Summary
Hydrolethalus syndrome is a recessively inherited lethal malformation syndrome characterized by hydrocephaly with absent midline structures of the brain, micrognathia, polydactyly, and several other abnormalities, mostly in the midline structures. Hydrolethalus syndrome was described in 1981 in Finland, where the incidence is 1:20,000. Only a few cases have been reported elsewhere, and the pathogenesis has remained unknown. Here we report the assignment of the hydrolethalus syndrome locus to chromosome 11q23-25 in Finnish families. The initial genome scan was performed using DNA samples from only 15 affected individuals. In the next step, the hydrolethalus syndrome locus was assigned to an 8.5-cM interval between markers D11S4144 and D11S1351 by linkage analysis in eight families. Finally, the critical locus could be restricted by linkage disequilibrium and haplotype analyses to a 0.5–1-cM region between markers D11S933 and D11S934. Genealogical studies performed in 40 families affected by hydrolethalus revealed no regional clustering, suggesting a relatively early introduction of the disease mutation into the Finnish population and the spreading of the mutation with the inhabitation of the late-settlement area.
Introduction
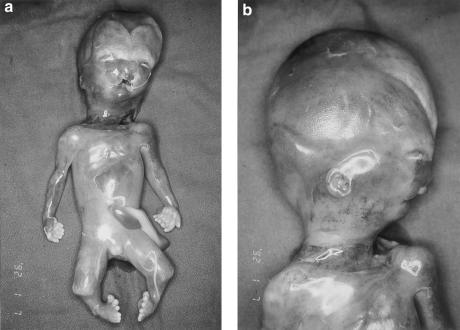
Hydrolethalus syndrome (MIM 236680) is a multiple midline malformation syndrome leading to stillbirth or death during the first day of life. The hallmark features include external hydrocephaly with absent midline structures of the brain, polyhydramnions, and micrognathia. Most patients also have polydactyly, anomalous eyes and nose, and a keyhole-shaped defect of the occipital bone. A cleft lip or palate, anomalous or low-set ears, abnormal larynx or trachea, defective lobulation of the lungs, congenital heart defect, abnormal genitalia, and club feet are often present (fig. 1). The syndrome shows an autosomal recessive mode of inheritance (Salonen et al. 1981; Salonen and Herva 1990).
Figure 1.
Male fetus with hydrolethalus syndrome (case 1a) after termination of pregnancy at 18 wk. Note hydrocephaly, small and wide-spaced eyes, anomalous bifid nose, bilateral cleft lip, micrognathia, low-set ears, broad neck, polydactyly in both hands and feet, and shortened lower limbs with club-foot deformity.
Hydrolethalus syndrome was first described in Finland (Salonen et al. 1981), where it occurs much more frequently than elsewhere in the world. Several hydrolethalus cases have been reported in other populations, but they may not all represent the same phenotypic entity. However, some reports reveal a clinical phenotype similar to Finnish cases and could represent true hydrolethalus cases (Muenke et al. 1991; Sharma et al. 1992; Pryde et al. 1993; Norgard et al. 1996). Since 1970, 57 families with 81 children affected with hydrolethalus syndrome have been identified in Finland, and the incidence in this population has been approximated to be at least 1:20,000 (Salonen and Herva 1990). Hydrolethalus syndrome thus belongs to the Finnish disease heritage (Norio et al. 1973), which consists of about 35 monogenic diseases, which have been intensively studied. Almost all Finnish disease loci have been mapped to well-defined chromosomal regions, and, in about half of the diseases, the causative genes have been identified (Peltonen 1997).
Here we report the assignment of the hydrolethalus syndrome locus to 11q23-25 by use of linkage, linkage-disequilibrium, and haplotype analyses in Finnish families. We also performed extensive genealogical studies that complement the genealogical data published earlier (Salonen et al. 1981).
Subjects and Methods
Family Material, Controls, DNA Samples, and Genealogy
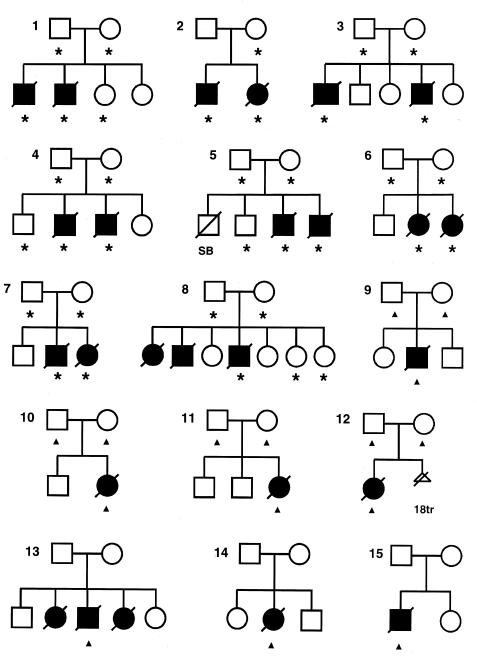
Fifteen affected individuals and 20 healthy family members from eight families affected by hydrolethalus were included in the linkage studies. In addition, seven affected individuals from different families and the parents of four of these families were included in the linkage-disequilibrium studies. The pedigrees are shown in figure 2. No relationships could be established between these families, but the parents of family 8 were first cousins.
Figure 2.
Pedigrees used in the linkage and linkage-disequilibrium studies. Asterisks and triangles indicate individuals from whom DNA samples were available. Individuals marked with asterisks were used in linkage and linkage-disequilibrium analyses, and those marked with triangles provided information for linkage-disequilibrium studies only. Individuals affected with hydrolethalus syndrome are marked with blackened symbols. In most cases, the pregnancy was terminated, after prenatal diagnosis by ultrasound. Only the first affected children in families 3, 8, 13, and 14 were delivered. SB=stillborn (hydrolethalus-syndrome status not known), 18tr=termination of a pregnancy with trisomy 18 but not hydrolethalus syndrome.
Two of the 22 affected individuals were born, in 1981 and 1991, and the remaining 20 were fetuses aborted between 1982 and 1998 as a result of ultrasonography diagnosis (gestational age 10–23 weeks). All the families included represented very typical cases, and all the diagnoses were confirmed by one clinician (R.S.). All the cases exhibited hydrocephaly and several other malformations typical of hydrolethalus syndrome. The phenotypic features of the affected individuals included in the linkage and linkage-disequilibrium studies are presented in table 1.
Table 1.
Findings for the 22 Affected Individuals Included in the Linkage and Linkage-Disequilibrium Studies[Note]
|
Family |
||||||||||||||||||||||
| Malformation | 1a | 1b | 2a | 2b | 3a | 3b | 4a | 4ba | 5a | 5b | 6a | 6b | 7a | 7b | 8c | 9 | 10 | 11 | 12 | 13b | 14 | 15 |
| Hydrocephalus | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Occipital bone defect | ? | + | ? | ? | + | + | + | + | + | + | + | + | + | + | ? | + | + | − | + | + | + | + |
| Micrognathia | + | + | + | + | + | + | + | ? | + | + | + | + | + | + | + | + | + | + | + | + | ? | + |
| Cleft lip/palate | + | − | + | + | + | + | + | ? | + | + | + | − | − | − | + | + | − | + | − | + | + | − |
| Anomalous nose | + | + | + | ? | + | + | + | ? | + | + | + | + | − | + | + | + | + | + | + | − | ? | ? |
| Small/deep-set eyes | + | + | ? | ? | + | + | + | ? | + | − | + | + | + | + | ? | + | + | + | + | + | + | ? |
| Anomalous/low-set ears | + | + | + | ? | + | + | + | ? | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Abnormal larynx/trachea | + | − | − | − | + | + | + | ? | − | − | − | − | + | − | + | − | + | + | − | − | + | − |
| Defective lobation of the lungs | + | + | + | + | + | + | + | ? | + | − | + | − | + | − | + | + | + | + | + | + | − | + |
| Congenital heart defect | + | − | + | + | + | + | + | ? | − | − | − | − | − | − | + | − | − | − | + | ? | + | − |
| Urinary-tract anomalies | + | − | − | + | + | + | − | ? | + | − | − | − | − | − | − | − | − | − | − | − | + | − |
| Abnormal genitalia | + | − | − | + | + | − | + | ? | − | ? | − | + | + | − | + | + | − | − | + | − | + | − |
| Uterus duplexb | m | m | m | − | m | m | m | m | m | m | − | + | m | − | m | m | − | − | + | m | + | m |
| Polydactyly: | + | − | + | + | + | + | + | + | + | − | − | − | − | + | + | − | + | + | + | + | + | + |
| Of fingers | + | − | + | + | + | + | − | + | + | − | − | − | − | − | + | − | + | + | − | + | − | + |
| Of toes | + | − | + | + | + | + | + | + | + | − | − | − | − | + | + | − | + | + | + | + | + | − |
| Duplicated big toe | + | − | + | ? | + | + | + | ? | + | − | − | − | − | − | + | − | + | + | + | + | + | − |
| Club feet | + | + | + | + | + | − | + | ? | + | + | + | + | − | + | − | + | + | ? | + | + | ? | − |
| Short limbs | + | + | ? | + | + | + | + | ? | + | + | + | − | − | + | − | − | − | + | + | + | ? | − |
Note.—+ = Malformation present; − = malformation absent; ? = status not known.
After suction curettage, at 10+5 wk of gestation, a thorough examination of the fetus was not possible.
m=Male individual.
Forty-one parental DNA samples collected for the mapping of infantile-onset spinocerebellar ataxia (IOSCA) and a new lethal metabolic syndrome with iron accumulation were used as controls for the linkage-disequilibrium analyses. These diseases also belong to the Finnish disease heritage and are regionally distributed in a similar way to hydrolethalus syndrome (Varilo et al. 1996; Fellman et al. 1998). We have recently used this principle successfully to select controls for the mapping of the new Finnish metabolic syndrome with iron accumulation (Visapää et al. 1998).
DNA from the affected individuals was extracted from fresh tissue samples obtained at autopsy (placenta, liver) or from cultured fibroblasts, in accordance with standard procedures (Maniatis et al. 1982), except for the affected individuals of family 2, for whom only paraffin-embedded tissue samples were available. DNA from the paraffin-embedded tissues was extracted as described elsewhere (Isola et al. 1994). DNA from the healthy family members and controls was extracted from peripheral blood leukocytes, in accordance with standard procedures.
The genealogical study was performed in accordance with published criteria (Varilo et al. 1996). The names, dates, and places of birth of the patients' parents were used to trace ancestors of the 40 families affected by hydrolethalus back to the 1800s, from local church registers. Eighteen of the core families have been described before (Salonen et al. 1981). This study was approved by the Ethical Committee of the Department of Obstetrics and Gynecology at Helsinki University Central Hospital.
Marker Genotyping, Radiation-Hybrid Mapping, and Statistics
A slightly modified Weber screening set (Sheffield et al. 1995) of 380 polymorphic microsatellite markers was used in the initial genome screen. In the denser mapping of the critical region on chromosome 11, the Généthon marker set (Dib et al. 1996) was used. To define the marker order and distances, radiation-hybrid (RH) mapping of the markers in the critical region was performed with the Stanford medium resolution (G3) radiation-hybrid panel (Research Genetics). Markers D11S4094, D11S4464, D11S1752, D11S933, D11S4158, and D11S1896 were typed twice in the G3 panel. With markers D11S1353, D11S1328, D11S934, D11S4110, D11S1351, and D11S912, raw hybrid data for the same panel were obtained from the Stanford Human Genome Center Internet pages. The Genome Database was used to identify the chromosomal band where the critical markers were located.
The markers were amplified for genotyping by PCR with fluorescently labeled primers, and the PCR products were electrophoretically separated with an ABI 377 sequencer (PE Biosystems). The genotype data were processed with the Genotyper computer program, version 2.0 (PE Biosystems). For the typing of the RH panel, the markers were amplified by PCR, and the products were separated by electrophoresis on 1.5% agarose gels stained with ethidium bromide.
Two-point linkage analyses were performed using the MLINK option of the LINKAGE package computer programs, version FASTLINK 2.2 (Lathrop and Lalouel 1984), assuming autosomal recessive inheritance, complete penetrance, and no phenocopies. The allele frequencies of the polymorphic markers were determined on the basis of the control samples used for the linkage-disequilibrium analyses. The consanguinity of the parents of family 8 was taken into account in the linkage analysis. Two-point and multipoint linkage-disequilibrium analyses were performed using the DISLAMB and DISMULT computer programs, respectively (Terwilliger 1995). A disease-allele frequency of .01 was assumed in the analyses.
The RH mapping data were analyzed with version 3.0 of the FORTRAN program RHMAXLIK of the RHMAP package (Boehnke et al. 1991), using the branch and bound ordering option. This program orders markers and calculates marker distances in centirays (cR). The marker distances were converted from centirays to centimorgans (cM) by dividing the known total distance (in centimorgans, in accordance with Dib et al. 1996) of the region proportional to the marker distances in centirays.
Results
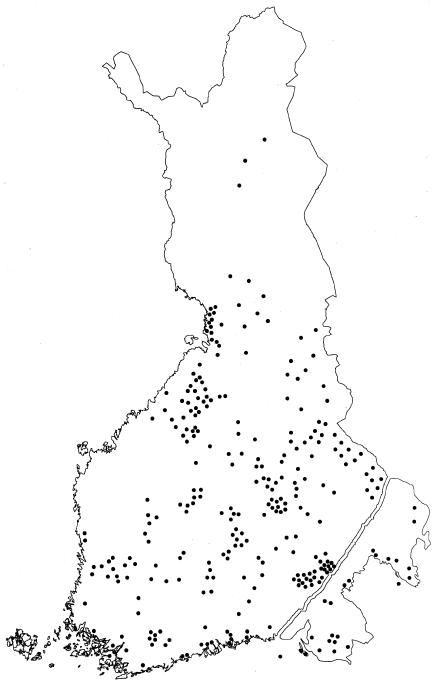
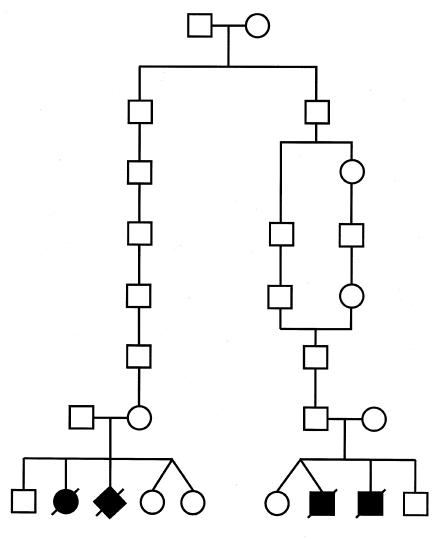
Genealogical studies of all the available 40 families affected by hydrolethalus, going back 3–10 generations, revealed a similar general distribution of ancestors, as reported elsewhere with a smaller number of families (Salonen et al. 1981). The birthplaces of the great-grandparents were distributed over a relatively wide geographical area in late-settlement eastern and central Finland, outside the densely populated early-settlement area of southern and western Finland (fig. 3). Six parents were found to be related. The earliest identified link between ancestors was seven generations (i.e., 220 years) old (fig. 4), and the most recent link was found in family 8, where the parents were first cousins.
Figure 3.
Birthplaces of the great-grandparents of 40 families affected by hydrolethalus on the map of Finland. The birthplaces are mostly distributed in the late-settlement area, outside the densely populated southern and western parts of Finland.
Figure 4.
Pedigree of the oldest traced family connection of the Finnish families affected by hydrolethalus. Their common ancestors originated from the parish of Ruokolahti, in eastern Finland, and were born in 1761 and 1764.
The initial genome scan was performed using the DNA of 15 individuals—seven affected sibpairs and one affected individual in family 8. Since the parents in family 8 were first cousins, we assumed that the affected individual would be homozygous with the markers flanking the disease locus. In the first stage of the genome scan, the genotypes were analyzed simply by comparing the genotypes of each sibpair, searching for shared alleles, and monitoring for homozygosity of the affected individual in family 8. Several markers revealed shared genotypes between six of the seven sibpairs, but those regions were not analyzed further because the adjacent markers (on average, 10 cM apart) did not show any tendency to allele sharing. After we had analyzed 270 markers, one marker, D11S4464, revealed identical genotypes in all seven sibpairs. With marker D11S1998, 8 cM centromeric from D11S4464, four sibpairs also shared identical genotypes, and with the next telomeric marker, D11S912, 11 cM from D11S4464, five sibpairs carried identical genotypes. All three markers also revealed homozygosity of the affected individual in family 8. These three markers were then genotyped in the complete family material. No obligatory recombinations were identified between hydrolethalus syndrome and marker D11S4464, which provided a LOD score of 3.77 at recombination fraction (θ) 0. The maximum LOD score with marker D11S912 was 2.04 at θ=.05, whereas marker D11S1998 was uninformative.
In the next phase, we analyzed several markers around marker D11S4464 in the complete family material and constructed haplotypes spanning the region. Several markers within an 8.5-cM region on chromosome 11q23-25, flanked by markers D11S4144 and D11S1351, provided some evidence of linkage. There were positive LOD scores (2.7–4.7) with nine markers over this interval at θ=0 (table 2). The disease chromosomes of the parents of family 8 revealed identical haplotypes over the complete region.
Table 2.
Pairwise LOD Scores between Hydrolethalus Syndrome and 13 Marker Loci on Chromosome 11q23-25 in Eight Families[Note]
| LOD Score at θ = |
|||||
| Locus | 0 | .01 | .05 | .10 | .20 |
| D11S1353 | −∞ | 2.44 | 2.60 | 2.26 | 1.40 |
| D11S4144 | −∞ | 2.45 | 2.68 | 2.40 | 1.58 |
| D11S4094 | 3.25 | 3.15 | 2.75 | 2.26 | 1.36 |
| D11S4464 | 3.77 | 3.67 | 3.26 | 2.75 | 1.76 |
| D11S1328 | 4.66 | 4.53 | 4.00 | 3.34 | 2.10 |
| D11S1752 | 2.71 | 2.64 | 2.33 | 1.95 | 1.23 |
| D11S933 | 3.08 | 2.99 | 2.61 | 2.16 | 1.32 |
| D11S4158 | 3.70 | 3.60 | 3.17 | 2.63 | 1.64 |
| D11S1896 | 3.91 | 3.80 | 3.37 | 2.83 | 1.80 |
| D11S934 | 4.57 | 4.45 | 3.94 | 3.31 | 2.10 |
| D11S4110 | 4.03 | 3.91 | 3.45 | 2.89 | 1.81 |
| D11S1351 | −∞ | 0.66 | 1.10 | 1.07 | 0.75 |
| D11S912 | −∞ | 1.15 | 2.04 | 2.01 | 1.40 |
Bold type indicates the maximum LOD score for each marker.
To restrict the critical chromosomal region, we monitored for linkage disequilibrium in the alleles of the nine markers revealing linkage (table 3). Thirty hydrolethalus chromosomes and 105 control chromosomes, including 23 chromosomes of the healthy parents in families affected by hydrolethalus and 41 DNA samples from regionally selected controls, were used in these analyses. In two-point linkage-disequilibrium analyses, two markers provided highly significant P values. With marker D11S1896, all the hydrolethalus chromosomes carried the same allele 4, which was present in 33% of the control chromosomes, and, with marker D11S4158, 77% of the disease chromosomes carried allele 1, which was present in only 16% of the control chromosomes. The corresponding P values were 4×10−13 for marker D11S1896 and 2×10−9 for marker D11S4158.
Table 3.
Allele Frequencies in Hydrolethalus and Control Chromosomes and Two-Point Linkage-Disequilibrium Analyses of Nine Marker Loci on 11q23-25[Note]
|
Percentage of Hydrolethalus Chromosomes (n=30) /Percentage of Control Chromosomes (n=105) for Allele |
||||||||||||||||
| Locus | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | χ2 LRTa | P | λb |
| D11S4094 | 7/17 | 7/13 | 7/12 | 3/10 | 73/35 | 0/6 | 0/7 | 3/0 | … | … | … | … | … | 10.9 | .0005 | .56 |
| D11S4464 | 3/6 | 53/35 | 23/30 | 13/18 | 7/7 | 0/5 | … | … | … | … | … | … | … | 1.0 | .2 | .26 |
| D11S1328 | 40/7 | 30/29 | 13/31 | 3/8 | 3/1 | 10/20 | 0/4 | 0/1 | … | … | … | … | … | 12.0 | .0003 | .35 |
| D11S1752 | 10/31 | 7/13 | 83/57 | … | … | … | … | … | … | … | … | … | … | 6.6 | .005 | .61 |
| D11S933 | 0/3 | 17/9 | 0/1 | 63/53 | 7/21 | 3/2 | 0/1 | 10/9 | 0/1 | 0/1 | … | … | … | .0 | .4 | .06 |
| D11S4158 | 77/16 | 0/3 | 3/7 | 7/45 | 13/24 | 0/2 | 0/1 | 0/1 | 0/1 | 0/1 | … | … | … | 35.2 | <.0001 | .72 |
| D11S1896 | 0/14 | 0/10 | 0/43 | 100/33 | … | … | … | … | … | … | … | … | … | 51.2 | <.0001 | 1.00 |
| D11S934 | 0/1 | 13/22 | 0/3 | 0/11 | 40/41 | 0/4 | 0/4 | 3/1 | 0/2 | 3/4 | 33/2 | 3/6 | 3/0 | 13.6 | .0001 | .31 |
| D11S4110 | 10/10 | 0/1 | 20/32 | 0/3 | 20/21 | 33/20 | 17/12 | 0/1 | … | … | … | … | … | .0 | .5 | .00 |
Bold type indicates alleles showing strong association with hydrolethalus syndrome.
χ2 statistic of the likelihood-ratio test (Terwilliger 1995).
Proportion of excess of a certain allele in the hydrolethalus chromosomes.
To confirm the marker order, for multipoint linkage-disequilibrium analysis and for reliable construction of haplotypes of disease chromosomes, we performed RH mapping of the linked markers to define their order. The most probable marker order and the approximate marker distances (converted to centimorgans) over the critical region were: cen-D11S4094-0.6 cM-D11S4464-1.3 cM-D11S1328-0.3 cM-D11S1752-0.1 cM-D11S933-0.8 cM-D11S4158-0.3 cM-D11S1896-0.1 cM-D11S934-1.9 cM-D11S4110-tel.
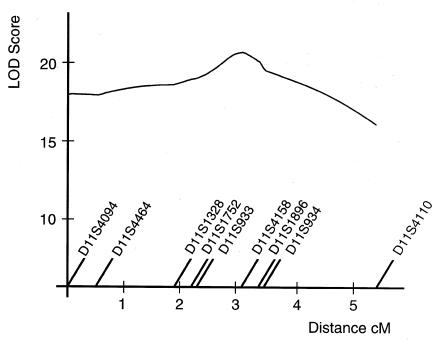
Multipoint linkage-disequilibrium analysis was performed with the critical markers (fig. 5). The maximum LOD score of 21.1 was obtained at the position of marker D11S4158. A LOD score >20 was obtained between markers D11S933 and D11S934. To identify the most probable location of the disease gene, we monitored for a shared ancestral haplotype between the disease chromosomes (table 4). Based on haplotype sharing, it was concluded that the hydrolethalus locus lay in the vicinity of markers D11S4158 and D11S1896. Every disease chromosome carried the same allele 4 with marker D11S1896, and 23 of 30 disease chromosomes carried the same allele 1 with marker D11S4158. It is probable that the hydrolethalus locus lies in the vicinity of marker D11S1896 and that the ancient recombinations of marker D11S4158 limit the locus on the centromeric side, but since allele 4 with marker D11S1896 was very common in the control chromosomes, the centromeric border of the hydrolethalus locus cannot be confirmed with complete certainty as lying at marker D11S4158. Markers D11S933 on the centromeric and D11S934 on the telomeric side limit the possible gene locus, there being several ancient recombinations.
Figure 5.
Multipoint linkage-disequilibrium analysis of nine markers in the linkage region with 30 hydrolethalus chromosomes and 105 control chromosomes
Table 4.
Haplotypes of the Hydrolethalus-Carrying Chromosomes with Nine Marker Loci[Note]
| Familya | D11S4094 | D11S4464 | D11S1328 | D11S1752 | D11S933 | D11S4158 | D11S1896 | D11S934 | D11S4110 |
| 1P | 5 | 2 | 3 | 3 | 8 | 5 | 4 | 12 | 3 |
| 1M | 2 | 3 | 2 | 3 | 4 | 1 | 4 | 5 | 6 |
| 2P | 5 | 3 | 2 | 2/3b | 2 | 1/4b | 4 | 11* | 5* |
| 2M | 5 | 5 | 2 | 2/3b | 4 | 1/4b | 4 | 5 | 5 |
| 3P | 5 | 4 | 6 | 3 | 8 | 5 | 4 | 11* | 7 |
| 3M | 5 | 3 | 5 | 3 | 4 | 3 | 4 | 5 | 3 |
| 4P | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 2 | 6 |
| 4M | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 11* | 1 |
| 5P | 5 | 2 | 1 | 3 | 4/5b | 1 | 4 | 5 | 1 |
| 5M | 5 | 2 | 1 | 3 | 4/5b | 1 | 4 | 11* | 5* |
| 6P | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 11* | 5* |
| 6M | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 10 | 6 |
| 7P | 5 | 2 | 1 | 3 | 5 | 1 | 4 | 5 | 1 |
| 7M | 5 | 5 | 2 | 2 | 4 | 1 | 4 | 5 | 6 |
| 8P | 5 | 4 | 2 | 3 | 2 | 1 | 4 | 5 | 7 |
| 8M | 5 | 4 | 2 | 3 | 2 | 1 | 4 | 5 | 7 |
| 9P | 2 | 3 | 1 | 1 | 6 | 4 | 4 | 5 | 3 |
| 9M | 5 | 4 | 2 | 3 | 2 | 1 | 4 | 5 | 6 |
| 10P | 8 | 1 | 2 | 3 | 4 | 1 | 4 | 5 | 6 |
| 10M | 1 | 3 | 6 | 3 | 2 | 1 | 4 | 11* | 5* |
| 11P | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 11* | 6 |
| 11M | 3 | 2 | 3 | 1 | 4 | 1 | 4 | 11* | 3 |
| 12P | 5 | 2 | 3 | 1 | 4 | 5 | 4 | 2 | 7 |
| 12M | 5 | 2 | 1 | 3 | 4 | 1 | 4 | 11* | 7 |
Note.—Underlining indicates alleles which are part of the most common haplotype, and asterisks indicate an alternative telomeric end of that haplotype.
P=paternal disease chromosome; M=maternal disease chromosome.
Not possible to determine which of the alleles originates from the paternal and which from the maternal disease chromosome.
Construction of the haplotype by use of markers D11S4158, D11S1896, and D11S934 was possible in 22 disease chromosomes (families 1 and 3–12, for which DNA samples from both parents were available). With these three markers, two ancient core haplotypes were observed: 8 of the 22 disease chromosomes carried haplotype 1–4–5, and seven chromosomes carried haplotype 1–4–11. The distribution of ancestors' birthplaces for these haplotypes was monitored, but neither group of great-grandparents revealed any obvious regional clustering. Since allele 1 with marker D11S4158 was equally present in chromosomes carrying allele 5 or 11 with marker D11S934, we consider it improbable that these two ancestral haplotypes represent two different founding mutations. It is more likely that ancient recombination events have taken place between markers D11S1896 and D11S934.
Discussion
When attempting to identify the locus for a monogenic disease with no known biochemical abnormalities, it is essential to have sufficient family material with reliable diagnoses. Differential diagnosis between hydrolethalus syndrome and other malformation syndromes has proven to be difficult—hydrolethalus syndrome as a disease entity was actually discovered in the course of a study of another lethal malformation syndrome, Meckel syndrome, in Finland (Salonen et al. 1981). The difficulties of differential diagnostics in multiple midline malformation syndromes have been discussed in several publications. In 1991, Hennekam et al. described a sibpair with multiple symptoms characteristic of hydrolethalus syndrome but concluded that they represented a separate entity, pseudotrisomy 13 syndrome. In 1993, Pryde et al. discussed the problems of prenatal differential diagnosis between hydrolethalus, severe Smith-Lemli-Opitz, orofaciodigital type IV and VI, pseudotrisomy 13, and Pallister-Hall syndromes and concluded that it was unclear whether similar or related pathogenetic defects lie behind these syndromes or whether they represent similar phenotypic expressions of several unique molecular defects. In 1995, Verloes compared these syndromes, using numerical syndromology, and concluded that the five syndromes are clearly independent phenotypic entities. Later, mutations in the Δ7-sterol reductase gene (DHCR7, chr 11q13) have been proven to cause Smith-Lemli-Opitz syndrome (Cormier-Daire et al. 1996; Fitzky et al. 1998; Wassif et al. 1998; Waterham et al. 1998) and mutations in the GLI3 gene (chr 7p13) to cause Pallister-Hall syndrome (Kang et al. 1997a, 1997b). Meckel syndrome has proven to be a heterogeneous disease and has been mapped to chromosome 17q22 in the Finnish population and to 11q13 in Middle Eastern and North African families (Paavola et al. 1995; Roume et al. 1998; Salonen and Paavola 1998). So far, the molecular defects behind orofaciodigital syndrome types IV and VI and pseudotrisomy 13 syndrome are not known.
Although the diagnosis of hydrolethalus syndrome is problematic, we are convinced that the family material used in our linkage and linkage-disequilibrium studies consists of true hydrolethalus cases. The syndrome is common enough in Finland to be recognized by specialists, and we included only highly typical cases in this study. The genealogical studies also suggested that, as in several other Finnish diseases (Peltonen et al. 1995), one major founding mutation could be predicted. Thus, we considered the family material sufficient for a linkage-based genomewide scan. The results of the linkage and linkage-disequilibrium analyses revealed that all our study families were linked to the same locus and that every disease chromosome carried the same allele 4 with marker D11S1896, demonstrating locus and allelic homogeneity in hydrolethalus syndrome in Finland. However, since 33% of the control chromosomes carried the same allele 4 with marker D11S1896, we cannot be totally convinced that there is only one ancestral mutation before we have a yet denser marker map and can find a haplotype specific to hydrolethalus-carrying chromosomes with several markers.
This mapping project is—like many other mapping projects of the Finnish disease heritage—a good example of the power of linkage disequilibrium in disease-gene mapping in genetic isolates. Using linkage-disequilibrium analyses, it is possible to limit the critical chromosomal region after conventional linkage analysis before starting the construction of a physical map or candidate-gene analyses; in this study, the critical genetic interval decreased from an initial 8.5 cM to 0.5–1 cM. When the family material available for linkage studies is limited, linkage disequilibrium also confirms the results of linkage analysis. In this study, the two-point LOD scores of 2.7–4.7, obtained with nine markers, were satisfactory, but the result of multipoint linkage-disequilibrium analysis, providing a LOD score of 21.1, left no room for doubt about the locus assignment.
The regional distribution of the ancestors of families affected by hydrolethalus in the late-settlement region of Finland resembles those of infantile-onset spinocerebellar ataxia (IOSCA) (Varilo et al. 1996) and the new Finnish metabolic syndrome with iron accumulation (Visapää et al. 1998). These disease mutations were most probably introduced into the Finnish population around the same time, some 30–40 generations ago. No obvious clustering of ancestors was observed, nor does the distribution of the ancestors' birthplaces reflect the distribution of the current Finnish population. As in IOSCA and the new metabolic syndrome, the distribution resembles the routes of the internal migration in the 1500s, from the southeastern county of Savo, and suggests that the disease alleles were introduced into subpopulations during this inland inhabitation process (Norio 1981). Compared to IOSCA and the new metabolic syndrome, the distribution of ancestors stretches somewhat further into the area of early settlement, suggesting that this mutation was introduced somewhat earlier. That there are fewer identified consanguineous families than in the case of IOSCA lends further support to this theory. The size of the common core haplotype and the interval revealing very significant linkage disequilibrium in two-point analysis (P< .0001) spans 2 cM in IOSCA (Nikali et al. 1995; Varilo et al. 1996), 1 cM in the new metabolic syndrome (Visapää et al. 1998), and 0.5 cM in hydrolethalus syndrome, further supporting the hypothesis that the hydrolethalus mutation is the oldest of the three. In fact, considering the similarity of the distribution of the ancestors in these three diseases, this chromosomal interval is surprisingly small in hydrolethalus syndrome, but this could also be due to inaccuracies in the genetic and RH maps.
There are 10 cloned genes and 80 expressed sequence tags in the 9-cM interval between D11S1353 and D11S1351 in the New Gene Map of the Human Genome of the National Center for Biotechnology Information. Some of the genes can be excluded as candidate genes for hydrolethalus syndrome on the basis of their known specific function or expression in only a few tissues—for example, two testis-specific genes, ZNF202 and ACRV1 (Golden et al. 1993; Monaco et al. 1998). However, many of the genes encode proteins involved in very basic cell functions—for example, potassium channel KCNJ1, sialyltransferase SIAT4C, and signal-recognition particle receptor SRPR. Although these genes are not directly known to be involved in embryogenesis, they cannot be excluded as candidate genes for hydrolethalus syndrome in view of their important role in cell functions. Which of the regional genes are located in the restricted critical chromosomal region and, thus, deserve closer consideration and analysis as candidate genes, remains to be analyzed when a complete physical map of the region has been constructed.
Linkage analysis can now be provided to families with a previous child with hydrolethalus syndrome to confirm the prenatal diagnosis as early as possible. At present prenatal diagnosis of hydrolethalus syndrome is done ultrasonographically (Hartikainen-Sorri et al. 1983; Ämmälä and Salonen 1995). For pregnancies with a known risk for hydrolethalus syndrome, a vaginal ultrasound examination can be performed at 11–12 wk of pregnancy, when the earliest signs of the syndrome can be detected. The diagnosis can be confirmed at 13–15 wk by an experienced clinician who knows the typical ultrasonographic features of the syndrome. Besides the diagnostics of Finnish cases, from now on, the segregation-based haplotype analysis can be used to address the issue of locus heterogeneity in non-Finnish cases. Most importantly, the precise positioning of the hydrolethalus-syndrome locus in the genome will now allow the disease-causing gene to be cloned and the molecular mechanisms behind this severe developmental disturbance to be revealed.
Acknowledgments
We are grateful to the Finnish families affected by hydrolethalus syndrome for providing data for this study. Drs. Pertti Aula, Riitta Herva, Riitta Karikoski-Leo, and Kalle Simola kindly provided samples from patients and families, and Drs. Vineta Fellman, Tuula Lönnqvist, and Kaisu Nikali provided control samples. We would like to thank Dr. Reijo Norio for his genealogical collaboration. Anne Jokiaho, Mira Kyttälä, and Pekka Ellonen are thanked for their excellent technical assistance. This study was supported by the Academy of Finland and the Ulla Hjelt Fund of the Foundation for Pediatric Research.
Electronic-Database Information
URLs for data in this article are as follows:
- Généthon, http://www.genethon.fr (for marker maps)
- Genome Database, http://gdbwww.gdb.org/ (for the cytogenetic localization of the critical markers)
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/genemap98 (for the New Gene Map of the Human Genome)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
- Stanford Human Genome Center, http://shgc-www.Stanford.edu/ (for raw RH data)
References
- Ämmälä P, Salonen R (1995) First-trimester diagnosis of hydrolethalus syndrome. Ultrasound Obstet Gynecol 5:60–62 [DOI] [PubMed]
- Boehnke M, Lange K, Cox DR (1991) Statistical methods for multipoint radiation hybrid mapping. Am J Hum Genet 49:1174–1188 [PMC free article] [PubMed]
- Cormier-Daire V, Wolf C, Munnich A, Le Merrer M, Nivelon A, Bonneau D, Journel H, et al (1996) Abnormal cholesterol biosynthesis in the Smith-Lemli-Opitz and the lethal acrodysgenital syndromes. Eur J Pediatr 155:656–659 [DOI] [PubMed]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed]
- Fellman V, Rapola J, Pihko H, Varilo T, Raivio KO (1998) Iron-overload disease in infants involving fetal growth retardation, lactic acidosis, liver haemosiderosis, and aminoaciduria. Lancet 351:490–493 [DOI] [PubMed]
- Fitzky BU, Witsch-Baumgartner M, Erdel M, Lee JN, Paik Y-K, Glossmann H, Utermann G, et al (1998) Mutations in the delta-7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc Natl Acad Sci USA 95:8181–8186 [DOI] [PMC free article] [PubMed]
- Golden WL, von Kap-herr C, Kurth B, Wright RM, Flickinger CJ, Eddy R, Shows T, et al (1993) Refinement of the localization of the gene for human intraacrosomal protein SP-10 (ACRV1) to the junction of bands q23àq24 of chromosome 11 by nonisotopic in situ hybridization. Genomics 18:446–449 [DOI] [PubMed]
- Hartikainen-Sorri AL, Kirkinen P, Herva R (1983) Prenatal detection of hydrolethalus syndrome. Prenat Diagn 3:219–224 [DOI] [PubMed]
- Hennekam RCM, van Noort G, de la Fuente AA (1991) Familial holoprosencephaly, heart defects, and polydactyly. Am J Med Genet 41:258–262 [DOI] [PubMed]
- Isola J, DeVries S, Chu L, Ghazvini S, Waldman F (1994) Analysis of changes in DNA sequence copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol 145:1301–1308 [PMC free article] [PubMed]
- Kang S, Allen J, Graham JM, Grebe T, Clericuzio C, Patronas N, Ondrey F, et al (1997a) Linkage mapping and phenotypic analysis of autosomal dominant Pallister-Hall syndrome. J Med Genet 34:441–446 [DOI] [PMC free article] [PubMed]
- Kang S, Graham JM, Olney AH, Biesecker LG (1997b) GLI3 frameshift mutations cause autosomal dominant Pallister-Hall syndrome. Nat Genet 15:266–268 [DOI] [PubMed]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed]
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Monaco C, Helmer Citterich M, Caprini E, Vorechovsky I, Russo G, Croce CM, Barbanti-Brodano G, et al (1998) Molecular cloning and characterization of ZNF202: a new gene at 11q23.3 encoding testis-specific zinc finger proteins. Genomics 52:358–362 [DOI] [PubMed]
- Muenke M, Ruchelli ED, Rorke LB, McDonald-McGinn DM, Orlow MK, Isaacs A, Craparo FJ, et al (1991) On lumping and splitting: a fetus with clinical findings of the oral-facial-digital syndrome type VI, the hydrolethalus syndrome, and the Pallister-Hall syndrome. Am J Med Genet 41:548–556 [DOI] [PubMed]
- Nikali K, Suomalainen A, Terwilliger J, Koskinen T, Weissenbach J, Peltonen L (1995) Random search for shared chromosomal regions in four affected individuals: the assignment of a new hereditary ataxia locus. Am J Hum Genet 56:1088–1095 [PMC free article] [PubMed]
- Norgard M, Yankowitz J, Rhead W, Kanis AB, Hall BD (1996) Prenatal ultrasound findings in hydrolethalus: continuing difficulties in diagnosis. Prenat Diagn 16:173–179 [DOI] [PubMed]
- Norio R (1981) Diseases of Finland and Scandinavia. In: Rothschild HR (ed) Biocultural aspects of disease. Academic Press, New York, pp 359–414 [Google Scholar]
- Norio R, Nevanlinna HR, Perheentupa J (1973) Hereditary diseases in Finland: rare flora in rare soil. Ann Clin Res 5:109–141 [PubMed]
- Paavola P, Salonen R, Weissenbach J, Peltonen L (1995) The locus for Meckel syndrome with multiple congenital anomalies maps to chromosome 17q21-q24. Nat Genet 11:213–215 [DOI] [PubMed]
- Peltonen L (1997) Molecular background of the Finnish disease heritage. Ann Med 29:553–556 [DOI] [PubMed]
- Peltonen L, Pekkarinen P, Aaltonen J (1995) Messages from an isolate: lessons from the Finnish gene pool. Biol Chem Hoppe Seyler 376:697–704 [DOI] [PubMed]
- Pryde PG, Qureshi F, Hallak M, Kupsky W, Johnson MP, Evans MI (1993) Two consecutive hydrolethalus syndrome-affected pregnancies in a nonconsanguinous black couple: discussion of problems in prenatal differential diagnosis of midline malformation syndromes. Am J Med Genet 46:537–541 [DOI] [PubMed]
- Roume J, Genin E, Cormier-Daire V, Ma HW, Mehaye B, Attie T, Razavi-Encha F, et al (1998) A gene for Meckel syndrome maps to chromosome 11q13. Am J Hum Genet 63:1095–1101 [DOI] [PMC free article] [PubMed]
- Salonen R, Herva R, Norio R (1981) The hydrolethalus syndrome: delineation of a "new," lethal malformation syndrome based on 28 patients. Clin Genet 19:321–330 [DOI] [PubMed]
- Salonen R, Herva R (1990) Hydrolethalus syndrome. J Med Genet 27:756–759 [DOI] [PMC free article] [PubMed]
- Salonen R, Paavola P (1998) Meckel syndrome. J Med Genet 35:497–501 [DOI] [PMC free article] [PubMed]
- Sharma AK, Phadke S, Chandra K, Upreti M, Khan EM, Naveed M, Agarwal SS (1992) Overlap between Majevski and hydrolethalus syndromes: a report of two cases. Am J Med Genet 43:949–953 [DOI] [PubMed]
- Sheffield VC, Weber JL, Buetow KH, Murray JC, Even DA, Wiles K, Gastier JM, et al (1995) A collection of tri and tetranucleotide repeat markers used to generate high quality, high resolution human genome-wide linkage maps. Hum Mol Genet 4:1837–1844 [DOI] [PubMed]
- Terwilliger J (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed]
- Varilo T, Nikali K, Suomalainen A, Lönnqvist T, Peltonen L (1996) Tracing an ancestral mutation: genealogical and haplotype analysis of the infantile onset spinocerebellar ataxia locus. Genome Res 6:870–875 [DOI] [PubMed]
- Verloes A (1995) Numerical syndromology: a mathematical approach to the nosology of complex phenotypes. Am J Med Genet 55:433–443 [DOI] [PubMed]
- Visapää I, Fellman V, Varilo T, Palotie A, Raivio KO, Peltonen L (1998) Assignment of the locus for a new lethal neonatal metabolic syndrome to 2q33-37. Am J Hum Genet 63:1396–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassif CA, Maslen C, Kachilele-Linjewile S, Lin D, Linck LM, Connor WE, Steiner RD, et al (1998) Mutations in the human sterol delta7-reductase gene at 11q12-13 cause Smith-Lemli-Opitz syndrome. Am J Hum Genet 63:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Wijburg FA, Hennekam RC, Vreken P, Poll-The BT, Dorland L, Duran M, et al (1998) Smith-Lemli-Opitz syndrome is caused by mutations in the 7-dehydrocholesterol reductase gene. Am J Hum Genet 63:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]