Natl Acad Sci USA. 1975;72:678–682. doi: 10.1073/pnas.72.2.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E Blackburn, J Gall. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- M Melek, D Shippen. Chromosome healing: spontaneous and programmed de novo telomere formation by telomerase. Bioessays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- Wright AD, D Lynn. Maximum ages of ciliate lineages estimated using a small subunit rRNA molecular clock: Crown eukaryotes date back to the paleoproterozoic. Arch Protistenkd. 1997;148:329–341. [Google Scholar]
- A Zahler, D Prescott. Telomere terminal transferase activity in the hypotrichous ciliate Oxytricha nova and a model for replication of the ends of linear DNA molecules. Nucleic Acids Res. 1988;16:6953–6972. doi: 10.1093/nar/16.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Lingner, L Hendrick, TR Cech. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–98. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- M Horvath, V Schweiker, J Bevilacqua, J Ruggles, S Schultz. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/S0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- R Coyne, D Chalker, Yao MC. Genome downsizing during ciliate development: Nuclear division of labor through chromosome restructuring. Annu Rev Gene. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- G Herrick. Germline-soma relationships in ciliated protozoa: the inception and evolution of nuclear dimorphism in one-celled animals. Sem Dev Biol. 1994;5:3–12. [Google Scholar]
- J Ward, M Davis, C Allis, G Herrick. Effects of nullisomic chromosome deficiencies on postzygotic conjugation events in Tetrahymena thermophila: insufficiency of the parental macronucleus to direct postzygotic development. Genetics. 1995;140:989–1005. doi: 10.1093/genetics/140.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Ward, G Herrick. Gene expression inhibitor studies establishing the requirement of zygotic gene expression for execution of post-fertilization events of conjugation in Tetrahymena thermophila. Dev Biol. 1996;173:174–184. doi: 10.1006/dbio.1996.0015. [DOI] [PubMed] [Google Scholar]
- M Davis, J Ward, G Herrick, C Allis. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Develop Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-Z. [DOI] [PubMed] [Google Scholar]
- M Kloc, B Zagrodzinska. Chromatin elimination – an oddity or a common mechanism in differentiation and development? Differentiation. 2001;68:84–91. doi: 10.1046/j.1432-0436.2001.680202.x. [DOI] [PubMed] [Google Scholar]
- M Sadofsky. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Ammermann, Steinbrück G, Lv Berger, W Hennig. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974;45:401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- K Klobutcher, D Prescott. The special case of the hypotrichs. In: J Gall, editor. The Molecular Biology of Ciliated Protozoa. NY: Academic Press; 1986. pp. 111–154. [Google Scholar]
- C Jahn, L Klobutcher. Genome remodeling in ciliated protozoa. Ann Rev Microbiol. 2002. [DOI] [PubMed]
- D Hoffman, R Anderson, M DuBois, D Prescott. Macronuclear gene-sized molecules of hypotrichs. Nucleic Acids Res. 1995;23:1279–1283. doi: 10.1093/nar/23.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Seegmiller, K Williams, G Herrick. Two two-gene macronuclear chromosomes of the hypotrichous ciliates Oxytricha fallax and O. trifallax generated by alternative processing of the 81 locus. Dev Genet. 1997;20:348–357. doi: 10.1002/(SICI)1520-6408(1997)20:4<348::AID-DVG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- S Cartinhour, G Herrick. Three different macronuclear DNAs in Oxytricha fallax share a common sequence block. Mol Cell Biol. 1984;4:931–938. doi: 10.1128/mcb.4.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G Herrick, D Hunter, K Williams, K Kotter. Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Develop. 1987;1:1047–1058. doi: 10.1101/gad.1.10.1047. [DOI] [PubMed] [Google Scholar]
- L Klobutcher, M Huff, G Gonye. Alternative use of chromosome fragmentation sites in the ciliated protozoan Oxytricha nova. Nucleic Acids Res. 1988;16:251–264. doi: 10.1093/nar/16.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Baroin, A Prat, F Caron. Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res. 1987;15:1717–1728. doi: 10.1093/nar/15.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Forney, E Blackburn. Developmentally controlled telomere addition in wild-type and mutant Paramecia. Mol Cell Biol. 1988;8:251–258. doi: 10.1128/mcb.8.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- F Caron. A high degree of macronuclear chromosome polymorphism is generated by variable DNA rearrangements in Paramecium primaurelia during macronuclear differentiation. J Mol Biol. 1992;225:661–678. doi: 10.1016/0022-2836(92)90393-X. [DOI] [PubMed] [Google Scholar]
- D Dawson, G Herrick. Rare internal C4A4 repeats in micronuclear genome of Oxytricha fallax. Mol Cell Biol. 1984;4:2661–2667. doi: 10.1128/mcb.4.12.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L Klobutcher, C Jahn, D Prescott. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell. 1984;36:1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- Yao M-C, C Yao, B Monks. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- R Coyne, D Chalker, Yao MC. Genome downsizing during ciliate development: Nuclear division of labor through chromosome restructuring. Annu Rev Gene. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Q Fan, Yao M-C. A long stringent sequence signal for programmed chromosome breakage in Tetrahymena thermophila. Nucleic Acids Res. 2000;28:895–900. doi: 10.1093/nar/28.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S Baird, L Klobutcher. Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev. 1989;3:585–597. doi: 10.1101/gad.3.5.585. [DOI] [PubMed] [Google Scholar]
- L Klobutcher, S Gygax, J Podoloff, J Vermeesch, C Price, C Tebeau, Jahn C. Conserved DNA sequences adjacent to chromosome fragmentation and telomere addition sites in Euplotes crassus. Nucleic Acids Res. 1998;26:4230–4240. doi: 10.1093/nar/26.18.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L Klobutcher. Characterization of in vivo developmental chromosome fragmentation intermediates in E. crassus. Mol Cell. 1999;4:695–704. doi: 10.1016/S1097-2765(00)80380-9. [DOI] [PubMed] [Google Scholar]
- Jönsson F, Steinbrück G, H Lipps. Both subtelomeric regions are required and sufficient for specific DNA fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol. 2001;2:1–11. doi: 10.1186/gb-2001-2-2-research0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G Yu, E Blackburn. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-C. [DOI] [PubMed] [Google Scholar]
- J Bednenko, M Melek, E Greene, D Shippen. Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J. 1997;16:2507–2518. doi: 10.1093/emboj/16.9.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M Melek, E Greene, D Shippen. Processing of nontelomeric 3' ends by telomerase: default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H Wang, E Blackburn. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K Collins, CW Greider. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7B:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- E Greene, D Shippen. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes Dev. 1998;12:2921–2931. doi: 10.1101/gad.12.18.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- H Niu, J Xia, N Lue. Characterization of the interaction between the nuclease and reverse transcriptase activity of the yeast telomerase complex. Mol Cell Biol. 2000;20:6806–6815. doi: 10.1128/MCB.20.18.6806-6815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M-C, Yao C-H, B Monks. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- Q Fan, Yao M-C. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol. 1996;16:1267–1274. doi: 10.1128/mcb.16.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L Harrington, C Greider. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- D Gilley, J Preer, K Aufderheide, B Polisky. Autonomous replication and addition of telomere-like sequences to DNA microinjected into Paramecium tetraurelia macronuclei. Mol Cell Biol. 1988;8:4765–4772. doi: 10.1128/mcb.8.11.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllenbeck M, LA Klobutcher. De novo telomere addition to spacer sequences prior to their developmental degradation in Euplotes crassus. Nucleic Acids Res. 2002;30:523–531. doi: 10.1093/nar/30.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M-C, S Duharcourt, D Chalker. Genome-wide rearrangements of DNA in ciliates. In: N Craig, R Craigie, M Gellert, editor. Mobile DNA II. Washington, D.C.: ASM Press; 2002. pp. 730–758. [Google Scholar]
- K Williams, Herrick G. Expression of the gene encoded by a family of macronuclear chromosomes generated by alternative DNA processing in Oxytricha fallax. Nucleic Acids Res. 1991;19:4717–4724. doi: 10.1093/nar/19.17.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- P Mueller, B Wold. Ligation-mediated PCR: applications to genomic footprinting. PCR Methods. 1991;2:20–31. [Google Scholar]
- L Klobutcher, M Swanton, P Donini, D Prescott. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3' terminus. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JM Clark. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Preer. Genetics of the protozoa. In: T-T Chen, editor. Research in Protozoology. Oxford: Pergamon Press; 1969. pp. 133–278. [Google Scholar]
- H-W Kuhlmann, K Heckmann. Nuclear processes in Euplotes octocarinatus during conjugation. Europ J Protistol. 1991;26:370–386. doi: 10.1016/S0932-4739(11)80158-6. [DOI] [PubMed] [Google Scholar]
- B Spear, M Lauth. Polytene chromosomes of Oxytricha: Biochemical and morphological changes during macronuclear development in a ciliated protozoan. Chromosoma. 1976;54:1–13. doi: 10.1007/BF00331828. [DOI] [PubMed] [Google Scholar]
- Jareño M. Études sur les chromosomes polytenes de Stylonychia mytilus. Protistologica. 1974;10:527–532. [Google Scholar]
- Wu C-t, J Morris. Transvection and other homology effects. Curr Opin Genet Dev. 1999;9:237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]
- D Witherspoon, T Doak, K Williams, A Seegmiller, J Seger, G Herrick. Selection on the protein-coding genes of the TBE1 family of transposable elements in the ciliates Oxytricha fallax and O. trifallax. Mol Biol Evol. 1997;14:696–706. doi: 10.1093/oxfordjournals.molbev.a025809. [DOI] [PubMed] [Google Scholar]
- S Ghosh, J Jaraczewski, L Klobutcher, C Jahn. Characterization of transcription initiation, translation initiation, and poly(A) addition sites in the gene-sized macronuclear DNA molecules of Euplotes. Nucleic Acids Res. 1994;22:214–221. doi: 10.1093/nar/22.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A Seegmiller, K Williams, R Hammersmith, T Doak, T Messick, Witherspoon D, L Storjohann, G Herrick. Internal eliminated sequences interrupting the Oxytricha 81 locus: allelic divergence, conservation, conversions, and possible transposon origins. Mol Biol Evol. 1996;13:1351–1362. doi: 10.1093/oxfordjournals.molbev.a025581. [DOI] [PubMed] [Google Scholar]
- A Pingoud, A Jeltsch. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- K Kramer, J Haber. New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- F Paques, J Haber. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D Dawson, G Herrick. Micronuclear DNA sequences of Oxytricha fallax homologous to the macronuclear inverted terminal repeat. Nucleic Acids Res. 1982;10:2911–2924. doi: 10.1093/nar/10.9.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- F Jönsson, HJ Lipps. The Biology of Telomeres in Hypotrichous Ciliates. In: G Krupp, R Parwaresch, editor. Telomeres and Telomerases: Cancer and Biology. Landes Bioscience; 2002. [Google Scholar]