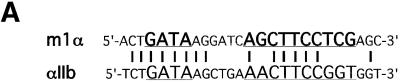Abstract
The transcription factor GATA-1 and its cofactor FOG-1 are essential for the normal development of erythroid cells and megakaryocytes. FOG-1 can stimulate or inhibit GATA-1 activity depending on cell and promoter context. How the GATA-1–FOG-1 complex controls the expression of distinct sets of gene in megakaryocytes and erythroid cells is not understood. Here, we examine the molecular basis for the megakaryocyte-restricted activation of the αIIb gene. FOG-1 stimulates GATA-1-dependent αIIb gene expression in a manner that requires their direct physical interaction. Transcriptional output by the GATA-1–FOG-1 complex is determined by the hematopoietic Ets protein Fli-1 that binds to an adjacent Ets element. Chromatin immunoprecipitation experiments show that GATA-1, FOG-1 and Fli-1 co-occupy the αIIb promoter in vivo. Expression of several additional megakaryocyte-specific genes that bear tandem GATA and Ets elements in their promoters also depends on the physical interaction between GATA-1 and FOG-1. Our studies define a molecular context for transcriptional activation by GATA-1 and FOG-1, and may explain the occurrence of tandem GATA and Ets elements in the promoters of numerous megakaryocyte-expressed genes.
Keywords: Ets/Fli-1/FOG/GATA/megakaryocyte
Introduction
Development of hematopoietic cell lineages is controlled by both tissue-restricted and widely expressed transcription factors. The related erythroid and megakaryocytic cell lineages are derived from a common progenitor cell and express an overlapping set of lineage-restricted transcription factors, including GATA-1 and the GATA-1 cofactor, Friend of GATA-1 (FOG-1). However, how these factors control the expression of distinct sets of genes in different lineages is an unresolved question.
GATA-1 is a zinc finger transcription factor that is expressed in erythroid cells, megakaryocytes, mast cells and eosinophils (Weiss and Orkin, 1995). Functional GATA elements are present in the proximal promoters of virtually all erythroid- and megakaryocyte-restricted genes examined. Gene targeting studies revealed that GATA-1 is required for the normal maturation of both erythroid and megakaryocytic cells (Pevny et al., 1991, 1995; Fujiwara et al., 1996; Shivdasani et al., 1997). FOG-1 was identified based on its ability to specifically bind the N-terminal zinc finger of GATA-1 (Tsang et al., 1997). The expression pattern of FOG-1 resembles that of GATA-1, with the highest levels observed in erythroid cells and megakaryocytes. Mice lacking FOG-1 display an erythroid differentiation block similar to that observed in GATA-1-deficient mice (Tsang et al., 1998), providing in vivo evidence that these factors function in the same transcriptional pathway. While FOG-1 deficiency virtually ablates the development of the megakaryocytic lineage (Tsang et al., 1998), megakaryocytes lacking GATA-1 are increased in number, but do not differentiate normally (Shivdasani et al., 1997; Vyas et al., 1999). Recent studies showed that the FOG-1-interacting transcription factor GATA-2 can partially compensate for the loss of GATA-1, thus explaining the less dramatic phenotype in GATA-1 null megakaryocytes (Chang et al., 2002). The importance of direct physical interaction between GATA-1 and FOG-1 is illustrated by the observation that point mutations in the N-terminal zinc finger of GATA-1 that disrupt FOG-1 binding lead to defective erythropoiesis and megakaryopoiesis (Crispino et al., 1999; Nichols et al., 2000; Chang et al., 2002). In transient transfection assays, FOG-1 can repress or activate GATA-1 activity depending on cell and promoter context. For example, while FOG-1 stimulates GATA-1 activity on the p45 NF-E2 gene promoter, which is active in erythroid cells and megakaryocytes (Tsang et al., 1997), it represses GATA-1 activity on the erythroid-specific EKLF and transferrin receptor II promoters, as well as on a synthetic GATA-1-dependent promoter (Fox et al., 1999; Kawabata et al., 2001). There are also examples where other GATA-dependent promoters are repressed by members of the FOG family that are present in various tissues and diverse organisms (Fossett and Schulz, 2001).
The requirement for GATA-1 and FOG-1 for normal erythroid and megakaryocytic differentiation and the dependence of their function on their direct interaction appear at odds with the observation that FOG-1 represses GATA-1 activity on numerous promoters in transfection-based assays. The studies presented here using the megakaryocyte-restricted αIIb gene promoter reveal a molecular context that specifies synergistic gene activation by GATA-1 and FOG-1.
The αIIb gene, which encodes the α integrin chain of the platelet fibrinogen receptor αIIb/β3, has long served as a model for understanding the molecular basis of high level, megakaryocyte-specific gene expression. Transient transfection and transgenic mice studies showed that ∼900 bp of αIIb promoter upstream sequence are sufficient to direct megakaryocyte-specific expression of a linked reporter (Uzan et al., 1991; Prandini et al., 1992; Block et al., 1994; Tronik-Le Roux et al., 1995). Two pairs of GATA and Ets consensus binding motifs in the 5′-flanking region of the αIIb gene contribute to high level, tissue-specific expression (Uzan et al., 1991; Prandini et al., 1992). Neighboring GATA and Ets elements have been identified in numerous megakaryocyte-specific regulatory regions and are viewed as hallmarks of megakaryocyte-expressed genes. Yet, how these elements function in concert is not understood. While GATA-1 can bind both αIIb GATA elements in vitro (Romeo et al., 1990) and, besides GATA-2, is probably the major GATA-binding activity in these cells, the nature of the Ets-binding proteins is more complex. Thus, Ets-1, Ets-2 and Fli-1, but not PU.1 (Spi-1), can bind in vitro to the Ets site closest to the transcriptional start site (TSS), whereas PU.1, but not Fli-1 or Ets-1, can bind to the distal Ets element (Lemarchandel et al., 1993; Zhang et al., 1993; Doubeikovski et al., 1997). Transient expression studies in non-megakaryocytic cell lines have shown that GATA-1 can stimulate the expression of a reporter gene containing 75 bp of the proximal αIIb promoter (Lemarchandel et al., 1993). More recently, it was shown that stably expressed GATA-1 and FOG-1 cooperated during the activation of the αIIb promoter in heterologous cells (Gaines et al., 2000). However, it remained unresolved whether activation resulted from direct occupation of the αIIb regulatory elements by both GATA-1 and FOG-1, and whether it required physical interaction between them.
To elucidate the basis for megakaryocyte-specific expression of a GATA-1-dependent gene, our studies focused on the regulation of the αIIb promoter. We found that activation of this promoter by GATA-1 and FOG-1 is dependent on their direct physical interaction and requires the presence of a specialized Ets element. This Ets element, when placed next to an isolated GATA site, enabled GATA-1/FOG-1 synergy. The hematopoietic Ets protein Fli-1 binds to this site and stimulates GATA-1/FOG-1-dependent transcription. Finally, in vivo expression of several megakaryocyte-restricted genes, whose promoters contain GATA and Ets elements, depends on direct interaction between GATA-1 and FOG-1. These studies suggest that cooperativity between GATA-1, FOG-1 and Fli-1 is important for establishing and/or maintaining the megakaryocytic cell lineage.
Results
Physical interaction between GATA-1 and FOG-1 is required for αIIb promoter activation
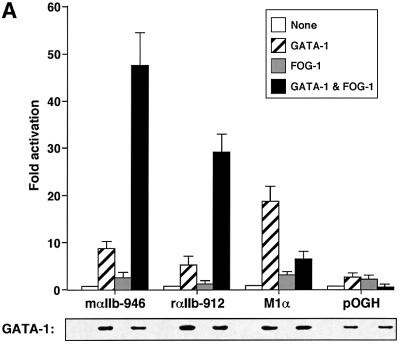
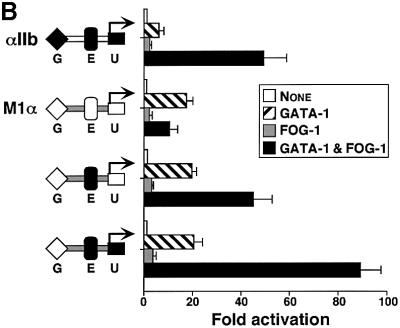
To examine the molecular mechanism by which GATA-1 and FOG-1 activate expression of the megakaryocyte-restricted αIIb gene, we performed transient transfection studies using the αIIb promoter fused to the human growth hormone reporter gene. Expression of GATA-1 alone led to 5- and 8-fold activation of the murine and rat αIIb promoters, respectively (Figure 1A), consistent with previous reports (Lemarchandel et al., 1993; Gaines et al., 2000). While expression of FOG-1 alone had little or no effect on reporter activity, co-expression of GATA-1 and FOG-1 activated these promoters 45- (murine) and 30-fold (rat) (Figure 1A). Control western blots showed that FOG-1 did not alter the expression of GATA-1, indicating that the effects of GATA-1 and FOG-1 are due to functional synergy at the αIIb promoter (Figure 1A). When assayed on a synthetic reporter gene construct that contains a single GATA element (M1α; Martin and Orkin, 1990), FOG-1 repressed GATA-1 activity ∼3-fold, similar to the findings of a previous report (Fox et al., 1999). These results suggest that the αIIb promoter contains a functional element(s) that leads to stimulation of GATA-1 activity by FOG-1 and is absent in M1α.
Fig. 1. Dependence of αIIb promoter activation on physical interaction between GATA-1 and FOG-1. (A) Activities of mouse (m, 946 bp) and rat (r, 912 bp) αIIb promoters in transiently transfected NIH-3T3 cells. GATA-1 and FOG-1 were co-expressed as indicated. Promoterless human growth hormone (GH) reporter and the synthetic GATA-1-dependent M1α reporter served as controls. Means ± SE are shown. Lower panel: anti-GATA-1 western blot. (B) Experiments were performed as in (A) using the mouse αIIb promoter.
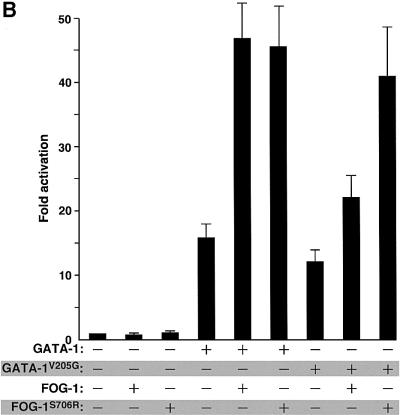
To determine whether FOG-1 function is mediated directly through GATA-1 or through other transcription factors bound at the αIIb promoter, we examined whether the observed functional synergy requires direct interaction between GATA-1 and FOG-1. To this end, we utilized mutant versions of GATA-1 and FOG-1 that alter their association. GATA-1V205G contains a point mutation in the N-terminal zinc finger of GATA-1 that diminishes FOG-1 binding without affecting its ability to bind DNA (Crispino et al., 1999). This mutation impairs the ability of GATA-1 to trigger terminal differentiation of GATA-1-dependent proerythroblasts (Crispino et al., 1999). A mutation at the same residue of GATA-1 was identified in male patients with X-linked dyserythroblastic anemia and thrombocytopenia (Nichols et al., 2000). FOG-1S706R contains a single amino acid substitution in its sixth zinc finger that restores binding to GATA-1V205G. Expression of FOG-1S706R in erythroid cells expressing GATA-1V205G rescues GATA-1-dependent erythroid differentiation (Crispino et al., 1999). Both wild-type GATA-1 and GATA-1V205G alone activated the αIIb promoter with similar efficiencies (Figure 1B). However, FOG-1 stimulated the activity of GATA-1V205G substantially less than wild-type GATA-1, indicating that physical interaction between GATA-1 and FOG-1 is required for their synergy. To rule out the possibility that GATA-1V205G has additional defects unrelated to its impaired FOG-1 interaction, we examined the effects of co-expressed FOG-1S706R on αIIb promoter activity. The results show that FOG-1S706R displayed strong transcriptional synergy with GATA-1V205G, comparable to that seen with their wild-type counterparts (Figure 1B). These findings demonstrate the importance of direct interaction between GATA-1 and FOG-1 during activation of the αIIb gene promoter in vivo.
FOG-1 and GATA-1 occupy the proximal αIIb promoter region in vivo
The above studies indicate that GATA-1 and FOG-1 regulate the expression of the αIIb gene by binding to its promoter region. To demonstrate that both GATA-1 and FOG-1 occupy this region in vivo in an appropriate cellular context, we performed chromatin immunoprecipitation (ChIP) assays, using the murine megakaryocytic cell line Y10, which expresses endogenous αIIb, GATA-1 and FOG-1 (Ishida et al., 1993; data not shown). Antibodies against GATA-1 and FOG-1, but not isotype-matched control antibodies, immunoprecipitated chromatin that was enriched for proximal αIIb promoter sequences (–110 bp to +226 bp) (Figure 2). As a negative control, a more distal αIIb promoter domain, between –3.3 and –3.6 kb upstream of the αIIb TSS which does not contain a GATA-1-binding site, was not enriched. As an additional control, the promoter region of the housekeeping gene GAPDH, which is not regulated by GATA-1 (Tsang et al., 1998), was not bound by GATA-1 or FOG-1 (Figure 2). These results show that GATA-1 and FOG-1 specifically contact the domain of the αIIb promoter that contains a functionally important GATA element, supporting the hypothesis that transcriptional activation of the αIIb gene is mediated directly by a GATA-1–FOG-1 complex in vivo.
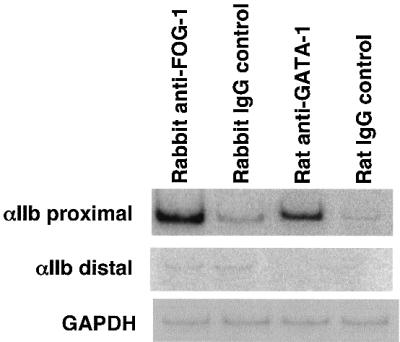
Fig. 2. Occupancy of the αIIb promoter by GATA-1 and FOG-1 in vivo. ChIP assays using the murine megakaryocytic cell line Y10, anti-FOG-1 and anti-GATA-1 antibodies or isotype-matched control antibodies. Purified DNA fragments were amplified by PCR with primers specific for the proximal mouse αIIb promoter (–110 to +226 bp) and, as control, a distal region lacking GATA and Ets elements (–3.3 to –3.6 kb). The GATA-1-independent housekeeping gene GAPDH served as control. One of three independent experiments is shown.
60 bp of αIIb upstream sequence are sufficient to mediate GATA-1/FOG-1 synergy
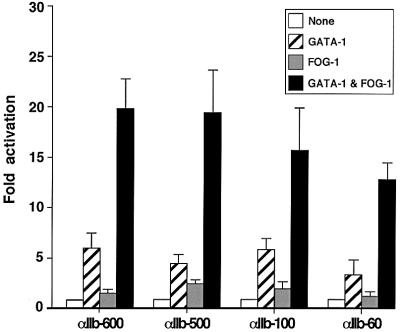
To identify the minimal region of the αIIb promoter sufficient for mediating synergistic activation by GATA-1 and FOG-1, we examined a series of 5′ to 3′ deletion constructs in transiently transfected NIH-3T3 cells. The results revealed that a construct spanning from –60 to +32 bp displayed GATA-1/FOG-1 synergy comparable with that observed with the full-length promoter (Figure 3). This region contains functional, phylogenetically conserved GATA and Ets elements, suggesting that these sites might be sufficient for transcriptional synergy by GATA-1 and FOG-1.
Fig. 3. Sixty base pairs of upstream region are sufficient for GATA-1/FOG-1 synergy. Promoter constructs containing the indicated lengths of upstream region were analyzed as in Figure 1.
Specificity of the Ets element determines transcriptional output by GATA-1 and FOG-1
To determine the cis-acting elements that confer activation by GATA-1 and FOG-1, we compared the αIIb promoter with the M1α promoter, where FOG-1 inhibits GATA-1 activity (Figure 1A; Fox et al., 1999). We noted that the M1α promoter contains an Ets element positioned at the same distance from the GATA site as that found in the αIIb promoter (Figure 4A). However, the M1α and αIIb Ets elements differ from each other in the nucleotides flanking the core 5′-GGA-3′ sequence (Figure 4A). These nucleotide differences are predicted to alter Ets binding specificity (Graves and Petersen, 1998). To examine whether the αIIb Ets motif determines GATA-1/FOG-1 activity, we replaced the M1α Ets element with that derived from the αIIb gene. Remarkably, the presence of the αIIb Ets element converted FOG-1 from an inhibitor to an activator of GATA-1 activity (Figure 4B). Substitution of the M1α 5′-untranslated region (5′-UTR) with that derived from αIIb further increased GATA-1 and FOG-1 synergy (Figure 4B). Thus, these data suggest that the αIIb Ets site is sufficient to convert FOG-1 from a GATA-1 repressor into a co-activator, and that additional sequences in the αIIb regulatory region contribute to maximal activation of GATA-1 by FOG-1.
Fig. 4. The αIIb Ets element can mediate GATA-1/FOG-1 synergy on a synthetic promoter construct. (A) Sequence alignment of αIIb in the 60 bp construct and in the M1α GATA and Ets elements. (B) Insertion of the αIIb Ets element into M1α leads to activation of GATA-1 by FOG-1. G, GATA-1-binding site; E, Ets-binding site; U, 5′-UTR. Means ± SE are shown.
Fli-1 can bind to the αIIb Ets element in vitro and in vivo
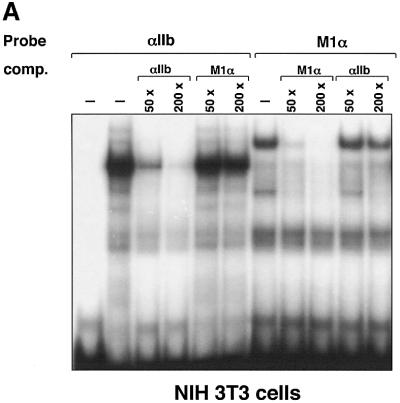
Given the importance of the Ets element during GATA-1- and FOG-1-mediated transcription, we used electrophoretic mobility shift assays (EMSAs) to characterize the protein(s) that bind to the αIIb and M1α Ets elements. Using nuclear extracts from NIH-3T3 cells, protein complexes with distinct mobilities were observed that showed little or no cross-competition (Figure 5A), suggesting that these elements bind different members of the Ets protein family. When Y10 cell extracts were used for the EMSA, an additional, faster moving complex was detected with the αIIb Ets probe (Figure 5B). This complex was undetectable with NIH-3T3 cell extracts (Figure 5A) or when the M1α Ets element was used as probe (Figure 5B). This suggests that at least one Ets factor binds in both a site- and a tissue-specific manner.
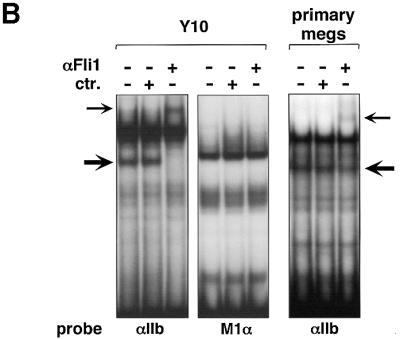
Fig. 5. Selective binding of Fli-1 to the αIIb Ets element. (A) αIIb and M1α Ets elements bind distinct Ets proteins. EMSA using nuclear extracts from NIH-3T3 cells. Note that there is little or no cross-competition between the αIIb and M1α Ets elements. (B) Nuclear extracts from Y10 cells (left panel) and fetal liver-derived primary megakaryocytes (right panel) were used. Anti-Fli-1 (αFli-1), but not control (ctr), antibodies reacted with a band (thick arrow), resulting in a supershift (thin arrow).
The Ets family protein Fli-1 is a likely candidate for this Ets-binding activity since recombinant Fli-1 can bind the proximal αIIb Ets element in vitro, and this element matches a consensus Fli-1-binding site (Zhang et al., 1993; Mao et al., 1994; Szymczyna and Arrowsmith, 2000). Moreover, Fli-1 is essential for the normal development of the megakaryocytic lineage (Hart et al., 2000; Spyropoulos et al., 2000; Kawada et al., 2001). To test directly whether Fli-1 is the megakaryocyte-specific Ets-binding protein, we added anti-Fli-1 antibodies to the EMSA reaction. The results show that anti-Fli-1 antibodies supershifted the megakaryocyte-specific band, but not the other Ets-binding activities (Figure 5B). In contrast, anti-Ets-1, anti-Ets-2 and anti-PU.1 antibodies did not alter the mobility of this band (data not shown). To determine whether Fli-1 DNA-binding activity is present in primary megakaryocytes, fetal liver cells were expanded in culture in the presence of thrombopoietin (TPO), leading to enrichment of megakaryocytes up to 80% of the total population of cells as determined by acetylcholinesterase (AChE) staining (data not shown). Nuclear extracts from these cells yielded protein complexes similar to those found in Y10 cells, and included a protein that specifically reacted with anti-Fli-1 antibodies (Figure 5B). However, anti-Fli-1 antibodies consistently supershifted only a fraction of this complex, suggesting that other megakaryocyte-expressed Ets family members also bind the αIIb Ets element (Figure 5B).
To determine whether Fli-1 occupies the αIIb promoter in vivo, we performed ChIP assays, using Y10 cells. Two anti-Fli-1 antibodies, but not isotype-matched control antibodies, immunoprecipitated chromatin that was enriched for proximal αIIb promoter sequences (–110 bp to +226 bp) (Figure 6). In contrast, a more distal domain, between –3.3 and –3.6 kb upstream of the αIIb transcription initiation site, was not enriched. These results show that Fli-1 binds in vivo to the proximal αIIb promoter and suggest that Fli-1 is a strong candidate Ets factor that mediates transcriptional synergy by GATA-1 and FOG-1.
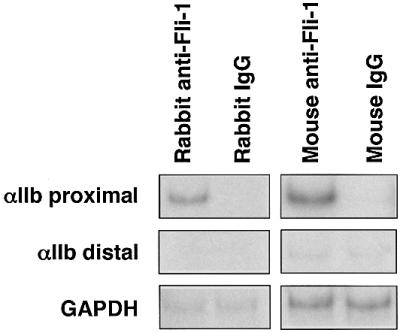
Fig. 6. Occupancy of the αIIb promoter by Fli-1 in vivo. ChIP assays using Y10 cells, rabbit and mouse anti-Fli-1 antibodies or isotype-matched control antibodies. Primer pairs were the same as in Figure 2. The GAPDH gene served as control.
Fli-1 mediates GATA-1/FOG-1 synergy at the αIIb promoter
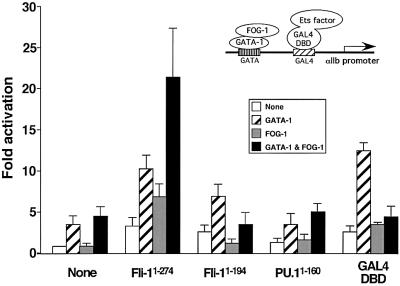
To determine whether Fli-1 can mediate transcriptional synergy by GATA-1 and FOG-1, we performed transient transfection assays but, to avoid interference by endogenous Ets proteins, we used an αIIb reporter gene construct in which the Ets element had been replaced with a GAL4-binding site (pGL2-αIIb100). While GATA-1 alone weakly activated this construct, FOG-1 co-expression failed to augment GATA-1 activity, consistent with a requirement for an Ets protein activity for αIIb gene expression (Figure 7). We then generated a construct in which the N-terminal 274 amino acids of Fli-1, lacking the DNA-binding domain, were fused to the DNA-binding domain of the yeast transcription factor GAL4 (GAL4-Fli-1). When GAL4-Fli-1 was co-expressed together with GATA-1 and FOG-1, transcriptional activation was restored (Figure 7). In contrast, expression of only the GAL4 DNA-binding domain failed to activate this reporter in the presence of GATA-1 and FOG-1. Control western blots showed that neither FOG-1 nor GAL-4 fusion proteins altered GATA-1 expression (data not shown). These results suggest that Fli-1 is sufficient to mediate transcriptional activation by GATA-1 and FOG-1. The effects of Fli-1 were specific since a construct containing only amino acids 1–194, which lacks an important protein interaction domain required for Fli-1 activity (Watson et al., 1997) was ineffective (Figure 7). To assess whether other Ets family proteins can substitute for Fli-1, we examined a construct in which the N-terminal 160 amino acids of the myeloid/lymphoid Ets protein PU.1 were fused to GAL4. The GAL4-PU.1 construct is functional, since it was used previously to activate gene expression in a myeloid-specific fashion (Maitra and Atchison, 2000). In addition, we generated a construct in which GAL4 was fused to the lymphoid Ets factor Spi-B. Both GAL4-PU.1 and GAL4-Spi-B failed to activate the αIIb promoter in the presence of GATA-1 and FOG-1, indicating that the effects of Fli-1 are specific (Figure 7; data not shown).
Fig. 7. Fli-1 mediates GATA-1/FOG-1 synergy on the αIIb promoter. Inset: schematic of the assay. The reporter construct contained 100 bp of αIIb promoter upstream region in which a GAL4-binding site was substituted for the Ets element. GAL4 fusion constructs were co- expressed as indicated.
Interaction between GATA-1 and FOG-1 is required for the expression of multiple megakaryocyte-specific genes in vivo
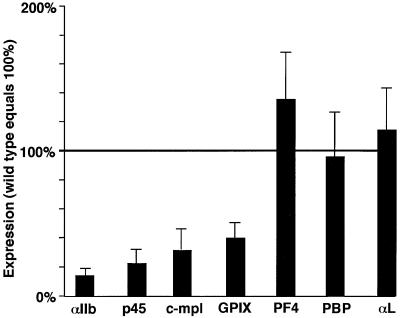
The frequent presence of tandem GATA- and Ets-binding sites in the regulatory regions of megakaryocyte-expressed genes has been noted previously (reviewed by Kaluzhny et al., 2001). To determine whether the expression of other megakaryocyte-specific genes also depends on a physical interaction between GATA-1 and FOG-1, we analyzed megakaryocytes derived from in vitro differentiated murine embryonic stem (ES) cells and compared them with those derived from ES cells in which the GATA-1 gene had been replaced with the FOG-1-binding-defective GATA-1V205G through homologous recombination (Chang et al., 2002). ES cells were differentiated into embryoid bodies (EBs) in the presence of TPO to increase the number of megakaryocytes. AChE staining demonstrated that ∼15% of the cells were megakaryocytes in both wild-type and mutant cultures (data not shown). Total RNA from these EBs was analyzed for αIIb gene expression by quantitative RT–PCR. The results show that αIIb levels in GATA-1V205G-containing megakaryocytes were reduced to 16% when compared with their wild-type counterparts, consistent with a requirement for the GATA-1–FOG-1 interaction for αIIb gene expression in vivo (Figure 8). To determine whether other megakaryocyte-restricted genes similarly depend on the GATA-1–FOG-1 complex, we measured mRNA levels of the c-mpl, p45 NF-E2, GPIX, PF4 and PBP genes. Remarkably, GATA-1V205G-containing megakaryocytes displayed a substantial reduction in the mRNA levels of p45 NF-E2, c-mpl and GPIX, but not of PF4 and PBP (Figure 8), indicating that the GATA-1–FOG-1 interaction is required for the expression of many, but not all, megakaryocyte-specific genes. Normal expression of the megakaryocyte-specific chemokines PF4 and PBP demonstrates that the GATA-1V205G mutation did not simply abrogate megakaryocyte development. Instead, these results indicate that a subset of megakaryocyte-restricted genes is independent of a GATA-1–FOG-1 complex.
Fig. 8. In vivo dependence of megakaryocyte-specific gene expression on physical interaction between GATA-1 and FOG-1. Total RNA was isolated from megakaryocyte-enriched EBs derived from in vitro differentiated wild-type or GATA-1V205G mutant ES cells. mRNA levels of the indicated genes were determined using quantitative RT–PCR. Expression levels in GATA-1V205G mutant cells were normalized for HPRT expression and plotted as a percentage of wild-type levels. The myeloid-expressed integrin gene αL served as additional control.
Activity of GATA-1 and FOG-1 on megakaryocytic gene promoters
Previous studies have shown that GATA-1 and FOG-1 synergistically activate the p45 NF-E2 promoter, similar to what we observed with the αIIb promoter (Tsang et al., 1997). Therefore, we wanted to determine whether additional megakaryocytic gene promoters were also targets of GATA-1 and FOG-1. Specifically, we asked whether promoters of genes that are affected by the GATA-1V205G mutation are activated by GATA-1 and FOG-1, and, conversely, whether genes that are expressed normally in GATA-1V205G-containing megakaryocytes are independent of FOG-1. The c-mpl gene promoter has functionally important GATA- and Ets-binding elements within its first 100 bp of upstream sequence (Deveaux et al., 1996). Therefore, we transfected a luciferase reporter gene construct driven by 100 bp of murine c-mpl promoter sequence into NIH-3T3 cells together with plasmids expressing GATA-1 and/or FOG-1. While GATA-1 alone activated the reporter gene 7-fold, co-expression of FOG-1 increased promoter activity up to 20-fold (Figure 9). In contrast, 222 bp of the murine PF4 promoter (Ravid et al., 1991; Minami et al., 1998), containing important GATA- and Ets-binding elements, showed activation by GATA-1, and but no further activation upon FOG-1 co-expression (Figure 9). Instead, FOG-1 repressed GATA-1 activity 6-fold, similar to what has been observed with the M1α reporter gene. Preliminary results (not shown) further suggest that the PBP promoter is also not activated by GATA-1 and FOG-1. Together with the results obtained using the αIIb and p45 NF-E2 promoters, these data establish a correlation between gene activation by GATA-1 and FOG-1 in transfection-based assays and sensitivity to the GATA-1V205G mutation in megakaryocytes.
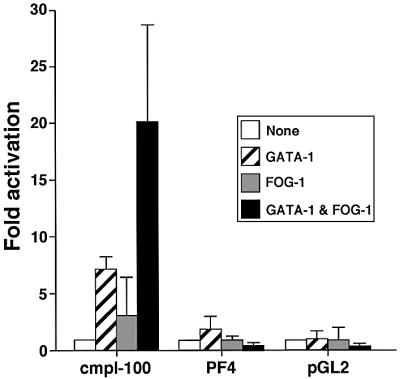
Fig. 9. GATA-1- and FOG-1-dependent activation of select megakaryocytic gene promoters. Transient transfection studies were performed as in Figure 1, with reporter gene constructs containing 100 bp of the murine c-mpl promoter or 222 bp of murine PF4 promoter. A pro moterless (pGL2) vector served as negative control.
Discussion
GATA-1 and FOG-1 cooperate during the differentiation of megakaryocytes and erythroid cells (Tsang et al., 1997; Crispino et al., 1999; Nichols et al., 2000; Chang et al., 2002). FOG-1 binds to the N-terminal zinc finger of GATA-1, thereby modulating GATA-1 activity. The effects of FOG proteins on the transcriptional activities of GATA factors are determined by the cell type and the context of cis-regulatory elements present at a given promoter. To understand better the molecular basis for cooperative transcriptional activation by GATA-1 and FOG-1, we examined the effects of GATA-1 and FOG-1 on the αIIb gene promoter. We found that FOG-1 strongly increases GATA-1 activity on the αIIb promoter in a fashion that requires direct physical interaction. ChIP assays showed that GATA-1 and FOG-1 co-occupy this promoter in megakaryocytes in vivo, indicating that the actions of GATA-1 and FOG-1 are direct. Deletion analysis showed that 60 bp of the proximal promoter plus sequences containing the 5′-UTR were sufficient for full activation by GATA-1 and FOG-1 in NIH-3T3 cells. Remarkably, gain-of-function experiments, using synthetic promoter constructs, revealed that the mere presence of a specialized Ets element is sufficient to convert FOG-1 from a GATA-1 inhibitor into a co-activator. These findings provide a molecular explanation for the frequent occurrence of neighboring GATA and Ets elements in megakaryocyte-restricted genes.
Fli-1 probably functions at the proximal αIIb Ets element in vivo for several reasons. First, recombinant and cellular Fli-1 proteins bind to this site in vitro (Zhang et al., 1993) (Figure 5B), consistent with the observation that the sequences flanking the proximal core Ets ele ment match the Fli-1 consensus binding site perfectly (Zhang et al., 1993; Mao et al., 1994; Szymczyna and Arrowsmith, 2000). Secondly, ChIP experiments showed that Fli-1 binds the αIIb promoter in vivo in a megakaryocytic cell line (Figure 7). Thirdly, when fused to GAL4, Fli-1 but not PU.1 and Spi-B can mediate transcriptional activation by GATA-1 and FOG-1. Fourthly, mice homozygous for a Fli-1 null mutation display altered megakaryocytic proliferation and differentiation (Hart et al., 2000; Spyropoulos et al., 2000; Kawada et al., 2001). Finally, forced expression of Fli-1 in K562 cells augments αIIb expression (Athanasiou et al., 1996), and Fli-1 can activate the αIIb promoter in transient transfection assays (Bastian et al., 1999). However, it is likely that other Ets proteins might function at this site as well since anti-Fli-1 antibodies supershifted only part of the Ets protein complex that co-migrated with Fli-1 in primary megakaryocytes. Furthermore, transcriptional activation of the αIIb promoter by FOG-1 and GATA-1 was observed in NIH-3T3 cells, which do not express detectable amounts of Fli-1. Thus, select members of the large and diverse Ets protein family might substitute for Fli-1 function in non-hematopoietic tissues. Moreover, functional redundancy among Ets family proteins might explain the normal expression in Fli-1 null mice of several megakaryocytic genes that contain functional Ets elements in the regulatory regions, including c-mpl and αIIb (Hart et al., 2000).
Through analysis of various Ets elements, we note that even when sequences do not match the Fli-1 consensus sequence, they might still be able to mediate activation by Fli-1. For example, based on sequence alone, the Ets sites in the GPIbα and GPIX promoters would be predicted to be poor binding sites for Fli-1. Yet both promoters are activated by Fli-1 and, in the case of GPIX, activation has been shown to depend on its Ets element (Bastian et al., 1999). Therefore, promoter architecture appears to be critical for mediating transcriptional activation by Fli-1. In accordance with this interpretation, recruitment of Fli-1 but not PU.1 to the αIIb promoter via GAL4-binding sites led to activated transcription together with GATA-1 and FOG-1. These results suggest that megakaryocyte-specific promoter activity is determined by both the nucleotide sequence of the Ets-binding site and distinct domains within the bound Ets factor.
The above studies indicate that the proximal promoter region is sufficient for GATA-1- and FOG-1-dependent transcription, but they raise questions regarding the role of the distal conserved GATA and Ets sites between positions –457 and –506 bp upstream of the TSS. While both tandem GATA/Ets sites have enhancer activity, the distal enhancer is active in both erythroid and megakaryocytic lineages (Prandini et al., 1992), indicating that it is not the critical determinant for megakaryocyte-specific expression of the αIIb gene. Furthermore, the distal Ets element does not conform to the Fli-1 consensus binding site. Indeed, PU.1, which has a substantially diverged DNA-binding domain and different binding site preference, has been shown to bind this Ets element (Doubeikovski et al., 1997). Megakaryocytes derived from PU.1-deficient ES cells expressed the αIIb gene normally, showing that PU.1 is not essential for αIIb expression (Zhang et al., 1997). As appears to be the case for the proximal Ets site at –35 bp, it is possible that multiple Ets family members can bind and promote αIIb expression through the distal Ets element. The identity of these Ets proteins remains to be determined.
Direct physical interaction between GATA-1 and FOG-1 is required for αIIb expression in transfected cells and in ES cell-derived megakaryocytes, since a point mutation in GATA-1 that disrupts FOG-1 binding leads to loss of gene activation. Megakaryocytes bearing the GATA-1V205G mutation express reduced levels of additional megakaryocyte-restricted genes, including c-mpl, p45 NF-E2 and GPIX, indicating that these genes are also regulated by the GATA-1–FOG-1 complex in vivo. In the cases of c-mpl and p45NF-E2, we and others (Tsang et al., 1997) have shown that GATA-1 and FOG-1 can activate these genes in transfection-based assays, supporting a model in which GATA-1 and FOG-1 control these genes directly. It is important to note that the PF4 and PBP genes, which are expressed at late stages of megakaryocytic differentiation (Lepage et al., 2000), were unaffected by the GATA-1V205G mutation. This indicates that reduced expression of αIIb, c-mpl, p45 NF-E2 and GPIX is not simply the result of failed megakaryopoiesis, but instead reflects differences in GATA-1/FOG-1 dependence between distinct sets of genes. It is of note that the Ets elements in the upstream region of the c-mpl and GPIX genes have been shown to bind to Fli-1 in vitro (Deveaux et al., 1996; Bastian et al., 1999). Our observation that PF4 expression is independent of the GATA-1–FOG-1 interaction suggests alternative mechanisms of PF4 gene regulation. It is worth pointing out that the rat and human proximal PF4 promoters differ, with the human gene lacking a GATA-1 consensus element (Eisman et al., 1990). Furthermore, the PF4 Ets elements do not conform to Fli-1 consensus sites, suggesting that other Ets proteins might regulate this gene. It is also possible that GATA-1 performs FOG-1-independent functions at this gene, or that GATA-2, which is expressed in megakaryocytes and also binds FOG-1 (Tsang et al., 1997), might substitute for GATA-1. At the PBP gene, no functional GATA-1-binding sites have been identified, and GATA-1 did not activate the PBP promoter in transient transfection assays (M.Poncz, unpublished observation). Remarkably, all examined megakaryocyte-specific genes that were affected by the GATA-1V205G mutation had regulatory regions that could be activated by GATA-1 and FOG-1 in transfection assays. In contrast, the promoters of the PF4 and PBP genes that were insensitive to the GATA-1V205G mutation could not be activated by GATA-1 and FOG-1.
Our results are consistent with findings in patients with mutations in the N-terminal finger GATA-1 residues V205, G208 or D218 (Nichols et al., 2000; Freson et al., 2001; Mehaffey et al., 2001). These patients exhibit different degrees of macrothrombocytopenia, bleeding diathesis and dyserythropoiesis. Among these GATA-1 mutations, V205M is the most severe. Patients carrying the V205M mutation were anemic and severely thrombocytopenic. Their bone marrows contained numerous dysplastic megakaryocytes, consistent with an essential role for the GATA-1–FOG-1 complex in megakaryo poiesis and platelet production. Since these patients had undergone bone marrow transplantation, the levels of various megakaryocyte-specific genes could not be measured.
It is instructive to compare our results with those obtained from megakaryocytes that express markedly reduced levels of wild-type GATA-1 as a result of a targeted mutation in the GATA-1 regulatory region (Shivdasani et al., 1997; Vyas et al., 1999). While the GATA-1V205G cells showed widespread, but not universal defects in megakaryocytic gene expression, the GATA-1-deficient megakaryocytes displayed reduced expression of all genes examined, including c-mpl, p45 NF-E2, αIIb and PF4. This more generalized decrease could be interpreted to reflect a general defect of megakaryocytic maturation. Alternatively, genes whose expression is diminished in the GATA-1-deficient but not in the GATA-1V205G cells, might be regulated in a GATA-1-dependent, but FOG-1-independent, fashion. In addition, the degree to which GATA-2 might compensate for impaired GATA-1 acti vity might vary between GATA-1V205G-containing and GATA-1-deficient megakaryocytes.
The frequent presence of GATA and Ets elements in megakaryocyte-expressed genes is indicative of the generality of our observations. This list of genes with functional GATA and Ets sites in their regulatory regions includes αIIb, c-mpl, GPIbα, GPV and GPIX, some of which are early markers of megakaryocytic development (Lepage et al., 2000). Thus, we speculate that the GATA-1, FOG-1 and Fli-1 cooperativity is critical during the formation and/or maintenance of the megakaryocytic lineage. However, in the case of the p45 NF-E2 gene, it remains an open question whether GATA-1/FOG-1-mediated activation requires the presence of an Ets element, since transactivation experiments were performed on reporter gene constructs containing 7 kb of upstream region (Tsang et al., 1997).
The mechanism by which Fli-1 exerts its effects on GATA-1 and FOG-1 remains to be determined. It is possible that Fli-1 forms specific contacts with FOG-1, thereby altering its conformation or promoting or interfering with additional protein contacts. It is also possible that Fli-1 communicates directly with GATA-1, although published studies showed relatively poor Fli-1–GATA-1 interaction in vitro (Rekhtman et al., 1999). We found that amino acids 194–274 of Fli-1 were essential for its function in the GAL4 recruitment assays. This region contains a domain from amino acids 231 to 248 that is similar to the B-box present in select Ets proteins, including SAP1a and ELK1, but not in Ets-1, Ets-2 and PU.1 (Watson et al., 1997). This domain has been shown to mediate contacts with other proteins to form higher order protein complexes on certain regulatory regions (Watson et al., 1997). It is possible that other Ets proteins containing this domain might also be able to mediate GATA-1/FOG-1 synergy. This domain might aid in positioning the GATA-1–FOG-1 complex at the promoter in a way that is favorable for transcriptional activation. Alternatively, it might aid in replacing co-repressor complexes similar to what is observed in nuclear hormone receptors upon ligand binding (McKenna and O’Malley, 2002).
In summary, our work sheds new light on the mechanism by which megakaryocyte-specific gene expression is accomplished, providing new insights into the GATA–Ets signature motif found in the promoter regions of numerous megakaryocytic genes. In addition, we identified Fli-1 as a tissue-specific Ets-binding protein that converts FOG-1 from a repressor into a GATA-1 co-activator. The implications of these findings might extend to genes controlled by GATA and FOG proteins in diverse tissues, including erythroid cells, lymphoid cells and the heart. Thus, we speculate that transcriptional regulators that cooperate with GATA factors in non-megakaryocytic cells might serve the same function as Fli-1 in megakaryocytes, which is to determine transcriptional output of GATA– FOG complexes.
Materials and methods
Plasmid constructs
The 912 bp rat αIIb promoter–growth hormone reporter construct has been described (Block et al., 1994). Mouse αIIb promoter constructs were generated by PCR, and introduced between BglII and HindIII sites of the luciferase reporter vector pGL2-basic (Promega). The 946 bp mouse αIIb promoter was cloned into the human growth hormone-based vector pOGH (Nichols Institute Diagnostics) using the same restriction sites. The M1α promoter (Martin and Orkin, 1990) was introduced into pGL2. Segments of 100 and 222 bp of the murine c-mpl and PF4 promoters, respectively, were subcloned into pGL2. In the pGL2-αIIb100 bp construct, the proximal Ets element between –32 and –50 bp was substituted with a single GAL4-binding site (5′-GCGGAGTACTGTCCTCCGA-3′) by overlapping PCR (Block et al., 1994). All PCR-based constructs were sequenced.
pXM-GATA-1, pXM-GATA-1V205G, pMT2-FOG-1, pMFG-FOG-1S706R, GAL4-PU.11–160 and pCMX-GAL4 have been described (Martin and Orkin, 1990; Tsang et al., 1997; Crispino et al., 1999; Maitra and Atchison, 2000). FOG-1S706R was subcloned into pMT2 at the EcoRI site. pCMX-GAL4-FLI-11–274 and pCMX-GAL4-FLI-11–194 were made by PCR and introduced into the GAL4 fusion expression vector pCMX.
Cell lines
NIH-3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM). Murine Y10 cells (Ishida et al., 1993) were maintained in F-12 Nutrient Mixture (Invitrogen). All media contained 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM l-glutamine and 10% fetal calf serum.
DNA transfection, reporter gene assay and western blotting
Cells were transfected with the calcium phosphate precipitation method. The total amount of transfected DNA was kept constant in all samples. After 48 h, luciferase activity was determined using commercial reagents (Promega). Growth hormone activity was assayed with a Nichols Diagnostic Institute kit.
In vitro ES cell differentiation, RNA extraction and semi-quantitative RT–PCR analysis
Wild-type and mutant TL-1 ES cell lines containing the GATA-1V205G substitution have been described (Chang et al., 2002). ES cells were differentiated into EBs as previously described using 100 ng/ml recombinant mouse TPO (R&D Systems) (Zhang et al., 1997). Megakaryocytes in the EBs were identified by their distinct morphology and by their staining with AChE. Total cellular RNA was prepared using RNA STAT-60 reagent (TEL-TEST). The following sets of murine sense/antisense primers were used for RT–PCR: αIIb (GGCTGGAGCACACCTATGAGCT; GCTCAACCTTGGGAGGCT); p45 NF-E2 (ACGTGGACATGTACCCAGTGG; GCCACCTTGTTCTTGCCCCGT); c-mpl (ACCAAGGTCCCTGGAGCG; AGGAGGCTGGGTTCCACTT); GPIX (AGGCCCTGTACCTGCCAGTCC; GCCCAGCTCATAACCTGTCAGCT); PF4 (GTCCAGTGGCACCTCTTGA; AATTGACATTTAGGCAGCTGA); PBP (GCCTGCCCACTTCATAACCTC; GGGTCCAGGCACGTTTTTTG); integrin αL (GATCTGTACTACCTCATGGATCTC; GCAACTTGCATTATGGCATCCAGC); and HPRT (TCCAGAACTAGGACACCTGC; GCTGGTGAAAAGGACCTCT). PCRs were performed in the presence of 1 µCi of [α-32P]dCTP, separated on a polyacrylamide gel, and band intensities measured by Phosphorimager analysis.
Chromatin immunoprecipitation assays
ChIP assays were performed as described (Forsberg et al., 2000) using anti-GATA-1 (N6, Santa Cruz), mouse anti-Fli-1 (C-19, Santa Cruz), rabbit anti-Fli-1 (PharMingen) and affinity-purified rabbit anti-FOG-1 serum raised against amino acids 11–25 of mouse FOG-1. The following modifications were carried out: DNA–protein–antibody complexes were recovered by eluting twice with 100 µl of 0.1 M NaHCO3 and diluted with TE buffer (10 mM Tris–HCl pH 7.4, 1 mM EDTA) to 400 µl total volume. PCRs were performed for 26 cycles at 94°C for 20 s, 52°C for 30 s, 72°C for 45 s in the presence of [α-32P]dCTP using the following primer pairs: αIIb –110 bp to +226 bp region (GTCGACGTCTAGAGGCTATTG; CTCTTAACGCCCATATGTCCT); αIIb –3.3 to –3.6 kb region (TGTGAGTCCCTGCCTGCCATT; TCTAGAGCAGGTTAAGCCCAG); and GAPDH (ACCACAGTCCATGCCATCAC; TCCACCACCCTGTTGCTGTA). PCR products were separated on a 5% TBE polyacrylamide gel and quantitated by phosphorimager analysis.
Fetal liver cultures
Day 13 fetal livers from C57Blk6 mice were differentiated into morphologically mature megakaryocytes as described (Vyas et al., 1999). AchE staining was performed after 5 days in culture with 100 ng/ml recombinant mouse TPO; cells were stained for AChE.
Electrophoretic mobility shift assays
A 0.2 ng aliquot (∼105 c.p.m.) of a [γ-32P]dATP- (Amersham) labeled probe was incubated with 10 µg of nuclear extract. The sequence of the αIIb Ets probe was TAAGCTGAAACTTCCGGTGGTGGGAAC, and that of the M1αEts probe was TAAGGATCAGCTTCCTCGAGCGACCTT. A 2 µg aliquot of the indicated antibodies: anti-FLI-1 (C19), anti-Ets-1 (N-276), anti-Ets-2 (C-20) and anti-PU.1 (T-21) (Santa Cruz Biotechnology) was added to nuclear extracts for 20 min prior to the addition of the probe.
Acknowledgments
Acknowledgements
We thank Stuart Orkin for the GATA-1V205G ES cells, Mike Atchinson for the GAL4-PU.1 construct, and Merlin Crossley and Mitch Weiss for helpful discussions and critical comments on the manuscript. This work was supported by in part by grants PO1 HL40387 (M.P.) and RO1 DK54937 (G.A.B.), and gifts from the Schulman Foundation (M.P.) and the Plummer Family (M.P.).
References
- Athanasiou M., Clausen,P.A., Mavrothalassitis,G.J., Zhang,X.K., Watson,D.K. and Blair,D.G. (1996) Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ., 7, 1525–1534. [PubMed] [Google Scholar]
- Bastian L.S., Kwiatkowski,B.A., Breininger,J., Danner,S. and Roth,G. (1999) Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood, 93, 2637–2644. [PubMed] [Google Scholar]
- Block K.L., Ravid,K., Phung,Q.H. and Poncz,M. (1994) Characterization of regulatory elements in the 5′-flanking region of the rat GPIIb gene by studies in a primary rat marrow culture system. Blood, 84, 3385–3393. [PubMed] [Google Scholar]
- Chang A.N., Cantor,A.B., Fujiwara,Y., Lodish,M.B., Droho,S., Crispino,J.D. and Orkin,S.H. (2002) GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl Acad. Sci. USA, 99, 9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino J.D., Lodish,M.B., MacKay,J.P. and Orkin,S.H. (1999) Use of altered specificity mutants to probe a specific protein–protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell, 3, 219–228. [DOI] [PubMed] [Google Scholar]
- Deveaux S., Filipe,A., Lemarchandel,V., Ghysdael,J., Romeo,P.H. and Mignotte,V. (1996) Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood, 87, 4678–4685. [PubMed] [Google Scholar]
- Doubeikovski A., Uzan,G., Doubeikovski,Z., Prandini,M.-H., Porteu,F., Gisselbrecht,S. and Dusanter-Fourt,I. (1997) Thrombopoietin-induced expression of the glycoprotein IIb gene involves the transcription factor PU.1/Spi-1 in UT7-Mpl cells. J. Biol. Chem., 272, 24300–24307. [DOI] [PubMed] [Google Scholar]
- Eisman R., Surrey,S., Ramachandran,B., Schwartz,E. and Poncz,M. (1990) Structural and functional comparison of the genes for human platelet factor 4 and PF4alt: characterization of the human β-thromboglobulin gene. Comparison with the gene for platelet factor 4. Blood, 76, 336–344. [PubMed] [Google Scholar]
- Forsberg E.C., Downs,K.M., Christensen,H.M., Im,H., Nuzzi,P.A. and Bresnick,E.H. (2000) Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA, 97, 14494–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N. and Schulz,R.A. (2001) Conserved cardiogenic functions of the multitype zinc-finger proteins: U-shaped and FOG-2. Trends Cardiovasc. Med., 11, 185–190. [DOI] [PubMed] [Google Scholar]
- Fox A.H., Liew,C., Holmes,M., Kowalski,K., Mackay,J. and Crossley,M. (1999) Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J., 18, 2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freson K. et al. (2001) Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood, 98, 85–92. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Browne,C.P., Cunniff,K., Goff,S.C. and Orkin,S.H. (1996) Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA, 93, 12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines P., Geiger,J.N., Knudsen,G., Seshasayee,D. and Wojchowski, D.M. (2000) GATA-1-and FOG-dependent activation of mega karyocytic αIIb gene expression. J. Biol. Chem., 275, 34114–34121. [DOI] [PubMed] [Google Scholar]
- Graves B.J. and Petersen,J.M. (1998) Specificity within the Ets family of transcription factors. Adv. Cancer Res., 75, 1–55. [DOI] [PubMed] [Google Scholar]
- Hart A., Melet,F., Grossfeld,P., Chien,K., Jones,C., Tunnacliffe,A., Favier,R. and Bernstein,A. (2000) Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity, 13, 167–177. [DOI] [PubMed] [Google Scholar]
- Ishida Y., Levin,J., Baker,G., Stenberg,P.E., Yamada,Y., Sasaki,H. and Inoue,T. (1993) Biological and biochemical characteristics of murine megakaryoblastic cell line L8057. Exp. Hematol., 21, 289–298. [PubMed] [Google Scholar]
- Kaluzhny Y., Ravid,K. and Poncz,M. (2001) Transcription factors involved in lineage-specific gene expression during megakaryo poiesis. In Ravid,K. and Licht,J. (eds), Transcription Factors: Normal and Malignant Development of Blood Cells. Wiley-Liss, New York, NY, pp. 31–49.
- Kawabata H., Germain,R.S., Ikezoe,T., Tong,X., Green,E.M., Gombart,A.F. and Koeffler,H.P. (2001) Regulation of expression of murine transferrin receptor 2. Blood, 98, 1949–1954. [DOI] [PubMed] [Google Scholar]
- Kawada H., Ito,T., Pharr,P.N., Spyropoulos,D.D., Watson,D.K. and Ogawa,M. (2001) Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int. J. Hematol., 73, 463–468. [DOI] [PubMed] [Google Scholar]
- Lemarchandel V., Ghysdael,J., Mignotte,V., Rahuel,C. and Romeo,P.H. (1993) GATA and ETS cis-acting sequences mediate megakaryocyte-specific gene expression. Mol. Cell. Biol., 13, 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage A., Leboeuf,M., Cazenave,J.P., de la Salle,C., Lanza,F. and Uzan,G. (2000) The α(IIb)β(3) integrin and GPIb-V–IX complex identify distinct stages in the maturation of CD34(+) cord blood cells to megakaryocytes. Blood, 96, 4169–4177. [PubMed] [Google Scholar]
- Maitra S. and Atchison,M. (2000) BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell. Biol., 20, 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Miesfeldt,S., Yang,H., Leiden,J.M. and Thompson,C.B. (1994) The FLI-1 and chimeric EWS–FLI-1 oncoproteins display similar DNA binding specificities. J. Biol. Chem., 269, 18216–18222. [PubMed] [Google Scholar]
- Martin D.I. and Orkin,S.H. (1990) Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev., 4, 1886–1898. [DOI] [PubMed] [Google Scholar]
- McKenna N.J. and O’Malley,B.W. (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell, 108, 465–474. [DOI] [PubMed] [Google Scholar]
- Mehaffey M.G., Newton,A.L., Gandhi,M.J., Crossley,M. and Drachman,J.G. (2001) X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood, 98, 2681–2688. [DOI] [PubMed] [Google Scholar]
- Minami T., Tachibana,K., Imanishi,T. and Doi,T. (1998) Both Ets-1 and GATA-1 are essential for positive regulation of platelet factor 4 gene expression. Eur. J. Biochem., 258, 879–889. [DOI] [PubMed] [Google Scholar]
- Nichols K.E., Crispino,J.D., Poncz,M., White,J.G., Orkin,S.H., Maris,J.M. and Weiss,M.J. (2000) Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet., 24, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L., Simon,M.C., Robertson,E., Klein,W.H., Tsai,S.-F., D’Agati,V., Orkin,S.H. and Costantini,F. (1991) Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature, 349, 257–260. [DOI] [PubMed] [Google Scholar]
- Pevny L., Lin,C.S., D’Agati,V., Simon,M.C., Orkin,S.H. and Costantini,F. (1995) Development of hematopoietic cells lacking transcription factor GATA-1. Development, 121, 163–172. [DOI] [PubMed] [Google Scholar]
- Prandini M.H., Uzan,G., Martin,F., Thevenon,D. and Marguerie,G. (1992) Characterization of a specific erythromegakaryocytic enhancer within the glycoprotein IIb promoter. J. Biol. Chem., 267, 10370–10374. [PubMed] [Google Scholar]
- Ravid K., Doi,T., Beeler,D.L., Kuter,D.J. and Rosenberg,R.D. (1991) Transcriptional regulation of the rat platelet factor 4 gene: interaction between an enhancer/silencer domain and the GATA site. Mol. Cell. Biol., 11, 6116–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhtman N., Radparvar,F., Evans,T. and Skoultchi,A.I. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev., 13, 1398–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo P.-H., Prandini,M.-H., Joulin,V., Mignotte,V., Prenant,M., Vainchenker,W., Marguerie,G. and Uzan,G. (1990) Megakaryocytic and erythrocytic lineages share specific transcription factors. Nature, 344, 447–449. [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A., Fujiwara,Y., McDevitt,M.A. and Orkin,S.H. (1997) A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J., 16, 3965–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos D.D., Pharr,P.N., Lavenburg,K.R., Jackers,P., Papas,T.S., Ogawa,M. and Watson,D.K. (2000) Hemorrhage, impaired hematopoiesis and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol., 20, 5643–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna B.R. and Arrowsmith,C.H. (2000) DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein–DNA recognition. J. Biol. Chem., 275, 28363–28370. [DOI] [PubMed] [Google Scholar]
- Tronik-Le Roux D., Roullot,V., Schweitzer,A., Berthier,R. and Marguerie,G. (1995) Suppression of erythro-megakaryocytopoiesis and the induction of reversible thrombocytopenia in mice transgenic for the thymidine kinase gene targeted by the platelet glycoprotein αIIb promoter. J. Exp. Med., 181, 2141–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang A.P., Visvader,J.E., Turner,C.A., Fujiwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- Tsang A.P., Fujiwara,Y., Hom,D.B. and Orkin,S.H. (1998) Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev., 12, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzan G., Prenant,M., Prandini,M., Martin,F. and Marguerie,G. (1991) Tissue-specific expression of the platelet GPIIb gene. J. Biol. Chem., 266, 8932–8939. [PubMed] [Google Scholar]
- Vyas P., Ault,K., Jackson,C.W., Orkin,S.H. and Shivdasani,R.A. (1999) Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood, 93, 2867–2875. [PubMed] [Google Scholar]
- Watson D.K., Robinson,L., Hodge,D.R., Kola,I., Papas,T.S. and Seth,A. (1997) FLI1 and EWS–FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene, 14, 213–221. [DOI] [PubMed] [Google Scholar]
- Weiss M.J. and Orkin,S.H. (1995) GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol., 23, 99–107. [PubMed] [Google Scholar]
- Zhang C., Gadue,P., Scott,E., Atchison,M. and Poncz,M. (1997) Activation of the megakaryocyte-specific gene platelet basic protein (PBP) by the Ets family factor PU.1. J. Biol. Chem., 272, 26236–26246. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lemarchandel,V., Romeo,P.H., Ben-David,Y., Greer,P. and Bernstein,A. (1993) The Fli-1 proto-oncogene, involved in erythroleukemia and Ewing’s sarcoma, encodes a transcriptional activator with DNA-binding specificities distinct from other Ets family members. Oncogene, 8, 1621–1630. [PubMed] [Google Scholar]