Abstract
The heterodimeric nuclear cap-binding complex (CBC) binds to the 5′ cap structure of RNAs in the nucleus and plays a central role in their diverse maturation steps. We describe the crystal structure at 2.1 Å resolution of human CBC bound to an m7GpppG cap analogue. Comparison with the structure of uncomplexed CBC shows that cap binding induces co-operative folding around the dinucleotide of some 50 residues from the N- and C-terminal extensions to the central RNP domain of the small subunit CBP20. The cap-bound conformation of CBP20 is stabilized by an intricate network of interactions both to the ligand and within the subunit, as well as new interactions of the CBP20 N-terminal tail with the large subunit CBP80. Although the structure is very different from that of other known cap-binding proteins, such as the cytoplasmic cap-binding protein eIF4E, specificity for the methylated guanosine again is achieved by sandwiching the base between two aromatic residues, in this case two conserved tyrosines. Implications for the transfer of capped mRNAs to eIF4E, required for translation initiation, are discussed.
Keywords: cap-binding complex/m7G cap/MIF4G domain/RNA maturation/RNP domain
Introduction
The nuclear cap-binding complex (CBC) is a conserved eukaryotic protein complex which plays a central role in the maturation of pre-mRNA and uracil-rich small nuclear RNA (U snRNA). It is a functional heterodimer, comprising a small (CBP20) and large (CBP80) subunit and binds with high affinity to the 5′ cap structure [7-methyl-G(5′)ppp(5′)N or m7GpppN, where N is any nucleotide] of nascent RNA polymerase II transcripts. CBC enhances the efficiency of pre-mRNA splicing (Izaurralde et al., 1994) and polyadenylation (Flaherty et al., 1997) in the nucleus and is co-exported with mRNA to the cytoplasm (Visa et al., 1996; Shen et al., 2000) where it is exchanged for the eIF4E cap-binding component of the translation initiation complex. In metazoans, CBC is essential for the nuclear export of U snRNAs (Izaurralde et al., 1995), an obligatory step in the assembly of U snRNPs, through the interaction of the CBC–U snRNA complex with PHAX, a phosphorylated adaptor protein, which binds via a nuclear export sequence to the nuclear export receptor Crm1-RanGTP (Ohno et al., 2000).
Previously, we have described the 2 Å resolution crystal structure of a trypsinated form of human CBC (hCBC) which consists of the central protease-resistant core of CBP20 tightly bound to essentially intact CBP80 (Mazza et al., 2001). hCBP80 (790 residues) comprises three tandem MIF4G-like (‘middle domain of eIF4G’) domains each of which is made up of five or six successive helical hairpins. MIF4G domains are found in a number of proteins involved in RNA processing and maturation (SMART; Schultz et al., 2000; Marcotrigiano et al., 2001). The central part of hCBP20 (residues 37–119 out of a total of 156) comprises a canonical RNP domain and binds via its helical face to the second and third MIF4G domains leaving exposed the putative RNA-binding, β-sheet surface. Differential effects of CBP20 point mutants on competitive discrimination between capped RNA and various cap analogues allowed us to identify four conserved residues from the solvent-exposed face of the RNP domain, Tyr43, Phe83, Phe85 and Asp116, that are essential for cap binding. In two other structurally unrelated cap-binding proteins, vaccinia virus nucleoside-2′-O-methyltransferase VP39 (Hodel et al., 1997) and the initiation factor eIF4E (Marcotrigiano et al., 1997; Matsuo et al., 1997), specific recognition of the methyl ated guanosine (m7G) is achieved by sandwiching the base between two aromatic residues (Quiocho et al., 2000). We proposed that Tyr43 might form the bottom of such a sandwich in CBC but were unable to identify the putative top component, suggesting that it might occur on either the N- or C-terminal extensions to the RNP domain that are not present in the structure of trypsinated CBC.
Here we present two new high resolution structures which reveal how CBC binds capped RNAs. Preparation, crystallization and data collection of these complexes are described elsewhere (Mazza et al., 2002). First, we have determined the structure of intact apo-CBC. This turns out to be similar to that of trypsinated CBC and shows that the N- or C-terminal extensions to the RNP domain of CBP20 are disordered in the absence of cap. Secondly, using an internal deletion mutant of CBP80, we were able to crystallize CBC in the presence of the cap analogue m7GpppG in two crystal forms. In these structures, the cap is tightly bound by CBP20, with essential interactions coming from the fully ordered N- and C-terminal extensions to the RNP domain.
Results and discussion
Involvement of the N- and C-terminal extension of CBP20 in cap binding
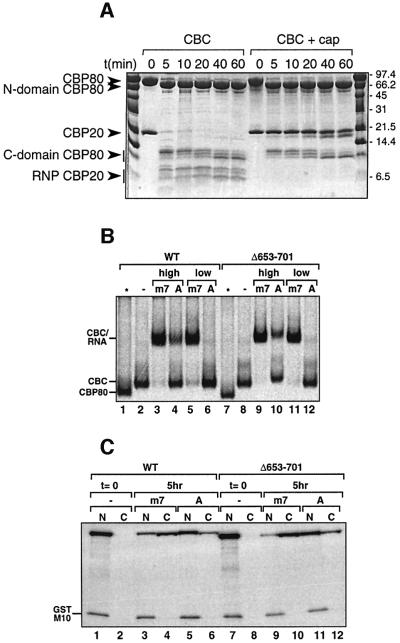
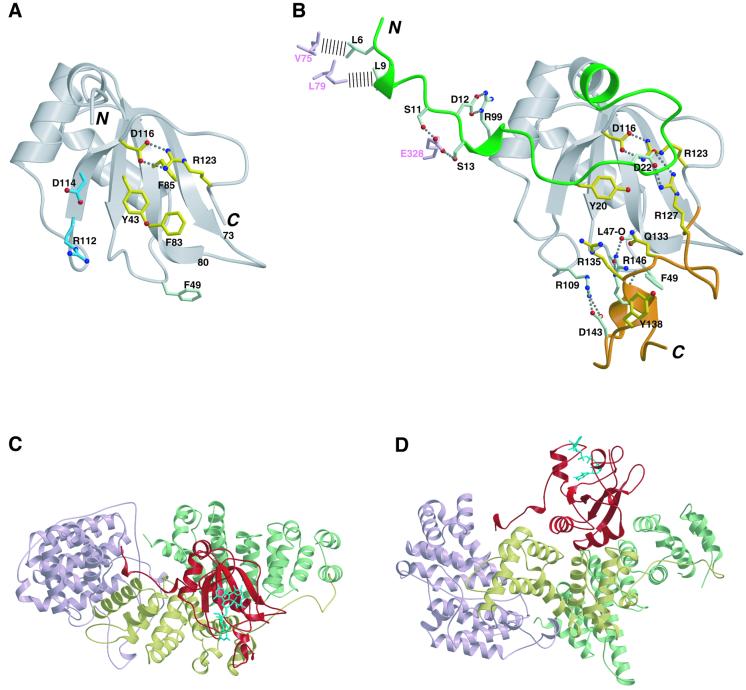
In our previous work (Mazza et al., 2001), we found that mild trypsination of apo-CBC leads to degradation of the N- (up to residue 22) and C- (from residue 120) terminal extensions to the central RNP domain of CBP20, as well as a cleavage in an internal loop (residues 76–79). Furthermore, trypsinated CBC is unable to bind cap. However, when CBC is pre-bound to the m7GpppG cap analogue, CBP20 is strongly protected from trypsination (Figure 1A). Neither the degradation of the N- and C-terminal extensions nor the internal cleavage within the RNP domain occurs, although the cleavage in the loop of the long coiled coil of CBP80 remains. These observations suggest that the N- and C-terminal extensions of CBP20 might be critical for cap binding. We have now determined the structure at 2.0 Å resolution of the intact apo-complex using a CBP80 construct (CBP80ΔNLS) in which the 19 N-terminal residues, including the nuclear localization sequence (NLS), have been deleted (Table I). Although full-length CBP20 is present in the crystal, the intact CBCΔNLS structure is very similar to that of the trypsinated complex. Only an additional eight and seven residues are visible at the N- and C-terminal ends, respectively, of the RNP domain of CBP20, and residues 1–29 and 126–156 remain disordered (Figure 2A). The internal loop of the RNP domain (residues 73–80) is now intact, leading to a native configuration of the RNA-binding surface as predicted (Mazza et al., 2001), whereas previously it had been distorted by the trypsin cleavage. Taken together, these results strongly suggest that the terminal regions of CBP20 are structured and stabilized upon cap binding.
Fig. 1. (A) Time course of limited proteolysis. A 42 µg aliquot of CBC was incubated at room temperature with 210 ng of trypsin in the absence or presence of 10 mM cap analogue m7GpppG in a total volume of 60 µl. Aliquots of 10 µl were taken off every 0, 5, 10, 20, 40 and 60 min, denatured by 3 µl of denaturing buffer (125 mM Tris pH 6.8, 260 mM DTT, 30% glycerol, 10% SDS and 0.025% Coomassie Blue) and loaded onto a 13.5% Tricine SDS–polyacrylamide gel. (B) Cap-binding activity of CBP80 Δ653–701. [35S]Methionine-labelled wild-type or mutant CBP80 was incubated for 30 min at room temperature in the absence (–) or presence of either 1.7 µM (high) or 53 nM (low) m7GpppG-capped (m7) or ApppG-capped (A) unlabelled U1ΔSm RNAs. The samples were fractionated by native 6% PAGE followed by fluorography. Free CBP80, CBC and the CBC–RNA complex are indicated. In the lanes indicated by an asterisk, the corresponding CBP80 proteins were loaded without CBP20. (C) Shuttling activity of CBP80ΔNLSΔ653–701 in Xenopus oocytes. [35S]methionine-labelled CBP80ΔNLS2 or CBP80ΔNLS2Δ653–701 were injected together with [35S]methionine-labelled GST–M10 into Xenopus oocyte nuclei either (1) alone (lanes 1 and 2, and 7 and 8), (2) together with m7GpppG-capped unlabelled U1ΔSm RNAs (lanes 3 and 4, and 9 and 10) or (3) together with ApppG-capped U1ΔSm RNAs (lanes 5 and 6, and 11 and12). Oocytes were dissected either immediately (lanes 1 and 2, and 7 and 8) or 5 h after injection (lanes 3–6 and 9–12) and the proteins analysed by SDS–PAGE followed by fluorography. GST–M10 is a mutant of HIV Rev with a non-functional nuclear export signal used as a negative control. See Ohno et al. (2000) or Segref et al. (2001) for more details.
Table I. Refinement statistics.
| CBCΔNLS | CBCΔCC + cap form 1 | CBCΔCC + cap form 2 | |
|---|---|---|---|
| Space group | C2 | P3121 | P212121 |
| Cell dimensions (Å) | a = 264.14, b = 59.60, c = 75.43, β = 99.52 | a = b = 112.78, c = 158.31, γ = 120 | a = 111.84, b = 125.72, c = 188.76 |
| Resolution range (Å) | 25–2.0 | 20–2.15 | 20–2.4 |
| Completeness (%) | 79.5 | 99.8 | 89.1 |
| Rmerge (%) | 5.3 | 7.9 | 11.4 |
| No. of working (test) reflections | 59 304 (3112) | 60 513 (3204) | 91 470 (4794) |
| R-factor (%) | 21.2 | 23.0 | 19.4 |
| Rfree (%) | 24.7 | 26.6 | 24.5 |
| R.m.s.d. from ideal bond length (Å) | 0.0057 | 0.006 | 0.006 |
| R.m.s.d. from ideal bond angles (°) | 1.1 | 1.1 | 1.1 |
| Ramachandran plot (%) | |||
| Favoured | 93.3 | 91.1 | 91.8 |
| Additional | 6.6 | 8.8 | 8.0 |
| Generous | 0.1 | 0.1 | 0.1 |
| Disallowed | 0.0 | 0 | 0.1 |
| No. of water molecules | 294 | 365 | 1426 |
| No. of non-hydrogen atoms (chain) | CZ | CZT | AXT BYU |
| CBP80 | 5905 | 5746 | 5765 5762 |
| CBP20 | 762 | 1193 | 1214 1202 |
| Cap analogue | – | 52 | 52 52 |
| Average B-factor (Å2) | |||
| CBP80 | 51.45 | 45.14 | 37.32 36.3 |
| CBP20 | 51.21 | 48.17 | 41.03 35.82 |
| Cap analogue | – | 61.06 | 40.35 46.50 |
| Anisotropic B-factor correction (Å2) | B11 = –14.53, B22 = 0.55, B33 = 13.98, B13 = –6.17, B12 = B23 = 0 | B11 = 3.79, B22 = 3.79, B33 = –7.58, B12 = 0.27, B13 = B23 = 0 | B11 = 5.44, B22 = –14.56, B33 = 9.12, B12 = B13 = B23 = 0 |
R = Σ(|Fobs| – k|Fcal|)/Σ|Fobs|
Rfree is calculated using 5% of the reflections.
Fig. 2. (A) Structure of CBP20 in the CBCΔNLS complex. Ribbon representation of the cap-free conformation of CBP20 showing residues 30–125 (grey). Residues involved in cap binding are shown in yellow (already in their cap-bound conformation) and blue (undergo a conformational change to interact with the cap) (compare with B). Phe49 (light green) changes conformation to help stabilize the C-terminal domain (compare with B). Salt bridges and hydrogen bonds are indicated by dashed lines. (B) Stabilization of the N- and C-terminal extensions of CBP20 upon cap binding. Ribbon representation of cap-bound CBP20 showing residues 32–125 (already ordered in the cap-free form) in grey and the N- and C-terminal extensions that fold upon cap binding in green and orange, respectively. Yellow residues stabilize the folded conformation of these two extensions through interactions with the cap. This stabilization is reinforced by protein–protein interactions involving the light green residues. D116, R123 and R127 take part to both kinds of contacts. CBP80 residues interacting with residues 5–13 from CBP20 are depicted in pink. Hydrogen bonds and salt bridges are represented as dashed lines, and the hydrophobic contacts as dashed bars. (C and D) Two views of CBC bound to the cap analogue m7GpppG. The three MIF4G domains of CBP80 are represented in pink, yellow and green for domains 1, 2 and 3, respectively. CBP20 is depicted in red, and the cap analogue m7GpppG in cyan. (A), (B) and (C) were generated with Molscript (Kraulis, 1991) and Render (Merritt and Murphy, 1994).
Crystal structure determination of CBC with the m7GpppG cap analogue
Attempts to crystallize the cap-bound wild-type complex have not been successful. To proceed, we hypothesized that truncation of the long protruding coiled coil located in the third MIF4G domain of CBP80 might result in a more readily crystallizable complex (Mazza et al., 2002). We thus re-cloned CBP80ΔNLS to replace 49 residues of the coiled coil by a glycine (Val652-Gly-Ala702, denoted CBP80ΔNLSΔCC). Figure 1B and C shows that this deletion mutant is still active in CBP20 binding, capped RNA binding and U snRNA export. We co-crystallized reconstituted CBCΔNLSΔCC with the cap analogue m7GpppG, obtaining two crystal forms which were solved by molecular replacement to give structures at 2.15 and 2.3 Å resolution, respectively (Mazza et al., 2002) (Table I). In crystal form 1, there is one complex in the asymmetric unit (denoted by the chain designations as CZT, for CBP80, CBP20 and cap analogue, respectively) whereas in crystal form 2 there are two complexes in the asymmetric unit (denoted by the chain designations as AXT and BYU). The conformation of the m7GTP moiety of the cap analogue is the same in all three independent examples of the complex, while the conformation of the second guanosine is dependent on crystal packing (see below and Materials and methods).
Co-operative folding of the N- and C-terminal domain upon cap binding
Almost the entire N- and C-terminal extensions of CBP20, disordered in the CBCΔNLS structure, as well as the m7GpppG cap analogue, which binds exclusively to CBP20, could be built into positive difference electron density. The highly hydrophilic extensions fold co-operatively around the cap analogue in an intricate hydrogen-bonded network which has little regular secondary structure apart from a few helical turns (Figures 2B, D and 5). The m7G is sandwiched between Tyr43, from the RNP2 motif, on the bottom, and Tyr20, from the N-terminal extension, on the top (Figures 3B and 4C). Folding and stabilization of the arginine–glycine–tyrosine-rich C-terminal domain of CBP20 is achieved mainly by interaction of five residues with the cap analogue and reinforced by intradomain interactions (Figure 2B). This necessitates rearrangement of some RNP core residues, for instance Phe49, which reorientates upon cap binding to pack between the backbones of residues 146–147 and 81–82 (compare Figure 2A and B). Some residues, such as Asp116, Arg123 and Arg127, are involved in both cap and intraprotein interactions. The N-terminal domain conformation is stabilized by a salt bridge between Arg127 and Asp22 which ties together the C- and N-terminal extensions. Tyr20 makes critical interactions with the cap analogue, not only forming the top of the m7G sandwich but also interacting with the ribose and β-phosphate (Pβ). Stabilization of residues 14–29 is buttressed further by the interaction via both hydrophobic interactions and hydrogen bonds of the extreme N-terminal residues 5–13 in a groove between the MIF4G domains 2 and 3 of CBP80 (Figure 2C and D). There are no significant changes to the structure of CBP80 or the CBP20–CBP80 interface upon cap binding.
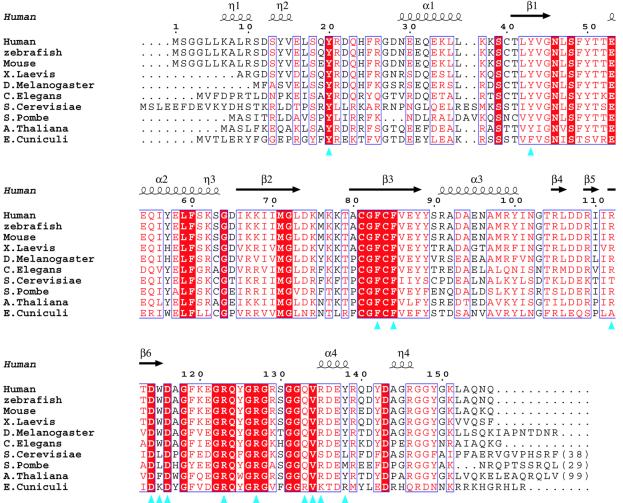
Fig. 5. Sequence alignment of the CBP20s. Human (P52298), zebrafish (AAM28218), mouse (NP_080830), Drosophila melanogaster (CAB53185), Xenopus laevis (P52299), Caenorhabditis elegans (NP_492130), Saccharomyces cerevisiae (NP_015147), Schizosaccharomyces pombe (NP_596414), Arabidopsis thaliana (NP_199233) and Encephalitozoon cuniculi (NP_597585). Residues that are 100% conserved are in red boxes. Homology >70% (based on Risler et al., 1988) is depicted in red. Blue triangles indicate residues involved in cap binding. The secondary structure of the human CBP20 is in black (α, α-helices; π, 310-helices; β, β-strand). The figure was generated with CLUSTALW (Thompson et al., 1994) and ESPript (Gouet et al., 1999).
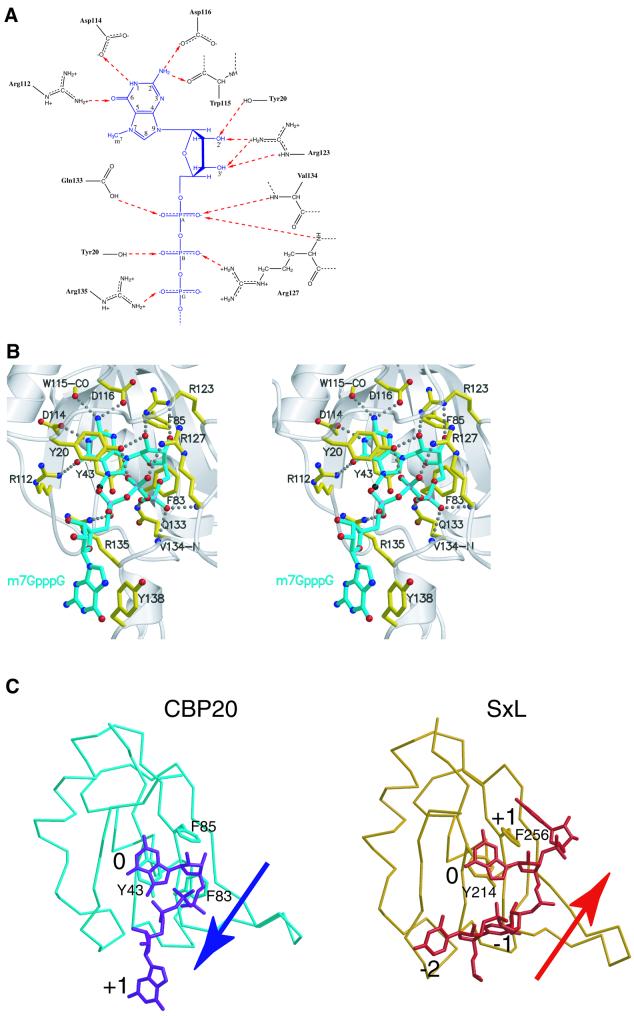
Fig. 3. (A) Diagram of the polar interactions between m7GTP and CBP20. m7GTP is represented in blue, and the amino acids involved in its stabilization in black. Dashed red arrows indicate the hydrogen bonds between donors and acceptors. (B) Stereoscopic view of the cap-binding site. The m7GpppG cap analogue is depicted in cyan and the CBP20 residues involved in its stabilization in yellow. Oxygen, nitrogen and phosphor atoms are in red, blue and pink, respectively, and the methyl group on the N7 position is in black. Hydrogen bonds and salt bridges are represented as dashed lines. (C) Comparison of the mode of RNA binding to the RNP domains of CBP20 and sex-lethal (SxL) protein. The complex between the CBP20 RNP domain (residues 38–118 in light blue) and the cap analogue m7GpppG (dark blue) is compared with the complex between the SxL RNP domain 2 (residues 209–289 in orange) and its cognate RNA (bases 2–5 in red). 0 represents the conserved base stacking on to the aromatic residue from the RNP2 motif. The –2, –1 and +1 base positions are 5′ to 3′ relative to the 0 position. The arrows indicate the direction (5′ to 3′) of the RNA relative to the RNA-binding domain. (A) was generated with ChemDraw, and (B) and (C) were generated with Molscript (Kraulis, 1991) and Render (Merritt and Murphy, 1994).
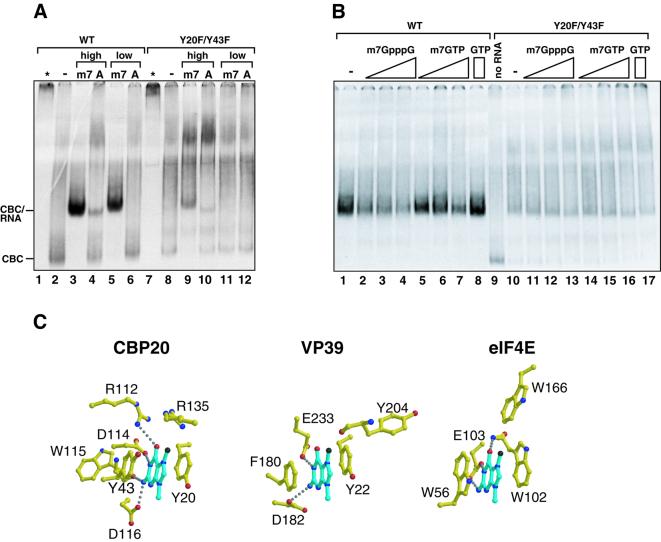
Fig. 4. (A) Reduced cap-binding activity of the CBP20 double mutant Y20F/Y43F. The same as Figure 1B except that CBP20 was labelled with [35S]methionine and incubated with recombinant CBP80ΔNLS2 and 3.3 µM RNA was used in the high concentrations. (B) A competition binding experiment was performed with 3.3 µM m7GpppG-capped U1ΔSm RNAs in the absence (–) or presence of m7GpppG (1.3, 4 or 12 mM) or m7GTP (4.3, 12 or 24 mM) or GTP (24 mM). In the lane where ‘no RNA’ is indicated, the corresponding CBC was loaded alone. The film was exposed twice longer for the mutant compared with the wild-type. (C) Comparison of the mode of m7G binding to CBP20, VP39 and eIF4E. Residues involved in the stabilization of the methylated guanosine (blue) in CBP20, VP39 and eIF4E are depicted in yellow. Oxygen and nitrogen atoms are in red and blue, respectively. Hydrogen bonds are represented as dashed lines. The figure was generated with Molscript (Kraulis, 1991) and Render (Merritt and Murphy, 1994).
Recognition of the cap analogue
Details of the interaction of the m7GpppG cap analogue with CBP20 are shown in Figure 3. There are direct hydrogen bonds to almost all of the possible acceptor or donor groups on the m7Gppp moiety of the ligand, including four to the Watson–Crick positions of the base, three to the 2′ and 3′ hydroxyl groups of the ribose, three to the α-phosphate, two to Pβ and one to Pγ. Guanosine specificity is ensured by hydrogen bonds from Arg112 and Asp114 to the O6 and N1 positions, respectively, of the base and two hydrogen bonds from the N2 position to the carbonyl group of Trp115 and OD2 of Asp116 (Figure 3A and B). A D116A mutant exhibits a reduction in RNA binding by a factor of at least 100 and is not able to distinguish between m7G-capped-RNA and A-capped-RNA (Mazza et al., 2001). In contrast, the binding is only reduced by a factor of eight in the D114A mutant and unchanged in the R112A mutant, and both retain specificity for the m7G base (Mazza et al., 2001; Table II). Therefore, Asp116 is certainly the key residue in determining the specificity of CBC for a guanine cap residue. Mono- or di-methylation of the N2 position would result in the loss of hydrogen bonds with Asp116 and Trp115 and an unfavourable environment for the methyl groups. This is consistent with the observation that di- or tri-methylated cap analogues (m2,7GpppG and m2,2,7GpppG, the latter being the mature form of the U snRNA cap) are 1000 times less efficient than the mono-methylated cap in competing with m7G-capped-RNA for CBC binding (Izaurralde et al., 1992).
Table II. Site-directed mutagenesis of CBP20 residues involved in cap binding.
| CBP20 mutants | Relative affinity for m7G-capped RNA (%)a | Relative affinity for m7GTP–Sepharose (%)b |
|---|---|---|
| Wild type | 100 (13 nM) | 100 |
| Y20A | 3.3 | 0 |
| Y20F | 25 | 0 |
| Y43A | 1 | 0 |
| Y43F | 100 | 62 |
| R112A | 100 | 58 |
| R112T | 100 | 103 |
| Y138A | 100 | 89 |
| Y20F/Y43F | 1 | Not done |
aRelative affinity of CBP20 mutants for m7G-capped RNA: same conditions as in Figure 4A.
bRelative affinity of CBP20 mutants for m7GTP–Sepharose: measured as described in Materials and methods.
The exact conformation of the second guanosine of m7GpppG is dependent on crystal packing. In two (CZT and BYU) of the three independent examples of the complex, the second guanosine stacks on Tyr138; in the third example (AXT), this region is distorted by crystal contacts, and both Tyr138 and the second guanosine stack separately on a neighbouring molecule. We believe stacking with Tyr138 represents the native conformation of the cap since the fact that the binding affinity for CBC is 100 times stronger for m7GpppG than for m7GTP (Izaurralde et al., 1992) strongly suggests a direct interaction of CBC with the second nucleotide. However, the full explanation for this substantially increased affinity for m7GpppG is not readily apparent since a Y138A mutant does not have a detectable effect on CBC affinity for capped RNA (see below and Table II). The second guanosine base is not specifically recognized within the complex, consistent with it being equivalent to the first, arbitrary nucleotide of a longer capped RNA. However, in both the CZT and BYU complexes, the base makes hydrogen bonds with a neighbouring protein in the crystal. The ribose of the second guanosine, which in the natural cap structure is 2′-O-methylated, is poorly defined, and the temperature factors of the base and the extreme C-terminal region of CBP20 to which it binds are relatively high, indicating residual flexibility of this region. It is possible that subsequent nucleotides of a longer capped RNA could interact non-specifically with CBC. Indeed, in the case of U snRNAs, binding of PHAX to proximal regions of the RNA via a novel non-specific binding domain enhances the stability of the CBC–PHAX–U snRNA complex (Ohno et al., 2000; Segref et al., 2001). Interestingly, in contrast to CBC, which binds m7GpppG 100-fold more strongly than m7GTP, eIF4E binds m7GpppG 20-fold less strongly than m7GTP (Niedzwiecka et al., 2002). In the recent X-ray structure of eIF4E complexed with m7GpppG, the second guanosine is not in contact with the protein and not visible in the electron density (Niedzwiecka et al., 2002).
An unusual orientation of the RNA on an RNP domain: comparison of the RNP–cap complex with other RNP–nucleic acid complexes
The orientation of the methylated guanylate (m7GMP) on the β-sheet surface of the CBP20 RNP domain is very similar to that of the central nucleotide (denoted position 0) in several other known RNP domain structures in complex with their cognate RNA, such as sex lethal protein (SxL; Handa et al., 1999) (Figure 3C). Both the m7G and Gua-4 stack onto the absolutely conserved aromatic residue from the RNP2 motif (Tyr43 in CBP20 and Tyr214 in SxL), and base-specific interactions are provided by residues from the C-terminal strand of the β-sheet and its extension. Previously we have shown that single mutations to alanine of either Tyr43 (RNP2), Phe83 or Phe85 (both RNP1 motif) reduce cap binding by factors of 100, 100 and 25, respectively (Mazza et al., 2001). The important role of Phe83 in making van der Waals contacts with the m7G ribose is now clear, equivalent interactions being observed commonly in other such systems. However, the role of Phe85 in cap binding differs from the usual function of this conserved aromatic residue in other known complexes where it is invariably found to stack with the +1 (rarely the +2) base. Instead, in CBP20, Phe85 is situated under the guanidinium group of Arg123, which makes the crucial salt bridge with Asp116 (Figure 3B). It is thus important in stabilizing the conformation of the 116–123 loop whose position blocks any possibility of base stacking on Phe85. Consequently, rather than the canonical mode of RNA binding, crossing the β-sheet surface diagonally from β1/RNP2 to β2/RNP1 in the 5′ to 3′ direction, the cap structure, with its unique 5′–5′ connection and tri-phosphate bridge, goes in the opposite direction (Figure 3C). If 0 is the position of the m7G stacking onto Tyr43 from RNP2, then the next guanosine (+1), which actually can be any base, roughly occupies the position equivalent to –2 in the SxL protein–RNA complex (Handa et al., 1999) (Figure 3C).
Specific recognition of the methylated base: comparison with other cap-binding proteins
The extensive hydrogen bonding interactions between the m7G and the RNP core are critical for guanosine specificity, but not sufficient to explain the strong preference of CBC for the N7-methylated cap. Indeed, the m7GpppG cap analogue is found to bind to CBC with a Kd of ∼13 nM (Mazza et al., 2001), some 100-fold better than unmethylated GpppG. Extensive structural and biochemical work on VP39 (Hodel et al., 1997; Hsu et al., 2000; Quiocho et al., 2000), eIF4E (Marcotrigiano et al., 1997; Niedzwiecka et al., 2002) and small molecule systems (Ueda et al., 1991; Stolarski et al., 1996; Ishida et al., 1988) has shown that m7G specificity is achieved principally by parallel stacking of the methylated base between two aromatic residues. The rationale for this is that the positive charge arising from methylation is delocalized on the base in its cationic form (as opposed to the zwitterionic form; Stolarski et al., 1996) and enhances the interactions with the π-electrons of the stacked aromatic rings. It is also plausible that the positive charge of the methylated base provides electrostatic reinforcement to the hydrogen bond interactions made by the acidic residues (e.g. Asp114 and Asp116 in CBC) to the N1 and N2 positions. In VP39, the two stacking aromatic residues are Tyr22 and Phe180, and in eIF4E they are Trp56 and Trp102 (Figure 4C). Van der Waals or weak polar interactions with the methyl group itself (with Trp166 in eIF4E and with the carbonyl group of Tyr204 in VP39) appear to play a minor role in m7G specificity. In CBP20, the m7G is sandwiched between Tyr43 and Tyr20, and individual mutation of either of these to alanine abolishes binding to m7GTP (Mazza et al., 2001; Table II). The closest similarity in binding configuration is between CBC and VP39, with the sandwiching aromatic rings being orientated in almost exactly the same way (Figure 4C). Also, in both cases, two acidic side chains are implicated in the guanosine specificity. In CBC, a possible van der Waals contact of the methyl group with Arg135, however, would leave room for an ethyl-substituted base which has been shown to be an equally good ligand for CBC (Izaurralde et al., 1992). Despite the relatively high resolution of the structure, we see no water molecules contributing to the CBP20–cap interface. In contrast, for instance in the case of eIF4E, several ordered water molecules interact notably with the α-phosphate (Niedzwiecka et al., 2002).
In early work, it was proposed from chemical considerations that the stacking ability of aromatic amino acids with m7G would decrease in the order tryptophan, tyrosine, phenylalanine (Ishida et al., 1988). Recently, mutational studies on VP39 and eIF4E combined with more accurate measurements of binding constants, led to the hypothesis that various combinations of aromatics could be sufficient for m7G discrimination provided at least one tyrosine or tryptophan was present (Hsu et al., 2000). In the case of CBC, a Y43F mutation still binds capped RNA and m7GTP well (Table II), consistent with the fact that a phenylalanine is found at this position in the eukaryote parasite Encephalitozoon cuniculi CBP20 (Katinka et al., 2001) (Figure 5). A Y20F mutation leads to a 75% reduction in capped RNA binding, whereas the double mutant Y20F/Y43F is completely deficient (Figure 4A; Table II). This would appear to support the hypothesis of Hsu et al. (2000), although the importance of the absolutely conserved Tyr20 is likely to be enhanced by the additional interactions of its hydroxyl group with the 2′OH of the ribose and the β-phosphate (Figure 3B). Note that in Table II we show relative binding efficiencies of CBC mutants to both capped RNAs and m7GTP– Sepharose, the differences between the results being indicative of additional interactions with CBC by the longer RNAs. Clearly, more precise measurements of the relative binding constants of methylated and unmethylated cap structures, as has been done recently with eIF4E (Niedzwiecka et al., 2002), are required to strengthen these conclusions.
A phylogenetically conserved mode of cap binding
All 14 residues directly contacting the cap are absolutely conserved in higher eukaryotes and, with a few exceptions, also in the lower eukaryotes, Saccharomyces cerevisiae, Schizosaccharomyces pombe and E.cuniculi (Figure 5). One variable position is Arg112, which interacts with the O6 of the methylated base; in S.cerevisiae, it is a threonine. Arg135, which interacts with the γ-phosphate and weakly with the methyl group of the m7G, is exceptionally a serine in S.cerevisiae, rather than a conserved basic residue. Other phylogenetic variations would affect mainly interactions with the second base. In particular, Tyr138 is replaced by either a leucine, methionine or arginine, all of which could conserve a non-specific base stacking function, as observed in other protein–RNA complexes. However, individual mutations of Tyr138 and Arg135 to alanine as well as R112T in hCBP20 do not significantly reduce the binding affinity of the human complex for capped RNA (Table II and data not shown), indicating that the contribution of each of these residues to the global binding is small. We are thus unable at this stage to provide an explanation for the lack of discrimination between methylated and non-methylated guanosine observed for yeast CBC (Gorlich et al., 1996).
These observations suggest that the mode of cap binding to CBC is evolutionarily highly conserved. The high conservation of the CBP20 subunit, which binds a universally conserved chemical entity, the cap, is in strong contrast to the very low overall conservation of CBP80 (Mazza et al., 2001). Presumably, CBP80 has diverged through co-evolution with its other protein-binding partners, this being evident even, for instance, in the interactions with the N-terminal tail of CBP20.
Conclusions
Our results show that cap binding by CBC is a co-operative, induced fit process involving ordering and stabilization of some 50 residues from the C- and N-termini of CBP20 around the dinucleotide. Comparison of the cap-bound and unbound structures enables a possible pathway for this process to be envisaged. Residues Tyr43, Phe85 and Phe83 and the salt bridge between Arg123 and Asp116, which are unchanged in conformation between the two structures, are plausibly the anchoring points for the initial binding of the cap (compare Figure 2A with B). They provide a bottom platform for the m7G and make specific interactions with the N2 and ribose hydroxyl groups. Re-orientation of Asp114 and Arg112 would then reinforce the specific interactions with the guanosine base (compare Figure 2A with B). Folding of the C-terminal domain could start with the interaction of the Arg127 and Val134 main chain amides, as well as Gln133, with Pα and then the side chain of Arg127 with Pβ. Correct positioning of Arg127 could then initiate folding of the N-terminal domain through formation of the interdomain salt link with Asp22 (Figure 2B). This would facilitate the crucial stacking of Tyr20, which interacts additionally with the ribose and β-phosphate, on top of the methylated guanosine. Final steps would be the parallel folding of the extreme C-terminal residues of CBP20 and stabilization of the second nucleotide and the interaction of the CBP20 N-terminal tail with CBP80 (Figure 2B–D). As there is no structure for apo-eIF4E, the exact extent of the suspected conformational changes (Niedzwiecka et al., 2002) in the protein upon cap binding is unknown, although there is no evidence for large unfolded regions as in CBP20. In VP39, comparison of structures of the apo and cap-bound protein shows that there is very little structural perturbation upon cap binding (Hu et al., 1999). Thus the large-scale induced fit recognition of cap by CBC is exceptional. It is interesting to note that conformational ordering on this scale may result in a significant entropic cost in the overall free energy of cap binding to CBC, which must be offset by the large number of new protein–protein and protein–ligand interactions.
Once in the cytoplasm, efficient mRNA translation requires that the cap be bound by eIF4F which recruits the ribosome to the translation start site. eIF4F is a translation initiation complex made up of the three proteins, eIF4E, eIF4A and eIF4G. The mechanism by which the capped mRNA is transferred from CBC to the cap-binding eIF4E subunit of eIF4F is still unclear. This could occur by simple dissociation from CBC and rebinding to eIF4E, but the complex and tight binding of cap to CBC suggests that a specific mechanism may be required to destablize the CBC complex with capped RNA. Recent studies suggest that there is an interaction between eIF4G and CBP80 in both yeast (Fortes et al., 2000) and mammals (McKendrick et al., 2001). In yeast, CBP80 has been found to interact directly with a domain of eIF4G located between the eIF4E-binding site and the MIF4G domain (Fortes et al., 2000; Marcotrigiano et al., 2001). Even though the corresponding domain in mammals shows low homology with yeast, this interaction is thought to be evolutionarily conserved (McKendrick et al., 2001). In a plausible model for assisted transfer, the scaffold protein eIF4G would simultaneously bind the nuclear (CBC) and cytoplasmic (eIF4E) cap-binding proteins in close proximity. mRNA could then be transferred from the CBP20 cap-binding site, which might be destabilized by the interaction with eIF4G, to eIF4E whose capped mRNA-binding affinity is significantly enhanced by binding to eIF4G (Haghighat and Sonenberg, 1997).
Figure 2D shows in a striking fashion that only CBP20 has a direct role in binding cap and that CBP80 is unlikely to make any contacts with capped RNAs. What then is the role of CBP80? CBP20 cannot bind to the cap on its own (Izaurralde et al., 1995), suggesting that CBP80 induces a conformational change in CBP20 that allows cap interaction. In fact, preliminary NMR data suggest that CBP20 is unstructured in solution (our unpublished data). Whether or not CBP20 is entirely unstructured without CBP80, this induced structural change is the mechanistic reason why cap binding absolutely requires the formation of the CBC heterodimer. The second function of CBP80 is to provide a large surface for binding to multiple partner proteins involved in different aspects of capped RNA maturation, such as PHAX and eIF4G. Future work should be directed at completing identification of these partners and mapping their interaction sites on CBC. It is interesting to speculate that some of these partners might make use of the additional binding surfaces offered by the folded N- and C-terminal extensions of CBP20 for cap-dependent CBC interactions.
Materials and methods
Limited proteolysis
Time course proteolysis was performed on CBC in the presence or absence of 10 mM m7GpppG cap analogue. The experiment was carried out at room temperature, using a trypsin/CBC ratio of 1/200 (w/w). Reactions were stopped by the addition of denaturing buffer and analysed by SDS–PAGE. See also Figure 1A.
Mutagenesis of CBP20, and cap binding and competition band shift assays
Alanine mutagenesis of CBP20 was performed using a pT77-vector with the QuickChange Site-Directed Mutagenesis kit (Stratagene). Band shift experiments were performed as described in Mazza et al. (2001). See also Figures 1B, 4A and B.
Nuclear injection experiments
Experiments were performed as described previously (Ohno et al., 2000). See also Figure 1C.
Binding of CBC mutants to m7GTP–Sepharose
35S-labelled CBP20 was incubated with 42 ng/µl recombinant CBP80NLS2 overnight at 4°C to form active CBC, or with phosphate-buffered saline (PBS) for control samples. m7GTP (a gift from Edward Darzynkiewicz) was pre-incubated for 20 min at 4°C in binding buffer [20 mM HEPES pH 7.9, 0.2 mM EDTA, 100 mM KCl, 1 mM dithiothreitol (DTT), 1 mg/ml bovine serum albumin (BSA), 0.1% NP-40, 5% glycerol, 1× protease inhibitor cocktail]. For the binding studies, 5 µg of m7GTP beads were diluted in 50 µl of binding buffer together with 2 µl of labelled CBC or CBP20 as background control, and incubated for 30 min at room temperature while being shaken continuously. Subsequently, the supernatant was collected, the beads washed several times and the bound proteins eluted with SDS in sample buffer. Fifty percent of the bound and 10% of the unbound fraction were analysed by SDS–PAGE followed by autoradiography. The affinity was determined using a phosphoimager (FLA 2000) by taking the percentage of the bound fraction after background subtraction.
Structure determination of CBCΔNLS
The CBCΔNLS structure was determined at 2.0 Å resolution from a single monoclinic crystal (C2, a = 264.1 Å, b = 59.6 Å, c = 75.4 Å, β = 99.5°) that grew from a 2-year-old precipitate. Crystallization conditions and full data collection statistics are given elsewhere (Mazza et al., 2002; see also Table I). The structure was solved by molecular replacement using as search model trypsinated CBC (PDB entry 1h6k). CBP20 residues 30–37, 72–81 and 117–125, disordered or cleaved in trypsinated CBC, could be built into the extra electron density. No electron density could be observed for residues 1–29 and 126–156. Refinement was carried out using CNS (Brünger et al., 1998) and model building with O (Jones and Kjeldgaarg, 1997). Water molecules were added using ARP-wARP (CCP4, 1994). The final model has an R-factor (Rfree) of 21.2% (24.7%) and contains one complex (CBP80 = chain C, CBP20 = chain Z) and 294 water molecules (Table I).
Structure determination of CBCΔNLSΔCC complexed with the cap analogue
In the CBCΔNLSΔCC complex, there are two deletions in CBP80: 19 residues, including the NLS, were removed at the N-terminus, as well as residues 653–701 which were replaced by a glycine. Two different crystal forms were obtained by co-crystallization with the cap analogue m7GpppG (Mazza et al., 2002): form 1 (P3121, a = b = 112.8 Å, c = 158.3 Å) and form 2 (P212121, a = 111.8 Å, b = 125.7 Å, c = 188.8 Å). Crystallization conditions and full data collection statistics are given elsewhere (Mazza et al., 2002; see also Table I). The structures were solved by molecular replacement using as search model trypsinated CBC (PDB entry 1h6k). Additional CBP20 residues 5–29 and 126–152 could be built into the electron density map as well as the cap analogue (for experimental electron density for the cap see Mazza et al., 2002). The structures were refined with CNS to a final R-factor (Rfree) of 23.0% (26.6%) at 2.15 Å resolution (form 1) and 19.4% (24.5%) at 2.4 Å resolution (form 2). Form 1 contains in the asymmetric unit one ternary complex comprising CBP80 (chain C), CBP20 (chain Z), m7GpppG (chain T) and 365 water molecules. Form 2 contains two complexes per asymmetric unit comprising chains A, X, T and B, Y, U for CBP80, CBP20 and m7GpppG, and a total of 1426 water molecules (Table I).
The conformation of the second guanosine of the m7GpppG is dependent on crystal packing. In both form 1 and complex BYU from form 2, the second guanine stacks on Tyr138 from the C-terminal domain of CBP20 and is stabilized by hydrogen bonding and/or hydrophobic interactions between the base and crystal symmetry-related molecules. In the AXT complex from form 2, Tyr138 makes crystal contacts with residues 26–28 from a distinct CBP20 molecule (chain Y) which prevents the stacking of the second guanine upon it. The base is displaced from its presumed native conformation and stabilized by Watson–Crick interactions with crystal symmetry-related residues Gln19 and Asn29 from chain Y. We believe this conformation is a crystal packing artefact as it would suggest that the second base makes no interaction with the complex. Above we only describe the cap conformation observed in the complex CZT from crystal form 1 and BYU from form 2.
Acknowledgments
Acknowledgements
We thank members of the EMBL–ESRF Joint Structural Biology Group for access to ESRF beamline ID14, and Kornelius Zeth for assistance in data processing. Co-ordinates and structure factors have been deposited in the PDB with codes 1h2v for the full-length CBC, and 1h2t and 1h2u for the form 1 and form 2 complex, respectively, of CBC with the cap analogue.
References
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Flaherty S.M., Fortes,P., Izaurralde,E., Mattaj,I.W. and Gilmartin,G.M. (1997) Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl Acad. Sci. USA, 94, 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes P., Inada,T., Preiss,T., Hentze,M.W., Mattaj,I.W. and Sachs,A.B. (2000) The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell, 6, 191–196. [PubMed] [Google Scholar]
- Gorlich D., Kraft,R., Kostka,S., Vogel,F., Hartmann,E., Laskey,R.A., Mattaj,I.W. and Izaurraide,E. (1996) Importin provides a link between nuclear protein import and U snRNA export. Cell, 87, 21–32. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignements in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Haghighat A. and Sonenberg,N. (1997) eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem., 272, 21677–21680. [DOI] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Hodel A.E., Gershon,P.D., Shi,X., Wang,S.M. and Quiocho,F.A. (1997) Specific protein recognition of an mRNA cap through its alkylated base. Nat. Struct. Biol., 4, 350–354. [DOI] [PubMed] [Google Scholar]
- Hsu P.C., Hodel,M.R., Thomas,J.W., Taylor,L.J., Hagedorn,C.H. and Hodel,A.E. (2000) Structural requirements for the specific recognition of an m7G mRNA cap. Biochemistry, 39, 13730–13736. [DOI] [PubMed] [Google Scholar]
- Hu G., Gershon,P.D., Hodel,A.E. and Quiocho,F.A. (1999) mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl Acad. Sci. USA, 96, 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Doi,M. and Inoue,M. (1988) A selective recognition mode of a nucleic acid base by an aromatic amino acid: l-phenylalanine-7-methylguanosine 5′-monophosphate stacking interaction. Nucleic Acids Res., 16, 6175–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Stepinski,J., Darzynkiewicz,E. and Mattaj,I.W. (1992) A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol., 118, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis,J., McGuigan,C., Jankowska,M., Darzynkiewicz,E. and Mattaj,I.W. (1994) A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell, 78, 657–668. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Lewis,J., Gamberi,C., Jarmolowski,A., McGuigan,C. and Mattaj,I.W. (1995) A cap-binding protein complex mediating U snRNA export. Nature, 376, 709–712. [DOI] [PubMed] [Google Scholar]
- Jones T.A. and Kjeldgaarg,M. (1997) Electron-density map interpretation. Methods Enzymol., 277, 173–208. [DOI] [PubMed] [Google Scholar]
- Katinka M.D. et al. (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature, 414, 450–453. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures’. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Marcotrigiano J., Gingras,A.C., Sonenberg,N. and Burley,S.K. (1997) Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell, 89, 951–961. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J., Lomakin,I.B., Sonenberg,N., Pestova,T.V., Hellen,C.U. and Burley,S.K. (2001) A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell, 7, 193–203. [DOI] [PubMed] [Google Scholar]
- Matsuo H., Li,H., McGuire,A.M., Fletcher,C.M., Gingras,A.C., Sonenberg,N. and Wagner,G. (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol., 4, 717–724. [DOI] [PubMed] [Google Scholar]
- Mazza C., Ohno,M., Segref,A., Mattaj,I.W. and Cusack,S. (2001) Crystal structure of the human nuclear cap binding complex. Mol. Cell, 8, 383–396. [DOI] [PubMed] [Google Scholar]
- Mazza C., Segref,A., Mattaj,I.W. and Cusack,S. (2002) Co-crystallisation of the human nuclear cap-binding complex with a m7GpppG cap analogue using protein engineering. Acta Crystallogr. D, in press. [DOI] [PubMed] [Google Scholar]
- McKendrick L., Thompson,E., Ferreira,J., Morley,S.J. and Lewis,J.D. (2001) Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol., 21, 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A.M. and Murphy,E.P. (1994) Raster3D version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Niedzwiecka A. et al. (2002) Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol., 319, 615–635. [DOI] [PubMed] [Google Scholar]
- Ohno M., Segref,A., Bachi,A., Wilm,M. and Mattaj,I.W. (2000) PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell, 101, 187–198. [DOI] [PubMed] [Google Scholar]
- Quiocho F.A., Hu,G. and Gershon,P.D. (2000) Structural basis of mRNA cap recognition by proteins. Curr. Opin. Struct. Biol., 10, 78–86. [DOI] [PubMed] [Google Scholar]
- Risler J.L., Delorme,M.O., Delacroix,H. and Henaut,A. (1988) Amino acid substitutions in structurally related proteins. A pattern recognition approach. Determination of a new and efficient scoring matrix. J. Mol. Biol., 204, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Mattaj,I.W. and Ohno,M. (2001) The evolutionarily conserved region of the U snRNA export mediator PHAX is a novel RNA-binding domain that is essential for U snRNA export. RNA, 7, 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E.C., Stage-Zimmermann,T., Chui,P. and Silver,P.A. (2000) The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem., 275, 23718–23724. [DOI] [PubMed] [Google Scholar]
- Stolarski R., Sitek,A., Stepinski,J., Jankowska,M., Oksman,P., Temeriusz,A., Darzynkiewicz,E., Lonnberg,H. and Shugar,D. (1996) 1H-NMR studies on association of mRNA cap-analogues with tryptophan-containing peptides. Biochim. Biophys. Acta, 1293, 97–105. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Iyo,H., Doi,M., Inoue,M. and Ishida,T. (1991) Cooperative stacking and hydrogen bond pairing interactions of fragment peptide in cap binding protein with mRNA cap structure. Biochim. Biophys. Acta, 1075, 181–186. [DOI] [PubMed] [Google Scholar]
- Visa N., Izaurralde,E., Ferreira,J., Daneholt,B. and Mattaj,I.W. (1996) A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol., 133, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]