Abstract
Caspases play a critical role in the execution of metazoan apoptosis and are thus attractive therapeutic targets for apoptosis-associated diseases. Here we report that baculovirus P49, a homolog of pancaspase inhibitor P35, prevents apoptosis in invertebrates by inhibiting an initiator caspase that is P35 insensitive. Consequently P49 blocked proteolytic activation of effector caspases at a unique step upstream from that affected by P35 but downstream from inhibitor of apoptosis Op-IAP. Like P35, P49 was cleaved by and stably associated with its caspase target. Ectopically expressed P49 blocked apoptosis in cultured cells from a phylogenetically distinct organism, Drosophila melanogaster. Furthermore, P49 inhibited human caspase-9, demonstrating its capacity to affect a vertebrate initiator caspase. Thus, P49 is a substrate inhibitor with a novel in vivo specificity for a P35-insensitive initiator caspase that functions at an evolutionarily conserved step in the caspase cascade. These data indicate that activated initiator caspases provide another effective target for apoptotic intervention by substrate inhibitors.
Keywords: apoptosis/baculovirus P49/caspase inhibitor P35/Drosophila melanogaster/IAP
Introduction
Apoptosis is a widely conserved genetic program that deletes unwanted, abnormal or diseased cells in multicellular organisms (Jacobson et al., 1997; Vaux and Korsmeyer, 1999). Disruption of normal apoptotic regulation causes adverse effects that are correlated with tumorigenesis, immunodeficiency and degenerative disorders (Thompson, 1995; Yuan and Yankner, 2000). The caspase family of cysteine-dependent aspartate-specific proteases are essential components in the execution of apoptosis (Salvesen and Dixit, 1997; Cryns and Yuan, 1998; Thornberry and Lazebnik, 1998; Earnshaw et al., 1999; Chang and Yang, 2000). By selective proteolysis of cellular substrates, the caspases catalyze the biochemical events that lead to cellular disassembly, including DNA fragmentation, chromatin condensation and plasma membrane blebbing. Because of their pivotal role in the commitment to cell death, these proteases are important targets in therapeutic strategies for treating apoptosis-associated diseases (Jacobson, 1998; Nicholson, 2000; Yuan and Yankner, 2000).
Caspases are activated from dormant proenzymes by an ordered series of proteolytic cleavages. Initiator caspases are activated by mechanisms that include signal-induced interactions of specific adaptor proteins with the long N-terminal prodomain, which promotes caspase proteolytic processing (Earnshaw et al., 1999; Chang and Yang, 2000). The initiators subsequently activate the short prodomain-containing effector caspases by proteolysis. In the vertebrate mitochondrion/cytochrome c pathway, initiator caspase-9 is processed upon recruitment of its CARD-containing prodomain by adaptor Apaf-1 bound to cytochrome c released from mitochondria during apoptotic signaling (Li et al., 1997; Zou et al., 1997; Srinivasula et al., 1998). From this apoptosome complex, caspase-9 proteolytically activates effector proteases, including caspase-3 (Adrain and Martin, 2001). In invertebrates, a comparable apoptosome-mediated activation of initiator caspases is likely since initiator caspase candidates DREDD and DRONC of Drosophila melanogaster interact with the Apaf-1 homolog designated Ark (White, 2000). The existence of diverse pro- and anti-apoptotic factors that affect initiator caspase activation argues that this process is a key regulatory step in initiation of apoptosis (Chang and Yang, 2000; Adrain and Martin, 2001).
Viruses have evolved novel mechanisms to prevent apoptosis of their host cell and thereby promote virus multiplication (O’Brien, 1998; Roulston et al., 1999). To date, two distinct types of apoptotic suppressor, represented by P35 and the inhibitors of apoptosis (IAPs), have been identified in the invertebrate baculoviruses (Clem, 2001). Baculovirus IAPs were the first members of the IAP protein family to be discovered (Deveraux and Reed, 1999; Miller, 1999). The best-studied viral IAP, Op-IAP, prevents apoptosis in insects and mammals by a mechanism that includes interaction with itself and pro-apoptotic proteins like Reaper, HID and GRIM of Drosophila (Birnbaum et al., 1994; Duckett et al., 1996; Hawkins et al., 1996; Vucic et al., 1997, 1998; Hozak et al., 2000). In SF21 cells from Spodoptera frugiperda (Order Lepidoptera), a model system for insect apoptosis, Op-IAP blocks proteolytic activation of the principal effector caspase, Sf-caspase-1 (Seshagiri and Miller, 1997; LaCount et al., 2000). In contrast, caspase inhibitor P35 blocks apoptosis by targeting active Sf-caspase-1 (Bertin et al., 1996; Ahmad et al., 1997; Manji et al., 1997; LaCount et al., 2000). P35 is a pancaspase inhibitor in which cleavage of its solvent-exposed reactive-site loop (RSL) leads to formation of a stoichiometric complex with the caspase target (Bump et al., 1995; Zhou et al., 1998; Fisher et al., 1999; Xu et al., 2001). Despite its broad-spectrum anti-caspase activity, P35 fails to block proteolytic activation of pro-Sf-caspase-1, suggesting the existence of a novel P35-insensitive initiator caspase (LaCount et al., 2000; Manji and Friesen, 2001). This uncharacterized caspase is designated Sf-caspase-X.
Here, we describe a third type of baculovirus apoptotic suppressor, P49, that is distinguished by its capacity to inhibit the Spodoptera initiator caspase unaffected by P35. The p49 gene from baculovirus SlNPV was identified by its capacity to block apoptosis and restore replication of a p35-deletion virus (Du et al., 1999). P49 is 49% identical to P35 but is unrelated to any known cellular protein. It is characterized by its larger size (446 residues), the presence of a 120 residue insertion absent in P35 (299 residues) and a significantly different sequence (TVTD94↓G) at its predicted cleavage site (Figure 1A). These dissimilarities suggest that if P49 functions as a caspase inhibitor, it may exhibit a unique caspase specificity or target a distinct step in the death pathway. To test these possibilities, we explored the anti-caspase potential of P49 and defined the in vivo apoptotic step affected.
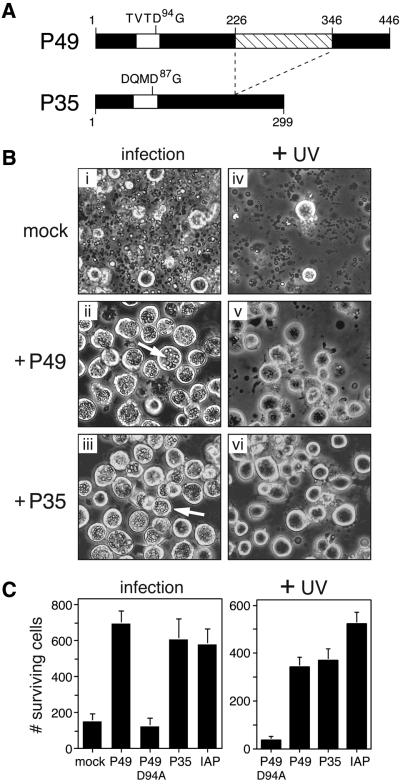
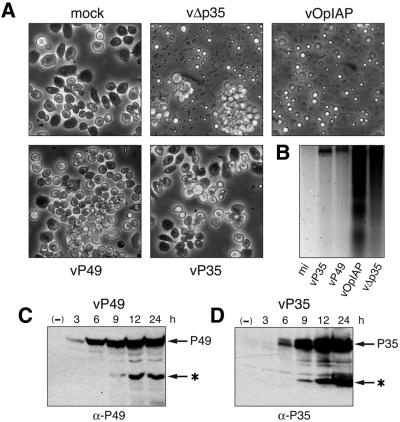
Fig. 1. P49 blocks apoptosis induced by diverse signals. (A) P49 and P35 structure. Amino acid similarity between P49 and P35 is colinear with the exception of a 120 residue insert (crosshatched) within P49. The caspase cleavage site within the predicted RSL (open) is indicated. (B) Virus- and UV radiation-induced apoptosis. SF21 cells were mock-transfected or transfected with plasmids encoding P49 or P35 and subsequently infected with apoptosis-inducing virus vΔp35 or irradiated with UV-B (125 mJ/cm2). Photographs (magnification, 100×) were taken 48 or 24 h after infection (i–iii) or UV irradiation (iv–vi), respectively. Arrows depict occluded virus particles in non-apoptotic cells. A representative experiment is shown. (C) Cell survival. SF21 cells transfected with plasmids encoding wild-type P49, D94A-mutated P49, P35 or Op-IAP were infected or UV-irradiated as described in (B). Survival was quantified by computer-aided microscopy and is reported as the average number of surviving, non-apoptotic cells ± standard deviation.
We report here that P49 is a caspase substrate inhibitor with a P35-like mechanism. However, unlike P35, P49 functions at an upstream step to inactivate the caspase that proteolytically activates effector caspases of Spodoptera cells. Thus, P49 exhibits a distinct in vivo target specificity for a novel P35-insensitive initiator caspase. These data indicate that despite comparable structures and mechanisms, caspase inhibitors P49 and P35 have a unique specificity for initiator or effector caspases in the context of the apoptotic cell. Our finding that P49 also has the capacity to block apoptosis in cultured cells of D.melanogaster and is a potent inhibitor of human initiator caspase-9 suggests that P49 functions at a highly conserved step in the caspase cascade and should therefore prove useful in delineating cell death pathways in many organisms.
Results
Baculovirus P49 blocks apoptosis induced by diverse stimuli
The high sequence similarity with P35 and the presence of a caspase-like cleavage site (Figure 1A) suggested that P49 functions as a caspase inhibitor. Thus, to test the capacity of P49 to block caspase-mediated apoptosis, we expressed p49 ectopically in cultured SF21 cells that were subsequently induced to undergo apoptosis. Upon plasmid transfection, P49 blocked apoptosis triggered by infection with baculovirus mutant vΔp35, which lacks apoptotic suppressors (Figure 1B). P49 prevented premature cell death and promoted virus replication as indicated by the accumulation of intranuclear virus particles. The level of p49-mediated protection was comparable to that conferred by p35 and baculovirus inhibitor of apoptosis Op-iap (Figure 1C). Similarly, P49 was a potent suppressor of UV radiation-induced apoptosis (Figure 1B). Upon transfection, P49 was as effective as P35 in protecting SF21 cells from a dose of UV radiation that caused 95% lethality (Figure 1C). Op-IAP was comparably protective.
To assess the contribution of the P49 predicted cleavage site (TVTD94↓G) to anti-apoptotic activity, Asp94 was substituted with alanine. Although readily synthesized in transfected cells (see below), D94A-mutated p49 failed to block apoptosis induced by infection or UV irradiation (Figure 1C). The loss of p49 function was confirmed by marker rescue assays (Table I) in which the anti-apoptotic activity of a plasmid-borne gene was measured by the extent to which it restored multiplication of a p35-deficient baculovirus (Bertin et al., 1996; Zoog et al., 1999). In contrast to wild-type P49, D94A-mutated P49 failed to rescue virus replication. As expected, wild-type P35 restored replication, but D87A cleavage site-mutated P35 did not (Table I). We concluded that P49 blocks apoptosis induced by distinct death signals through a mechanism requiring an aspartate residue at the predicted caspase cleavage site.
Table I. Marker rescue of vΔp35 mutant replication.
| Transfected gene | Virus yielda (× 103 p.f.u./ml) | Anti-apoptotic activity (%) |
|---|---|---|
| wt p49 |
125 ± 18 |
93.3 |
| D94A-p49 |
0.0 |
0.0 |
| wt p35 |
134 ± 18 |
100 |
| D87A-p35 | 0.4 ± 0.1 | 0.3 |
aVirus yields were determined by plaque assay using apoptosis-sensitive SF21 cells. Percentage anti-apoptotic activity is reported as the ratio of non-apoptotic, lacZ-expressing plaques produced by transfection of the gene indicated to that of wild-type p35. Values shown are the average ± standard deviation of triplicate transfections.
P49 is a substrate inhibitor of caspases
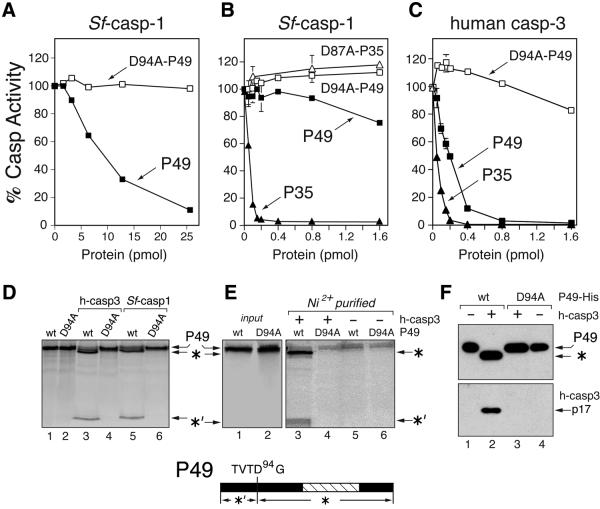
To determine whether P49 is a direct inhibitor of caspases, we generated C-terminal His6-tagged P49. When tested in dose-dependent assays that used the tetrapeptide DEVD-amc as substrate, the principal effector caspase of SF21 cells, Sf-caspase-1, was inhibited by purified P49-His6, but not D94A-mutated P49-His6 (Figure 2A). Nonetheless, P49-His6 was ∼100 times less effective than P35-His6, which is a stoichiometric inhibitor of Sf-caspase-1 (Figure 2B). In contrast, P49-His6 was a potent inhibitor of human caspase-3 (Figure 2C). In these assays, P49-His6 was nearly as effective as P35-His6, which is also a stoichiometric inhibitor of caspase-3 (Bump et al., 1995; Bertin et al., 1996; Zhou et al., 1998; Fisher et al., 1999). D94A-mutated P49-His6 had little effect on human caspase-3 over the range of protein concentrations tested (Figure 2C).
Fig. 2. P49 is a substrate inhibitor. (A and B) Inhibition of Sf-caspase-1. Purified recombinant Sf-caspase-1 (0.1 pmol) was incubated with the indicated amounts of purified P49-His6 (filled square), D94A-mutated P49-His6 (open square), P35-His6 (filled triangle) or D87A-mutated P35-His6 (open triangle). After 30 min, residual caspase activity was measured by using DEVD-amc as substrate. Plotted values are the average ± standard deviation of triplicate assays and are expressed as a percentage of uninhibited caspase activity. (A) and (B) differ in the range of P49 used. (C) Inhibition of human caspase-3. Purified recombinant caspase-3 (0.1 pmol) was incubated with the indicated amounts of purified P49-His6 (filled square), D94A- mutated P49-His6 (open square) or P35-His6 (filled triangle) and assayed as described above. (D) P49 cleavage. In vitro synthesized, 35S-radiolabeled, wild-type (wt) or D94A-mutated P49 was mixed with buffer (lanes 1 and 2), human caspase-3 (lanes 3 and 4) or Sf-caspase-1 (lanes 5 and 6) and analyzed by SDS–PAGE. The 40 kDa (*) and 9 kDa (*′) cleavage fragments are indicated (bottom). (E) P49–caspase-His6 complexes. Radiolabeled untagged wild-type or D94A-mutated P49 was mixed with buffer or His6-tagged human caspase-3 (300 pmol). Complexes recovered by Ni2+ affinity chromatography in the presence (lanes 3 and 4) or absence (lanes 5 and 6) of caspase-3 were analyzed by SDS–PAGE. Uncleaved P49 (input) was included (lanes 1 and 2). (F) P49-His6–caspase complexes. Purified wild-type (wt) or D94A-mutated P49-His6 was mixed with buffer or untagged caspase-3. Recovered Ni2+-bound complexes were subjected to immunoblot analysis by using α-P49 (top) or caspase-3 large subunit α-P17 (bottom) antisera.
Wild-type, but not D94A-mutated P49, was cleaved by human caspase-3 and Sf-caspase-1 to produce 40 and 9 kDa fragments (Figure 2D). Amino acid sequence analysis revealed that the N-terminus of the larger 40 kDa fragment was GGGAD (data not shown), indicating that cleavage occurred between P49 residues Asp94 and Gly95 within the predicted cleavage site TVTD94↓ GGGAD. To determine whether the P49 cleavage fragments stably associate with the targeted caspase in a manner analogous to that of P35, we used His6-tagged caspase-3 in Ni2+ affinity pull-down assays. Upon recovery of His6-tagged caspase-3 from cleavage reactions containing untagged P49, both 40 and 9 kDa fragments of wild-type P49 (Figure 2E, lane 3), but not D94A-mutated P49 (lane 4) were detected. Neither wild-type nor mutated P49 was recovered in the absence of His6-tagged caspase. In reciprocal assays, where the association of untagged caspase-3 with P49-His6 was tested, caspase-3 interacted stably with cleaved P49-His6 (Figure 2F, lane 2). In contrast, D94A-mutated P49 was neither cleaved nor associated with caspase-3 (lane 3). Collectively, these data indicate that caspase inhibition in vitro includes the formation of a stable P49–caspase complex, which requires cleavage at Asp94.
P49 functions upstream of P35 but downstream of Op-IAP
Although P49 and P35 function similarly in vitro, the structural deviations of these two caspase inhibitors suggested that they act differently in vivo, possibly by targeting distinct caspases. This notion was supported by the 100-fold difference in P49 and P35 inhibition of Sf-caspase-1 (Figure 2B). To investigate potential differences, we first defined the apoptotic step affected by P49. To this end, we constructed a recombinant baculovirus (vP49) that encodes P49 as its singular apoptotic inhibitor. By inoculating SF21 cultures with this virus, P49 was synthesized in every cell that received an apoptotic stimulus. Importantly, P49 activity could therefore be monitored in the context of physiologically relevant levels of death effectors.
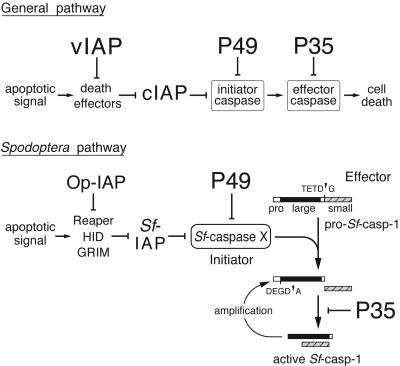
As expected, apoptosis was fully blocked in SF21 cells infected with vP49. Directed by the immediate early ie-1 promoter, P49 was synthesized early during infection (Figure 3A). Our α-P49 antiserum recognized full-length P49 and its 40 kDa fragment (*) but not the 9 kDa N-terminal fragment as demonstrated by comparing purified P49-His6 partially cleaved by caspase (Figure 3A). P49 cleavage was first detected between 6 and 12 h and thus coincided with virus-induced caspase activation (LaCount and Friesen, 1997; LaCount et al., 2000). To confirm the identity of the 40 kDa cleavage fragment that was present at low levels, we compared cells transfected with wild-type or mutated p49. The 40 kDa fragment was detected only in the presence of wild-type P49, not cleavage-impaired D49A-mutated P49 (Figure 3B, lanes 5 and 6). Furthermore, generation of the cleavage fragment required apoptotic signaling, since it was present in virus-infected cells (lane 5) but not mock-infected cells (lane 2). Another P49-related protein (Figure 3B, filled diamond) detected early in vP49 infection and upon transfection with wild-type and D49A-mutated P49 is a likely internal initiation product.
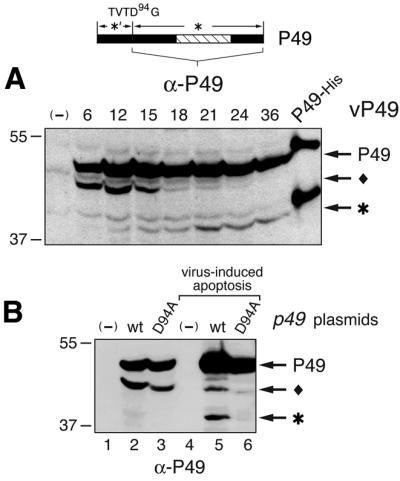
Fig. 3. In vivo P49 cleavage. (A) Time course. SF21 cell lysates prepared at the indicated hours after infection with vP49 were subjected to immunoblot analysis using α-P49, which recognized full-length P49 and the 40 kDa cleavage fragment (top), but not the N-terminal 9 kDa fragment. Due to the absence of His tags, in vivo P49 and its 40 kDa cleavage fragment (*) are smaller than P49-His6, which was partially cleaved by caspase (right lane). Another P49-related protein (filled diamond) is indicated. (B) Asp94-dependent P49 cleavage. SF21 cells were mock-transfected (–) or transfected with plasmid encoding wild-type (wt) or D94A-mutated P49. After mock infection (lanes 1–3) or infection with p35– vΔp35 to induce apoptosis (lanes 4–6), cell lysates (5 × 105 cell equiv) were prepared and subjected to α-P49 immunoblot analysis.
To define the apoptotic step affected by P49, we determined where P49 functions relative to P35 by testing the effect of P49 on in vivo cleavage of P35, a sensitive indicator of effector caspase activity (Bertin et al., 1996; Manji et al., 1997). Upon infection with a p35-carrying virus (p35+), activated caspase(s) readily cleaved P35 to generate the 25 kDa cleavage (*) fragment (Figure 4A, lane 2). However, upon coinfection with vP49, P49 blocked the cleavage of P35 (lane 3). Conversely, P49 was cleaved normally even in the presence of abundant P35 (Figure 4B, lane 3). Taking into account the reduced P49 synthesis due to p35+ virus coinfection, P49 cleavage was comparable to that in cells infected with vP49 alone (lane 2). We concluded that P49 directly or indirectly inhibits the caspase(s) responsible for P35 cleavage and thus functions upstream of P35.
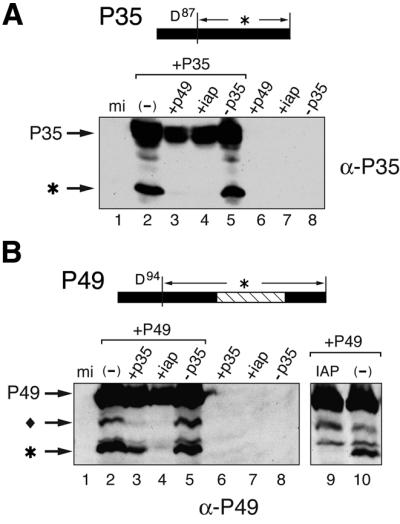
Fig. 4. P49 functions upstream of P35 but downstream of Op-IAP. (A) P49 inhibits P35 cleavage. Sf21 cells were mock-infected (mi) or infected with viruses vP49 (+p49), vOp-IAP (+iap) or vΔp35 (–p35) with (lanes 2–5) or without (lanes 6–8) wild-type p35+ virus (+P35). Lysates (3 × 105 cell equiv) prepared 24 h later were subjected to immunoblot analysis using α-P35. Top: P35 and its C-terminal cleavage product (*) are indicated. (B) Op-IAP inhibits P49 cleavage. Lysates (106 cell equiv) prepared 24 h after infection with the viruses described in (A) with (lanes 2–5) or without (lanes 6–8) vP49 (+P49) were subjected to immunoblot analysis with α-P49. SF21 cells constitutively expressing Op-iap (IAP) were included (lane 9). (Top) P49 and its C-terminal cleavage fragment (*) are indicated. The P49-related protein (filled diamond) is marked.
To determine where P49 functions relative to an IAP, we also assessed the effect of Op-IAP on P49 cleavage. In cells coinfected with vP49 and an Op-iap+ virus (vOp-IAP), Op-IAP blocked P49 cleavage (Figure 4B, lane 4). To confirm this finding, we infected SF21 cells that constitutively express Op-iap with vP49. In contrast to parental (Op-iap–) cells, P49 cleavage was fully blocked in Op-iap+ cells (Figure 4B, compare lanes 9 and 10). As expected, Op-IAP also prevented P35 cleavage (Figure 4A, lane 4) confirming its upstream function relative to P35 (Manji et al., 1997). These data indicated that Op-IAP functions at or upstream of the caspase that cleaves P49, which is upstream of the caspase(s) inhibited by P35.
P49, but not P35, prevents in vivo proteolytic processing of effector caspases
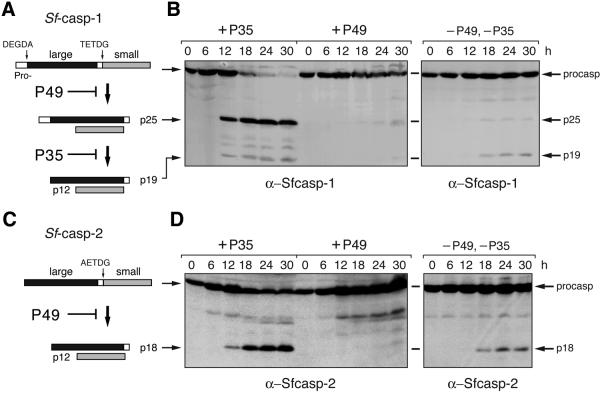
During apoptosis of SF21 cells, pro-Sf-caspase-1 (Figure 5A) and pro-Sf-caspase-2 (Figure 5C) are proteolytically processed (Ahmad et al., 1997; LaCount, 1998; LaCount et al., 2000). This signal-induced activation is initiated by a P35-insensitive caspase (LaCount et al., 2000; Manji and Friesen, 2001). Sf-caspase-1 undergoes a second cleavage, which removes the prodomain mediated by a P35-inhibitable protease, probably Sf-caspase-1 itself (LaCount et al., 2000). To determine whether P49 affects the caspase responsible for processing these effector caspases, we compared Sf-caspase activation in the presence of P49 and P35. Upon apoptotic signaling caused by p35+ virus infection, pro-Sf-caspase-1 was processed to its large subunit fragments p25 and p19 (Figure 5B). Thus, cleavage of pro-Sf-caspase-1 occurred at TETD↓G despite an abundance of P35. In contrast, P49 blocked the processing of pro-Sf-caspase-1 during infection with vP49 (Figure 5B). Levels of pro-Sf-caspase-1 were unchanged and cleavage products, including p25, were not detected until very late. In a similar pattern, P49 prevented proteolytic processing of pro-Sf-caspase-2, whereas P35 did not (Figure 5D). The large subunit p18 of Sf-caspase-2 accumulated in the presence of P35 but not P49. These data confirmed that P49 acts at a distinct step upstream from P35 in the apoptotic pathway of these lepidopteran cells. Furthermore, P49 likely targets the P35-insensitive initiator caspase responsible for activation of pro-Sf-caspase-1 and -2.
Fig. 5. P49 inhibits effector caspase proteolytic processing. (A and C) Processing of Sf-caspase-1 and -2. Activation of pro-Sf-caspase-1 initiates with cleavage at TETD↓G to generate intermediate p25, which is subsequently cleaved at DEGD↓G to generate the mature p19 and p12 subunits. Pro-Sf-caspase-2 is cleaved at AETD↓G to generate p18 and p12 subunits. (B and D) Effect of P49 on Sf-caspase-1 and -2 processing. SF21 lysates (3.8 × 105 cell equiv) prepared at the hours (h) indicated after infection with wild-type p35+ virus (+P35), vP49 (+P49) or vΔp35 (–P49, –P35) were subjected to immunoblot analysis using α-Sfcasp1 (B) or α-Sfcasp2 (D) antiserum.
Does the P49 cleavage site confer caspase specificity?
The similarity between the P49 cleavage site TVTD↓G (Figure 6A) and the initial processing sites TETD↓G and AETD↓G of pro-Sf-caspase-1 and -2, respectively, suggested that the apical caspase selectivity of P49 is conferred by its RSL cleavage residues. We hypothesized that the in vivo selectivity of P49 and P35 could be switched by exchanging cleavage site residues. Thus, we substituted the P4–P1 residues of P49 and P35 and determined the effect of these swaps by monitoring Sf-caspase activation in transfected cells.
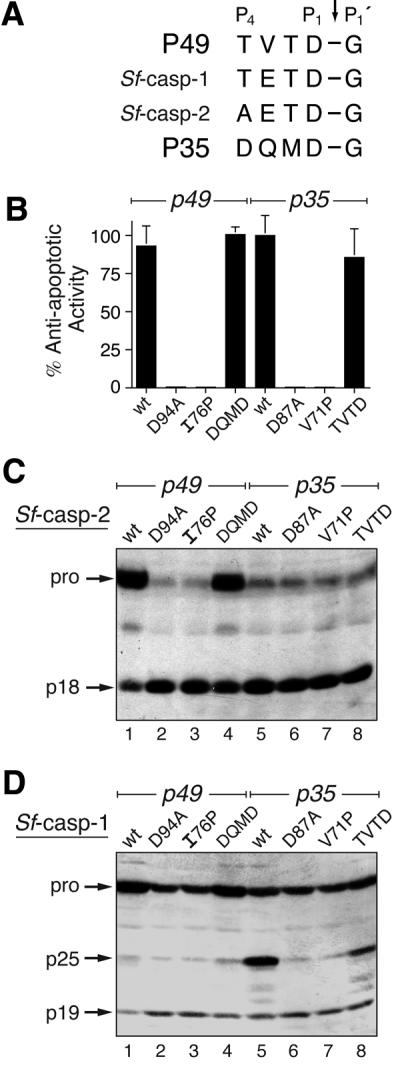
Fig. 6. In vivo caspase selectivity by P49 and P35. (A) Cleavage sites. The P4–P1′ residues of P49 and P35 are compared with the initial processing sites of pro-Sf-caspase-1 and -2. (B) Anti-apoptotic activity. The capacity of plasmid-borne wild-type (wt) or mutated p49 and p35 to block virus-induced apoptosis was determined by marker rescue. The average ± standard deviation of triplicate transfections is reported as a percentage of the capacity of wild-type P35 to restore virus replication. (C and D) Processing of Sf-caspases. SF21 cells were transfected with plasmids encoding wt or mutated P49 (lanes 1–4) or wt or mutated P35 (lanes 5–8), and infected 24 h later to induce apoptosis. Lysates (3.8 × 105 cell equiv) prepared 24 h after infection were subjected to immunoblot analysis with α-Sfcasp2 (C) or α-Sfcasp1 (D).
The exchange of P49 or P35 cleavage sites had no effect on the anti-apoptotic activity of either caspase inhibitor. Marker rescue assays demonstrated that DQMD94G-substituted P49 and TVTD87G-substituted P35 were as effective as the wild-type proteins in blocking virus-induced apoptosis (Figure 6B). In contrast, loss-of- function D94A- or I76P-mutated P49 and D87A- or V71P-mutated P35 failed to block apoptosis. Our previous studies indicated that substitution V71P disrupted the P35 RSL and thus caused loss of function (Fisher et al., 1999; Zoog et al., 1999). Although fully functional, the cleavage site substitutions failed to alter the effect of P49 or P35 on procaspase processing during virus infection. Both wild-type and DQMD94G-substituted P49 prevented pro-Sf-caspase-2 processing (Figure 6C, lanes 1 and 4), whereas loss-of-function D94A- or I76P-mutated P49 did not (lanes 2 and 3). The higher p18 levels (lanes 1 and 4) were attributed to those infected cells that failed to produce P49 because they were not transfected (transfection efficiencies ranged from 75 to 90%). Furthermore, neither wild-type (lane 5) nor TVTD87G-P35 (lane 8) blocked pro-Sf-caspase-2 processing. As expected, loss-of-function D87A- and V71P-mutated P35 (lanes 6 and 7) failed to block processing. Likewise, wild-type and DQMD94G-P49 prevented pro-Sf-caspase-1 processing (Figure 6D, lanes 1 and 4). TVTD87G-P35 exhibited a slightly increased capacity to prevent processing compared with wild-type P35 (Figure 6D, lanes 5 and 8). Overall, the exchange of cleavage sites had a minimal effect on processing of pro-Sf-caspase-1 and -2. We concluded that the P4–P1 residues are not sufficient to confer P49 or P35 target specificity in vivo. Thus, specificity is conferred by a determinant other than or in addition to the cleavage site residues.
P49 is a substrate inhibitor of human initiator caspase-9
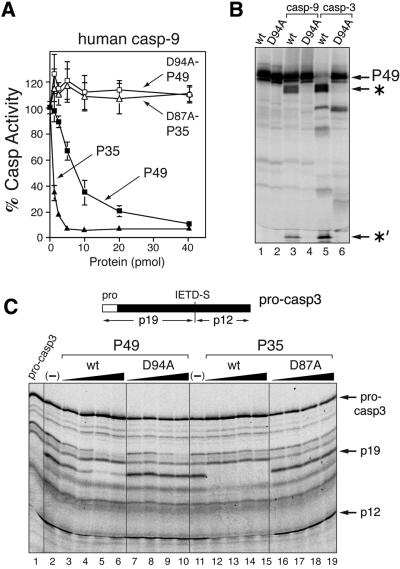
Our data suggested that P49 functions in vivo to block activity of an initiator caspase. Therefore, to determine whether P49 is capable of inhibiting a well characterized initiator caspase, we tested the effect of P49 on purified human caspase-9, which functions in the mitochondrion/cytochrome c death pathway of mammals (Earnshaw et al., 1999; Chang and Yang, 2000; Adrain and Martin, 2001). When tested in dose-dependent assays that used the tetrapeptide LEHD-afc as substrate, purified P49-His6 effectively inhibited caspase-9 but D94A-mutated P49-His6 did not (Figure 7A). Interestingly, wild-type but not D87A-mutated P35-His6 inhibited caspase-9. Under identical conditions, baculovirus vector purified P49 was ∼8-fold less potent than Escherichia coli-produced P35. Wild-type but not D94A-mutated P49 was cleaved by caspase-9 into 40 and 9 kDa fragments that were electrophoretically indistinguishable from those generated by human caspase-3 (Figure 7B). Collectively, these data indicate that P49 acts as a substrate inhibitor of caspase-9. To further determine whether P49 blocks the capacity of caspase-9 to proteolytically process and thus activate human effector caspase-3, we tested the effect of increasing concentrations of P49 on cleavage of pro-caspase-3 by active caspase-9 in vitro. As expected, caspase-9 cleaved pro-caspase-3 at IETD↓S between its large and small subunits to generate the p19 and p12 subunits (Figure 7C, lanes 2 and 11). However, this cleavage was blocked in a dose-dependent manner by P49-His6 (lanes 3–6) but not D94A-mutated P49-His6 (lanes 7–10). P35-His6 exhibited a similar pattern of inhibition (Figure 7C, lanes 12–19). We concluded that P49 has the capacity to inhibit the activity of caspase-9 that is required for proteolytic activation of human effector caspase-3.
Fig. 7. P49 is a substrate inhibitor of human caspase-9. (A) Dose-dependent inhibition. Purified recombinant caspase-9 (40 pmol) was incubated with the indicated amounts (pmol) of purified P49-His6, D94A-mutated P49-His6, P35-His6 or D87A-mutated P49-His6. After 30 min, residual caspase activity was measured by using the substrate LEHD-afc as described in the legend to Figure 2. (B) P49 cleavage. In vitro synthesized, 35S-radiolabeled, wild-type (wt) or D94A-mutated P49 was incubated with buffer alone (lanes 1 and 2), caspase-9 (lanes 3 and 4) or human caspase-3 (lanes 5 and 6) and analyzed by SDS–PAGE. The 40 kDa (*) and 9 kDa (*′) cleavage fragments are indicated. (C) Inhibition of procaspase-3 processing. Purified caspase-9 was mixed with buffer alone (–), wild-type (wt) or D94A-mutated P49-His6 (lanes 3–11), or wild-type or D87A-mutated P49-His6 (lanes 12–19). After 30 min, in vitro synthesized, 35S-labeled, procaspase-3 was added, incubated for 6 h and analyzed by SDS–PAGE. Input procaspase-3 is shown (lane 1). Caspase-3 p19 and p12 subunits are indicated (top).
P49 prevents apoptosis in Drosophila
The capacity of P49 to inhibit vertebrate caspases in vitro suggested that P49 would suppress apoptosis in a phylogenetically distinct organism. To explore this possibility, we tested the anti-apoptotic potential of P49 in DL-1 cells of D.melanogaster (Order: Diptera). To this end, we used recombinant baculoviruses simultaneously to deliver apoptotic regulatory genes and to induce apoptosis. Although DL-1 cells fail to support productive baculovirus infection (Morris and Miller, 1993), virus entry and gene expression are efficient. Upon inoculation with the p35-deletion mutant vΔp35, DL-1 cells underwent widespread apoptosis (Figure 8A). Likewise, vOp-IAP caused apoptosis that affected >95% of the DL-1 culture. Both viruses caused extensive apoptotic body formation (Figure 8A) and intracellular DNA fragmentation (Figure 8B). Consistent with caspase activation, the effector caspase drICE (Fraser et al., 1997) was proteolytically processed and DEVD-amc cleavage activity was readily detected (E.Lannan, S.Perry, J.Theisen and P.Friesen, unpublished data). Thus, Op-IAP failed to block apoptosis in DL-1 cells despite abundant synthesis (not shown). This finding suggests a fundamental difference between Op-IAP and the other IAPs that regulate apoptosis in Drosophila (Miller, 1999).
Fig. 8. P49 blocks baculovirus-induced apoptosis in Drosophila DL-1 cells. (A) Virus-induced apoptosis. Drosophila Line-1 (DL-1) cells were photographed 24 h after mock infection (mock) or inoculation with p35– vΔp35, Op-iap+ vOp-IAP, p49+ vP49 or wild-type p35+ virus (vP35). Magnification 100×. (B) DNA fragmentation. Low molecular weight DNA was selectively extracted from equivalent numbers of DL-1 cells and associated apoptotic bodies. Only fragmented DNA was visualized by agarose gel electrophoresis since intact cellular DNA was excluded. (C and D) P49 synthesis and cleavage. DL-1 lysates (3 × 106 cell equiv) prepared at the times indicated after inoculation with vP49 or vP35 were subjected to immunoblot analysis with α-P49 (C) or α-P35 (D). Full-length and C-terminal cleavage (*) proteins are indicated.
In contrast, P49 and P35 blocked virus-induced apoptosis of DL-1 cells when expressed from vP49 or p35+-virus vP35, respectively. Inoculated cells showed no signs of membrane blebbing (Figure 8A) or DNA fragmentation (Figure 8B). A majority of the inoculated cells detached from the monolayer, but remained intact for several days. Confirming that these cells were infected, P49 (Figure 8C) and P35 (Figure 8D) were synthesized in abundance. Furthermore, P49 was cleaved to generate the 40 kDa fragment (*) indicative of intracellular caspase activity. P49 cleavage was first detected 9–12 h after inoculation (Figure 8C), coinciding with caspase activation in these cells (E.Lannan, S.Perry, J.Theisen and P.Friesen, unpublished data). Previously shown to block apoptosis in Drosophila (Hay et al., 1994; White et al., 1996; Jiang et al., 1997), P35 was cleaved with kinetics similar to those of P49 (Figure 8D). These hallmark caspase-mediated cleavages argue that the anti-apoptotic activity of P49 in Drosophila, like that of P35, is due to caspase inactivation by direct substrate inhibition.
Discussion
P49 blocks a central caspase-mediated initiation step in apoptosis
Our study indicates that P49 is a potent apoptotic suppressor that functions as a substrate inhibitor of caspases from invertebrates and vertebrates. Although related to pancaspase inhibitor P35, P49 is distinguished by its unique in vivo capacity to inhibit an initiator caspase that is insensitive to P35. As such, P49 is the third distinct apoptotic regulator encoded by the invertebrate baculoviruses (Figure 9). The finding that P49 prevents apoptosis induced by diverse signals in cells from S.frugiperda and D.melanogaster suggests that this apoptotic regulator acts at a phylogenetically conserved step in the death pathway. In SF21 cells, P49 blocked proteolytic processing of effector caspases Sf-caspase-1 and -2 (Figure 5) by preventing cleavage at (T/A)ETD↓G between the large and small subunits. Thus, P49 likely inhibits the P35-insensitive initiator caspase, designated Sf-caspase-X, responsible for activation of effector caspases in Spodoptera (LaCount et al., 2000; Manji and Friesen, 2001). Consistent with the capacity of P49 to block activation of caspases that are targeted by P35, P49 prevented in vivo cleavage of P35 by these downstream caspases (Figure 4). Conversely, P35 had no effect on in vivo cleavage of P49. Thus, P49 functions at a distinct step upstream from and unaffected by P35 (Figure 9). In support of this conclusion, in vitro assays demonstrated that P49 is a poor inhibitor of active Sf-caspase-1 when compared with P35, which stoichiometrically inhibits this effector protease (Figure 2).
Fig. 9. Cell death regulation by baculovirus apoptotic suppressors. Top: general pathway. Baculovirus IAP (vIAP) and caspase inhibitors P49 and P35 prevent signal-induced apoptosis at distinct steps relative to endogenous cellular IAP (cIAP). A linear pathway is shown but converging paths are possible. Bottom: model for caspase activation in Spodoptera. Upon apoptotic signaling, prodeath effectors (Reaper, HID or GRIM homologs) trigger activation of initiator Sf-caspase-X by inhibition of Sf-IAP. Sf-caspase-X proteolytically activates downstream effector caspases, including Sf-caspase-1, by cleavage at the large– small subunit junction. Active effector caspases may amplify processing. Distinct steps are inhibited by P49 and P35. Op-IAP functions upstream by association with death effectors, Sf-caspase-X or both.
Collectively, our data support a model for cell death in Spodoptera (Figure 9) wherein P49 targets initiator Sf-caspase-X, which functions to activate effector caspases, including Sf-caspase-1. Sf-caspase-X is likely regulated by endogenous Sf-IAP, a Spodoptera IAP capable of inhibiting human initiator caspase-9 (Huang et al., 2000). Consistent with such IAP regulation, Op-IAP blocked in vivo caspase-mediated cleavage of P49 (Figure 4B). Thus, this viral IAP acts upstream to inhibit the activation or activity of Sf-caspase-X. P35 functions furthest downstream by inhibiting active effector caspases and blocking their maturation. Thus, initiator and effector steps are selectively targeted by these baculovirus caspase inhibitors (Figure 9).
P49 inhibits vertebrate initiator and effector caspases in vitro
Our finding that P49 inhibited invertebrate initiator caspase activity in vivo predicted that it would also affect other initiator caspases, including human caspase-9, which is well characterized. Indeed, tetrapeptide cleavage by recombinant caspase-9 was potently inhibited by P49 through a substrate inhibitor mechanism (Figure 7A). Furthermore, P49 blocked proteolytic processing of effector caspase-3 by caspase-9 (Figure 7C). These findings suggest that P49 has the capacity to inhibit in vivo activity of caspase-9, which has been reported to be insensitive to P35 (Vier et al., 2000). In a purified in vitro system, P49 also inhibited human effector caspase-3 (Figure 2). P49 was nearly as potent as P35, a stoichiometric inhibitor with a subnanomolar Ki (Bump et al., 1995; Bertin et al., 1996; Zhou et al., 1998). In addition, P49 inhibited effector Sf-caspase-1 but much less effectively than P35. Nonetheless, P49 may have the capacity to target both initiator and effector caspases in vivo. Thus, P49’s expanded potential to target multiple classes of caspases may confer a selective advantage to its virus during infection of specific tissues or animal hosts. The molecular mechanism determining in vivo target selectivity of P49 and P35 is of significant interest (see below).
P49 is a substrate inhibitor with a P35-like mechanism
Cleavage at TVTD94↓G was required for P49’s anti-caspase activity in vitro (Figures 2 and 7). Like P35, P49’s two cleavage fragments remained stably associated with the inhibited caspase (Figure 2E and F). Confirming the mechanistic significance of cleavage, D94A-mutated P49 was not cleaved in vivo and failed to prevent apoptosis (Figures 1 and 3). Cleavage-defective P49 also failed to block proteolytic processing of Sf-caspase-1 and -2 (Figure 6). The P35 cleavage site DQMD87↓G is located at the apex of a RSL that must be properly anchored to the main protein core for anti-caspase activity (Fisher et al., 1999; Zoog et al., 1999). The sequence similarity of P49 and P35 (Figure 1A) suggests that P49 has an RSL and a P35-like fold, despite its larger size. Supporting this model, the P49 substitution I76P, which disrupted the predicted α-helix anchoring the RSL, caused loss of function (Figure 6). Substitution of the analogous residue (Val71) in the α-helix of P35 also disrupted function (Fisher et al., 1999; Zoog et al., 1999). Upon cleavage, the P35 RSL undergoes a conformational rearrangement and forms a stable thioester linkage with the active site of the caspase (de la Cruz et al., 2001; Xu et al., 2001). Thus, on the basis of structural similarity and the capacity to form a stable inhibitor–caspase complex, it is likely that P49 uses a similar, if not identical, mechanism for suicide inhibition.
P4–P1 cleavage residues are insufficient for in vivo caspase selectivity
Although P49 and P35 use similar mechanisms, what accounts for the difference in their in vivo specificity? Since cleavage is critical for anti-caspase activity, we tested the contribution of cleavage site residues to caspase targeting. When the P4–P1 residues (TVTD94↓G) of P49 were swapped with those of P35 (DQMD87↓G), the exchange had no significant effect on specificity, since inhibition of Sf-caspase-1 and -2 processing by DQMD94G-substituted P49 was comparable to that by wild-type P49 (Figure 6). Likewise, the inability of P35 to block processing of both effector caspases was unaffected by substituting its cleavage site with that of P49, even though the TVTD87G-modified P35 was fully functional. In contrast to P49 and P35, alterations of the caspase cleavage residues affected the specificity and potency of CrmA (Xue and Horvitz, 1995; Ekert et al., 1999). Although we have not ruled out the possibility that additional RSL residues contribute to selectivity, our data suggest that P49 is not simply a larger version of P35 with a different cleavage site specificity. Rather, the caspase selectivity of P49 is conferred by a determinant absent in P35. An intriguing possibility is that the unique 120 residue insertion of P49 (Figure 1A) is involved.
P49 blocks virus-induced apoptosis in Drosophila
The capacity of P49 to inhibit both invertebrate and vertebrate caspases in vitro suggested that P49 should function in phylogenetically diverse organisms. Indeed, P49 blocked virus-induced apoptosis in cultured cells from Drosophila (Figure 8). Apoptosis of DL-1 cells was also suppressed by P35, which has been shown to prevent programmed cell death in invertebrates and vertebrates (Hay et al., 1994; Sugimoto et al., 1994; Grether et al., 1995; Izquierdo et al., 1999; Hisahara et al., 2000; Viswanath et al., 2000). Since P49 and P35 were proteolytically cleaved in Drosophila cells (Figure 8), a substrate inhibitor mechanism for both apoptotic suppressors is likely. Our preliminary experiments (E.Lannan, S.Perry, J.Theisen and P.Friesen, unpublished data) indicated that P49 and P35 have different targets among the seven Drosophila caspases identified to date (Kumar and Doumanis, 2000; Vernooy et al., 2000). An obvious Drosophila target for P49 is DRONC, a CARD-containing caspase that is resistant to P35 inhibition and likely functions as an initiator in Drosophila (Hawkins et al., 2000; Meier et al., 2000). However, the low in vitro activity of recombinant DRONC (Dorstyn et al., 1999) has precluded direct tests of P49 inhibition of DRONC (data not shown). Consequently, alternative approaches to iden tify the Drosophila targets of P49 and P35 are underway. We predict that like human initiator caspase-9, DRONC will be inhibited by P49.
Our study also demonstrates for the first time that it is possible to trigger uniform apoptosis in cultured Droso phila cells by using baculovirus vectors. Inoculation of DL-1 cells with moderate levels [∼10 plaque-forming units (p.f.u.) per cell] of AcMNPV recombinants that lack functional apoptotic suppressors caused complete apoptosis within 24 h. Moreover, the use of recombinant viruses allowed simultaneous expression of functional anti-apoptotic genes to block apoptosis at different steps in the death pathway. The capacity to deliver such apoptotic regulators with near 100% efficiency to cells that are normally refractive to standard transfection methods should facilitate studies on apoptotic pathways in Drosophila, a model organism for studies on programmed cell death (Bergmann et al., 1998; Abrams, 1999). In particular, P49 promises to be useful in defining the molecular steps of caspase activation in flies and other organisms.
Materials and methods
Cells and viruses
Spodoptera frugiperda IPLB-SF21 (Vaughn et al., 1977) and Drosophila Schneider DL-1 (Schneider, 1972) cell lines were propagated at 27°C in TC100 and Schneider’s growth medium, respectively (Gibco-BRL). For infection, cell monolayers were exposed to ≥15 p.f.u./cell. Wild-type AcMNPV (p35+, iap–) and AcMNPV recombinants vΔp35 (p35–, iap–), vΔp35/lacZ and vOp-IAP (p35–, iap+) have been described (Hershberger et al., 1992; Manji et al., 1997). Recombinant vP49 (p49+, p35–, iap–) was generated by allelic replacement (Hershberger et al., 1992) whereby the polyhedrin gene of parent vΔp35 was substituted with SlNPV p49 (Du et al., 1999) fused to the AcMNPV ie-1 promoter and linked to a lacZ reporter. To overproduce P49-His6, an AcMNPV expression vector was constructed with the pFastBac1 donor plasmid (BRL Life Technologies) using standard procedures.
Plasmids
pIE1-P49 was constructed by inserting the p49 open reading frame from plasmid pES2 (Du et al., 1999) downstream from the ie-1 promoter of pIE1hr/PA at an NdeI site introduced at p49 nucleotide position –1. p49 and nucleotide substitutions thereof were generated by using overlap extension PCRs and verified by nucleotide sequencing. Plasmids expressing AcMNPV p35 and OpMNPV Op-iap using the ie-1 promoter have been described (Bertin et al., 1996; Manji et al., 1997; Zoog et al., 1999).
Cell survival and marker rescue assays
SF21 monolayers were transfected with plasmid DNAs by using cationic liposomes as described previously (LaCount et al., 2000). After infection or UV irradiation, viable and apoptotic cells were distinguished by computer-aided microscopy as described previously (Manji and Friesen, 2001). The mean ± standard deviation was calculated from the number of surviving cells in five evenly distributed fields of view. For marker rescue assays (Bertin et al., 1996), SF21 cells were inoculated with vΔp35/lacZ 16 h after transfection with plasmid DNAs. The resulting extracellular virus was quantified by plaque assay using SF21 cells and X-gal to identify rescued (non-apoptotic) virus.
In vitro caspase inhibition assays
P35-His6 and D87A-mutated P35-His6 were obtained from E.coli (Fisher et al., 1999), but similar attempts to produce P49 in E.coli yielded only insoluble protein. Thus, wild-type and D94A-mutated P49-His6 were purified by Ni2+ affinity chromatography (Zoog et al., 1999) after production in SF21 cells by AcMNPV expression vectors. P49 preparations were judged ≥95% pure by electrophoretic analysis. Purified His6-tagged Sf-caspase-1 (LaCount et al., 2000), His6-tagged human caspase-3 (Fisher et al., 1999) or recombinant human caspase-9 (Calbiochem) was mixed with increasing quantities of P49- or P35-His6 using reaction conditions described previously (Bertin et al., 1996; Fisher et al., 1999). After 30 min, residual caspase activity was measured by using substrates Ac-DEVD-amc or Ac-LEHD-afc. Reported values are the average of triplicate assays in which the rate of product formation was obtained from the linear portion of the reaction curves within the first 10% of substrate depletion.
P49 cleavage and caspase association
Untagged, [35S]methionine-labeled P49 from coupled transcription– translation reactions (Promega) was mixed with purified His-tagged Sf-caspase-1 or human caspase-3 for 30 min in 10 mM HEPES pH 7.5–0.1% CHAPS–10% sucrose. Conversely, purified wild-type or D94A-mutated P49-His6 (5 µg) was mixed with E.coli extract containing untagged human caspase-3. After Ni2+ affinity purification (Fisher et al., 1999), proteins were subjected to SDS–PAGE and immunoblot analyses. For N-terminal sequence analysis, the large and small fragments of P49-His6 generated by cleavage with human caspase-3 or Sf-caspase-1 were separated electrophoretically and subjected to Edman degradation (Midwest Analytical, Inc.). For cleavage by human caspase-9, reactions containing [35S]methionine-labeled procaspase-3 in 50 mM HEPES pH 7.2, 50 mM NaCl, 10 mM DTT, 10 mM EDTA, 0.1% CHAPS, 5% glycerol were incubated at 37°C for the times indicated. The identity of each caspase was confirmed by use of caspase-specific tetrapeptide inhibitors (data not shown).
Immunoblot analysis and antisera
For P49-specific antiserum (α-P49), insoluble E.coli-produced P49-His6 was used as antigen for immunization of New Zealand white rabbits (University of Wisconsin Medical School Polyclonal Antibody Service). Immunoblot analyses were conducted using the following antisera: α-P49, α-p17 of human caspase-3 (Fisher et al., 1999), α-Sfcasp1 (LaCount et al., 2000), α-Sfcasp2 (LaCount, 1998) and monoclonal α-P35 (gift from Y.Lazebnik).
Acknowledgments
Acknowledgements
We thank Doug LaCount for preparation of α-Sfcasp2 antiserum and his efforts in the discovery and characterization of Sf-caspase-2. We also thank Yuri Lazebnik (Cold Spring Harbor Laboratory) for the gift of P35 monoclonal antibody, and Pascal Meier and Gerard Evans (Imperial Cancer Research Fund, London) for drICE antisera. This work was supported in part by Public Health Service grant AI40482 from the National Institute of Allergy and Infectious Diseases (P.D.F.), NIH Predoctoral Traineeship GM07215 (S.J.Z.) and NIH Postdoctoral Traineeship CA09075 (J.J.S.).
References
- Abrams J. (1999) An emerging blueprint for apoptosis in Drosophila. Trends Cell Biol., 9, 435–440. [DOI] [PubMed] [Google Scholar]
- Adrain C. and Martin,S.J. (2001) The mitochondrial apoptosome: a killer unleashed by the cytochrome seas. Trends Biochem. Sci., 26, 390–397. [DOI] [PubMed] [Google Scholar]
- Ahmad M., Srinivasula,S.M., Wang,L.J., Litwack,G., Fernandes-Alnemri,T. and Alnemri,E.S. (1997) Spodoptera frugiperda caspase-1, a novel insect death protease that cleaves the nuclear immunophilin FKBP46, is the target of the baculovirus antiapoptotic protein P35. J. Biol. Chem., 272, 1421–1424. [DOI] [PubMed] [Google Scholar]
- Bergmann A., Agapite,J. and Steller,H. (1998) Mechanisms and control of programmed cell death in invertebrates. Oncogene, 17, 3215–3223. [DOI] [PubMed] [Google Scholar]
- Bertin J., Mendrysa,S.M., LaCount,D.J., Gaur,S., Krebs,J.F., Armstrong,R.C., Tomaselli,K.J. and Friesen,P.D. (1996) Apoptotic suppression by baculovirus P35 involves cleavage by and inhibition of a virus-induced CED-3/ICE-like protease. J. Virol., 70, 6251–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum M.J., Clem,R.J. and Miller,L.K. (1994) An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol., 68, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bump N.J. et al. (1995) Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science, 269, 1885–1888. [DOI] [PubMed] [Google Scholar]
- Chang H.Y. and Yang,X. (2000) Proteases for cell suicide: functions and regulation of caspases. Microbiol. Mol. Biol. Rev., 64, 821–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem R.J. (2001) Baculoviruses and apoptosis: the good, the bad and the ugly. Cell Death Differ., 8, 137–143. [DOI] [PubMed] [Google Scholar]
- Cryns V. and Yuan,J. (1998) Proteases to die for. Genes Dev., 12, 1551–1571. [DOI] [PubMed] [Google Scholar]
- de la Cruz W.P., Friesen,P.D. and Fisher,A.J. (2001) Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage. J. Biol. Chem., 276, 32933–32939. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L. and Reed,J.C. (1999) IAP family proteins—suppressors of apoptosis. Genes Dev., 13, 239–252. [DOI] [PubMed] [Google Scholar]
- Dorstyn L., Colussi,P.A., Quinn,L.M., Richardson,H. and Kumar,S. (1999) DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl Acad. Sci. USA, 96, 4307–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Lehavi,D., Faktor,O., Qi,Y. and Chejanovsky,N. (1999) Isolation of an apoptosis suppressor gene of the Spodoptera littoralis nucleopolyhedrovirus. J. Virol., 73, 1278–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett C.S., Nava,V.E., Gedrich,R.W., Clem,R.J., Van Dongen,J.L., Gilfillan,M.C., Shiels,H., Hardwick,J.M. and Thompson,C.B. (1996) A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J., 15, 2685–2694. [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Martins,L.M. and Kaufmann,S.H. (1999) Mammalian caspases: structure, activation, substrates and functions during apoptosis. Annu. Rev. Biochem., 68, 383–424. [DOI] [PubMed] [Google Scholar]
- Ekert P.G., Silke,J. and Vaux,D.L. (1999) Inhibition of apoptosis and clonogenic survival of cells expressing crmA variants: optimal caspase substrates are not necessarily optimal inhibitors. EMBO J., 18, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.J., Cruz,W., Zoog,S.J., Schneider,C.L. and Friesen,P.D. (1999) Crystal structure of baculovirus P35: role of a novel reactive site loop in apoptotic caspase inhibition. EMBO J., 18, 2031–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A.G., McCarthy,N.J. and Evan,G.I. (1997) DrlCE is an essential caspase required for apoptotic activity in Drosophila cells. EMBO J., 16, 6192–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M.E., Abrams,J.M., Agapite,J., White,K. and Steller,H. (1995) The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev., 9, 1694–1708. [DOI] [PubMed] [Google Scholar]
- Hawkins C.J., Uren,A.G., Hacker,G., Medcalf,R.L. and Vaux,D.L. (1996) Inhibition of interleukin 1β-converting enzyme-mediated apoptosis of mammalian cells by baculovirus IAP. Proc. Natl Acad. Sci. USA, 93, 13786–13790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C.J., Yoo,S.J., Peterson,E.P., Wang,S.L., Vernooy,S.Y. and Hay,B.A. (2000) The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID and GRIM. J. Biol. Chem., 275, 27084–27093. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff,T. and Rubin,G.M. (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development, 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- Hershberger P.A., Dickson,J.A. and Friesen,P.D. (1992) Site-specific mutagenesis of the 35-kilodalton protein gene encoded by Autographa californica nuclear polyhedrosis virus: cell line-specific effects on virus replication. J. Virol., 66, 5525–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisahara S. et al. (2000) Targeted expression of baculovirus p35 caspase inhibitor in oligodendrocytes protects mice against autoimmune-mediated demyelination. EMBO J., 19, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak R.R., Manji,G.A. and Friesen,P.D. (2000) The BIR motifs mediate dominant interference and oligomerization of inhibitor of apoptosis Op-IAP. Mol. Cell. Biol., 20, 1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Deveraux,Q.L., Maeda,S., Salvesen,G.S., Stennicke,H.R., Hammock,B.D. and Reed,J.C. (2000) Evolutionary conservation of apoptosis mechanisms: lepidopteran and baculoviral inhibitor of apoptosis proteins are inhibitors of mammalian caspase-9. Proc. Natl Acad. Sci. USA, 97, 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo M., Grandien,A., Criado,L.M., Robles,S., Leonardo,E., Albar,J.P., de Buitrago,G.G. and Martinez,A.C. (1999) Blocked negative selection of developing T cells in mice expressing the baculovirus p35 caspase inhibitor. EMBO J., 18, 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M.D. (1998) Anti-apoptosis therapy: a way of treating neural degeneration? Curr. Biol., 8, R418–R421. [DOI] [PubMed] [Google Scholar]
- Jacobson M.D., Weil,M. and Raff,M.C. (1997) Programmed cell death in animal development. Cell, 88, 347–354. [DOI] [PubMed] [Google Scholar]
- Jiang C., Baehrecke,E.H. and Thummel,C.S. (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development, 124, 4673–4683. [DOI] [PubMed] [Google Scholar]
- Kumar S. and Doumanis,J. (2000) The fly caspases. Cell Death Differ., 7, 1039–1044. [DOI] [PubMed] [Google Scholar]
- LaCount D.J. (1998) Virus replication events and host factors involved in baculovirus-induced apoptosis. PhD thesis, University of Wisconsin-Madison, WI.
- LaCount D.J. and Friesen,P.D. (1997) Role of early and late replication events in induction of apoptosis by baculoviruses. J. Virol., 71, 1530–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D.J., Hanson,S.F., Schneider,C.L. and Friesen,P.D. (2000) Caspase inhibitor P35 and inhibitor of apoptosis Op-IAP block in vivo proteolytic activation of an effector caspase at different steps. J. Biol. Chem., 275, 15657–15664. [DOI] [PubMed] [Google Scholar]
- Li P., Nijhawan,D., Budihardjo,I., Srinivasula,S.M., Ahmad,M., Alnemri,E.S. and Wang,X. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell, 91, 479–489. [DOI] [PubMed] [Google Scholar]
- Manji G.A. and Friesen,P.D. (2001) Apoptosis in motion. An apical, P35-insensitive caspase mediates programmed cell death in insect cells. J. Biol. Chem., 276, 16704–16710. [DOI] [PubMed] [Google Scholar]
- Manji G.A., Hozak,R.R., LaCount,D.J. and Friesen,P.D. (1997) Baculovirus inhibitor of apoptosis functions at or upstream of the apoptotic suppressor P35 to prevent programmed cell death. J. Virol., 71, 4509–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P., Silke,J., Leevers,S.J. and Evan,G.I. (2000) The Drosophila caspase DRONC is regulated by DIAP1. EMBO J., 19, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.K. (1999) An exegesis of IAPs: salvation and surprises from BIR motifs. Trends Cell Biol., 9, 323–328. [DOI] [PubMed] [Google Scholar]
- Morris T.D. and Miller,L.K. (1993) Characterization of productive and non-productive AcMNPV infection in selected insect cell lines. Virology, 197, 339–348. [DOI] [PubMed] [Google Scholar]
- Nicholson D.W. (2000) From bench to clinic with apoptosis-based therapeutic agents. Nature, 407, 810–816. [DOI] [PubMed] [Google Scholar]
- O’Brien V. (1998) Viruses and apoptosis. J. Gen. Virol., 79, 1833–1845. [DOI] [PubMed] [Google Scholar]
- Roulston A., Marcellus,R.C. and Branton,P.E. (1999) Viruses and apoptosis. Annu. Rev. Microbiol., 53, 577–628. [DOI] [PubMed] [Google Scholar]
- Salvesen G.S. and Dixit,V.M. (1997) Caspases: intracellular signaling by proteolysis. Cell, 91, 443–446. [DOI] [PubMed] [Google Scholar]
- Schneider I. (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol., 27, 353–365. [PubMed] [Google Scholar]
- Seshagiri S. and Miller,L.K. (1997) Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc. Natl Acad. Sci. USA, 94, 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula S.M., Ahmad,M., Fernandes-Alnemri,T. and Alnemri,E.S. (1998) Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell, 1, 949–957. [DOI] [PubMed] [Google Scholar]
- Sugimoto A., Friesen,P.D. and Rothman,J.H. (1994) Baculovirus p35 prevents developmentally programmed cell death and rescues a ced-9 mutant in the nematode Caenorhabditis elegans. EMBO J., 13, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.B. (1995) Apoptosis in the pathogenesis and treatment of disease. Science, 267, 1456–1462. [DOI] [PubMed] [Google Scholar]
- Thornberry N.A. and Lazebnik,Y. (1998) Caspases: enemies within. Science, 281, 1312–1316. [DOI] [PubMed] [Google Scholar]
- Vaughn J.L., Goodwin,R.H., Thompkins,G.L. and McCawley,P. (1977) The establishment of two insect cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae). In Vitro, 13, 213–217. [DOI] [PubMed] [Google Scholar]
- Vaux D.L. and Korsmeyer,S.J. (1999) Cell death in development. Cell, 96, 245–254. [DOI] [PubMed] [Google Scholar]
- Vernooy S.Y., Copeland,J., Ghaboosi,N., Griffin,E.E., Yoo,S.J. and Hay,B.A. (2000) Cell death regulation in Drosophila: conservation of mechanism and unique insights. J. Cell Biol., 150, F69–F75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vier J., Furmann,C. and Hacker,G. (2000) Baculovirus P35 protein does not inhibit caspase-9 in a cell-free system of apoptosis. Biochem. Biophys. Res. Commun., 276, 855–861. [DOI] [PubMed] [Google Scholar]
- Viswanath V., Wu,Z., Fonck,C., Wei,Q., Boonplueang,R. and Andersen,J.K. (2000) Transgenic mice neuronally expressing baculoviral p35 are resistant to diverse types of induced apoptosis, including seizure-associated neurodegeneration. Proc. Natl Acad. Sci. USA, 97, 2270–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D., Kaiser,W.J., Harvey,A.J. and Miller,L.K. (1997) Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl Acad. Sci. USA, 94, 10183–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D., Kaiser,W.J. and Miller,L.K. (1998) Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell. Biol., 18, 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K. (2000) Cell death: Drosophila Apaf-1—no longer in the (d)Ark. Curr. Biol., 10, R167–169. [DOI] [PubMed] [Google Scholar]
- White K., Tahaoglu,E. and Steller,H. (1996) Cell killing by the Drosophila gene reaper. Science, 271, 805–807. [DOI] [PubMed] [Google Scholar]
- Xu G., Cirilli,M., Huang,Y., Rich,R.L., Myszka,D.G. and Wu,H. (2001) Covalent inhibition revealed by the crystal structure of the caspase-8/p35 complex. Nature, 410, 494–497. [DOI] [PubMed] [Google Scholar]
- Xue D. and Horvitz,H.R. (1995) Inhibition of the Caenorhabditis elegans cell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature, 377, 248–251. [DOI] [PubMed] [Google Scholar]
- Yuan J. and Yankner,B.A. (2000) Apoptosis in the nervous system. Nature, 407, 802–809. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Krebs,J.F., Snipas,S.J., Price,A., Alnemri,E.S., Tomaselli,K.J. and Salvesen,G.S. (1998) Interaction of the baculovirus anti-apoptotic protein p35 with caspases. Specificity, kinetics and characterization of the caspase/p35 complex. Biochemistry, 37, 10757–10765. [DOI] [PubMed] [Google Scholar]
- Zoog S.J., Bertin,J. and Friesen,P.D. (1999) Caspase inhibition by baculovirus P35 requires interaction between the reactive site loop and the β-sheet core. J. Biol. Chem., 274, 25995–26002. [DOI] [PubMed] [Google Scholar]
- Zou H., Henzel,W.J., Liu,X.S., Lutschg,A. and Wang,X.D. (1997) Apaf-1, a human protein homologous to C. elegans ced-4, participates in cytochrome c-dependent activation of caspase-3. Cell, 90, 405–413. [DOI] [PubMed] [Google Scholar]