Summary
Fibrodysplasia ossificans progressiva (FOP) is a severely disabling, autosomal-dominant disorder of connective tissue and is characterized by postnatal progressive heterotopic ossification of muscle, tendon, ligament, and fascia and by congenital malformation of the great toes. To identify the chromosomal location of the FOP gene, we conducted a genomewide linkage analysis, using four affected families with a total of 14 informative meioses. Male-to-male transmission of the FOP phenotype excluded X-linked inheritance. Highly polymorphic microsatellite markers covering all human autosomes were amplified by use of PCR. The FOP phenotype is linked to markers located in the 4q27-31 region (LOD score 3.10 at recombination fraction 0). Crossover events localize the putative FOP gene within a 36-cM interval bordered proximally by D4S1625 and distally by D4S2417. This interval contains at least one gene involved in the bone morphogenetic protein–signaling pathway.
Introduction
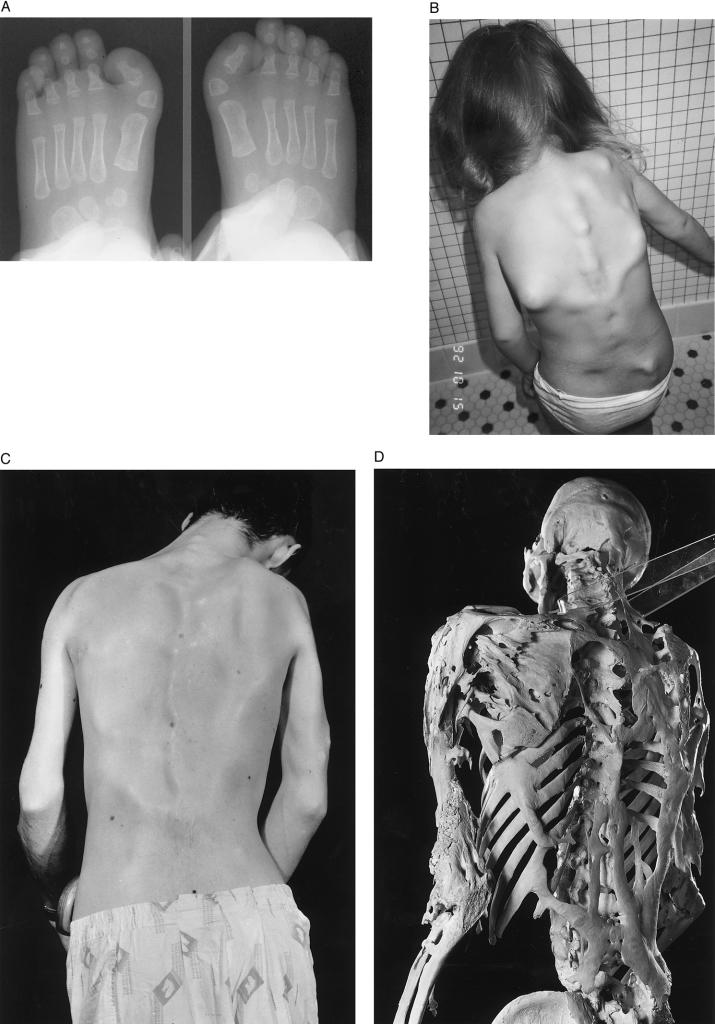
Fibrodysplasia ossificans progressiva (FOP [MIM 135100]) is a rare, severely disabling, autosomal-dominant disorder characterized by progressive postnatal heterotopic ossification and by congenital malformation of the great toes (fig. 1A and C; Schroeder and Zasloff 1980; Connor and Evans 1982a, 1982b; Connor et al. 1993; Kaplan et al. 1993a, 1998; Smith 1998). Frequently, the first sites of heterotopic bone formation in children occur within fibroproliferative lesions on the upper back and neck (fig. 1B). Heterotopic ossification in patients with FOP occurs in predictable anatomic patterns (Kaplan et al. 1990; Cohen et al. 1993; Rocke et al. 1994) and leads to extra-articular bony ankylosis of nearly all joints of the axial and appendicular skeleton (Connor and Evans 1982a; Cohen et al. 1993; Kaplan et al. 1993b; Rocke et al. 1994; Shah et al. 1994), which renders movement impossible (fig. 1C). Death results most often from complications of severe restrictive–chest-wall disease (Connor and Evans 1982a; Kussmaul et al. 1998).
Figure 1.
Clinical findings in FOP. A, Radiographs of the feet at age 4 mo in a child with FOP. The soft-tissue shadows clearly outline the short in-turned great toes due to malformed delta-shaped proximal phalanges (arrowheads), one of the common variations of great-toe malformations seen in FOP. B, Multiple variable-size soft-tissue nodules (preosseous fibroproliferative lesions) on the back of a 4-year-old child with FOP. These fibroproliferative lesions are frequently the first indication of heterotopic ossification in a child. C and D, Clinical appearance and skeleton of a man with FOP. The rigid posture in this 25-year-old man was due to ankylosis of the spine, shoulders, and elbows. He died of pneumonia at age 40 years. Plates and ribbons of ectopic bone contour the skin over the back and arms (C) and can be seen directly on the skeleton (D). (Courtesy of the Mutter Museum, College of Physicians of Philadelphia; reproduced with permission from Shafritz et al. [1996].)
Heterotopic ossification in FOP is induced by minor injury to the soft tissues but can occur without detectable trauma (Connor and Evans 1982a; Janoff et al. 1994; Luchetti et al. 1996; Glaser et al. 1998). The earliest pathological finding is a perivascular lymphocytic infiltration into normal-appearing skeletal muscle (Gannon et al. 1998). The early and transient inflammatory phase is followed by muscle-cell degeneration and by highly vascular fibroproliferative soft-tissue swelling that is indistinguishable histologically from aggressive juvenile fibromatosis (Gannon et al. 1998). The fibroproliferative lesions evolve, through an endochondral process, into mature lamellar bone with marrow elements (Kaplan et al. 1993b; Gannon et al. 1998).
Endochondral bone formation during embryogenesis is first evidenced by the condensation of mesenchymal cells, followed by the differentiation of these cells into chondrocytes and, subsequently, by the replacement of cartilage by bone. Recent investigations have identified transcription factors—such as Cbfa1 (Ducy et al. 1997; Komori et al. 1997; Mundlos et al. 1997; Otto et al. 1997) and Sox9 (Bi et al. 1999)—that direct bone and cartilage formation. Little is known, however, about the signals that initiate the differentiation of cells toward a chondrogenic or osteogenic pathway. The genetic defect in FOP is likely to be such an inductive signal.
The molecular cause and pathogenesis of FOP remain unknown. Overexpression of bone morphogenetic protein 4 (BMP-4) in immortalized lymphoblastoid cells (Shafritz et al. 1996; Lanchoney et al. 1998) and in lesional fibroproliferative cells (Gannon et al. 1997) has recently been associated with the condition. The elevation of steady-state levels of BMP-4 in FOP is due to an increased rate of BMP-4 transcription (Olmsted et al. 1996). However, BMP-4 expression is not elevated above normal levels in all cell lines from patients with FOP (Shafritz et al. 1996; Virdi et al. 1999), and mutations in the BMP-4 gene have not been identified in patients with FOP (Xu and Shore 1998), indicating that the molecular cause lies elsewhere, perhaps in a BMP-4 signaling–pathway component that stimulates expression of the BMP-4 gene or that impairs the action of an inhibitor.
FOP is a rare condition; the prevalence is ∼0.6/1 million live births (Connor and Evans 1982b; Delatycki and Rogers 1998). Reproductive fitness is low, and most cases appear to arise by spontaneous mutation (Kaplan et al. 1993a). The present study describes a genomewide linkage analysis in four small families containing individuals affected with FOP and demonstrates locus homogeneity and linkage of the FOP gene to a 36-cM segment on the long arm of human chromosome 4.
Subjects and Methods
Subjects
Four small multigenerational families with FOP participated in this study (pedigrees are shown in fig. 2): FOP 1 (Kaplan et al. 1993a), FOP 2 (Connor et al. 1993), and FOP 4 (Janoff et al. 1995) have been described elsewhere; FOP 3 is newly documented in the present report. All participants were evaluated clinically by one or more of the authors of the present report, and the FOP status was ascertained before molecular studies were initiated. Informed consent was obtained in accordance with the standards set by local institutional review boards.
Figure 2.
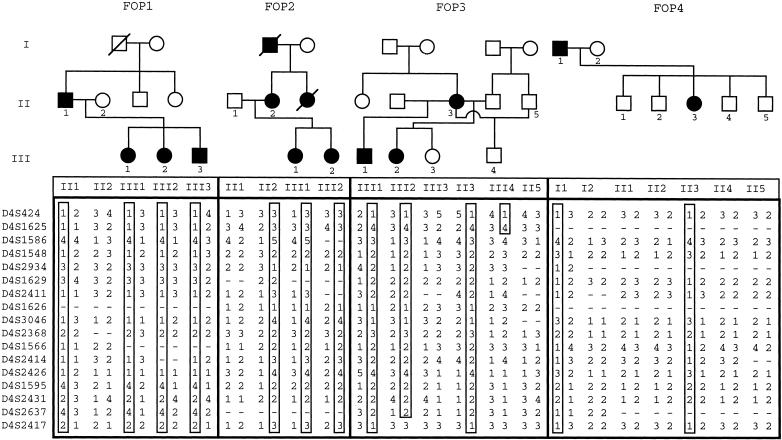
Pedigrees of families with FOP with genotypes for markers from chromosome 4q27-31. Standard pedigree notation is used: circles and squares denote females and males, respectively; blackened symbols indicate affected individuals; and generations are indicated by Roman numerals to the left of the pedigrees (only individuals from whom genotyping data was obtained are numbered). Haplotypes for the markers shown on the left are listed below the symbols for the family members. The haplotype cosegregating with the disorder in each family is boxed.
Genotyping
Peripheral blood was obtained from the subjects, and, by use of standard protocols (Sambrook et al. 1989), genomic DNA was isolated directly either from peripheral blood leukocytes or from Epstein-Barr virus–transformed lymphocyte cell lines. A set of 240 highly polymorphic microsatellite markers (Human MapPairs versions 3a and 8a; Research Genetics), spaced ⩽25 cM apart and spanning all human autosomes, were amplified in four families with FOP (families 1–4 in fig. 2). For fine mapping, 25 additional markers were identified from genetic maps of chromosome 4q (by use of the Research Genetics database) and were amplified. DNA samples were analyzed by PCR amplification using one to four primer pairs per reaction. Each reaction contained (in a final volume of 10 μl) 30 ng of genomic DNA, 10 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 0.01% gelatin, 200 μM each dTTP, dGTP, and dATP, 2.5 μM dCTP, 0.35 nCi α-[32P]dCTP, 1.5 mmol of each primer, and 0.15 U of Taq polymerase (PE Biosystems). Thermal cycling (Hybaid Omnigene thermal cycler) consisted of 27 cycles of 94°C for 30 s, 55°C for 75 s, and 72°C for 15 s. The amplified PCR products were denatured (at 95°C for 5 min) and were separated by sized markers on a 6% polyacrylamide 7.7-M urea gel. The gels were exposed to autoradiography film (XAR; Kodak) at −70°C, with intensifying screens, for 6–48 h. Autoradiograms were scored independently by at least two investigators.
Linkage Analysis
Two-point linkage analysis was performed by the MLINK program by use of data formatted by the Cyrillic program (Cherwell Scientific). On the basis of clinical experience with these families, penetrance of the FOP gene was assumed to be 100%. Multipoint analyses were conducted with the GENEHUNTER program (Kruglyak et al. 1996). Because of the consistent pattern of inheritance based on the pedigrees, a single model of a dominant highly penetrant trait was used. The disease-gene frequency was assigned a value of 6.0×10-7 on the basis of previous studies. Haplotypes were reconstructed from the genotype data, by use of the Cyrillic program.
Radiation-Hybrid Mapping
The Massachusetts Institute of Technology (MIT)/Whitehead Institute GeneBridge 4 radiation-hybrid map (Whitehead Institute) and National Center for Biotechnology Information (NCBI) databases were examined for previously localized positions of markers and candidate genes in the linked region. The positions of some additional markers and candidate genes were verified by use of the GeneBridge 4 Radiation Hybrid Panel (Research Genetics). Each 10-μl PCR contained 50 ng of hybrid template DNA, 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 μM each dNTP, 2.5 mM MgCl2, 3 × MasterAmp Enhancer (Epicenter Technologies), 300 mM each forward and reverse primer, and 0.3 U of Taq DNA polymerase (Life Technologies). Primers for SMAD1 were 5′-CTGCTGTCCTTTTGCATTTGGA (forward) and 5′-TTCATGGTGGC-TCTGAAGATCGG (reverse). PCR amplification used 36 cycles of 30 s at 94°C, 75 s at 58°C, and 15 s at 72°C, plus a final 6 min at 72°C, in an MJ Reseach PTC-200. Each reaction was electrophoresed through 1% agarose gels in TAE buffer (40 mM Tris-HCl [pH 7.5], 20 mM sodium acetate, 1 mM EDTA). Positive and negative amplification was scored for all 93 hybrid clones, and the data were submitted to the GeneBridge 4 mapping service at the Whitehead Institute.
Results
To identify the chromosomal locus of the FOP gene, a genomewide linkage analysis using four unrelated families with two or more affected individuals was conducted (fig. 2). In each family, penetrance was complete, and the inheritance pattern was consistent with autosomal-dominant transmission. Family 1 demonstrated male-to-male transmission of the FOP phenotype, thereby excluding X-linked transmission. Linkage data from polymorphic microsatellite markers for all autosomes showed that only markers in the 4q27-31 region segregated with the affected phenotype with no recombination in each of the four families.
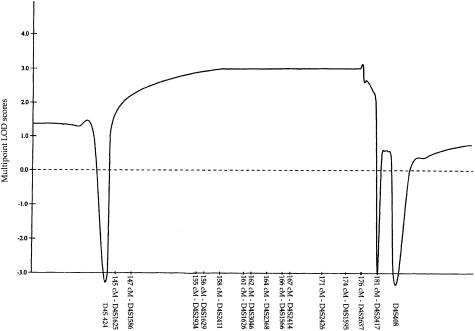
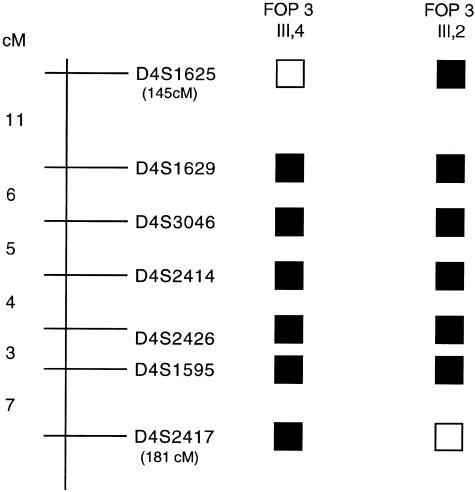
Fine mapping of the 4q27-31 region was conducted with use of additional polymorphic markers within this interval. The two-point LOD-score calculation for the combined family data (table 1) produced a maximum LOD score (Zmax) of 3.31 with D4S1548 and D4S2426 markers at a recombination fraction (θ) of 0. Multipoint analysis resulted in a multipoint Zmax of 3.10 within the linked interval (fig. 3). Boundaries of the linked region were established by crossover events for marker D4S1625 (map position 145 cM) in individual III-4 in family 3 and for marker D4S2417 (map position 181 cM) in individual III-2 in family 3, placing the putative FOP gene within a 36-cM interval on the long arm of human chromosome 4 (fig. 4).
Table 1.
Combined Two-Point LOD Scores for FOP and Chromosome 4 Markers
|
LOD Score at θ= |
|||||||||||||
| Markera | .00 | .05 | .10 | .15 | .20 | .25 | .30 | .35 | .40 | .45 | .50 | θmax | Zmax |
| D4S424 | −4.3 × 1019 | 1.16 | 1.21 | 1.12 | .98 | .80 | .60 | .39 | .20 | .05 | .00 | .10 | 1.21 |
| D4S1625 | −4.3 × 1019 | −.16 | .07 | .17 | .21 | .23 | .22 | .18 | .14 | .08 | .00 | .25 | .23 |
| D4S1586 | 2.41 | 2.19 | 1.95 | 1.70 | 1.44 | 1.17 | .88 | .59 | .32 | .11 | .00 | .00 | 2.41 |
| D4S1548 | 3.31 | 3.00 | 2.67 | 2.33 | 1.96 | 1.59 | 1.2 | .81 | .45 | .16 | .00 | .00 | 3.31 |
| D4S2934 | 1.51 | 1.33 | 1.15 | .97 | .78 | .60 | .43 | .27 | .13 | .03 | .00 | .00 | 1.51 |
| D4S1629 | 1.81 | 1.63 | 1.44 | 1.24 | 1.04 | .82 | .61 | .39 | .20 | .05 | .00 | .00 | 1.81 |
| D4S2411 | 2.11 | 1.91 | 1.70 | 1.47 | 1.24 | 1.00 | .75 | .51 | .28 | .10 | .00 | .00 | 2.11 |
| D4S1626 | .90 | .81 | .72 | .62 | .52 | .41 | .30 | .19 | .09 | .03 | .00 | .00 | .90 |
| D4S3046 | 3.01 | 2.72 | 2.42 | 2.10 | 1.76 | 1.42 | 1.07 | .72 | .41 | .15 | .00 | .00 | 3.01 |
| D4S2368 | .54 | .47 | .41 | .34 | .27 | .20 | .14 | .08 | .04 | .01 | .00 | .00 | .54 |
| D4S1566 | 2.11 | 1.91 | 1.70 | 1.47 | 1.24 | .99 | .73 | .48 | .24 | .07 | .00 | .00 | 2.11 |
| D4S2414 | 2.34 | 2.12 | 1.89 | 1.65 | 1.39 | 1.13 | .86 | .59 | .33 | .13 | .00 | .00 | 2.34 |
| D4S2426 | 3.31 | 3.00 | 2.67 | 2.33 | 1.96 | 1.59 | 1.20 | .81 | .45 | .16 | .00 | .00 | 3.31 |
| D4S1595 | 2.64 | 2.36 | 2.06 | 1.76 | 1.44 | 1.13 | .81 | .51 | .26 | .07 | .00 | .00 | 2.64 |
| D4S2431 | 2.71 | 2.44 | 2.16 | 1.87 | 1.56 | 1.23 | .90 | .58 | .29 | .80 | .00 | .00 | 2.71 |
| D4S2637 | −.16 | .21 | .30 | .31 | .28 | .23 | .17 | .10 | .05 | .01 | .00 | .15 | .31 |
| D4S2417 | −4.3 × 1019 | 1.38 | 1.43 | 1.31 | 1.16 | .97 | .76 | .54 | .32 | .12 | .00 | .10 | 1.43 |
Order shown is from centromere to telomere.
Figure 3.
Combined multipoint LOD scores for the markers in the 4q27-31 region in four families with FOP. Multipoint LOD scores were calculated for each of the markers indicated along the X-axis. The relative genetic distances of these markers, measured (in cM) from 4pter, were derived from the Research Genetics database.
Figure 4.
Localization of the FOP-linked interval on the long arm of human chromosome 4. Schematic representation of the 4q27-31 region shows some of the markers used in the genetic linkage analysis. The genetic distances between the markers are indicated and the cumulative genetic distances (in cM; pter is 0 cM) for the markers defining the proximal and distal boundaries of the linked interval are shown in parentheses, under the marker names. Blackened squares denote regions of allele sharing among affected individuals; open squares represent recombinant regions. Crossovers in individual III-4 from family 3 identified the proximal boundary, and those in individual III-2 from family 3 identified the distal boundary.
Examination of the 4q27-31 region identified several plausible FOP candidate genes previously localized within or near this interval, including SMAD1, IL-15, 15-hydroxyprostaglandin dehydrogenase (PGDH), LEF1, and a BMP receptor (BMPRIB). To determine whether these genes are within the linked interval, the markers at the proximal and distal borders of the linked region, D4S1625 and D4S2417, were localized on the physical radiation-hybrid map. The Genebridge 4 map from the Radiation Hybrid Mapping Web site of the MIT/Whitehead Institute and the NCBI GeneMap 1999 databases were used; unless indicated, cR values refer to the NCBI map. D4S1625 maps between 615.30 and 618.92 cR (669.6 cR on the MIT/Whitehead Institute map; 145 cM), whereas D4S2417 was identified at 676.27 cR (739.7 cR on the MIT/Whitehead Institute map; 181 cM). Radiation-hybrid mapping confirmed that SMAD1, a leading candidate gene, which encodes a critical intracellular component of the BMP-signaling pathway (Hoodless et al. 1996), is located within the FOP-linked interval of chromosome 4q at 623.30 cR (675.6 cR on the MIT/Whitehead Institute map; 145 cM), consistent with results reported elsewhere (Lechleider et al. 1996). PGDH has been localized close to the distal border within the linked region, at 671.36–673.33 cR (179 cM), and IL-15 has been mapped at 615.3 cR (142 cM) near the proximal border. BMPRIB and LEF1 map outside the linkage region, according to the GeneMap 1999 database. Note that actual ordering of genes is pending verification of radiation-hybrid mapping data.
Discussion
Linkage data from a genomewide screen support the localization of the gene that causes FOP to 4q27-31, a 36-cM interval on human chromosome 4. No recombination between markers in this region and the FOP phenotype was observed, and no genetic heterogeneity was observed in the four families studied, despite previous observation of clinical and biochemical heterogeneity (Janoff et al. 1995; Shafritz et al. 1996; Virdi et al. 1999). The catastrophic disability of FOP leads to low reproductive fitness and a paucity of multigenerational families (Connor and Evans 1982b; Delatycki and Rogers 1998; Kaplan et al. 1998). Although the number of informative meioses in this study was sufficient to demonstrate linkage (LOD score >3) of FOP within the 4q27-31 region, additional families that show crossovers within this region are needed in order to narrow the target interval before productive positional-cloning efforts can be accomplished, although candidate genes in the interval can be examined for mutations.
BMP-4, a powerful osteogenic morphogen and a member of the transforming-growth factor–β superfamily (Wozney et al. 1988; Massagué 1990; Kingsley 1994; Hogan 1996a, 1996b), has been proposed as a candidate gene for FOP (Kaplan et al. 1990), because it is selectively overexpressed in immortalized lymphoblastoid cell lines and in preosseous fibroproliferative cells from patients with FOP (Olmsted et al. 1996; Shafritz et al. 1996; Gannon et al. 1997; Lanchoney et al. 1998). Previous examination of the DNA sequence of the BMP-4 gene alleles of patients with FOP showed no FOP-associated mutations (Xu and Shore 1998). Furthermore, the present report excludes linkage of the FOP phenotype to chromosome 14, the location of the BMP-4 gene. However, SMAD1, a BMP pathway–specific gene involved in cytoplasmic and nuclear signal transduction (Kretzschmar and Massagué 1998), is located on human chromosome 4, within the FOP-linked interval and is a plausible candidate gene for FOP.
Since there are >100 anonymous transcripts in the 4q27-31–linked interval, the FOP gene may be an as-yet-undescribed gene in a pathway that influences BMP activity. However, in addition to SMAD1, several promising candidate genes in the linkage region warrant consideration. Histopathological examination of lesion biopsies from patients with FOP provides insight into the pathobiology of FOP and suggests a rationale for considering other plausible candidate genes within the linked region. The most prominent feature of an early FOP lesion is intense perivascular lymphocytic infiltration into normal-appearing skeletal muscle, followed by the appearance of fibroproliferative swellings with intense angiogenesis (Gannon et al. 1998). IL-15 (which maps near the proximal border of the FOP candidate region), a cytokine expressed in endothelial cells, has been shown to initiate angiogenesis and to play a role in T-cell and B-cell growth and differentiation (Kirman et al. 1998). NFκB, a transcription factor with a major role in BMP-4 repression during chick limb development, has an activating binding site on the IL-15 promoter (Azimi et al. 1998).
NAD-dependent 15-PGDH is a member of the short-chain alcohol dehydrogenase enzymes known to catalyze the reversible inactivation of prostaglandins, powerful mediators of the inflammatory response (Duax and Ghosh 1997; Pichaud et al. 1997b). This enzyme has recently been shown to play a role in vitamin D3–induced macrophage/osteoclast differentiation (Pichaud et al. 1997a) and is a plausible candidate gene for FOP, because of the known osteogenic effects of prostaglandins, specifically PGE-2, and because of the reported elevation of a PGE-2–like molecule in the serum of patients with FOP (Levitz et al. 1992).
In summary, we present evidence that the FOP gene is located on human chromosome 4, within the 4q27-31 region. The identification of the mutated gene that causes FOP will illuminate our understanding of the genetic, molecular, and cellular pathways that regulate osteoblast-cell differentiation and the de novo formation of bone and will, in addition, provide a foundation for development of treatments not only for FOP but also for the more common disorders of osteogenesis.
Acknowledgments
The authors gratefully acknowledge valuable scientific discussions with our colleagues Drs. Gregory Hahn, Victor McKusick, Richard Spielman, Jeffrey Tabas, and Michael Zasloff and the valuable technical contributions of Heather Mitchell and Mei-qi Xu. This work was supported in part by grants from the International Fibrodysplasia Ossificans Progressiva Association, the Ian Cali Fund for FOP Research, the Isaac and Rose Nassau Professorship of Orthopaedic Molecular Medicine, the National Institutes of Health (grant 2R01-AR41916-04), the European Neuromuscular Center, the Association Française Contre Les Myopathies, and the Medical Research Council of Great Britain.
Footnotes
These authors have contributed equally to this work.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- GeneMap 1999, http://www.ncbi.nlm.nih.gov/genemap/ (for localization of genes and expressed-sequence tags)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for FOP [MIM 135100]) [PubMed]
- Radiation Hybrid Mapping, http://carbon.mit.edu:8000/cgi-bin/contig/rhmapper.pl (for radiation-hybrid mapping)
- Research Genetics, Huntsville AL, http://www.resgen.com (for linkage analysis)
References
- Azimi N, Brown K, Bamford RN, Tagaya Y, Siebenlist U, Waldmann TA (1998) Human T cell lymphotropic virus type I Tax protein trans-activates interleukin 15 gene transcription through an NF-κB site. Proc Natl Acad Sci USA 95:2452–2457 [DOI] [PMC free article] [PubMed]
- Bi W, Deng JM, Zhang Z, Behringer RR, De Crombrugghe B (1999) Sox9 is required for cartilage formation. Nat Genet 22:85–89 [DOI] [PubMed]
- Cohen RB, Hahn GV, Tabas JA, Peeper J, Levitz CL, Sando A, Sando N, et al (1993) The natural history of heterotopic ossification in patients who have fibrodysplasia ossificans progressiva. A study of forty-four patients. J Bone Joint Surg Am 75:215–219 [DOI] [PubMed]
- Connor JM, Evans DAP (1982a) Fibrodysplasia ossificans progressiva: the clinical features and natural history of 34 patients. J Bone Joint Surg Br 64:76–83 [DOI] [PubMed]
- ——— (1982b) Genetic aspects of fibrodysplasia ossificans progressiva. J Med Genet 19:35–39 [DOI] [PMC free article] [PubMed]
- Connor JM, Skirton H, Lunt PW (1993) A three generation family with fibrodysplasia ossificans progressiva. J Med Genet 30:687–689 [DOI] [PMC free article] [PubMed]
- Delatycki M, Rogers JG (1998) The genetics of fibrodysplasia ossificans progressiva. Clin Orthop 346:15–18 [PubMed]
- Duax WL, Ghosh D (1997) Structure and function of steroid dehydrogenases involved in hypertension, fertility, and cancer. Steroids 62:95–100 [DOI] [PubMed]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747–754 [DOI] [PubMed]
- Gannon FH, Kaplan FS, Olmsted E, Finkel GC, Zasloff M, Shore E (1997) Bone morphogenetic protein (BMP) 2/4 in early fibromatous lesions of fibrodysplasia ossificans progressiva. Hum Pathol 28:339–343 [DOI] [PubMed]
- Gannon FH, Valentine BA, Shore EM, Zasloff MA, Kaplan FS (1998) Acute lymphocytic infiltration in extremely early lesions of fibrodysplasia ossificans progressiva. Clin Orthop 346:19–25 [PubMed]
- Glaser DL, Rocke DM, Kaplan FS (1998) Catastrophic falls in patients who have fibrodysplasia ossificans progressiva. Clin Orthop 346:110–116 [PubMed]
- Hogan BL (1996a) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 10:1580–1594 [DOI] [PubMed]
- ——— (1996b) Bone morphogenetic proteins in development. Curr Opin Genet Dev 6:432–438 [DOI] [PubMed]
- Hoodless PA, Haerry T, Abdollah S, Stapleton M, O'Connor MB, Attisano L, Wrana JL (1996) Madr1, a Mad-related protein that functions in Bmp2 signaling pathways. Cell 85:489–500 [DOI] [PubMed]
- Janoff HB, Tabas JA, Shore EM, Muenke M, Dalinka MK, Schlesinger S, Zasloff MA, et al (1995) Mild expression of fibrodysplasia ossificans progressiva: a report of 3 cases. J Rheumatol 22:976–978 [PubMed]
- Janoff HB, Zasloff M, Kaplan FS (1996) Submandibular swelling in patients with fibrodysplasia ossificans progressiva. Otolaryngol Head Neck Surg 114:599–604 [DOI] [PubMed]
- Kaplan FS, Delatycki M, Gannon FH, Rogers JG, Smith R, Shore EM (1998) Fibrodysplasia ossificans progressiva. In Emery, AEH (ed) Neuromuscular disorders: clinical and molecular genetics. John Wiley & Sons, Chichester, England, pp 289–321 [Google Scholar]
- Kaplan FS, McCluskey W, Hahn G, Tabas JA, Muenke M, Zasloff MA (1993a) Genetic transmission of fibrodysplasia ossificans progressiva: report of a family. J Bone Joint Surg Am 75:1214–1220 [DOI] [PubMed]
- Kaplan FS, Tabas JA, Gannon FH, Finkel G, Hahn GV, Zasloff MA (1993b) The histopathology of fibrodysplasia ossificans progressiva: an endochondral process. J Bone Joint Surg Am 75:220–230 [DOI] [PubMed]
- Kaplan FS, Tabas JA, Zasloff MA (1990) Fibrodysplasia ossificans progressiva: a clue from the fly? Calcif Tissue Int 47:117–125 [DOI] [PubMed]
- Kingsley DM (1994) The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev 8:133–146 [DOI] [PubMed]
- Kirman I, Vainer B, Nielsen OH (1998) Interleukin-15 and its role in chronic inflammatory diseases. Inflamm Res 47:285–289 [DOI] [PubMed]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, et al (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764 [DOI] [PubMed]
- Kretzschmar M, Massagué J (1998) SMADs: mediators and regulators of TGF-beta signaling. Curr Opin Genet Dev 8:103–111 [DOI] [PubMed]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed]
- Kussmaul WG, Esmail AN, Sagir Y, Ross J, Gregory S, Kaplan FS (1998) Pulmonary and cardiac function in advanced fibrodysplasia ossificans progressiva. Clin Orthop 346:104–109 [PubMed]
- Lanchoney TF, Olmsted EA, Shore EM, Gannon FH, Zasloff MA, Rosen V, Kaplan FS (1998) Characterization of bone morphogenetic protein-4 receptors in fibrodysplasia ossificans progressiva. Clin Orthop 346:38–45 [PubMed]
- Lechleider RJ, de Caestecker MP, Dehejia A, Polymeropoulos MH, Roberts AB (1996) Serine phosphorylation, chromosomal localization, and transforming growth factor-beta signal transduction by human bsp-1. J Biol Chem 271:17617–17620 [DOI] [PubMed]
- Levitz CL, Cohen RB, Zasloff MA, Kaplan FS (1992) The role of prostaglandins in the pathogenesis of fibrodysplasia ossificans progressiva. Calcif Tissue Int 50:3871571850
- Luchetti W, Cohen RB, Hahn GV, Rocke DM, Helpin M, Zasloff M, Kaplan FS (1996) Severe restriction in jaw movement after routine injection of local anesthetic in patients who have fibrodysplasia ossificans progressiva. Oral Surg Oral Med Oral Pathol Oral Radiol Endo 81:21–25 [DOI] [PubMed]
- Massagué J (1990) The transforming growth factor-beta family. Annu Rev Cell Biol 6:597–641 [DOI] [PubMed]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, et al (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773–779 [DOI] [PubMed]
- Olmsted EA, Liu C, Haddad JG, Shore EM, Kaplan FS (1996) Characterization of mechanisms controlling bone morphogenetic protein-4 message expression in fibrodysplasia ossificans progressiva. J Bone Miner Res 11:S164 [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, et al (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771 [DOI] [PubMed]
- Pichaud F, Delage-Mourroux R, Frenkian M, Frendo JL, Roux S, de Vernejoul MC, Jullienne A (1997a) Type-I 15-hydroxyprostaglandin dehydrogenase: role in macrophage/osteoclast differentiation. Adv Exp Med Bio 433:399–402 [DOI] [PubMed]
- Pichaud F, Delage-Mourroux R, Pidoux E, Jullienne A, Rousseau-Merck MF (1997b) Chromosomal localization of the type-I 15-PGDH gene to 4q34-q35. Hum Genet 99:279–281 [DOI] [PubMed]
- Rocke DM, Zasloff M, Peeper J, Cohen RB, Kaplan FS (1994) Age- and joint-specific risk of initial heterotopic ossification in patients who have fibrodysplasia ossificans progressiva. Clin Orthop 301:243–248 [PubMed]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Schroeder HW Zasloff M (1980) The hand and foot malformations in fibrodysplasia ossificans progressiva. Johns Hopkins Med J 147:73–78 [PubMed]
- Shafritz AB, Shore EM, Gannon FH, Zasloff MA, Taub R, Muenke M, Kaplan FS (1996) Overexpression of an osteogenic morphogen in fibrodysplasia ossificans progressiva. N Engl J Med 335:555–561 [DOI] [PubMed]
- Shah PB, Zasloff MA, Drummond D, Kaplan FS (1994) Spinal deformity in patients who have fibrodysplasia ossificans progressiva. J Bone Joint Surg Am 76:1442–1450 [DOI] [PubMed]
- Smith R (1998) Fibrodysplasia (myositis) ossificans progressiva: clinical lessons from a rare disease. Clin Orthop 346:7–14 [PubMed]
- Virdi AS, Shore EM, Oreffo ROC, Li M, Connor JM, Smith R, Kaplan FS, et al (1999) Phenotypic and molecular heterogeneity in fibrodysplasia ossificans progressiva. Calcif Tissue Int 65:250–255 [DOI] [PubMed]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, et al (1988) Novel regulators of bone formation: molecular clones and activities. Science 242:1528–1534 [DOI] [PubMed]
- Xu M, Shore EM (1998) Mutational screening of the bone morphogenetic protein 4 gene in a family with fibrodysplasia ossificans progressiva. Clin Orthop 346:53–58 [PubMed]