Abstract
The expression of FREQUENCY, a central component of the circadian clock in Neurospora crassa, shows daily cycles that are exquisitely sensitive to the environment. Two forms of FRQ that differ in length by 99 amino acids, LFRQ and SFRQ, are synthesized from alternative initiation codons and the change in their ratio as a function of temperature contributes to robust rhythmicity across a range of temperatures. We have found frq expression to be surprisingly complex, despite our earlier prediction of a simple transcription unit based on limited cDNA sequencing. Two distinct environmentally regulated major promoters drive primary transcripts whose environmentally influenced alternative splicing gives rise to six different major mRNA species as well as minor forms. Temperature-sensitive alternative splicing determines AUG choice and, as a consequence, the ratio of LFRQ to SFRQ. Four of the six upstream ORFs are spliced out of the vast majority of frq mRNA species. Alternative splice site choice in the 5′ UTR and relative use of two major promoters are also influenced by temperature, and the two promoters are differentially regulated by light. Evolutionary comparisons with the Sordariaceae reveal conservation of 5′ UTR sequences, as well as significant conservation of the alternative splicing events, supporting their relevance to proper regulation of clock function.
INTRODUCTION
Circadian rhythms are daily oscillations in gene expression and activity observed in virtually all groups of organisms. They are driven by an endogenous clock that integrates environmental input to allow the organism to synchronize its activities with the daily cycles of light and temperature (Dunlap et al., 2004). In many organisms studied to date, a core of the clock is composed of a negative feedback loop (Bell-Pedersen et al., 2005); in Neurospora, this loop comprises a negative element, the frequency (frq) gene, whose transcription is activated by the positive elements, the products of the wc-1 and wc-2 genes (Liu, 2003; Dunlap and Loros, 2004). FRQ in turn feeds back to repress its own synthesis by interacting with and influencing the phosphorylation status of the WC-1 and WC-2 proteins (Liu, 2003; Schafmeier et al., 2005). This transcription/translation-based negative feedback loop thus generates a self-sustaining oscillation in the components of the clock. The levels of both frq RNA and FRQ protein oscillate with a circadian period and this daily oscillation is necessary for the circadian clock (Aronson et al., 1994b). FRQ also exerts positive influences on the levels of WC-1 and WC-2 and these positive feedback loops interlock with the negative feedback loop to contribute to the robustness of the rhythm (Lee et al., 2000; Cheng et al., 2001, 2002).
Light and temperature are the major environmental stimuli that keep the clock in tune with the environment. Although light is primarily an entraining signal, temperature has a more complex and interesting relationship to the clock.
An organism experiences large variations in ambient temperature but the clock maintains a relatively constant period by a mechanism known as temperature compensation. Nevertheless, the clock can be reset by temperature steps and temperature pulses, and temperature can be a stronger entraining influence than light under certain experimental paradigms in Neurospora (Liu et al., 1998), whereas in many other systems light is predominant (Dunlap et al., 2004). Interestingly, the temperature range within which clocks can function is more restricted than the range within which the organism can thrive. These considerations led us to studies of the influence of temperature on the molecular mechanism of the oscillator (Liu et al., 1997, 1998).
Previous work (Garceau et al., 1997) revealed that FRQ protein is synthesized in two forms, LFRQ and SFRQ, which differ in length by 99 amino acids. The two proteins arise from the use of two alternative translation initiation sites. Additionally, these protein coding sequences are preceded by a long 5′ UTR containing several small upstream open reading frames (uORFs), and deletion of a subset of these increased the overall level of expression of total FRQ (Liu et al., 1997). This was consistent with actions of a leaky ribosome scanning mechanism and, interestingly, the relative use of the two start codons was seen to be dependent on temperature within the physiological range of clock function, such that the ratio of LFRQ to SFRQ increased as the temperature was raised from 20 to 30°C (Liu et al., 1997). The synthesis of both protein forms was necessary for robust clock function. Sequence analysis of five non-full-length cDNAs (Aronson et al., 1994a) was consistent with a model whereby frq was expressed from a simple transcription unit with a single promoter and no splicing. This led to the interpretation that the thermosensitive alternative translation initiation is due to direct effects of temperature on translation, for example modulation of the extent of leaky ribosome scanning (Liu et al., 1997; Kozak, 2002).
We sought to extend this work through use of strand-specific primers and polysomal RNA, but results soon indicated discrepancies with this model, and the identification of a full-length antisense transcript (Kramer et al., 2003) provided a partial explanation: The previously sequenced partial cDNAs had arisen from both the sense and antisense transcripts and the preliminary view of the frq transcription unit had perforce convolved these. For this reason, and to understand the basis for the temperature-dependent choice of initiation codon, we used strand-specific primers for RT-PCR and 5′ RACE (rapid amplification of cDNA ends), along with ribonuclease protection assays, to develop a detailed analysis of the structure of the 5′ region of frq transcripts, how they are regulated, and how the FRQ initiation codon is chosen. The resulting picture reveals one of the most intriguing and unexpectedly complex transcription units to be described in a lower eukaryote. Choice of the FRQ initiation codon is the result of a temperature-influenced alternative splicing event. Overall, the extent, variety and magnitude of the environmental influences on promoter choice and alternative splicing may reflect the importance of the expression of this key circadian clock gene. Additionally, they extend the domain of such complex transcriptional and posttranscriptional control to nonmulticellular organisms.
MATERIALS AND METHODS
Strains, Growth Conditions, and Race Tube Assay
Neurospora strains 87-3 (bd; a), KAJ120 (Aronson et al., 1994a), YL15, YL31 (Liu et al., 1997), and HVC16 (this study) were cultured, harvested, and analyzed on race tubes by standard methods (Davis and deSerres, 1970; Aronson et al., 1994b) and as previously described (Froehlich et al., 2003). Analysis of race tubes was done with Chrono (Roenneberg and Taylor, 2000).
Plasmids
pHVC16, harboring two point mutations in the 5′ splice site of intron 2, was made by site directed mutagenesis in the plasmid pUC19.frqHindIIIA described previously (Liu et al., 1997), using the Transformer Site-directed Mutagenesis kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions. The mutagenic primer was CACAAGCTGCTCGAACATGACTAGCATCGCAGGCTTC. The AflII/DraIII fragment from the resulting construct was subcloned into pKAJ120 digested with AflII and DraIII to create pHVC16. The mutation was verified by sequencing the plasmid DNA. pHVC16 was transformed into the his-3 locus of strain 93-4 as previously described (Bell-Pedersen et al., 1996). pYL6, used for the RPA experiments, contains the 637-base pair EcoRI/EcoNI fragment of the 5′ UTR of frq, followed by a T3 promoter for the in vitro transcription of antisense riboprobes.
RNA Analyses
Cultures were grown in liquid and harvested as described above. RNA for all analyses was made from 50 to 100 mg of frozen mycelium with Trizol (Invitrogen, Carlsbad, CA) following the manufacturer's recommendations including modifications specific for filamentous fungi: to remove cell wall debris, the frozen tissue was ground with 1 ml of Trizol in liquid nitrogen in a mortar and pestle, transferred to a microfuge tube, allowed to thaw, and centrifuged 10 min before the addition of chloroform. Subsequent steps were exactly as recommended. Before RT-PCR, the RNA was treated with amplification grade DNase (Invitrogen). The C.therm. One Step RT-PCR System (Roche, Indianapolis, IN) was used for RT-PCR analyses, according to the manufacturer's directions. Total DNase-treated RNA, 250 ng, was subjected to 25–28 cycles of PCR (which was determined to be in the linear range of the reaction) and 3.5 min as the initial extension time. For Figure 2B, 5 μCi 32P-dCTP was included in the PCR reaction and the products were separated on a denaturing polyacrylamide gel. For Figure 3A, one-fifth of the RT-PCR reaction was separated on a 5% nondenaturing polyacrylamide gel and stained with VistraGreen (Molecular Probes, Eugene, OR). Band intensities were determined on a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and normalized for fragment size to obtain the relative abundance of each band within a given sample. The primers used for these experiments were: CCGCACCCGCACTCACCTGACCG and GATATGGCTCTCGTCATGAAAGGC for Neurospora. For Sordaria, the primers used in the forward direction were as follows: GCTGCATCGCATCCACTTCGCACACC, GCAAAAACGGCATTGGATGA, and AACCAGAACGTAGCAGTGTGGC, and the primer for the reverse direction was GGATTCGGTCACTCCCAGTGCG.
Figure 2.
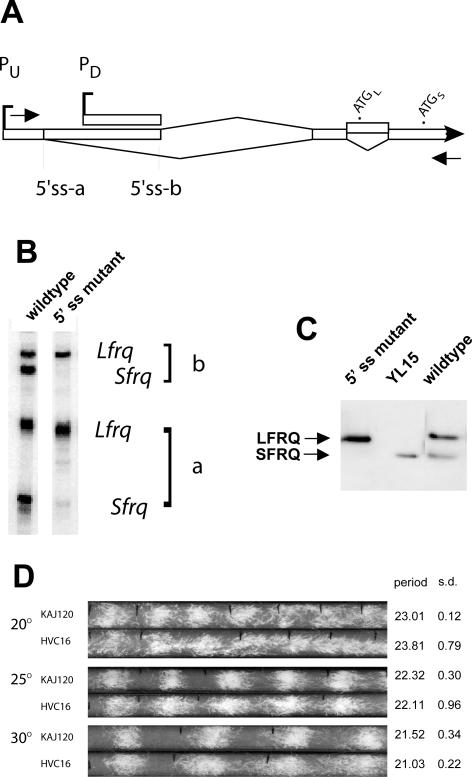
Splicing of intron 2 is necessary for translation to initiate at AUGS. (A) Schematic of the 5′ end of frq (see Figure 1) with primers for RT-PCR indicated by arrows. (B) RT-PCR products amplified from total RNA from a wild-type strain and a frq10 strain transformed with a frq construct (pHVC16) in which the 5′ splice site for intron 2 has been mutated (see text). The four bands correspond to the top four transcripts depicted in Figure 1; this primer pair would not amplify transcripts coming from PD. Note the absence of the bands (Sfrq) in the 5′ss mutant lane that correspond to splicing of intron 2; the two remaining bands reflect the use of alternative 5′ splice sites a and b in the large 5′ UTR intronic region, respectively. (C) Western analysis of FRQ protein made in three strains: wild-type, HVC16, and YL15 (a strain bearing a deletion of five uORFs, as well as AUGL, which makes only SFRQ; Liu et al., 1997). The 5′ splice site mutant protein shown was prepared from tissue grown at 30°C, but other temperatures give a similar pattern with lower expression. The other strains were grown at 25°C; all were harvested 16 h after transfer to darkness (DD16), in the subjective morning. The extracts were treated with lambda protein phosphatase before analysis to allow clear visualization of SFRQ and LFRQ (Garceau et al., 1997). (D) Race tube analysis of strain HVC16 compared with KAJ120 (a transformant bearing a complete wild-type copy of frq and surrounding sequences; Liu et al., 1997) at three temperatures. The average periods and SDs are shown on the right (n = 6).
Figure 3.
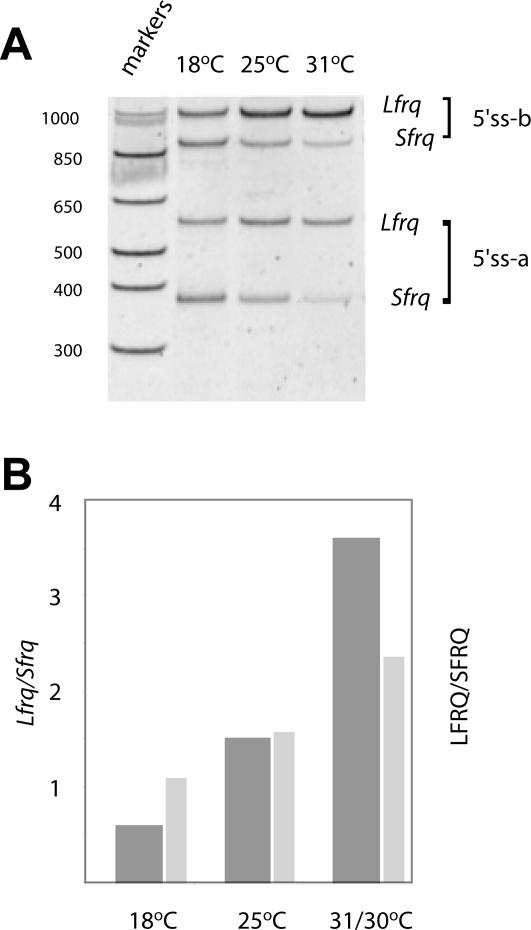
Levels of alternatively spliced products reflect the ambient temperature. (A) The gel shows RT-PCR products amplified from total RNA of strains grown at 18, 25, and 31°C in constant light (LL); the DNA was stained with VistraGreen and visualized on a Storm PhosphorImager. The primers and products are as in Figure 2. (B) The graph compares the ratios of the transcripts (dark bars), as determined from quantitation of the gel in A, with the ratios of the protein (light bars; Liu et al., 1997) at three temperatures. The absolute values of the ratios vary somewhat from experiment to experiment but their relationships are always similar to the representative sample shown here. Preliminary experiments were performed to determine conditions under which the RT-PCR was linear (unpublished data). Quantitative estimates are based on comparisons of bands obtained within each sample and not between samples.
5′ RACE was performed with the Gene Racer kit (Invitrogen) according to the manufacturer's instructions. Total RNA, 8 μg, prepared with Trizol from a wild-type strain grown at 20°C and harvested at DD19 was used, and the cDNA was generated with random primers. Several gene-specific primers were used to generate the final fragments for cloning and sequencing.
Real-time RT-PCR analyses were done as follows: the RT step used the TaqMan (Applied Biosystems, Foster City, CA) reverse transcription system and 200–300 ng RNA per 25 μl of reaction. The PCR was done with 2 μl of the RT reaction in a final volume of 25 μl, using the ABI PRISM 7700 Sequence Detection System with either the TaqMan SYBR Green PCR Core Reagents or the QuantiTect SYBR Green PCR Kit (Qiagen, Chatsworth, CA). Cycling conditions were as recommended by each manufacturer. The primers used for the forward direction were as follows: GGGTAGTCGTGTACTTTACACC for Ua and CCCCAGGAAAGCCCAGAAATCAAAGACACC for Ub; the reverse primer for both transcripts was CATGATACGGCTGTGTGCGTGTCC. For the data shown in Figure 4, raw data were obtained as CT values (cycle number at which the amplification curve crosses a given threshold value). The difference between the CT values for the two primer pairs for a given RNA (delta CT) was then compared with delta CT for other RNAs to obtain a value for the relative change in the ratio Ua/Ub at different temperatures. Two different types of validation experiments were performed: On the one hand, relative quantitation of each sample by use of a standard curve for each primer pair, followed by the determination and comparison of Ua/Ub ratios, gave results similar to the more direct delta CT method. On the other, samples with RNAs from deletion constructs that can generate only Ua or Ub transcripts were mixed in various ratios, subjected to real-time RT-PCR and analyzed by the delta CT method; the changes in Ua/Ub values determined experimentally corresponded closely to the known ratios in the input RNA mixtures.
Figure 4.
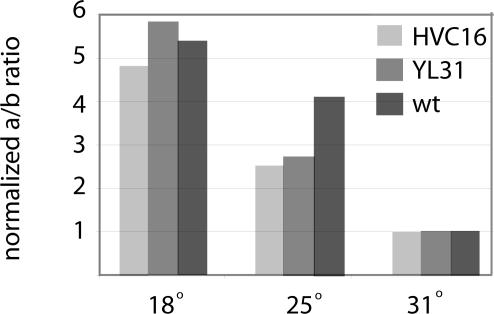
Choice of 5′ splice site for alternative splicing in the 5′ UTR is thermosensitive. Real-time RT-PCR was used to compare the ratios of Ua to Ub transcripts at different temperatures (see Materials and Methods). Ua/Ub at 31°C was set to 1. The three degrees of shading represent RNA from three different strains, all grown in LL for 22–24 h at the indicated temperatures: light gray is HVC16, the intron-2 5′ splice site mutant; medium gray is YL31 (Liu et al., 1997), which has mutations in AUGL and AUGS and makes an untranslatable frq transcript; and dark gray is wild-type.
Ribonuclease protection assays (RPA) were done with the HybSpeed RPA kit (Ambion, Austin, TX) according to the manufacturer's instructions. The probe was made by transcription from KpnI-digested pYL6 with T3 polymerase and 32P-labeled UTP, followed by DNase digestion and gel purification. The riboprobe thus generated was 321 nucleotides (nt) long. Growth conditions, temperature or light treatments, and amounts of RNA used are indicated in the legend to Figure 5. The protected fragments were separated on denaturing polyacrylamide gels and quantitated on a PhosphorImager.
Figure 5.
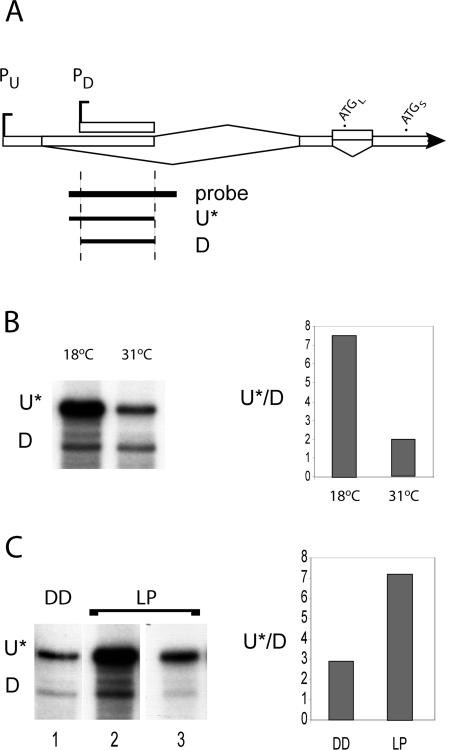
The two major promoters respond differentially to light and temperature. (A) Schematic of the frq 5′ end showing the probe used for ribonuclease protection assay (RPA) and expected protected products corresponding to transcripts from the two promoters, PU and PD. The probe is 455 nt long and would protect fragments of 300 (D, for PD) and 350 (U*, for PU*) nt. Note that the U* fragment represents only a fraction of the transcripts transcribed from the upstream start site PU, due to alternative splicing (see text). (B) Denaturing gels and quantitation by PhosphorImager of fragments protected in the RPA assay. RNA was made from a wild-type strain grown at the indicated temperatures in DD and collected at DD16 (for 18°C) or DD13 (for 31°C). Total RNA, 30 μg, was used for both samples. (C) RNA was made from a wild-type strain grown in constant darkness (DD) at 25°C for 22 h and subjected to a 5-min light pulse (LP) followed by 15 min in DD before harvesting. Ten micrograms of the LP sample and 30 μg of the DD control sample were used in the assay. Lanes 1 and 2 were exposed to the PhosphorImager screen for 3 d; lane 3 was exposed overnight. The quantitation is based on the gels shown, which are representative of four independent replications.
Western Blot Analysis
Protein extracts were prepared, treated with lambda phosphatase, and analyzed by Western blotting as described previously (Garceau et al., 1997). Growth conditions are as indicated in the legend for Figure 2.
RESULTS
frq Has a Complex Transcription Unit
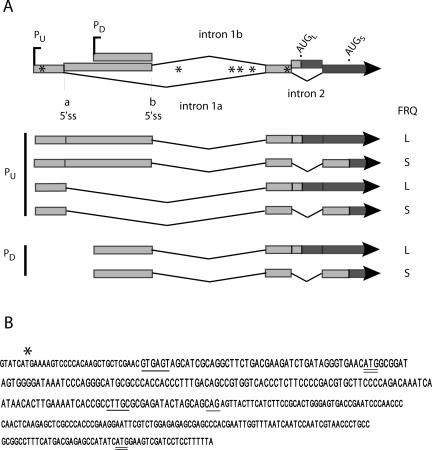
To understand the molecular basis for the temperature-dependent regulation of FRQ synthesis, we undertook a detailed analysis of the transcript structure upstream of and surrounding the LFRQ- and SFRQ-specific initiation codons. Figure 1A summarizes the results of numerous analyses that are described below and in Supplementary Figure S1, including RT-PCR, 5′ RACE and ribonuclease protection assays (RPA). The more distal 5′ end of the transcripts shown in Figure 1 is designated +1 as defined in previous experiments (Liu et al., 1997), although there is heterogeneity in the exact start of transcription in this region and others (see below). The translation initiation sites of the large and small FRQ proteins, AUGL and AUGS (formerly referred to as AUG#1 and AUG#3; Liu et al., 1997) are at positions 1519 and 1816, respectively. 5′ RACE and RPA analyses have confirmed the position of the major start site and revealed another promoter downstream. The vast majority of transcripts generated by the upstream promoter (PU) are alternatively spliced at two 5′ splice sites (5′ss-a and 5′ss-b) in the 5′ UTR and a common 3′ splice site, resulting in the removal of 1155 (intron 1a) or 500 nt (intron 1b), respectively. 5′ss-b is also used by most of the transcripts originating at the downstream promoter (PD). In all major transcripts, the various splicing events remove four out of the six uORFs (marked as asterisks in Figure 1A; Garceau, 1996; Liu et al., 1997). RT-PCR and 5′ RACE have also revealed the use of additional alternative 5′ and 3′ splice sites located within intron 1a in a small minority of transcripts (∼5%; Supplementary Figure S1), resulting in the retention of several uORFs (Supplementary Figure S1), as well as a minor population of transcripts that are not spliced in the 5′ UTR.
Figure 1.
Schematic of frq transcript structure. (A) This diagram is a compilation of the results from many experiments (see below), and represents the first ca. 1900 nt of the transcribed region of frq. The alternative start sites are designated PU and PD. The broad Vs represent introns. Alternative 5′ splice sites for the large upstream intronic region are called a and b. Intron 2 encompasses AUGL, as shown. The asterisks (*) represent upstream AUGs. The lighter shading indicates exon sequences, which vary among the transcripts, and the darker shading denotes coding sequences. (B) The sequence of intron 2 and adjacent regions is shown. Consensus sequences for splicing are underlined; the 5′ and 3′ splice sites were determined experimentally, but the branchpoint sequence shown, CTTGC, is the closest match to the known consensus for Neurospora (see Results). Initiation codons for LFRQ and SFRQ are doubly underlined, and the ATG for uORF 6 is indicated by an asterisk.
A downstream intron (intron 2) encompassing AUGL was also found by RT-PCR. Its sequence, as well as that of adjacent regions, is shown in Figure 1B. Alternative splicing of this 185-nt intron removes AUGL from a subpopulation of transcripts, leaving AUGS as the translation initiation site. Overall, six major transcripts arise from the major alternative splicing events in the 5′ UTR and the coding region. The only noncoding sequence common to all transcripts is the 141 nt exon directly upstream of intron 2. This exon contains the one consistently present uORF, whose AUG is 71 nt upstream of AUGL and 183 nt upstream of AUGS (when intron 2 is spliced out). Downstream of the common exon sequence the proximal 5′ UTR sequences of AUGL and AUGS are completely different and comprise 45 and 157 nt, respectively. The total lengths of the 5′ UTRs range from ∼363–975 nt, and the 5′ UTRs of the transcripts that encode LFRQ are 112 nt shorter than those of the SFRQ transcripts.
Temperature Regulation of Alternative Splicing of frq Can Account for Temperature Regulation of the FRQ Isoform Population
We have previously reported that two different FRQ proteins are generated by the temperature-dependent choice between two alternative AUGs (Liu et al., 1997). We proposed that leaky ribosome scanning (Kozak, 2002) led to the use of the downstream AUG, in a temperature-regulated manner, to produce SFRQ. This view was supported by data showing the expression of both LFRQ and SFRQ from an inducible construct in which the FRQ ORF was directly fused to the qa-2 promoter and 5′ UTR (Merrow et al., 1997). In the work reported here, we initially set out to confirm the mechanism (anticipated to be leaky scanning) by which the downstream AUG would be selected. However, results consistent with this model were not obtained. Instead, Figure 2, A and B, show the four products obtained by RT-PCR with the indicated primers; these correspond only to transcripts that start at PU. Sequencing of the products showed two major alternative splicing events, as described above: splicing in the 5′ UTR removes either intron 1a or 1b, and intron 2 is spliced in only some of the transcripts. Consistent results were seen from analysis of total RNA or from use of strand-specific primers with polysomal RNAs, and all revealed the presence of spliced products. Given this and in hindsight, we now view the appearance of LFRQ and SFRQ in the induced samples (Merrow et al., 1997) as an artifact deriving from the absence of 5′ UTR sequences sufficient to allow ribosomes to bind preferentially upstream of AUGL.
Importantly, this finding made the prediction that splicing alone could lead to initiation of SFRQ from AUGS, obviating the need to invoke translational mechanisms that would bypass the more-upstream AUGL in a temperature-dependent manner. To test that idea, we made two point mutations in the 5′ splice site consensus sequence to change it from GTGAGT (Figure 1B) to aTGAcT. These changes rendered the site nonfunctional as seen by the presence of only the unspliced products after RT-PCR (Figure 2, A and B). This mutation, in the context of an otherwise wild-type frq transcription unit, was transformed back into a frq deletion strain such that the only frq transcripts present arose from the construct. Consistent with this, when protein extracts from the mutant strain were analyzed, they showed that no SFRQ was produced (Figure 2C). Race tube analysis of mutant and wild-type transformed strains shows somewhat more diffuse banding in the mutant at all temperatures tested (Figure 2D). At 20°C, the rhythm is clearly present and displays a period length similar to wild type, confirming previous observations (Liu et al., 1997) that temperature compensation is still normal in a strain that cannot make SFRQ. The observed rhythm, however, is slightly less vigorous and is damped earlier in the mutant than in the wild type, consistent with the observation that the presence of both LFRQ and SFRQ contributes to the robustness of the rhythm at temperatures across the physiological range of the clock (Liu et al., 1997). At 30°C, the period of the rhythm is slightly shorter in both mutant and wild type. Altogether it is clear that, although alternative splicing determines the choice of AUGL versus AUGS and the subsequent synthesis of LFRQ versus SFRQ, this choice does not influence the temperature compensation of the clock.
As shown previously (Liu et al., 1997), the ratio of the proteins LFRQ and SFRQ changes with temperature. Our discovery of an intron whose splicing status determines AUG choice raised the possibility that splicing could be the thermosensitive event underlying the changing protein profile. We tested this hypothesis by performing quantitative RT-PCR assays of the extent of splicing of this intron as a function of temperature (Figure 3A). Within each lane, the relative levels of products were determined by quantitation of the DNA staining (see Materials and Methods); the ratios of the RNAs, Lfrq and Sfrq, are plotted in Figure 3B. It is clear that the proportion of spliced transcript (which can only produce SFRQ) indeed decreases as the temperature rises. The trend of the ratios of the S and L frq transcripts is quite similar to the trend of the protein ratios with increasing temperature (Liu et al., 1997), consistent with a model in which the protein population is largely the direct reflection of the RNA population, whose composition in turn reflects the ambient temperature.
Choice of Upstream 5′ Splice Sites Is Influenced by Temperature
Although it is clear that the temperature-influenced splicing of the second intron directs the type of FRQ protein made, the influence of temperature on frq expression extends beyond this. The RT-PCR results shown in Figure 3A further indicate that temperature also affects the relative levels of 5′ UTR splice variants. The top pair of bands corresponds to the use of the downstream 5′ splice site (5′ss-b; Figure 1A), resulting in products 655 nt longer (see above) than those corresponding to use of the upstream 5′ splice site (5′ss-a; Figure 1A). It is readily apparent that the ratio of the shorter pair to the longer pair (5′ss-a/5′ss-b) decreases with increasing temperature, indicating that as the temperature rises, the downstream 5′ splice site (5′ss-b) is increasingly preferred. This was confirmed by a real-time RT-PCR assay. Primers that span the two alternative exon-exon junctions were designed to allow only specific amplification of 5′ss-a or 5′ss-b products, respectively (see Materials and Methods). As shown in Figure 4, there is a fivefold higher 5′ss-a/5′ss-b ratio at 18 than at 31°C, indicating a shift in 5′ splice site choice in favor of the more downstream one with a rise in temperature. This shift results in a higher proportion of transcripts with much longer 5′ UTRs at the higher temperatures. Regardless of upstream 5′ splice site choice, however, intron 2 splicing remains temperature sensitive (see Figure 3A). Figure 4 also addresses a secondary issue raised by the observation that alternative splicing of the plant gene AtGRP7 is autoregulatory (Staiger et al., 2003). One of the strains used here (YL31) is mutated in both AUGL and AUGS and thus produces no FRQ protein. Splicing, however, is normally temperature regulated, negating any possible role of FRQ in affecting its own splicing.
Ambient Light and Temperature Conditions Influence the Choice between Alternative Promoters
Results to this point have documented the effect of ambient temperature on splicing of intron 2 and the choice of the 5′ splice site of intron 1, but temperature is only one of the two principal environmental influences on circadian rhythms; the other is light. Light has previously been shown to induce frq expression to high levels, an action that is the basis of light entrainment of the rhythm (Crosthwaite et al., 1995; Liu, 2003). Earlier experiments to determine the start of frq transcription pinpointed a start site that has been designated as +1 (Liu et al., 1997; Froehlich et al., 2002, 2003). However, the existence of more than one transcription start site has been suggested by the presence of two distinct size classes of frq transcripts as seen following lengthy exposures of Northerns of RNA isolated under some conditions (unpublished results). To investigate this possibility further, we performed a 5′ RACE analysis (see Materials and Methods). The results revealed a cluster of 5′ ends located at approximately +350, along with those mapping near +1 as expected (unpublished data). We confirmed the 5′ RACE results with RPA, using a probe that spanned the putative downstream promoter (Figure 5A). A major protected fragment of the predicted size (50 nt shorter than the PU-specific product) is seen, confirming the existence of the second transcription start site (Figure 5, B and C). Minor distinct fragments were also seen reproducibly and most likely reflect the heterogeneity of transcription start sites in this region, as suggested by the 5′ RACE results. Note that the absolute ratios of the protected major fragments do not directly reflect the relative abundance of transcripts originating from the two promoters, because the longer band represents only a subset of the upstream transcripts as a result of alternative splicing at 5′ splice sites 5′ss-a and 5′ss-b (see Figure 1). Any transcripts spliced at 5′ss-a would not be scored by this assay, so the data of Figure 5, B and C, underestimate the relative contributions of the stronger upstream promoter.
The RNAs used for the RPA came from strains that had been either grown at different temperatures in the dark or subjected to a light pulse. In addition to proving the existence of PD, the assays revealed that the in vivo conditions determine the relative use of the transcription start sites. Based on the RPA results, the ratio of upstream (PU) to downstream (PD) promoter use decreases with increasing temperature (Figure 5B). The RNAs used for these experiments were isolated at the time of peak expression of frq in constant darkness. From these data and others (unpublished data), we estimate that the downstream promoter contributes 5% of the total frq population at 18°C and at least 25% of the total at 31°C. As shown in Figure 5C, the ratio of upstream to downstream promoter use increases in response to a light pulse. As in Figure 5B, the ratio as shown on the graph is an underestimate because of the partial representation of PU activity by U*. Thus, under all conditions, but especially at lower temperatures or upon exposure to light, the use of PU predominates.
Complex Aspects of the frq Transcription Unit Are Phylogenetically Conserved
Previous work from this laboratory has shown that a frq homolog cloned from Sordaria fimicola could rescue clock function in a frq deletion strain, indicating conservation of both the protein and the regulation of the gene's expression (Merrow and Dunlap, 1994). Comparative sequencing showed considerable conservation in the coding region and relatively less in the noncoding region (5′ UTR), though portions of it were seen to have a surprising degree of conservation. In light of the complex environmental regulation of transcript processing that we have uncovered, we undertook a detailed analysis of transcript structure in Sordaria to gain more context for interpreting the significance of our findings with Neurospora frq. Among the primer pairs used for RT-PCR with Neurospora, several were found to work well with Sordaria RNA and these were used to characterize the Sordaria transcripts.
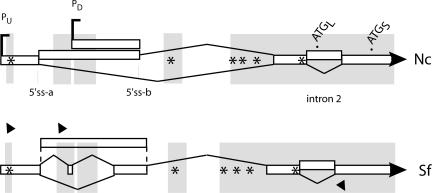
As seen in Figure 6, sequencing of the RT-PCR products reveals that the general features of alternative splicing of both the 5′ UTR and intron 2 are conserved. Even though the major 5′ splice sites in the 5′ UTR do not fall within highly conserved regions, their positions are reasonably well conserved. There is more diversity in the splicing of the Sordaria 5′ UTR: although with Neurospora we find very minor products that use additional sites within the 3′ half of the 5′ UTR (Figure 6 and Supplementary Figure S1), we have not seen products resembling the more complex pattern found in the 5′ half of the Sordaria 5′ UTR. Splicing of intron 2 was seen in some but not all Sordaria RT-PCR products, suggesting that it is regulated in a similar manner to that seen in Neurospora. Conserved regions in the 5′ UTR are clustered near the transcription start sites identified in the Neurospora gene. It is intriguing that the additional conserved regions correspond to the locations of the uORFs; the presence of six uORFs is conserved and the positions of four of them are strictly conserved within the highly similar regions near the 5′ end and just upstream of the bona fide coding region (Figure 6 and Supplementary Figure S1).
Figure 6.
Schematics of the structures of Neurospora and Sordaria frq transcripts. The symbols are the same as in Figure 1. The overall length of the region is slightly shorter in Sordaria, but RT-PCR analyses followed by sequencing of the products revealed a similar array of alternative splicing events. Primers (shown as filled triangles) were chosen after comparison of the sequences with pairwise BLAST. The regions thus designated as highly conserved are highlighted by the shaded rectangles. (Additional details about the BLAST parameters used, the extent of the conserved regions, and the data supporting the transcript processing pattern, are provided in Supplementary Figure S1.) The positions of four of the six uORFs, shown by asterisks, are strictly conserved (Supplementary Figure S1).
DISCUSSION
The work presented in this article describes the molecular basis for the temperature-dependent choice between two initiation codons to produce long or short FRQ protein, respectively (Liu et al., 1997). We have discovered a small intron encompassing AUGL; this intron is subject to alternative splicing and its retention results in the exclusive use of AUGL, whereas its removal makes AUGS the first bona fide start codon. Strains unable to splice this intron fail to produce SFRQ, indicating that leaky scanning makes no significant contribution to the production of SFRQ. Instead, the relative levels of the two alternatively spliced transcripts are thermosensitive and this temperature-modulated splicing underlies the temperature-dependent changes in the FRQ protein population. This choice of FRQ form has significance for the circadian system by extending the temperature range for vigorous rhythmicity, as previously noted (Liu et al., 1997).
Our data may reinforce a consensus regarding the nature of alternatively spliced introns, as well as contributing both a novel biological context and a genetically and molecularly tractable system in which to study this phenomenon. Regulated splicing is sometimes associated with weak consensus sequences such as those seen in intron 2 (Gao et al., 2004; Shin and Manley, 2004). The 3′ splice site consensus CAG is embedded in the sequence AGCAGCAGAG (Figure 1B) and may confound the factors responsible for selecting the 3′ splice site. The polypyrimidine tract usually found upstream of the 3′ splice site is absent. The branchpoint sequence is difficult to pinpoint unequivocally: the consensus for Neurospora is CTRAY (where R is a purine and Y is a pyrimidine; Kupfer et al., 2004). The closest match that is also conserved in Sordaria is CTTGC, located 16 nt upstream of the 3′ splice site CAG (see Figure 1B). If this is indeed the branchpoint sequence used, substitution of G for the almost invariant branchpoint A is striking and suggests that the very minor population of introns with a similarly nonconsensus branch-point (Kupfer et al., 2004) might also be highly regulated, perhaps by temperature. Efforts are underway to identify introns with similar features and test their splicing efficiencies as a function of temperature.
Organisms like plants and fungi are able to adapt to a wide range of temperatures, and the splicing machinery needs to work well over a broad temperature range. It is not surprising, therefore, that temperature-regulated alternative splicing is relatively rare (Sablowski and Meyerowitz, 1998), with only a few examples described in the literature. It is consistent with this that we have seen no evidence for temperature-dependent splicing in any other alternatively spliced Neurospora genes we have analyzed (including NCU2807.1, NCU08380.1, and NCU06702.1; unpublished data), again suggesting that the overall splicing machinery is designed to deliver consistent products across a temperature range. A perhaps noteworthy exception to this is an important thermosensitive splicing event in a circadian context previously noted in Drosophila. In pioneering work, Edery and coworkers (Cheng et al., 1998; Majercak et al., 1999) showed that a thermosensitive splicing event in the 3′ UTR of Drosophila per mRNA is important for advancing the phase of locomotor activity to the daytime on short, cold days. Their work indicates that it is the mere fact of increased splicing in the cold, rather than the identity of the products, that leads to the higher levels and phase advance of per mRNA accumulation. They proposed that the enhanced presence of the splicing machinery at the 3′ UTR could stimulate 3′ end processing, thus increasing per mRNA abundance. More recent work has further demonstrated that both clock factors and light influence the splicing event and that phospholipase C plays a role in communicating temperature information to the splicing machinery (Collins et al., 2004; Majercak et al., 2004). Similarly, the Arabidopsis homolog of the splicing factor SF2/ASF is itself subject to alternative selection of a 3′ splice site in one of its introns, and this choice is under temperature control (Lazar and Goodman, 2000). Several other known instances of splicing changes induced by temperature changes are due to actual mutations. For instance, a single-nucleotide polymorphism in a 5′ splice site of the waxy gene of rice results in less efficient splicing and a substantial sensitivity to temperature, such that there is more splicing at 18°C than at 25 or 32°C (Larkin and Park, 1999). Similarly, a mutation in a beta-globin gene creates aberrant splice sites whose use is dependent on temperature (Gemignani et al., 2002).
In addition to providing a molecular basis for the choice of the initiating AUG in frq, we have uncovered other details of frq transcript structure that are environmentally regulated. The 5′ UTR is spliced, and the choice between two major alternative 5′ splice sites varies with temperature. There is a minor transcription start site in addition to the major one reported previously; these start sites respond differently to temperature. Alternative initiation of transcription is also regulated by light. It should be noted that transcripts arising from use of the downstream transcription start site would not be shorter than the bulk of the frq transcripts (due to splicing of the 5′ UTR); therefore, the nature of the shorter transcript observed on Northern blots remains unclear. The general aspects of intron-exon structure, both in the 5′ UTR and surrounding AUGL, are conserved in Sordaria, though greater heterogeneity is seen. Although we have previously shown that highly conserved regions of the 5′ UTR are interspersed with divergent regions (Merrow and Dunlap, 1994), the expanded analysis reported here allows significance to be assigned to some of the conserved regions and suggests the presence of additional important regulatory features of this region at both the transcriptional and translational levels.
In addition to seasonal temperature differences, organisms are adapted to the daily cycle of temperature fluctuations. The temperature-regulated events we have uncovered would determine the population of transcripts at the pre-dawn temperature minimum in temperate latitudes, such that the clock is poised for a rapid response to light. When light pulses are given to Neurospora at different temperatures, the rise in frq RNA levels is significantly slower at low than high temperatures (HVC, unpublished results). Thus the specific transcripts available may need to be more efficient for translation. By analogy to the situation in flies, splicing of intron 2 could have this desired effect at least partly by virtue of its occurrence—it could lead to higher RNA levels and to transcripts that are more efficiently translated because they have undergone a second splicing event, in addition to splicing in the 5′ UTR. Splicing can lead to enhanced expression because of its stimulatory effect on transcription, RNA export, and translation (Le Hir et al., 2003; Kornblihtt et al., 2004).
We previously reported that strains harboring only the L or S form of FRQ were deficient in clock function at 18 and 30°C, respectively (Liu et al., 1997). In the present work, we did not find as large an effect on full clock function across this temperature range when a mutant frq gene unable to splice intron 2 (thus encoding only LFRQ) was transformed into a frq-deletion strain. This construct differs significantly from those used previously in a way that could not have been understood before the splicing events were recognized, and in this light the previous and present results can be rationalized. In previous work, we made mutations in the different AUGs to prevent synthesis of either long or short FRQ. However, the splicing of intron 2 would affect the final productive population of frq transcripts; for example, because more than half the transcripts are spliced at intron 2 at the cooler temperature, the effective concentration of translatable transcript would be greatly reduced for the ATGS-ablated form, because more than half of the RNAs (i.e., the spliced ones) would lack the remaining intact ATGL and would be untranslatable, leading to much lower levels of protein. We did report different phenotypes for different SFRQ strains, depending on the level of protein seen; i.e., the weakness of the rhythm observed at higher temperatures when only SFRQ is produced is partly abrogated by increasing its level (Liu et al., 1997). It is likely that the overall level of FRQ is at least as important as the S/L ratio for the robustness of the clock and that, as stated previously (Liu et al., 1997; Liu et al., 1998), higher threshold levels of protein are required at higher temperatures.
The location and regulation of the splicing events may shed light on the importance of the uORFs in the frq 5′ UTR. Specifically, alternative splicing events in the 5′ UTR remove five upstream AUGs within four uORFs from all major transcripts. Two AUGs remain; one is present near the 5′ end of the subset of transcripts initiated at PU and the other is in all coding transcripts. The uORF encoded by the latter is just upstream of intron 2, 71 nt away from AUGL and 183 nt away from AUGS. In addition, there is a minor heterogeneous population of frq mRNAs in which from three to six uORFs are retained (Supplementary Figure S1). uORFs have been implicated in tightly regulated translational control (Luo et al., 1995; Jin et al., 2003). The functional impact of upstream AUGs is thought to depend in part on their distance from the first bona fide AUG (Pesole et al., 2001; Kozak, 2002). Accordingly, Lfrq and Sfrq may be differentially regulated at the translational level by the retained uORFs, with Lfrq being more affected. uORFs can also target transcripts for nonsense-mediated decay (NMD), either as a way to remove improperly spliced transcripts or as a mechanism for quantitative control of gene expression (Wollerton et al., 2004). The presence of six uORF sequences is conserved between Neurospora and Sordaria (Figure 6 and Supplementary Figure S1), perhaps suggesting their importance in regulating frq expression from a subset of transcripts. In transcripts spliced for intron 2, the criteria are met for targeting any uORF-containing transcripts for NMD in that the premature stop codons are more than 55 nt upstream of an intron (Maquat, 2002). Note that the sixth uORF, which is retained in all transcripts, would not perform this function. In any case, although four uORFs are normally spliced out, some functional significance can be assigned to the remaining two, especially the final one that is retained all transcripts: constructs deleting all uORFs but the first (e.g., NG37 and YL15; Garceau, 1996; Liu et al., 1997) show elevated levels of FRQ expression, a result consistent with that recently reported by Diernfellner et al. (2005). It should be noted, however, that despite the effects on the levels of FRQ expression and even the complete loss of LFRQ (Liu et al., 1997), these strains have a normally temperature compensated circadian clock.
While this article was in review, similar findings were reported regarding the temperature dependent splicing of intron 2 (Diernfellner et al., 2005). In that publication, a strain bearing multiple mutations in the 5′ splice site, branchpoint region, and 3′ splice site of intron 2 (designated intron 6 in that publication) was used to block this splicing, and a temperature compensation defect in this multiply mutant strain was used to infer a role of the splicing in temperature compensation. The strain reported here, HVC16, used just two point mutations to similarly completely abrogate intron 2 splicing (Figure 2). However, this strain has completely normal temperature compensation, strongly suggesting that the temperature-dependent splicing of intron 2 cannot itself play a role in or be important for the temperature compensation mechanism of the clock. Likewise, because this strain does not produce any short FRQ (Figure 2C), the ratio of LFRQ to SFRQ per se cannot be the basis of temperature compensation. This is in agreement with results and conclusions previously reported by Liu et al. (1997).
By uncovering temperature-dependent splicing and transcription events that take place with the frequency transcript, we have elucidated novel aspects of temperature sensing by the circadian clock in Neurospora. Temperature influences circadian clocks in three different ways. First, abrupt changes in ambient temperature are sensed by the clock and are used as time cues to entrain the intracellular circadian clock to external time (Liu et al., 1998). It is likely that the temperature-influenced splicing described here plays a significant role in this temperature sensing, because the level of FRQ expression is known to be highly temperature-dependent (Liu et al., 1997, 1998). Further, we have previously suggested a more general role for the frq-based circadian oscillator in temperature sensing by the organism because, in a microarray study using a limited unigene set, strains lacking frq no longer showed cycles in gene expression in response to environmental temperature cycles (Nowrousian et al., 2003). A second facet of circadian temperature response is temperature compensation, the mechanism that acts to maintain a relatively constant cycle length across at least part of the physiological range of the organism. Our data suggest that temperature-influenced splicing does not play a role in temperature compensation, because strains making only LFRQ or SFRQ retain compensated clocks (Liu et al., 1997; unpublished data), as do strains that cannot splice intron 2 (Figure 2). We have previously suggested that temperature-influenced changes in the level of FRQ expression might contribute to the third facet of circadian temperature responses, the existence of physiological upper and lower limits for fully functional circadian regulation within the growth range of the organism (Liu et al., 1997). The involvement of alternative splicing in sensing temperature for proper clock function in both this organism and Drosophila suggests that this phenomenon may be more generally involved in circadian clocks and in other processes that require temperature information.
Supplementary Material
Acknowledgments
We thank Arun Mehra and Norman Garceau for access to unpublished results. This work was supported by the following grants from the National Institutes of Health: R37 GM34985 to J.C.D., MH44651 to J.C.D. and J.J.L. and National Science Foundation Grant MCB-0084509 to J.J.L.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0756) on September 29, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Aronson, B. D., Johnson, K. A., and Dunlap, J. C. (1994a). Circadian clock locus frequency: protein encoded by a single open reading frame defines period length and temperature compensation. Proc. Natl. Acad. Sci. USA 91, 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson, B. D., Johnson, K. A., Loros, J. J., and Dunlap, J. C. (1994b). Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen, D., Cassone, V. M., Earnest, D. J., Golden, S. S., Hardin, P. E., Thomas, T. L., and Zoran, M. J. (2005). Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen, D., Shinohara, M. L., Loros, J. J., and Dunlap, J. C. (1996). Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific. Proc. Natl. Acad. Sci. USA 93, 13096–13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, P., Yang, Y., Gardner, K. H., and Liu, Y. (2002). PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 22, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, P., Yang, Y., and Liu, Y. (2001). Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl. Acad. Sci. USA 98, 7408–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y., Gvakharia, B., and Hardin, P. E. (1998). Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol. Cell. Biol. 18, 6505–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B. H., Rosato, E., and Kyriacou, C. P. (2004). Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc. Natl. Acad. Sci. USA 101, 1945–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite, S. K., Loros, J. J., and Dunlap, J. C. (1995). Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Davis, R. L., and deSerres, D. (1970). Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 27A, 79–143. [Google Scholar]
- Diernfellner, A. C., Schafmeier, T., Merrow, M. W., and Brunner, M. (2005). Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 19, 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap, J. C., and Loros, J. J. (2004). The neurospora circadian system. J. Biol. Rhythms 19, 414–424. [DOI] [PubMed] [Google Scholar]
- Dunlap, J. C., Loros, J. J., and Decoursey, P. (2004). Chronobiology: Biological Timekeeping, Sunderland, MA: Sinauer Associates.
- Froehlich, A. C., Liu, Y., Loros, J. J., and Dunlap, J. C. (2002). White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297, 815–819. [DOI] [PubMed] [Google Scholar]
- Froehlich, A. C., Loros, J. J., and Dunlap, J. C. (2003). Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl. Acad. Sci. USA 100, 5914–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H., Gordon-Kamm, W. J., and Lyznik, L. A. (2004). ASF/SF2-like maize pre-mRNA splicing factors affect splice site utilization and their transcripts are alternatively spliced. Gene 339, 25–37. [DOI] [PubMed] [Google Scholar]
- Garceau, N. (1996). Identification and Regulation of a Circadian Clock Protein, FRQ, and a Clock-controlled Protein, CCG-1, in Neurospora crassa, Hanover, NH: Dartmouth College.
- Garceau, N. Y., Liu, Y., Loros, J. J., and Dunlap, J. C. (1997). Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Gemignani, F., Sazani, P., Morcos, P., and Kole, R. (2002). Temperature-dependent splicing of beta-globin pre-mRNA. Nucleic Acids Res. 30, 4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, X., Turcott, E., Englehardt, S., Mize, G. J., and Morris, D. R. (2003). The two upstream open reading frames of oncogene mdm2 have different translational regulatory properties. J. Biol. Chem. 278, 25716–25721. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A. R., de la Mata, M., Fededa, J. P., Munoz, M. J., and Nogues, G. (2004). Multiple links between transcription and splicing. RNA 10, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (2002). Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, C., Loros, J. J., Dunlap, J. C., and Crosthwaite, S. K. (2003). Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421, 948–952. [DOI] [PubMed] [Google Scholar]
- Kupfer, D. M., Drabenstot, S. D., Buchanan, K. L., Lai, H., Zhu, H., Dyer, D. W., Roe, B. A., and Murphy, J. W. (2004). Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3, 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, P. D., and Park, W. D. (1999). Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 40, 719–727. [DOI] [PubMed] [Google Scholar]
- Lazar, G., and Goodman, H. M. (2000). The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42, 571–581. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Nott, A., and Moore, M. J. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28, 215–220. [DOI] [PubMed] [Google Scholar]
- Lee, K., Loros, J. J., and Dunlap, J. C. (2000). Interconnected feedback loops in the Neurospora circadian system. Science 289, 107–110. [DOI] [PubMed] [Google Scholar]
- Liu, Y. (2003). Molecular mechanisms of entrainment in the Neurospora circadian clock. J. Biol. Rhythms 18, 195–205. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Garceau, N. Y., Loros, J. J., and Dunlap, J. C. (1997). Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 89, 477–486. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Merrow, M., Loros, J. J., and Dunlap, J. C. (1998). How temperature changes reset a circadian oscillator. Science 281, 825–829. [DOI] [PubMed] [Google Scholar]
- Luo, Z., Freitag, M., and Sachs, M. S. (1995). Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol. 15, 5235–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak, J., Chen, W. F., and Edery, I. (2004). Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol. 24, 3359–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majercak, J., Sidote, D., Hardin, P. E., and Edery, I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230. [DOI] [PubMed] [Google Scholar]
- Maquat, L. E. (2002). Nonsense-mediated mRNA decay. Curr. Biol. 12, R196–R197. [DOI] [PubMed] [Google Scholar]
- Merrow, M. W., and Dunlap, J. C. (1994). Intergeneric complementation of a circadian rhythmicity defect: phylogenetic conservation of structure and function of the clock gene frequency. EMBO J. 13, 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow, M. W., Garceau, N. Y., and Dunlap, J. C. (1997). Dissection of a circadian oscillation into discrete domains. Proc. Natl. Acad. Sci. USA 94, 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian, M., Duffield, G. E., Loros, J. J., and Dunlap, J. C. (2003). The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics 164, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole, G., Mignone, F., Gissi, C., Grillo, G., Licciulli, F., and Liuni, S. (2001). Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276, 73–81. [DOI] [PubMed] [Google Scholar]
- Roenneberg, T., and Taylor, W. (2000). Automated recordings of bioluminescence with special reference to the analysis of circadian rhythms. Methods Enzymol. 305, 104–119. [DOI] [PubMed] [Google Scholar]
- Sablowski, R. W., and Meyerowitz, E. M. (1998). Temperature-sensitive splicing in the floral homeotic mutant apetala3–1. Plant Cell 10, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier, T., Haase, A., Kaldi, K., Scholz, J., Fuchs, M., and Brunner, M. (2005). Transcriptional feedback of neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122, 235–246. [DOI] [PubMed] [Google Scholar]
- Shin, C., and Manley, J. L. (2004). Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell. Biol. 5, 727–738. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Zecca, L., Wieczorek Kirk, D. A., Apel, K., and Eckstein, L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 33, 361–371. [DOI] [PubMed] [Google Scholar]
- Wollerton, M. C., Gooding, C., Wagner, E. J., Garcia-Blanco, M. A., and Smith, C. W. (2004). Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell 13, 91–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.