Abstract
A method has been developed to follow fusion of individual pseudotyped virus expressing HIV-1 Env to cells by time-resolved fluorescence microscopy. Viral envelopes were labeled with a fluorescent lipid dye (DiD) and virus content was rendered visible by incorporating a Gag-GFP chimera. The Gag-GFP is naturally cleaved to the much smaller NC-GFP fragment in the mature virions. NC-GFP was readily released upon permeabilization of the viral envelope, whereas the capsid was retained. The NC-GFP thus provides a relatively small and mobile aqueous marker to follow viral content transfer. In fusion experiments, virions were bound to cells at low temperature, and fusion was synchronously triggered by a temperature jump. DiD transferred from virions to cells without a significant lag after the temperature jump. Some virions released DiD but retained NC-GFP. Surprisingly, the fraction of lipid mixing events yielding NC-GFP transfer was dependent on the type of target cell: of three infectable cell lines, only one permitted NC-GFP transfer within minutes of raising temperature. NC-GFP release did not correlate with the level of CD4 or coreceptor expression in the target cells. The data indicate that fusion pores formed by HIV-1 Env can remain small for a relatively long time before they enlarge.
INTRODUCTION
In the process of membrane fusion, two continuities are created: separate membranes join and distinct aqueous compartments become one. To identify the mechanisms of membrane fusion, both continuities should be followed and the temporal order in which they occur identified. In the case of the HIV-1 fusion protein Env, mechanistic studies have largely relied on fusion of cells expressing the protein to cells expressing CD4 and cognate chemokine receptors (e.g., Munoz-Barroso et al., 1998; Melikyan et al., 2000; Reeves et al., 2002; Abrahamyan et al., 2003; Gallo et al., 2003; Markosyan et al., 2003). But monitoring lipid dye spread in HIV-1 Env-mediated cell-cell fusion has not proved to be very useful in following membrane continuity, for several reasons. The dye does not reliably move between cells, and when it does move, the time course is slow, in large part because the dye segregates nonuniformly over the cell membrane. Also, the dye enters intracellular pools, making it difficult to follow transfer between fused membranes. Furthermore, in cell-cell fusion, the two bound cells are in contact over an appreciable area, so fusion could potentially occur at many sites (Frolov et al., 2000; Leikina and Chernomordik, 2000); the sites of origin for the observed lipid and aqueous dye spread could well be different.
By studying fusion of viral particles to cells, these problems can be overcome. The small size of virus minimizes the area of contact and accurately reflects the biological situation. Also, membrane retrieval processes that internalize lipid dye are absent in virions, and the viral envelope is simpler than a cell membrane. But until recently, suitable assays to follow both lipid and contents mixing and to time-order these movements in virus-cell fusion were not available. However, the mixing of lipid or contents, but not both, has often been measured. Viral envelopes have been labeled by fluorescent lipid dyes and release of the dye has been followed in population studies (Dimitrov et al., 1992; Stegmann et al., 1993; Chatterjee et al., 2000; Corver et al., 2000; Earp et al., 2003). When release of viral contents alone is measured, levels of infection, rather than fusion itself, have traditionally been measured. Because a multitude of steps occur downstream of the fusion step, only a small fraction of the fused virus particles may cause infection, and thus infection levels need not correspond to fusion levels (Tobiume et al., 2003; Daecke et al., 2005). In the past few years, assays have been developed that directly monitor the release of virus contents into a host cell subsequent to virus-cell fusion, and these provide a direct measure of fusion. For example, one can introduce β-lactamase into the viral core (Cavrois et al., 2002; Tobiume et al., 2003; Barnard et al., 2004; Wyma et al., 2004; Daecke et al., 2005) or associate luciferase with the viral envelope proteins (Kolokoltsov and Davey, 2004) and quantify fusion from measures of these enzymatic activities within cells. But these assays have limited temporal resolution, and the transfer of enzymes is not measured in real time. Methods to track the movements of fluorescently labeled single virions by fluorescence microscopy have succeeded. Using influenza virus with fluorescently labeled envelope, the viral pathway through cytosol, starting from the uptake into endosomes, followed by lipid mixing into the endosomal membrane, has been microscopically tracked (Lakadamyali et al., 2003). Individual HIV particles have been tracked, delineating reverse transcription steps and trafficking within a cell and between cells (McDonald et al., 2002). A means to study lipid and aqueous continuity between individual virions expressing Env of avian sarcoma and leukosis virus (ASLV) and cells has recently been developed (Melikyan et al., 2005), enabling the dissection of individual fusion events into discrete steps and identification of the sequential molecular processes involved. By combining these single particle approaches, we have developed a system to study fusion between individual HIV-1 Env-pseudotyped virions and target cells that allows lipid spread and contents mixing to be simultaneously resolved.
MATERIALS AND METHODS
Cells and Reagents
HEK 293T cells were obtained from ATCC (Rockville, MD) and maintained as described (Abrahamyan et al., 2003). HeLa JC-5.3, HOS.CD4.CCR5, and U87.CD4.CCR5 are stable cell lines expressing CD4 and CCR5. HeLa JC-5.3 line (hereafter designated JC-5.3) was a gift from Dr. D. Kabat (OHSU). The following cell lines were obtained from the NIH AIDS Research and Reference Reagent Program (NARRRP): HOS.CD4.CCR5 (hereafter referred to as HOS), provided by Dr. N. Landau (Deng et al., 1996); U87.CD4.CCR5 (referred to as U87), contributed by Drs. Deng and Littman (Bjorndal et al., 1997); and HeLaT4+ cells expressing CD4, contributed by Dr. R. Axel (Maddon et al., 1986). JC-5.3 cells were grown in DMEM medium supplemented with 10% fetal calf serum and penicillin/streptomycin. Growth media for HOS and U87 cells contained 10 μg puromycin and 1 μg puromycin/300 μg G418 per milliliter of medium, respectively.
Poly-l-lysine and n-propyl gallate were purchased from Sigma. The inhibitory peptide, C34 (Eckert and Kim, 2001; >95% purity) was synthesized by Macromolecular Resources (Fort Collins, CO). DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate) was purchased from Molecular Probes (Eugene, OR). The CD4 monoclonal antibody (mAb), SIM.2, was provided by Dr. J. Hildreth (McCallus et al., 1992) through the NARRRP. The CCR5 mAb, PA14, was a gift from Dr. W. Olson (Progenics Pharmaceuticals, Tarrytown, NY). The mAb CRL-1912 against the MLV capsid (p30) was purified from hybridoma cells (ATCC) and provided by Drs. J. Young and R. Barnard (Salk Institute, La Jolla, CA). All fluorescent secondary IgGs were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
HIV-1 JRFL Env with truncated cytoplasmic tail in a pCAGGS vector (Binley et al., 2003) was a gift from Dr. J. Binley (Torrey Pines Institute for Molecular Studies, San Diego, CA). Vectors expressing MLV Gag-pol, Gag-GFP, and LTR lacZ plasmids (Sherer et al., 2003) were kindly provided by Dr. W. Mothes (Yale University, New Haven, CT). The Gag-GFP chimera was constructed by inserting GFP in place of Pol, and this eliminated a viral protease cleavage site downstream of NC (Sherer et al., 2003). As a consequence, GFP was associated with NC within the virions, whereas GFP should have been unassociated if the cleavage site were still present (Andrawiss et al., 2003). HIV-1 Rev was expressed from a pCDNA3 plasmid provided by Dr. R. Doms (University of Pennsylvania, Philadelphia, PA).
Preparation of Pseudoviruses and Infectivity Assay
Viral particles pseudotyped with an MLV core and an HIV-1 Env were prepared by cotransfecting 293T cells with MLV Gag-Pol (18.5 μg), MLV Gag-GFP (6.2 μg), LTR lacZ (25 μg), HIV JRFL 140T Env (25 μg), and Rev (12.5 μg) plasmids per 10-cm culture dish, using a calcium phosphate method (Sherer et al., 2003; Melikyan et al., 2005). (For the control virus used in Western blotting, a pEYFP-MEM vector (Clontech, Palo Alto, CA) was used to incorporate palmitoylated YFP into the pseudoviral envelope.) Viral membranes were labeled with DiD, as described (Melikyan et al., 2005). Briefly, 293T cells were labeled with DiD on the day after transfection. DiD was injected from a stock solution in dimethyl sulfoxide into a serum-free medium to a final concentration of 2.5 μM and immediately applied to transfected cells. After a 3-h incubation at 37°C, the unincorporated dye was removed by washing, and cells were returned to regular growth medium. Two days after transfection, the extracellular medium was collected, centrifuged at low speed to remove cell debris, passed through a 0.45-μm filter, aliquoted, and stored at –80°C. The infectious titer of pseudoviruses was determined by an X-Gal assay (Sanes et al., 1986). Typically, these preparations contained 3·105–8·105 IU/ml. Only ∼25% of the Gag-GFP labeled virions also incorporated detectable levels of DiD. The lipid dye within the viral envelope did not appreciably self-quench (unpublished data), showing that DiD incorporation was low.
Western Blotting
Gag-GFP labeled virions (approx. 4·105 IU) were pelleted from culture medium using a benchtop centrifuge at 14,000 rpm for 90 min at 4°C. The pellet was resuspended in 40 μl medium and mixed with an equal volume of a 2 x SDS-PAGE reducing-loading buffer. One-half of the sample was loaded per well and resolved on a 12% gel. Proteins of resolved virus samples were electrotransferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA) as described in (Towbin et al., 1979). The transferred membrane was blotted for 5 min with phosphate-buffered saline (PBS) containing 0.05% Tween 20 and 5% skim milk (PBSTM) and was then incubated with (1:500) rabbit polyclonal anti-GFP serum (Novus Biologicals, Littleton, CO) in PBSTM at room temperature for 1 h. The membrane was washed 3 times with PBST and incubated with goat anti-rabbit IgG-HRP conjugate (1:1000) in PBSTM at room temperature for 1 h. Unbound second antibody was removed by washing, and the GFP bands were made visual by adding a metal enhanced DAB substrate (Pierce Biotechnology, Rockford, IL).
Permeabilization of Pseudoviruses
Poly-lysine coated no. 0 coverslips were placed in a six-well plate, overlaid with medium containing ∼105 IU of the pseudotyped virus, and centrifuged at 5000 × g at 4°C for 90 min in a Sorval Legend RT centrifuge. The coverslips were washed twice, and adhered viruses were permeabilized by treating with PBS containing either 100 μg/ml saponin (2–5 min, room temperature) or 0.5% Triton X-100 (10 min, room temperature). Saponin treatment of virions colabeled with Gag-GFP and DiD resulted in a steep reduction in the Gag-GFP signal (for many virions the fluorescence was reduced to background level) without appreciable changes in DiD fluorescence. By contrast, treatment with Triton X-100 led to detachment of almost all viruses from the coverslip and to the disappearance of both the DiD and Gag-GFP signals (unpublished data). To prevent detachment of viruses from coverslips when permeabilizing viral envelopes with Triton X-100, the viral particles were fixed with 2% paraformaldehyde for ∼5 min at room temperature before detergent exposure. The viruses remained attached to the coverslip and lost most of their Gag-GFP in the presence of Triton X-100 (e.g., see Figure 2); viruses fixed for 10 min partially retained their Gag-GFP (see Figure 2B). The loss of Gag-GFP fluorescence caused by detergent treatment was inversely proportional to the time of fixation. Because the membrane-impermeant paraformaldehyde was only present before the Triton X-100 treatment, a severe fixation of the envelope probably rendered detergent ineffective.
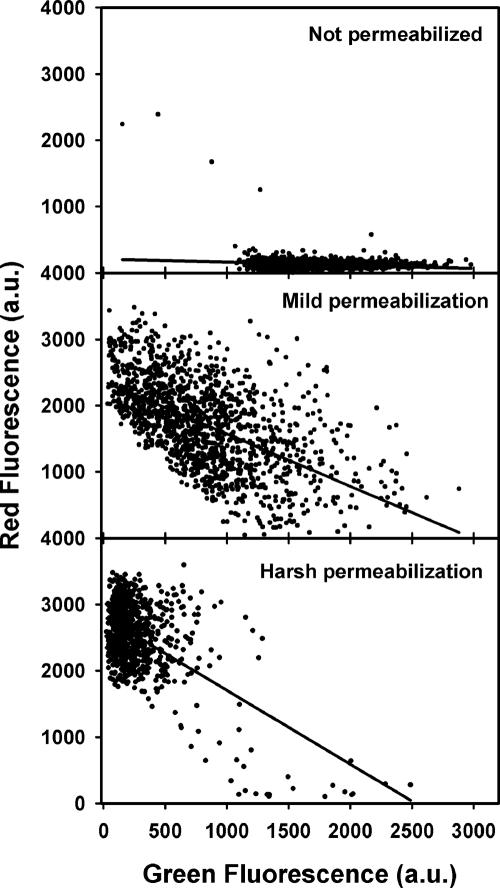
Figure 2.
Correlation between the degree of virus permeabilization (loss of MLV NC-GFP) and immunostaining of the capsid. Images of viral particles were analyzed to obtain the green and red fluorescence intensities for each viral particle. The two signals inversely correlated. The degree to which Triton X-100 permeabilized the envelopes was reduced by increasing the time of fixing with paraformaldehyde before adding detergent. Solid lines are linear regressions.
Immunostaining
Gag-GFP-labeled virions were adhered to coverslips, fixed with paraformaldehyde, and permeabilized by 0.5% Triton X-100 as described above. Permeabilized virions were incubated with undiluted hybridoma medium containing a rat anti-CA mAb, CRL-1912, for 1 h at 4°C. Virions were washed and incubated for 1 h at 4°C with Cy5-conjugated goat anti-rat IgG diluted 1:50 in PBS supplemented with 10% goat serum. Identically stained virus preparations that were not exposed to Triton X-100, as well as permeabilized virions that were incubated only with the secondary goat anti-rat antibody, served as controls. Saponin-permeabilized virions were not detectably stained by the anti-CA antibodies (unpublished data).
Virions were observed in a confocal microscope as described below. GagGFP (green) and CA (red, Cy5) fluorescence of individual viruses were acquired simultaneously, and images were analyzed off-line using IPLab software (Scanalytics, Fairfax, VA). Analysis of the cross-correlation (Figure 2) between the Gag-GFP signal and the extent of staining with anti-CA antibodies (red signal) was performed by creating image segments containing all fluorescent particles; the mean green and red fluorescence intensities were calculated for each segment.
Measurement of CD4 and CCR5 Expression Levels
The levels of expression of CD4 and CCR5 on target U87 and JC-5.3 cells were measured by flow cytometry as previously described (Markosyan et al., 2003). Briefly, cells were detached from culture dishes by incubating with a PBS/EDTA/EGTA buffer, followed by dislodging the cells by vigorous pipetting. Cells were then incubated either with anti-CD4 SIM.2 (1:10 dilution) or with anti-CCR5 PA-14 (2 μg/106 cells) mouse monoclonal antibodies. After 1 h at 4°C, the cells were washed and incubated for 1 h at the same temperature with FITC-conjugated goat anti-mouse IgG diluted 1:100 in PBS supplemented with 10% goat serum. Cells were washed and immediately analyzed with a flow cytometer (Guava EasyCyte, Guava Technologies, Hayward, CA). Target cells were treated with secondary antibodies (in the absence of the primary antibody) as negative controls.
Fusion Experiments
Viruses (approx. 1·105 IU) were spinoculated onto poly-lysine-coated 22-mm square coverslips, washed twice, and overlaid with ∼1.5·106 target cells in Hanks' balanced salt solution (Hanks' buffer) containing 10% fetal bovine serum. Cells were allowed to adhere to the substrate for 30 min at 18°C; these conditions prevented premature virus-cell fusion or significant virus internalization. The coverslips were washed once to remove loosely bound cells, broken into smaller pieces, and stored on ice before an experiment. The pieces of the coverslip were placed in a custom-made chamber that was mounted on a temperature-controlled microscope stage. Viruses were visualized by a laser scanning confocal Fluoview300 microscope (Olympus America, Melville, NY), using an UPlanApo 60X/1.20NA water-immersion objective. Gag-GFP and DiD were excited simultaneously with a 488-nm argon and a 632-nm HeNe laser, respectively. The light emitted by GFP and DiD was separated and detected by two PMTs, using standard EGFP and Cy5 filter sets. The transmitted light was collected by the third PMT to form DIC images. The lowest possible laser intensities were used and temporal resolution was killed in order to minimize photobleaching so that the time virions retained fusion activity was extended for as long as possible. Images were scanned every 10 s, taking 2.7 s to scan each image. To further reduce photobleaching, the experimental solution (Hanks' buffer) was supplemented with 1 mM n-propyl gallate.
A Peltier-based devise (20/20 Technology, Wilmington, NC) was used to cool the experimental chamber to 4°C. Because the water-immersion objective was not explicitly cooled, the local temperature around the imaged cells was ∼18–19°C as measured by a miniature thermocouple. This temperature was sufficiently low to prevent premature Env-mediated virus-cell fusion (unpublished data). Virus-cell fusion was triggered by locally raising the temperature to 37°C by focusing an IR laser diode onto the image field (Melikyan et al., 2000; Markosyan et al., 2003). Images were saved and analyzed off-line. Regions of interest (ROI) were drawn around the fusing virions, as determined by visual examination, and the changes in average fluorescence over time for these virions were analyzed using the Fluoview300 imaging software.
RESULTS
NC-GFP, But Not the Viral Capsid, Is Released from Permeabilized Virions
Our virions were prepared by pseudotyping HIV-1 Env with an MLV core. To label the viral interior, we used a Gag-GFP chimera with GFP attached to the C-terminus (as described in Sherer et al., 2003). We reasoned that if all the Gag polyprotein cleaved into matrix (MA), capsid (CA), and nucleocapsid (NC), all GFP would tag NC, creating “NC-GFP.” To test whether this was the case, we performed a Western blot analysis for GFP and found that only a ∼35-kDa band was prominent (Figure 1A, first lane). The absence of a high-molecular-weight band shows that the overwhelming majority of the Gag-GFP polyprotein was cleaved; the 35-kDa band is indeed consistent with the prediction that almost all GFP is in the form of NC-GFP (GFP ∼27 kDa, MLV NC ∼10 kDa). In a control (second lane), we incorporated palmitoylated YFP into the pseudovirions (Melikyan et al., 2005). It migrated as a 31-kDa band, showing that the GFP family of proteins migrates according to their expected molecular weights. Henceforth, we refer to the GFP probe as NC-GFP.
Figure 1.
Cleavage of Gag-GFP to NC-GFP. (A) A Western blot analysis of the culture medium of cells producing Gag-GFP labeled pseudoviruses (for details see Materials and Methods). The left lane is a product of Gag-GFP polyprotein cleavage. A band corresponding to palmitoylated YFP incorporated into pseudovirions (see Melikyan et al., 2005) is also shown for comparison (right lane). (B) Kinetics of NC-GFP fluorescence after addition of 100 μg/ml saponin. The total GFP-fluorescence in an image field, as shown in C, is plotted as a function of time. (C) Individual pseudovirions colabeled with Gag-GFP (green) and DiD (red) were adhered to a poly-lysine-coated coverslip and imaged using a laser scanning confocal microscope. Yellow particles correspond to viruses that contain both GFP and DiD. The same image field is shown before (left panel) and 2 min after (right panel) exposure to 100 μg/ml saponin at room temperature. Reduction in the number of green and yellow (arrowheads) particles occurs because of dispersal of Gag-GFP from the viruses. (D) Viruses labeled with Gag-GFP (but not with DiD) were fixed, permeabilized by Triton X-100, and immunostained by anti-CA antibodies (red), as indicated. Detergent treatment led to virtually complete loss of the NC-GFP signal (green) and to staining by the secondary IgG-Cy5 antibody (right). The core was not stained if virions were not permeabilized (left) or if only the secondary antibody was added (middle).
We expected that because GFP is considerably larger than NC, NC-GFP would not bind viral RNA and therefore would not pack into the viral core as effectively as NC. It is thought that NC peptides contribute to Gag-Gag interactions by binding to the viral genomic RNA, rather than by interacting among themselves (Coffin et al., 1997). Thus, NC-GFP should be monomeric and mobile. To test whether the NC-GFP is free to move while the virus retains its core, we attached virions to poly-lysine coated coverslips, created large pores in viral envelopes by treating with 100 μg/ml saponin, and recorded NC-GFP and DiD fluorescence intensities. The DiD signal did not change appreciably for any of the virions, showing that except for the creation of pores, the envelopes were intact (Figure 1C). In contrast, the NC-GFP signal quickly decayed for the majority of virions (Figure 1C, losses of NC-GFP from double-labeled virions are marked by arrowheads). From a field of view containing many virions, the average GFP fluorescence decayed rapidly (Figure 1B). The finding that NC-GFP quickly passes through the saponin pores indicates that the viral NC-GFP is not tightly associated with the core. Saponin-treated virions that retained their GFP fluorescence may have been immature, having not cleaved the Gag-polyprotein.
Our next step was to show that the core remained in the viral interior despite the presence of pores that allowed the escape of NC-GFP. If antibodies against the core had access to the viral interior, we could determine if the core was still inside. For saponin-permeabilized virions, the antibodies did not recognize the core (unpublished data). Therefore either the intact core passed out of the virions or the antibodies could not get inside. Because antibodies are significantly smaller than the core, it was most likely that the saponin-induced pores created in the viral envelope were too small to pass antibodies. We found this to be the case: in order to ensure entry of the anti-CA antibody and the Cy5-conjugated secondary antibody, the viral envelopes were severely permeabilized by using a harsher detergent, Triton X-100. (For these experiments, DiD was not incorporated into virions to avoid spectral interference between DiD and Cy5.) The addition of the detergent caused almost all of the virions to detach from their coverslips (unpublished data), so we attached the virions more securely to the coverslips by briefly treating with paraformaldehyde (see Materials and Methods). Before the detergent-treatment, the fixed virions were identified from their NC-GFP fluorescence (Figure 1D, left panel, green virions) and the antibodies did not bind to these virions (left panel, absence of red fluorescence). Triton X-100 treatment permeabilized most of the virions, as evidenced by the loss of the NC-GFP signal (middle panel). The addition of the antibodies after the Triton X-100 treatment led to a drastic increase in red staining (right panel), which shows that the viral particles retained their cores. These results show that NC-GFP was free to leave the viral interior after the envelope was permeabilized, but the core was too large to do so.
Having qualitatively shown that permeabilization of the viral envelope permitted NC-GFP to leave and antibodies to better stain the core, we established this quantitatively for individual virions by measuring for each viral particle its NC-GFP fluorescence (green) and core fluorescence (red). The green and red signals inversely correlated (Figure 2). As expected, without permeabilization, the red signal was absent because the antibodies did not have access to the viral interior and the NC-GFP signal was large (top panel). For mild permeabilization (middle panel), virions that better released NC-GFP were more strongly stained by the antibodies. For our most severe Triton X-100 treatment, most virions lost their NC-GFP and could be stained by the antibody (bottom panel). Thus it is certain that NC-GFP leaves the viral interior through pores too small to permit passage of the viral core. The relatively small NC-GFP thus provides a convenient marker for viral content mixing during virus-cell fusion.
Lipid and Aqueous Dye Redistribution Can Be Simultaneously Measured
Virions bearing R5-tropic HIV Env were adhered to a poly-lysine coated coverslip. Then, at low temperature (18–19°C), U87 cells expressing CD4 and CCR5 were laid on top of the immobilized virions and allowed to bind for 30 min (see Materials and Methods). Placing cells above virus greatly simplifies the experiments and the analyses: Without immobilization, virions on the cell surface constantly move, making it extremely difficult to unambiguously follow the location of an individual virion over time. Also, by adhering virions to a flat substrate, they are all within the same focal plane and this eliminates the need to scan different cell depths, and it allows all cell-bound virions to be continually observed. At 18–19°C, the virions neither fused nor became engulfed by the cells (by endosomal entry or other processes) during the binding time. For study, we confined our attention to virions that were stained by both the lipid dye, DiD, and NC-GFP.
After binding cells to viruses, microscopic observation was begun, and fusion was synchronously triggered by quickly raising temperature to 37°C through a temperature-jump procedure. The DiD (red) that labeled the viral envelope disappeared at varied times, indicating that these virions released their lipid dye into the cell membrane (Figure 3, arrowhead and arrow in the top panel). For virions that released NC-GFP, the fluorescence signal always decayed to the background level. Quantitative analysis of these events confirmed that both the DiD and then the NC-GFP signals quickly decayed to the background fluorescence levels (Figures 3, bottom panel, and 4B). But the decay of NC-GFP fluorescence was often not instantaneous (resolution, 10 s/frame). Rather it usually occurred over several frames. If NC-GFP had packed into the core in the same way as unlabeled CA and NC proteins, all NC-GFP would have simultaneously permeated the fusion pore. There were no indications that virions moved into the cell (depth of focus ∼2 μm), and so this could not be the reason for the gradual decline in GFP fluorescence. All virions that released NC-GFP dispersed its DiD, showing that NC-GFP passed through a fusion pore connecting virus and cell, rather than through a leak within the envelope. Thus, the observation that the decrease in GFP fluorescence was often gradual confirms that the NC-GFP was not tightly associated with the core. The gradual release of the relatively small aqueous marker NC-GFP also indicates that the pore did not enlarge quickly.
Figure 3.
Imaging and analysis of individual virus-cell fusion events. Top panel, virions colabeled with NC-GFP and DiD are colored yellow, and those that do not contain DiD are green; DiD-only labeled vesicles are red. The first image (time = 0) shows a U87 cell adhered to virions immobilized on a coverslip. Fusion was triggered immediately after the first frame by a temperature-jump to 37°C (see Materials and Methods). The virus marked by arrowhead transferred its lipid and content into the target cell (third and fourth frames, respectively). Two viral particles that exhibited only DiD mixing activity are marked by arrow and an asterisk. Bottom panel, the DiD (red circles) and NC-GFP (green circles) fluorescence signals of the fusing virus (top panel, arrowhead) as a function of time at 37°C. The fluorescence traces for the virus marked by arrow are shown in magenta (DiD) and dark cyan (NC-GFP).
Figure 4.
Examples of lipid mixing (dotted lines) and content mixing (solid lines) events between individual pseudovirions and U87 cells. (A) Only lipid dye spread was observed. (B) Both lipid mixing and content mixing occurred. (C) Analyses of the extent of lipid (open bars) and content (hatched bars) mixing between virions and U87 cells in the absence (first column, n = 40 experiments) and in the presence (second column, n = 4) of 200 nM C34. For comparison, fusion to HeLaT4+ cells expressing CD4 and CXCR4 is also shown (third column, n = 3). The fraction of double-labeled viruses that transfer their contents and/or lipid was determined for each experiment and the mean fraction (± SEM) was plotted.
The fraction of viruses that exchanged both their fluorescent lipid and their NC-GFP varied between experiments, and the time intervals between these two events were highly variable. For virions that did not release NC-GFP after the disappearance of DiD, the NC-GFP intensity remained constant for the observation time (up to 10 min) of the experiment (Figure 4A). On average, ∼14% of the virions labeled by both DiD and NC-GFP exhibited lipid mixing, but only a third of these particles (∼4.5% of the total number of virions) transferred NC-GFP into the target cells (Figure 4C, first column). Thus, the fraction of virions that exhibited NC-GFP release after DiD spread was relatively low. Because NC-GFP is relatively small in size, virions that released DiD and retained NC-GFP either hemifused (i.e., merged apposing, proximal lipid monolayers while maintaining noncontacting, distal monolayers) to the cell without proceeding further, or formed a small fusion pore with the cell that did not appreciably enlarge.
We used C34 to confirm that the observed dye losses were mediated by HIV-1 Env, rather than by nonspecific dye transfer. C34 peptide binds to an intermediate conformation of HIV-1 gp41 and blocks fusion by preventing Env from folding into a final six-helix bundle structure (Eckert and Kim, 2001). In the presence of high concentrations of C34, neither lipid nor contents transferred within 10 min at 37°C (Figure 4C, second column), showing that dye transfer was in fact mediated by Env. We verified the coreceptor specificity of the observed fluorescent dye redistribution by binding the virions, which bear R5-tropic Env, to cells (HelaT4+ cells) expressing CD4 and CXCR4. These cells did not support any lipid or contents redistribution (Figure 4C, third column), further demonstrating that the observed lipid and content mixing activity was promoted by HIV Env.
Six-helix Bundle-blocking Peptides Inhibit Contents Release at a Lower Concentration than Needed to Inhibit Lipid Redistribution
As a rule, conditions must be closer to optimal for full fusion to occur than for hemifusion (e.g., Chernomordik et al., 1998; Munoz-Barroso et al., 1998; Markosyan et al., 2000). Because conditions are made less optimal by lowering the density of fusion proteins, introducing deleterious mutations, or weakening the triggers for fusion, pore formation is more readily reduced and eliminated than is hemifusion. We made conditions less optimal by including C34 in the external solution. After adhering the target cells to the immobilized virions in the presence of varied concentrations of C34, we triggered fusion by raising temperature from 18 to 37°C. We quantified C34 inhibitions by taking the average extent of lipid and content mixing and normalizing that by the extent in the absence of peptide (Figure 5). Content mixing (○) was clearly more sensitive to C34 than was lipid mixing (•). For example, NC-GFP transfer was completely blocked by 100 nM C34, but the extent of DiD mixing was still 20% of maximum. This result is similar to that when cells expressing HIV Env were fused to target cells (Munoz-Barroso et al., 1998). It appears that fewer copies of Env are needed to induce lipid transfer than to induce aqueous transfer.
Figure 5.
Dose-dependence of inhibition of DiD (•) and NC-GFP (○) transfer between virions and U87 cells by C34 peptide. The average fraction of individual virions exhibiting lipid mixing alone or in association with content mixing was determined from multiple experiments and plotted against C34 concentration. The total number of double-labeled virions analyzed for each C34 concentration were as follows: 1658 particles in the absence of C34; 75 for 25 nM C34; 458 for 50 nM; 104 for 100 nM; and 610 for 200 nM.
Viral Content Mixing Occurs after Lipid Transfer
We compared the distribution of waiting times for lipid and content transfer into U87 cells by rank ordering these times after raising temperature to 37°C and then plotting cumulatively (Figure 6A). For virions that released both DiD and NC-GFP, the distribution of release times was somewhat shorter for DiD (○), but comparable to that of NC-GFP (•). By analyzing the delay times between lipid (L) and content (C) transfer for individual virions, TC – TL, it is clear that ∼70% of the NC-GFP transfer events were delayed compared with those of DiD mixing (Figure 6B). For the remaining 30% of the events, the mixing times were the same for lipid and contents. We consider it likely that movement of DiD before contents mixing could not be detected in this 30% because the time between lipid dye and aqueous dye movement was too short to be discerned from our limited temporal resolution (10 s per frame). For virions in which DiD spread without subsequent NC-GFP release, the distribution of times for the onset of lipid dye dispersal (Figure 6A, ▵) was similar to that when full fusion occurred (○). This similarity in times indicates that the pathways that lead to lipid dye spread alone, or to both lipid dye spread and content mixing, diverge subsequent to the spread of lipid. In other words, at the point lipid dye spreads, the events that determine whether aqueous contents will mix have not yet occurred.
Figure 6.
Kinetics of lipid mixing and content mixing (A) and the distribution of delay times between DiD and NC-GFP transfer for individual viral particles (B). (A) Waiting times between raising temperature to 37°C and DiD (▵ and ○) and NC-GFP (•) transfer were determined for each virion and plotted as cumulative distributions. The kinetics of lipid mixing for virions that did not transfer their contents is shown by open triangles. (B) The time interval between lipid (TL) and content (TC) mixing events for the same virion were calculated for 54 particles from data shown in B, ranked, and plotted as a cumulative distribution. Measurements were done with U87 cells as targets.
Whether NC-GFP Transfers after DiD Dispersal Depends on Target Cell Type
We used two additional cell lines (JC-5.3 and HOS), both of which express CD4 and CCR5, to test whether the observed dye spreads were only dependent on the levels of CD4 and CCR5, and independent of other properties of the target cells. The extent of lipid dye spread was considerably lower (by about a factor of 2) for these two cell lines (Figure 7A, second and third columns, open bars) than for U87 (first column). In striking contrast to U87, there was almost no content mixing for these two additional cell lines (hatched bars). For example, of the double-labeled virions adhered to JC-5.3 cells, ∼7% exchanged lipids, but only 0.2% delivered NC-GFP into target cells. In other words, virus became connected to JC-5.3 and HOS cells, via either hemifusion or a small fusion pore, at a reasonable frequency, but pore formation and/or growth was a rare event over our observation time (which ranged up to 10 min) after lipid dye redistribution.
Figure 7.
The extents of lipid and content mixing (A) and the kinetics of lipid mixing (B) between single virions and various target cells. (A) The fraction of viruses that transfer their contents (hatched bars) and/or lipid (open bars) was determined for each experiment and the mean fraction (±SEM) was plotted for U87 cells (first column, n = 40 experiments), JC-5.3 cells (second column, n = 11), and HOS cells (third column, n = 10). (B) The kinetics of HIV-1 Env-induced lipid mixing. Fusion between single virions and target cells was triggered by raising temperature to 37°C. The waiting times for the onset of lipid mixing was plotted as cumulative distributions for U87 (○), JC-5.3 (▵), and HOS (▪) cells.
The waiting times for virus dispersing its lipid dye did not depend strongly on the cell line that was used as target (Figure 7B). However, DiD spread exhibited a pronounced lag time for HOS cells, more than 1 min (▪), that was not seen with U87 and JC-5.3 cells. The kinetics of lipid dye spread was the same for JC-5.3 (▵) and U87 (○) cells, but the extents were greater for the U87 cells (Figure 7A). To determine whether large fusion pores formed for JC-5.3 and HOS cells at times greater than could be monitored by our fluorescence methods, we measured infectivity levels for the same batch of virions and found that in fact JC-5.3 cells showed higher (about fourfold) levels of infection than did U87. Thus, enlarged pores do form for JC-5.3 cells, and with a greater efficiency than U87 cells. But the times of pore opening and/or dilation are much greater for the JC-5.3 cells.
The finding that there was less fusion at the cell surface for JC-5.3 than for U87 cells over the time of our fluorescence experiments was surprising, because the JC-5.3 had been subcloned to yield high CCR5 expression levels (Kuhmann et al., 2000). We used flow cytometry to confirm that virtually all (∼90%) of the JC-5.3 cells expressed CCR5 at high density. In contrast, only 15% of the U87 cells did so. Also, for those cells that did express CCR5, the CCR5 density was greater for the JC-5.3 cells (Figure 8, bottom panel). Similarly, flow cytometry showed that CD4 was expressed at higher levels for JC-5.3 than U87 cells, both in percentage of cells and in density (Figure 8, top panels). Because we have found that CD4 and CCR5 densities are not the cause of greater fusion of virus to U87 than to JC-5.3 cells, we suggest that other factors in U87 cells are promoting the more rapid formation and/or dilation of the fusion pores.
Figure 8.
Flow cytometric analysis of CD4 (top) and CCR5 (bottom) expression in U87 (left) and JC-5.3 (right) cells. Cell surface expressions of CD4 and CCR5 were measured using SIM.2 and PA-14 antibodies, respectively (see Materials and Methods). Thin lines show background binding of fluorescently labeled secondary antibody in the absence of the primary antibody. Thick lines are the signals in the presence of both antibodies.
A Fast Rate of Virus-Cell Lipid Transfer Is Necessary But Is Not Sufficient for Content Mixing
We determined the average rate of DiD redistribution from virus to different target cell lines by aligning individual fluorescence traces from several viruses at the onset of lipid transfer. This analysis revealed that the rate of lipid transfer was significantly slower for JC-5.3 cells (Figure 9A, circles) than for U87 (triangles), or HOS (squares) cells. Lipid transfer into HOS cells was almost as fast as into U87 cells, and yet HOS cells did not support significant NC-GFP transfer, whereas U87 cells did. Thus lipid dye must quickly escape for enlarged pores to readily form, but fast escape does not guarantee this outcome.
Figure 9.
The rate of lipid and content transfer from viruses to target cells. (A) Lipid mixing profiles for U87 (triangles, n = 42), HOS (squares, n = 19), and JC-5.3 (circles, n = 28) cells are shown. The average rate of lipid mixing was determined by subtracting the background signal (fluorescence of an area of the target cell not containing fluorescent particles and adjacent to the region of interest, ∼20% of the total signal), normalizing the resulting DiD fluorescence to that at the onset of lipid mixing, and aligning fluorescence traces at that time point. (B) Analyses of lipid and content transfer from virions into U87 cells. Content (•, n = 22) and lipid (○, n = 22) transfer rates are shown for single virion events that exhibited both transfers. The rate of lipid mixing when content release did not occur is shown by open triangles (n = 20). (C) Lipid dye transfer events into JC-5.3 cells (shown in A by open circles, n = 28) were separated into fast (n = 9) and slow (n = 19) events and plotted as average DiD fluorescence versus time (• and ○, respectively). Lipid transfer to U87 cells in the presence (▵, n = 6) or absence (▴, n = 6) of 50 nM C34 was analyzed for events in which only DiD was observed to spread. Error bars, SEM.
We compared the rates of lipid mixing for cases when NC-GFP did (Figure 9B, ○) or did not (▵) transfer into U87 cells to determine whether the rate of lipid movement correlated with subsequent NC-GFP transfer. We found that the rate of lipid movement was the same for both cases. Thus, the rate of lipid transfer does not correlate with the propensity of fusion pores to quickly enlarge. We also found that the rates of DiD and NC-GFP mixing were identical (Figure 9B, ○ vs. •). That is, although the onset of NC-GFP transfer was usually delayed compared with DiD transfer (Figure 6B), and the two probes moved by very different routes (one through aqueous spaces, and the other confined to the membrane), their rates of loss were similar once each began to move. The comparable rates of mixing may result from the complete transfer of each marker within one frame for more than half of the events. Thus, quick pore enlargement after lipid dye movement was independent of the waiting times between the temperature jump and lipid dye spread (Figure 6A), and the rate of lipid dye transfer was independent of whether contents would eventually transfer (Figure 9B). These results strongly indicate that the event that determines whether a pore will form and/or enlarge occurs subsequent to lipid dye spread.
The times for lipid to transfer from virion to cell (tens of seconds) were much greater than would occur if lipid freely diffused (μs). Are complexes of Env proteins surrounding a fusion site responsible for this restriction, as has been proposed for influenza hemagglutinin-induced fusion (Chernomordik et al., 1998)? We reduced the number of fusion-competent Envs that could create a barrier around the fusion site by adding an intermediate concentration of C34 (50 nM) that did not eliminate lipid dye spread for all virions (Figure 5). We found that in the presence of C34, lipid moved more slowly into U87 cells (Figure 9C, ▵) than in the absence of the inhibitor (▴). Thus, when less Env was available due to C34 inhibition, virus that could still transfer lipid dye did so more slowly than in the absence of C34 (Figure 9C). In other words, lipid moved more readily into U87 cells when there were a greater number of fusion-active Envs. If this result was generally true, DiD should have transferred more quickly into JC-5.3 cells than U87 cells. But the opposite occurred (Figure 9A). Therefore, we conclude that restriction of DiD movement by Env is not responsible for slow DiD transfer into cells.
An examination of individual events showed that the rates of DiD transfer into U87 cells were always fast (unpublished data), but both slow and fast transfers occurred into JC-5.3 cells. By dividing records for JC-5.3 cells into fast and slow transfer events, we found that the average rate of the decay of DiD fluorescence exhibiting fast transfer into JC-5.3 cells (Figure 9C, •) was similar to that for U87 cells in the absence of C34 (▴); the slow transfer into JC-5.3 cells (○) occurred at a rate close to that for U87 cells in the presence of C34 (▵). That is, even when DiD transfer into JC-5.3 cells was fast (∼30% of the events), NC-GFP did not transfer. These results reinforce the finding that fast lipid transfer from virus into the target cell does not ensure rapid pore enlargement.
DISCUSSION
We have shown that membrane and aqueous continuity, resulting from fusion of individual pseudovirions expressing HIV Env to cell membranes, can be simultaneously monitored. We found that 1) The Gag-GFP chimera is cleaved to yield a relatively small probe that allows one to monitor fusion pores still small enough to prevent release of the viral core. 2) Aqueous contents never transfer before the spread of lipid dye, and the time between these transfers varies widely. 3) The ability of NC-GFP to transfer within minutes after lipid dye spread is surprisingly dependent on the target cell. 4) After binding virus to optimal target cells at low temperature, fusion proceeds without significant delay upon raising temperature.
The Pseudoviral-cell Fusion System
We used R5-tropic (JRFL) Env with its CT eliminated and an MLV core to produce pseudoviruses. It has been known that pseudotyping full-length HIV Env with an MLV core does not yield infectious particles, but Env with the CT deleted is fusogenic (Mammano et al., 1997). Deletion of HIV Env's CT increases expression levels in cells and somewhat increases cell-cell fusion, without affecting the fundamental properties of the process (Abrahamyan et al., 2005). HIV-1 Env pseudotyped against an HIV core led to the same level of infectivity as when the CT was deleted (Binley et al., 2003). In short, deleting the CT has minimal consequences on the fusion process.
Proper cleavage of the Gag-GFP chimera by MLV protease creates NC-GFP. Our data shows that NC-GFP is not tightly associated with the viral core. The NC-GFP diffuses through pores in the viral envelope, and its movement thus provides a convenient and sensitive means to monitor fusion of an individual HIV particle to a cell. It had previously been found that when creating pseudovirus with an MLV core containing a Gag-GFP chimera with a cleavage site downstream of NC still intact (see Materials and Methods), GFP alone was predominantly cleaved from a Gag-GFP. There were only minor amounts of unprocessed Gag-GFP and of NC-GFP (Andrawiss et al., 2003). Because GFP is slightly smaller than NC-GFP, this alternate Gag-GFP chimera may be somewhat preferable for future fusion experiments.
HIV Env-induced fusion of pseudovirions to cells was a relatively rare event. This is consistent with the very low ratio of infectious/noninfectious particles reported for HIV (Piatak et al., 1993; Gao and Goff, 1999). Our observation time (∼10 min) was limited by inactivation of fluorescently labeled virus caused by photobleaching after prolonged exposure to the laser beam and so our extent of fusion probably underestimates the actual extent. We began illumination simultaneously with raising temperature and DiD and NC-GFP mixing was completed within ∼5 min (Figure 6). A reporter gene assay has shown that fusion of virus does not plateau for several hours (Platt et al., 2005) and photoaffinity labeling of SIV surface proteins upon fusion with cells indicated that fusion did not saturate for at least tens of minutes (Raviv et al., 2002). The faster saturation we observed is likely due to photo-induced inactivation. Even if this is the case, our present methods would still be valid for the first few minutes after triggering fusion.
Our procedures definitely allow, for the first time, study of HIV Env-induced fusion kinetics at early times and at the level of individual virions. These advances depend critically on using temperature-jump methodology. By quickly raising temperature from a state in which virus and cells were bound at low temperature, fusion is synchronously initiated and this permits meaningful interpretation of lipid and content mixing time courses.
Factors Other than Env, CD4, and Chemokine Receptor Densities Can Affect the Rate of Lipid Mixing and the Extent of Content Transfer
JC-5.3 cells have much higher CD4 and CCR5 levels than U87 cells, and our pseudovirus yielded somewhat higher levels of infection for JC-5.3 cells than for U87 cells. Yet U87 cells supported content mixing whereas JC-5.3 cells only yielded lipid dye spread. It is possible that immobilizing virions on a coverslip disfavors pore dilation more for HeLa (JC-5.3) and HOS cells than for U87 cells. Or contributions to the infectious route via endocytosis may be more prominent for JC-5.3 and HOS cells than U87 cells, in which case viral fusion to the plasma membrane need not correlate with levels of infection. But what properties of the plasma membrane could account for the lack of correlation between CD4 and chemokine receptor levels with content release?
There is considerable evidence in the literature that molecules other than CD4 and chemokine receptors, such as ceramide (Finnegan et al., 2004), glycosphingolipids (Puri et al., 1999; Fantini et al., 2000), and protein disulfide isomerases (Fenouillet et al., 2001; Gallina et al., 2002; Barbouche et al., 2003; Markovic et al., 2004), affect efficiency of fusion and/or infection. Such molecules or other factors may strongly influence the transfer of aqueous contents from virus to the cell interior. Although it is possible that this influence over content release also affects the diffusional rate of lipid dye transfer, there is no reason, a priori, this should be the case. Experiment and theory have shown that membrane tension promotes pore formation (Cohen et al., 1982; Kozlovsky et al., 2002) and pore growth (Markosyan et al., 1999; Chizmadzhev et al., 2000). Perhaps U87 cells have higher membrane tensions than JC-5.3 or HOS cells.
Based on cell-cell fusion, it has been commonly thought that fusion proteins restrict the movement of lipid dye and higher densities of fusion proteins are needed for pores than for hemifusion, and so lipid movement is more severely restricted at hemifusion sites that lead to pores (Chernomordik et al., 1998). But our data for virus-cell fusion shows that conditions that reduce the probability of content mixing (e.g., moderate concentrations of C34) also lead to greater restriction of lipid dye movement during either hemifusion or during the small pore stage. Thus, fusion proteins do not appear to restrict lipid dye movement in HIV Env-mediated fusion to cells. Because the probability of content release after lipid mixing was independent of the rate of lipid dispersal (Figure 9), we suggest that the pathway leading to lipid mixing is the same regardless of whether content mixing actually occurs. The pathway's point of divergence occurs subsequent to formation of the structure through which lipids spread. It is likely that this structure is dependent on the number of Envs that actively participate in fusion.
The HIV Pore
Our findings that the Gag-GFP cleavage product, NC-GFP, does not pack into the viral core and is mobile suggest a new interpretation of recent work imaging fusion of individual MLV particles expressing ASLV Env (Melikyan et al., 2005). In the light of our findings, the Gag-YFP used to label the virus core in the prior work undoubtedly yielded a relatively small, diffusible NC-YFP chimera. But the NC-YFP did not transfer through fusion pores formed by ALSV Env (Melikyan et al., 2005). Slow pore growth may be common to ASLV and HIV Env-induced virus-cell fusion, as demonstrated by the inability of the relatively small NC-G(Y)FP to transfer subsequent to lipid dye dispersal.
The virus-cell fusion system developed in the present study allows simultaneous monitoring of both lipid and aqueous dyes for single viral fusion events. Many fundamental questions regarding HIV fusion can now be addressed directly, without the ambiguities that arise when inferences must be made from population studies. For example, how do cells control the enlargement of fusion pores necessary for the release of the viral core into cytosol? Fluorescently tagging viral core proteins (e.g., HIV Vpr; McDonald et al., 2002) could provide a means for following pore enlargement. Also, the intermediate states subsequent to lipid dye spread that we found to be long-lived for some target cells could be manipulated to make the states proceed on to enlarged pores. More generally, the time-resolved monitoring of lipid dye and NC-GFP movement through small pores will allow for precise single virus investigations of HIV Env's cell biological and biophysical mechanisms of inducing membrane continuity.
Acknowledgments
We thank Dr. Michael Leung for kindly performing the Western blot analysis of the Gag-GFP chimeric protein and Dr. Samvel Mkrtchyan for preparing Figure 8. We thank Drs. Levon Abrahamyan for help in making pseudoviruses, Walther Mothes for providing MLV Gag-GFP and Gag-Pol plasmids, David Kabat for providing JC-5.3 cells, and David McDonald for sharing his protocol for labeling virus with DiD. We are grateful to Drs. Richard Barnard and John Young for providing the anti-CA CRL-1912 antibody and to Dr. James Binley for the HIV JRFL Env expression vector. We thank the National Institutes of Health AIDS Research and Reference Reagent Program for cell lines and reagents. Supported by NIH Grants GM27367 and GM54787.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–06–0496) on September 29, 2005.
Abbreviations used: NC-GFP, Gag-GFP chimera cleaved to nucleocapsid-GFP; CA, capsid protein; MA, matrix protein; DiD, far-red fluorescent lipid analogue.
References
- Abrahamyan, L. G., Markosyan, R. M., Moore, J. P., Cohen, F. S., and Melikyan, G. B. (2003). Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77, 5829–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamyan, L. G., Mkrtchyan, S. R., Binley, J., Lu, M., Melikyan, G. B., and Cohen, F. S. (2005). The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J. Virol. 79, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrawiss, M., Takeuchi, Y., Hewlett, L., and Collins, M. (2003). Murine leukemia virus particle assembly quantitated by fluorescence microscopy: role of Gag-Gag interactions and membrane association. J. Virol. 77, 11651–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbouche, R., Miquelis, R., Jones, I. M., and Fenouillet, E. (2003). Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J. Biol. Chem. 278, 3131–3136. [DOI] [PubMed] [Google Scholar]
- Barnard, R. J., Narayan, S., Dornadula, G., Miller, M. D., and Young, J. A. (2004). Low pH is required for avian sarcoma and leukosis virus Env-dependent viral penetration into the cytosol and not for viral uncoating. J. Virol. 78, 10433–10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley, J. M., Cayanan, C. S., Wiley, C., Schulke, N., Olson, W. C., and Burton, D. R. (2003). Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77, 5678–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorndal, A., Deng, H., Jansson, M., Fiore, J. R., Colognesi, C., Karlsson, A., Albert, J., Scarlatti, G., Littman, D. R., and Fenyo, E. M. (1997). Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71, 7478–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrois, M., De Noronha, C., and Greene, W. C. (2002). A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20, 1151–1154. [DOI] [PubMed] [Google Scholar]
- Chatterjee, P. K., Vashishtha, M., and Kielian, M. (2000). Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74, 1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik, L. V., Frolov, V. A., Leikina, E., Bronk, P., and Zimmerberg, J. (1998). The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140, 1369–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizmadzhev, Y. A., Kuzmin, P. I., Kumenko, D. A., Zimmerberg, J., and Cohen, F. S. (2000). Dynamics of fusion pores connecting membranes of different tensions. Biophys. J. 78, 2241–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin, J. M., Highes, S. H., and Varmus, H. E. (1997). Retroviruses. New York: Cold Spring Harbor Laboratory Press. [PubMed]
- Cohen, F. S., Akabas, M. H., and Finkelstein, A. (1982). Osmotic swelling of phospholipid vesicles causes them to fuse with a planar phospholipid bilayer membrane. Science 217, 458–460. [DOI] [PubMed] [Google Scholar]
- Corver, J., Ortiz, A., Allison, S. L., Schalich, J., Heinz, F. X., and Wilschut, J. (2000). Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology 269, 37–46. [DOI] [PubMed] [Google Scholar]
- Daecke, J., Fackler, O. T., Dittmar, M. T., and Krausslich, H. G. (2005). Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J. Virol. 79, 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, H. et al. (1996). Identification of a major co-receptor for primary isolates of HIV-1. Nature 381, 661–666. [DOI] [PubMed] [Google Scholar]
- Dimitrov, D. S., Willey, R. L., Martin, M. A., and Blumenthal, R. (1992). Kinetics of HIV-1 interactions with sCD4 and CD4+ cells: implications for inhibition of virus infection and initial steps of virus entry into cells. Virology 187, 398–406. [DOI] [PubMed] [Google Scholar]
- Earp, L. J., Hernandez, L. D., Delos, S. E., and White, J. M. (2003). Receptor-activated binding of viral fusion proteins to target membranes. Methods Enzymol. 372, 428–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert, D. M., and Kim, P. S. (2001). Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810. [DOI] [PubMed] [Google Scholar]
- Fantini, J., Hammache, D., Pieroni, G., and Yahi, N. (2000). Role of glycosphingolipid microdomains in CD4-dependent HIV-1 fusion. Glycoconj. J. 17, 199–204. [DOI] [PubMed] [Google Scholar]
- Fenouillet, E., Barbouche, R., Courageot, J., and Miquelis, R. (2001). The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J. Infect. Dis. 183, 744–752. [DOI] [PubMed] [Google Scholar]
- Finnegan, C. M., Rawat, S. S., Puri, A., Wang, J. M., Ruscetti, F. W., and Blumenthal, R. (2004). Ceramide, a target for antiretroviral therapy. Proc. Natl. Acad. Sci. USA 101, 15452–15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov, V., Cho, M., Reese, T. S., and Zimmerberg, J. (2000). Both hemifusion and fusion pores are induced by GPI-linked influenza hemagglutinin. Traffic 1, 622–630. [DOI] [PubMed] [Google Scholar]
- Gallina, A., Mandel, R., Trahey, M., Broder, C. C., and Ryser, H. J. (2002). Inhibitors of protein disulfide isomerase (PDI) prevent cleavage of disulfide bonds in receptor-bound gp120 and prevent HIV-1 entry. J. Biol. Chem. 277, 50579–50588. [DOI] [PubMed] [Google Scholar]
- Gallo, S. A., Finnegan, C. M., Viard, M., Raviv, Y., Dimitrov, A., Rawat, S. S., Puri, A., Durell, S., and Blumenthal, R. (2003). The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta 1614, 36–50. [DOI] [PubMed] [Google Scholar]
- Gao, G., and Goff, S. P. (1999). Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol. Biol. Cell 10, 1705–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolokoltsov, A. A., and Davey, R. A. (2004). Rapid and sensitive detection of retrovirus entry by using a novel luciferase-based content-mixing assay. J. Virol. 78, 5124–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky, Y., Chernomordik, L. V., and Kozlov, M. M. (2002). Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys. J. 83, 2634–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann, S. E., Platt, E. J., Kozak, S. L., and Kabat, D. (2000). Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J. Virol. 74, 7005–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali, M., Rust, M. J., Babcock, H. P., and Zhuang, X. (2003). Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina, E., and Chernomordik, L. V. (2000). Reversible merger of membranes at the early stage of influenza hemagglutinin-mediated fusion. Mol. Biol. Cell 11, 2359–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddon, P. J., Dalgleish, A. G., McDougal, J. S., Clapham, P. R., Weiss, R. A., and Axel, R. (1986). The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47, 333–348. [DOI] [PubMed] [Google Scholar]
- Mammano, F., Salvatori, F., Indraccolo, S., De Rossi, A., Chieco-Bianchi, L., and Gottlinger, H. G. (1997). Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J. Virol. 71, 3341–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan, R. M., Cohen, F. S., and Melikyan, G. B. (2000). The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol. Biol. Cell 11, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan, R. M., Cohen, F. S., and Melikyan, G. B. (2003). HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell 14, 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosyan, R. M., Melikyan, G. B., and Cohen, F. S. (1999). Tension of membranes expressing the hemagglutinin of influenza virus inhibits fusion. Biophys. J. 77, 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic, I., Stantchev, T. S., Fields, K. H., Tiffany, L. J., Tomic, M., Weiss, C. D., Broder, C. C., Strebel, K., and Clouse, K. A. (2004). Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 103, 1586–1594. [DOI] [PubMed] [Google Scholar]
- McCallus, D. E., Ugen, K. E., Sato, A. I., Williams, W. V., and Weiner, D. B. (1992). Construction of a recombinant bacterial human CD4 expression system producing a bioactive CD4 molecule. Viral Immunol. 5, 163–172. [DOI] [PubMed] [Google Scholar]
- McDonald, D., Vodicka, M. A., Lucero, G., Svitkina, T. M., Borisy, G. G., Emerman, M., and Hope, T. J. (2002). Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan, G. B., Barnard, R. J., Abrahamyan, L. G., Mothes, W., and Young, J. A. (2005). Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. USA 102, 8728–8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan, G. B., Markosyan, R. M., Hemmati, H., Delmedico, M. K., Lambert, D. M., and Cohen, F. S. (2000). Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Barroso, I., Durell, S., Sakaguchi, K., Appella, E., and Blumenthal, R. (1998). Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J. Cell Biol. 140, 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatak, M., Jr., Saag, M. S., Yang, L. C., Clark, S. J., Kappes, J. C., Luk, K. C., Hahn, B. H., Shaw, G. M., and Lifson, J. D. (1993). High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259, 1749–1754. [DOI] [PubMed] [Google Scholar]
- Platt, E. J., Durnin, J. P., and Kabat, D. (2005). Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J. Virol. 79, 4347–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri, A., Hug, P., Jernigan, K., Rose, P., and Blumenthal, R. (1999). Role of glycosphingolipids in HIV-1 entry: requirement of globotriosylceramide (Gb3) in CD4/CXCR4-dependent fusion. Biosci. Rep. 19, 317–325. [DOI] [PubMed] [Google Scholar]
- Raviv, Y., Viard, M., Bess, J., Jr., and Blumenthal, R. (2002). Quantitative measurement of fusion of HIV-1 and SIV with cultured cells using photosensitized labeling. Virology 293, 243–251. [DOI] [PubMed] [Google Scholar]
- Reeves, J. D. et al. (2002). Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99, 16249–16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes, J. R., Rubenstein, J. L., and Nicolas, J. F. (1986). Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 5, 3133–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer, N. M., Lehmann, M. J., Jimenez-Soto, L. F., Ingmundson, A., Horner, S. M., Cicchetti, G., Allen, P. G., Pypaert, M., Cunningham, J. M., and Mothes, W. (2003). Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4, 785–801. [DOI] [PubMed] [Google Scholar]
- Stegmann, T., Schoen, P., Bron, R., Wey, J., Bartoldus, I., Ortiz, A., Nieva, J. L., and Wilschut, J. (1993). Evaluation of viral membrane fusion assays. Comparison of the octadecylrhodamine dequenching assay with the pyrene excimer assay. Biochemistry 32, 11330–11337. [DOI] [PubMed] [Google Scholar]
- Tobiume, M., Lineberger, J. E., Lundquist, C. A., Miller, M. D., and Aiken, C. (2003). Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 77, 10645–10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyma, D. J., Jiang, J., Shi, J., Zhou, J., Lineberger, J. E., Miller, M. D., and Aiken, C. (2004). Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78, 3429–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]