Abstract
Notch proteins are the receptors for an evolutionarily highly conserved signalling pathway that regulates numerous cell fate decisions during development. Signal transduction involves the presenilin-dependent intracellular processing of Notch and nuclear translocation of the intracellular domain of Notch, Notch-IC. Notch-IC associates with the DNA-binding protein RBP-Jκ/CBF-1 to activate transcription of Notch target genes. In the absence of Notch signalling, RBP-Jκ/CBF-1 acts as a transcriptional repressor through the recruitment of histone deacetylase (HDAC) corepressor complexes. We identified SHARP as an RBP-Jκ/CBF-1-interacting corepressor in a yeast two-hybrid screen. In cotransfection experiments, SHARP-mediated repression was sensitive to the HDAC inhibitor TSA and facilitated by SKIP, a highly conserved SMRT and RBP-Jκ-interacting protein. SHARP repressed Hairy/Enhancer of split (HES)-1 promoter activity, inhibited Notch-1-mediated transactivation and rescued Notch-1-induced inhibition of primary neurogenesis in Xenopus laevis embryos. Based on our data, we propose a model in which SHARP is a novel component of the HDAC corepressor complex, recruited by RBP-Jκ to repress transcription of target genes in the absence of activated Notch.
Keywords: corepressor/gene expression/Notch/RBP-Jκ/SHARP
Introduction
The differentiation of cells during development is often regulated by cell–cell interactions. An important function for Notch signalling is to control a mechanism called lateral inhibition, which ensures that two distinct cell types are produced in correct numbers from a population of initially homogenous cells. The number of neurons that develop from neural precursor cells is controlled in this way (for a review, see Beatus and Lendahl, 1998). During this process, neural precursors arise via the activity of basic helix–loop–helix (bHLH) transcriptions factors encoded by proneural genes, such as those in the Achaete-Scute (A-Sc) gene complex in Drosophila, and MASH in mammals. A second class of bHLH repressor proteins downstream of Notch, such as those encoded by the genes within the Enhancer of Split [E(Spl)] complex in Drosophila and HES (Hairy/Enhancer of Split) genes in mammals, appear to inhibit the formation of neural precursors. This mechanism ultimately restricts the number of cells that form the neural precursors.
Despite their diverse function in multiple developmental programs, Notch receptors and ligands are highly conserved. The mammalian family of Notch proteins consists of at least four transmembrane receptors (Notch-1 to Notch-4). The Notch ligands (Jagged-1, Jagged-2, Delta-1, Delta-2 and Delta-3) represent transmembrane proteins of the DSL (Delta, Serrate, and Lag-2) family that, like Notch, contain multiple epidermal growth factor (EGF)-like repeats in their extracellular domains (Egan et al., 1998). After ligand binding, an extracellular cleavage step mediated by TACE, an ADAM family protease, occurs (Brou et al., 2000) followed by a cleavage step near the transmembrane region of the C-terminal protein fragment. This final proteolytic cleavage, which has been linked to presenilins (De Strooper et al., 1999; Struhl and Greenwald, 1999), releases the intracellular domain of Notch (Notch-IC, activated Notch), which then translocates to the nucleus and associates with the ubiquitous DNA-binding protein RBP-Jκ/CBF-1, the mammalian homologue of Drosophila Suppressor of Hairless [Su(H)] (Schroeter et al., 1998; Struhl and Adachi, 1998). RBP-Jκ has been shown to act as a repressor of transcription (Dou et al., 1994; Oswald et al., 1998).
RBP-Jκ-mediated repression includes destabilization of the general transcription factor IID (TFIID)/TFIIA interaction (Olave et al., 1998) and recruitment of histone deacetylase (HDAC) corepressor complexes. Indeed, CIR-1, an RBP-Jκ-interacting corepressor protein, was shown to associate with a corepressor complex including SAP-30 and HDAC-2 (Hsieh et al., 1999). RBP-Jκ has also been shown to interact with the corepressor complex proteins SMRT and HDAC-1 (Kao et al., 1998). Moreover, the LIM protein KyoT2 negatively regulates transcription by association with RBP-Jκ (Taniguchi et al., 1998).
Here we present the identification and characterization of SHARP as an RBP-Jκ-interacting protein. SHARP, originally identified as an SMRT-associated protein in a yeast two-hybrid screen, was previously implicated in nuclear receptor signalling (Shi et al., 2001). We show that SHARP physically interacts with RBP-Jκ in vitro and in vivo. In cotransfection experiments, SHARP repressed transcription in an HDAC-dependent fashion and transactivation mediated by Notch-1 was inhibited by SHARP. In addition, overexpression of SHARP induced a neurogenic phenotype in Xenopus laevis embryos and rescued loss of primary neurogenesis resulting from overexpression of dominant active Notch-1. Our data suggest that SHARP is recruited by RBP-Jκ to a HDAC corepressor complex regulating the Notch signalling pathway.
Results
SHARP interacts with RBP-Jκ
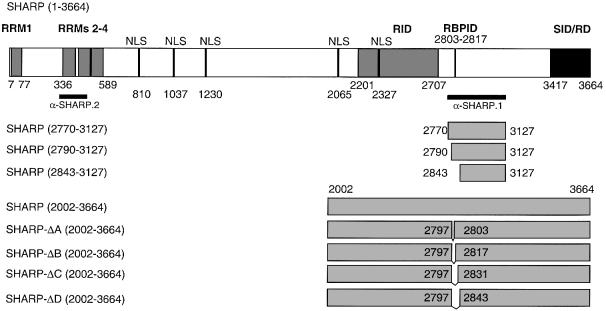
The RBP2N cDNA was fused to the Gal4 DNA-binding domain in pGBT9, and a yeast two-hybrid screen for RBP-Jκ-interacting proteins was performed using a human embryonic liver cDNA library (Stratagene). We identified an 1100 bp open reading frame having no significant homology to known sequences at the cDNA or predicted amino acid level. While this study was in progress, the corresponding full-length protein was identified as the SMRT-interacting corepressor, SHARP, implicated in nuclear receptor signalling (Shi et al., 2001). A schematic representation of the SHARP protein fragments used in this study is shown in Figure 1.
Fig. 1. Schematic representation of SHARP-specific expression constructs. Reported and putative functional domains are shown in the full-length construct. The protein contains four putative RRM and five putative nuclear localization signals (NLS). The receptor interaction domain (RID) and the SMRT interaction/repression domain (SID/RD) were characterized previously (Shi et al., 2001). The RBP interaction domain (RBPID) is characterized in this study. The black bars represent protein fragments used for antibody production. The first and last amino acids compared with the full-length protein are indicated in parentheses. The SHARP-Δ constructs represent in-frame deletions of the indicated amino acids.
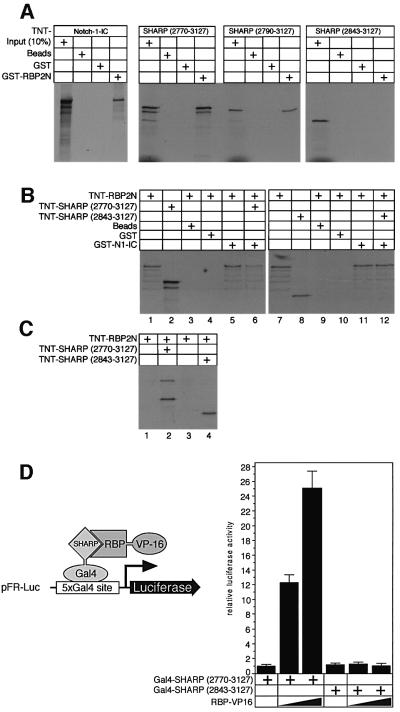
Interaction of SHARP with RBP-Jκ was verified in vitro by pull-down assays with glutathione S-tranferase (GST)– RBP2N. Glutathione–Sepharose beads were coated with bacterially expressed GST or GST–RBP2N proteins, and used as bait for cell-free synthesized and radiolabelled SHARP fragments (Figure 2A). Radiolabelled mNotch-1- IC protein was used as a positive control for RBP binding. As expected, a clear interaction of mNotch-1-IC with GST–RBP2N was observed (Figure 2A, left). No interaction could be detected with glutathione–Sepharose or with GST alone. Two fragments of SHARP (2770–3127 and 2790–3127) interacted with GST–RBP2N (Figure 2A, middle). No interaction was detected with a smaller SHARP fragment (2834–3127) (Figure 2A, right). These results show that SHARP physically interacts with GST–RBP2N, and that the region between amino acids 2790 and 2840 is required for interaction.
Fig. 2. SHARP interacts with RBP-Jκ. (A) Cell-free synthesized SHARP(2770–3127) and the N-terminal-deleted SHARP(2790–3127) bind specifically to GST–RBP2N (middle). A further N-terminal- truncated SHARP(2843–3127) construct fails to interact with GST–RBP2N (right). Interaction of GST–RBP2N with the intracellular form of Notch-1 (Notch-1-IC) was used as a control (left). GST–RBP2N was immobilized on Sepharose beads and incubated with in vitro translated, radiolabelled proteins. After extensive washing, proteins were eluted and separated on SDS–PAGE. (B) RBP-Jκ binding to GST–Notch-1-IC is reduced by the addition of SHARP. Cell-free synthesized proteins alone are shown in lanes 1, 2, 7 and 8. GST–Sepharose beads and GST protein alone were used as negative controls (lanes 3, 4, 9 and 10). RBP-Jκ binding in the absence (lanes 5 and 11) and presence of either SHARP (2770–3127, lane 6) or SHARP lacking the RBP-binding site (lane 12) is shown. (C) Supernatants from GST–Notch-1-IC pull-downs (B, lanes 5, 6, 11 and 12) were coimmuno precipated with anti-Flag-agarose (lanes 1–4, respectively). RBP2N was only immunoprecipitated with a SHARP protein containing the RBP-binding domain (compare lanes 2 and 4). (D) SHARP and RBP interact in the mammalian two-hybrid assay. HeLa cells were cotransfected with the indicated Gal4–SHARP constructs and increasing amounts of CMV–RBP–VP16 together with pFR-Luc. Fold-activation was determined by the relative luciferase activity after cotransfection of the Gal4–SHARP(2770–3127) construct alone. Luciferase activity was determined from 100 µg portions of total cell extract. Mean values and standard deviations from four experiments are shown.
Since RBP binds SHARP and Notch, the question arose whether both proteins could bind RBP simultaneously. Therefore, we performed a pull-down using GST– Notch-1-IC as bait (Figure 2B). Cell-free synthesized RBP2N (lane 1) did not bind to beads or GST protein alone (lanes 3 and 4), but bound specifically to GST–Notch-1-IC (lane 5). Addition of cell-free synthesized SHARP (2770–3217, lane 2) reduced the amount of RBP2N protein bound to Notch-1-IC (lane 6). A cell-free synthesized SHARP fragment lacking the RBP-binding site did not reduce RBP binding to Notch-1-IC (lane 12). The supernatants from Figure 2B, lanes 5, 6, 11 and 12 were used in an anti-Flag coimmunoprecitation (Figure 2C). The RBP protein displaced from GST– Notch-1-IC was bound to the SHARP protein (lane 2). These experiments suggest that RBP binds either Notch or SHARP exclusively.
To test SHARP RBP-Jκ interaction on a cellular background, we performed a mammalian two-hybrid assay. SHARP(2770–3127) and SHARP(2843–3127) were fused to the Gal4 DNA-binding domain. The SHARP–Gal4 fusion vectors were cotransfected into HeLa cells together with increasing amounts of CMV–RBP–VP16 and the reporter plasmid, pFR-Luc, containing five copies of the Gal4-binding site upstream of a luciferase gene. Luciferase activity was detected when RBP–VP16 was coexpressed with Gal4–SHARP(2770– 3127), but not after coexpression of RBP–VP16 with Gal4–SHARP(2843–3127, Figure 2D). As in the pull-downs, the N-terminally deleted SHARP(2843–3127) fragment had lost the capacity to interact with the RBP fusion protein in the mammalian two-hybrid assay.
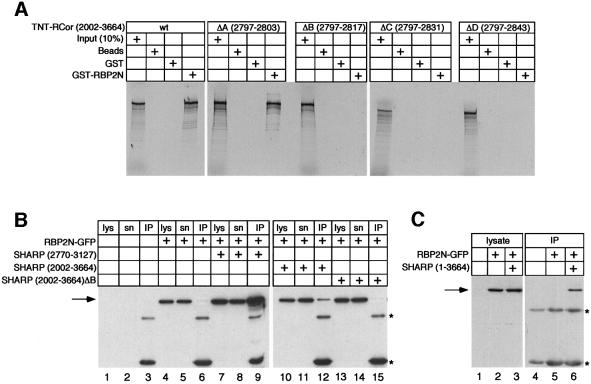
To characterize the RBP-Jκ-binding domain of SHARP in more detail, we performed additional pull-down assays using GST–RBP2N and the C-terminal part of SHARP, SHARP(2002–3664), as well as the in frame deletions, SHARP-ΔA to ΔD (see Figure 1). As shown in Figure 3A, cell-free synthesized SHARP(2002–3664) interacted specifically with GST–RBP2N. A strong interaction could also be detected with SHARP-ΔA containing a seven amino acid in-frame deletion. GST–RBP2N did not bind the SHARP-ΔB, -ΔC and -ΔD deletion constructs (Figure 3A). The results from these mapping experiments suggest that amino acids 2804–2816 within SHARP are required for interaction with RBP-Jκ.
Fig. 3. SHARP binds RBP in vivo and requires a 14 amino acid region. (A) Cell-free synthesized SHARP(2002–3664) binds specifically to GST–RBP2N. Deletion of seven amino acids within SHARP-ΔA had no effect on binding to GST–RBP2N. The in-frame deletion mutants SHARP-ΔB, -ΔC and -ΔD failed to bind to GST–RBP2N. (B) Expression of transfected RBP2N–GFP was detected in lysate (lys, lanes 4, 7, 10 and 13) as well as supernatant (sn, lanes 5, 8, 11 and 14) using an antibody against GFP. Coimmunoprecipitated RBP2N–GFP was detected in the IP fraction after cotransfection with SHARP(2770–3127) (IP, lane 9) and SHARP(2002–3664) (IP, lane 12). RBP2N was not coimmunoprecipitated after cotransfection of SHARP(2002–3664)-ΔB (IP, lane 15). (C) Full-length SHARP binds to RBP2N–GFP in vivo. Expression of transfected RBP2N–GFP was detected in the lysate (lanes 2 and 3). RBP2N–GFP was coimmunoprecipitated after cotransfection of SHARP (lane 6). HEK-293 cells were transiently transfected with an expression plasmid for RBP2N–GFP alone or together with expression plasmids for the indicated N-terminal Flag-tagged SHARP proteins. The SHARP proteins were immunoprecipitated using an antibody directed against the Flag-tag. Coimmunoprecipitated RBP2N–GFP proteins were detected by western blotting using an anti-GFP antibody. The asterisks indicate the heavy and light chains of the anti-Flag antibody.
In vivo interaction of RBP and SHARP was tested by coimmunoprecipitation assays. HEK-293 cells were transfected with an expression plasmid for RBP2N–green fluorescent protein (GFP) alone or together with expression plasmids for various N-terminally Flag-tagged SHARP proteins. The SHARP proteins were immuno precipitated using an antibody against the Flag tag. Coimmunoprecipitated RBP2N–GFP proteins were detected by western blotting using an anti-GFP antibody (Figure 3B). No cellular protein was detected by the anti-GFP antibody in either, the lysate, or supernatant after immunoprecipitation of untransfected HEK-293 cells (Figure 3B, lanes 1 and 2). After transfection of RBP2N– GFP alone, the GFP fusion protein was detected in the lysate (lane 4) and in the supernatant (lane 5), but not in the IP fraction (lane 6). The RBP2N–GFP fusion protein was coimmunoprecipitated from lysates of HEK-293 cells cotransfected with the SHARP(2770–3127) expression construct (lane 9), suggesting an interaction of SHARP and RBP2N in vivo. Interaction was also observed in lysates of cells cotransfected with SHARP (2202–3664, lane 12). In contrast, the RBP2N–GFP protein was not coimmunoprecipitated from lysates of cells cotransfected with the SHARP-ΔB expression plasmid (lane 15). Finally, the RBP protein was also coimmunoprecipitated after cotransfection of both RBP2N–GFP and the full-length SHARP construct (Figure 3C, lane 6). These results show that (i) full-length SHARP is able to interact with RBP2N–GFP in vivo, and (ii) amino acids 2804–2816 within SHARP are required for interaction.
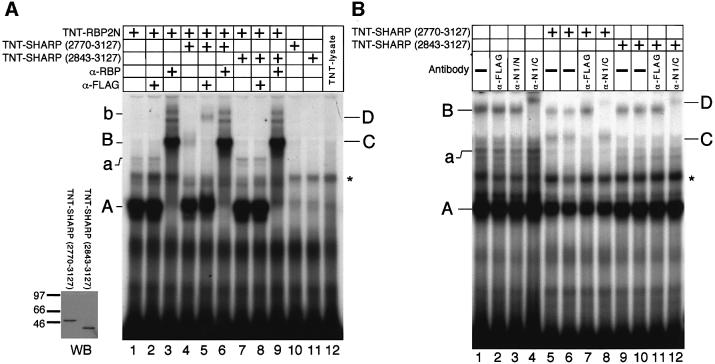
To investigate the formation of SHARP/RBP DNA-binding complexes, we performed band shift experiments using in vitro translated RBP2N (TNT–RBP2N) together with the in vitro translated SHARP fragments, TNT–SHARP(2770–3127) and TNT–SHARP(2843– 3127). Translation of TNT–SHARP proteins was controlled in the anti-Flag western blot shown in Figure 4A (WB). The RBP DNA-binding complexes A and a (lane 1) were supershifted after addition of an anti-RBP antibody to generate complexes B and b (lanes 3, 6 and 9). Addition of an anti-Flag antibody had no effect on the RBP DNA-binding complexes (lane 2). Addition of cell-free synthesized SHARP(2770–3127) to the reaction mixture resulted in the formation of a novel, higher order complex (complex C, lane 4). This novel complex disappeared after addition of an anti-Flag antibody to generate the supershifted complex D containing the flag-tagged SHARP (lane 5). No higher order complex was detected after addition of cell-free synthesized SHARP(2843– 3127) to the reaction mixture (lane 7). The RBP DNA-binding complexes A and a could also be detected in nuclear extracts from SUP-T1 cells (Figure 4B). Additionally, the slower migrating complex B was present, and this complex was supershifted with an antibody against the C-terminus of human Notch-1 (lane 4), indicating that complex B represents an RBP–Notch-1 complex. Addition of cell-free synthesized SHARP(2770– 3127) but not SHARP(2843–3127) lacking the RBP-binding site results in the formation of complex C at the expense of complexes A, a and interestingly also B. Again, complex C was supershifted with the anti-Flag antibody (lane 7). These results show that (i) SHARP(2770–3127) can form a DNA-binding complex with both cell-free RBP2N and endogenous RBP-Jκ, whereas SHARP lacking the RBP-binding site cannot, and (ii) the DNA-bound RBP–SHARP complex in SUP-T1 lysates is formed at the expense of both the DNA-bound RBP complex and the RBP–Notch-1 complex. These data combined with those shown in Figure 2B and C suggest that the presence of either SHARP or Notch in the DNA-bound RBP complex is exclusive.
Fig. 4. (A) Detection of RBP/SHARP DNA-binding complexes. The insert (left) shows the TNT–SHARP translation products visualized in an anti-Flag western blot. Cell-free synthesized RBP2N formed specific DNA-binding complexes (complexes A and a, lane 1). These complexes were supershifted after addition of an anti-RBP antibody to generate complexes B and b (lanes 3, 6 and 9). Addition of cell-free synthesized SHARP(2770–3127) resulted in the formation of a novel higher order complex C (lane 4), which was supershifted by an antibody directed against the N-terminal Flag tag on the SHARP protein to generate complex D (lane 5). Cell-free synthesized SHARP(2843–3127) failed to form a higher order complex with RBP (lane 7), and no supershifted complex could be detected after addition of the anti-Flag antibody (lane 8). (B) SHARP or Notch presence in the DNA-bound RBP complex is exclusive. Sup-T1 cell lysate formed specific DNA-binding complexes A, a and B (lane 1). Complex B was not supershifted with α-Flag (lane 2) or an antibody against the N-terminus of human Notch-1 (α-N1/N, lane 3), but was supershifted to complex D with α-N1/C, an antibody against the C-terminus of human Notch-1, (lanes 4, 8 and 12). Addition of cell-free synthesized SHARP(2770–3127) created an additional higher order complex, C (lanes 5 and 6), which could be supershifted with α-Flag (lane 7). Note that this complex migrates at the same position as complex B (compare to A, lane 5). No changes in the DNA-binding complexes A, a and B were observed with the addition of cell-free synthesized SHARP missing the RBP-binding site (lanes 9–12). The 32P-labelled oligonuleotide FO233 was used as a probe. The asterisk designates a non-specific complex.
Expression of SHARP mRNA in the mouse
SHARP is ubiquitously expressed in the embryonic and adult mouse. Hybridization of a SHARP riboprobe to 9.5 and 14.5 d.p.c. mouse embryo sections showed little differences in SHARP expression between organs, although expression in the liver and lung were slightly higher (see Supplementary data, available at The EMBO Journal Online). All adult mouse tissues examined expressed SHARP, although at variable levels according to real-time RT–PCR analysis (see Supplementary data). The highest expression was measured in the brain and testis. SHARP expression overlapped with, but was not restricted to, the expression pattern of Notch-1 during mouse development.
SHARP is a nuclear protein and colocalizes with RBP-Jκ and SKIP
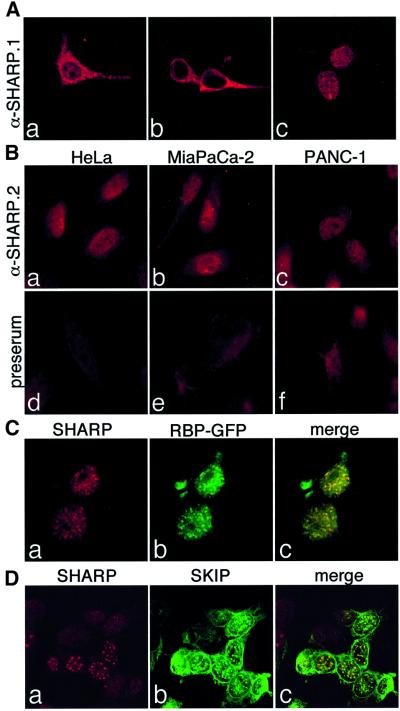
To test subcellular localization of SHARP proteins, we performed transient transfection experiments with plasmids expressing SHARP(2770–3127), SHARP(2002– 3664) and the full-length SHARP protein in HEK-293 cells. The expressed proteins were localized using the α-SHARP.1 rabbit polyclonal antiserum and confocal microscopy. Whereas SHARP(2770–3127) was localized in the cytoplasm as well as in the nucleus (Figure 5Aa), the C-terminal part of SHARP(2002–3664) was only found in the cytoplasm (Figure 5Ab). The full-length SHARP protein was localized exclusively in the nucleus where it was expressed in a speckled pattern (Figure 5Ac). Subcellular localization of endogenous SHARP protein from various human cell lines was investigated by immunofluorescence using a second polyclonal antiserum directed against the N-terminus of SHARP (α-SHARP.2, see also Figure 1). Again, the SHARP protein was located in the nuclei of HeLa as well as pancreatic carcinoma cell lines, MiaPaCa-2 and PANC-1 (Figure 5B). HEK-293 cells were transiently transfected with expression plasmids for SHARP and RBP2N–GFP. Immunofluorescence staining showed that RBP2N–GFP and SHARP were colocalized in speckles in the nuclei (Figure 5C).
Fig. 5. (A) Subcellular localization of SHARP constructs. HEK-293 cells were transfected with plasmids expressing N-terminal Flag-tagged SHARP(2770–3127) (a and d), SHARP(2002–3664) (b and e), and full-length SHARP(1–3664) (c and f). At 24 h after transfection, cells were fixed, permeabilized and immunostained with an antiserum directed against human SHARP (α-SHARP-1). (B) Nuclear localization of endogenous SHARP. The human cell lines HeLa (a and d), MiaPaCa-2 (b and e) and PANC-1 (c and f) were fixed and immunostained with a rabbit polyclonal antiserum (α-RCor.2) directed against human SHARP (upper) or the pre-immune serum from the same rabbit (lower). (C) SHARP colocalizes with RBP2N–GFP. HEK-293 cells were transiently transfected with an expression plasmid for SHARP and RBP2N–GFP. Cells were fixed 24 h after transfection, permeabilized and immunostained with the SHARP antiserum. The subcellular localization of SHARP (red, a) and RBP2N-GFP (green, b) was assayed by confocal microscopy. (D) SHARP colocalizes with SKIP. HEK-293 cells were transiently transfected with an expression plasmid for Flag-tagged human SKIP. Cells were fixed 24 h after transfection, permeabilized and immunostained with an antibody directed against the Flag epitope and with the SHARP antiserum. The subcellular localization of SHARP (red, a) and SKIP proteins (green, b) was assayed by confocal microscopy.
It was shown previously that RBP-Jκ acts as a transcriptional repressor by recruiting a corepressor complex involving SMRT and HDAC-1 (Kao et al., 1998). Originally identified as an RBP-Jκ-associated protein, SKIP (Zhou et al., 2000b) was shown to be a component of this corepressor complex (Zhou et al., 2001). To investigate the role of SHARP in this context, we performed transient transfection experiments in HEK-293 cells with expression plasmids for N-terminally Flag-tagged SKIP. Localization of SKIP and SHARP was assayed by immunofluorescence using the anti-Flag antibody and α-SHARP.2 antiserum. Overexpressed SKIP protein was localized in a halo around the nuclear membrane as well as in speckles in the nucleus (Figure 5Db). Interestingly, SHARP localized to these nuclear spots (Figure 5Da) resulting in a colocalized signal in the overlay (Figure 5Dc).
Transcriptional repression via SHARP depends on HDAC activity and is facilitated by SKIP
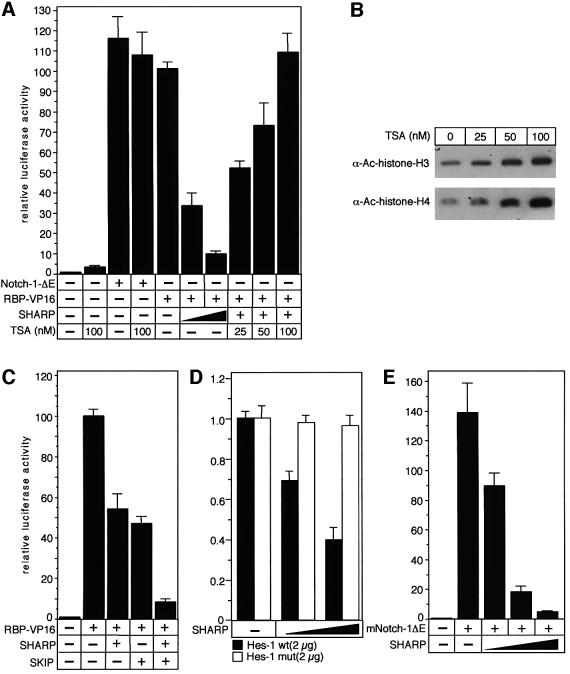
To characterize the function of SHARP in RBP-Jκ-mediated transcriptional regulation, we performed transient cotransfection experiments with expression plasmids for RBP–VP16 and full-length SHARP together with the RBP-Jκ responsive reporter construct, pGa981/6. This luciferase reporter plasmid carries six repeats of the EBNA-2 responsive element within the Epstein–Barr Virus (EBV) TP-1 promoter upstream of a minimal β-globin promoter. As shown previously (Oswald et al., 2001), cotransfection of a plasmid expressing RBP fused to the VP16 transactivation domain (RBP–VP16) into HeLa cells resulted in stimulation of luciferase activity (Figure 6A). Transactivation mediated by RBP–VP16 was gradually reduced after cotransfection of increasing amounts of SHARP suggesting that SHARP acts as an RBP-interacting corepressor. In addition, this repression was HDAC-dependent since incubation of the cells with increasing amounts of the HDAC inhibitor trichostatin A (TSA) for 6 h resulted in a complete loss of repression (Figure 6A). This appeared to be specific, since TSA could not further activate the reporter in the presence of Notch-1-ΔE, a dominant active form of Notch-1. Taken together, incubation of the cells with increasing amounts of TSA resulted in histone hyperacetylation and completely restored RBP–VP16-mediated activation (Figure 6A and B).
Fig. 6. (A) SHARP-mediated repression depends on HDAC activity. Portions (2 µg) of reporter construct pGa981/6 were transfected alone into HeLa cells or cotransfected with 100 ng of plasmids expressing RBP–VP16 together with increasing amounts (200 and 500 ng) of plasmid expressing SHARP. Cotransfected cells were incubated with increasing amounts (25, 50 and 100 nM) of TSA 6 h prior to harvesting. Cells transfected with reporter construct alone had 3-fold higher activation in the presence of 100 nM TSA. Cotransfection of Notch-1-ΔE with the reporter plasmid shows the activated condition, which could not be further increased with 100 mM TSA treatment. (B) Treatment of HeLa cells with TSA results in hyperacetylation of histones. Extracts (20 µg) from cells treated with the indicated amounts of TSA were used for western blotting to detect acetylated forms of H3 and H4. (C) SKIP facilitates SHARP-mediated repression. The pGa981/6 reporter construct (2 µg) was cotransfected into HeLa cells with 100 ng of plasmid expressing RBP–VP16 together with an SHARP expression plasmid (100 ng), or a SKIP expression plasmid (100 ng), or both. (D) SHARP represses HES promoter activity. Portions (2 µg) of the human HES-1-specific reporter constructs HES-1wt and HES-1mut were cotransfected into HeLa cells alone and with increasing amounts (200 and 500 ng) of SHARP expression plasmids. (E) SHARP represses Notch-1-mediated transactivation. The reporter construct pGa981/6 (2 µg) was cotransfected into HeLa cells with 20 ng of a dominant active Notch-1 expression plasmid (Notch-1ΔE) together with increasing amounts (100, 200 and 500 ng) of SHARP expression plasmid. Luciferase activity was determined from 100 µg portions of total-cell extracts, and normalized to the basal promoter activity of the reporter construct. Mean values and standard deviations from four independent experiments are shown.
To test the role of SKIP in the context of SHARP-mediated repression, we performed additional cotransfection experiments (Figure 6C). The RBP–VP16-mediated transactivation of the pGa981/6 reporter construct was again reduced after cotransfection of 100 ng of SHARP expression plasmid. In addition, cotransfection of a SKIP expression plasmid (100 ng) also resulted in ∼50% reduction of luciferase activity. Interestingly, cotransfection of both SHARP and SKIP resulted in a 90% reduction of RBP–VP16-mediated transactivation (Figure 6C). Therefore, SKIP not only colocalizes with, but enhances reduction of transcriptional activation by SHARP.
SHARP represses HES-1 promoter activity and Notch-1-mediated transactivation
HES-1 was identified as a Notch target gene in mammals (Jarriault et al., 1995, 1998; Kuroda et al., 1999). To test the effect of SHARP on a naturally existing Notch responsive promoter, we performed cotransfection experiments with the human HES-1 promoter fused to the luciferase gene. Cotransfection of increasing amounts of full-length SHARP resulted in a clear repression of basal promoter activity. No repression of luciferase activity was measured when the HES-1 promoter construct in which both RBP-Jκ-binding sequences were inactivated by point mutations was used (Figure 6D). These results demonstrate that SHARP represses promoter activity from a Notch responsive gene, and that this repression depends on functional RBP-Jκ-binding sites.
To test the role of SHARP in Notch-1-mediated transactivation, we transiently cotransfected expression plasmids for Notch-1ΔE together with SHARP and the luciferase reporter pGa981/6 into HeLa cells. Again, cotransfection of mNotch-1ΔE stimulated promoter activity. This Notch-1-mediated transactivation was abrogated by gradually increasing the amount of SHARP (Figure 6E). Therefore, SHARP antagonises Notch-1-mediated transactivation.
SHARP rescues a Notch-1-induced phenotype in Xenopus embryos
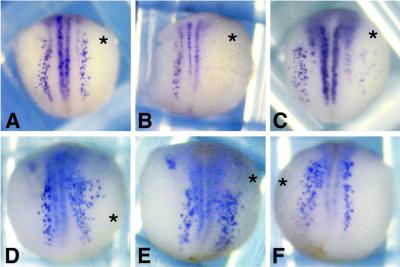
It was previously shown that activation of the Notch pathway by the Delta ligand expressed on a neighbouring cell suppresses neuronal differentiation in a process called lateral inhibition (Chitnis et al., 1995). To investigate whether SHARP can play a functional role in Notch signalling during X.laevis embryogenesis, we overexpressed the constitutively active Notch-1ΔE construct with or without full-length SHARP in one half of the embryo. Whole mount in situ hybridization for N-tubulin allowed identification of primary neurons in the X.laevis embryos at neurula stages 14–15 (Figure 7). Embryos injected with Notch-1ΔE RNA alone exhibited no lateral and in some cases also no intermediate primary neurons in the injected sides (38/56 = 68%, Figure 7B). However, coinjection of SHARP RNA (Figure 7C) rescued the formation of intermediate and lateral primary neurons (47/72 = 65%) although these stripes were somewhat broader and less organized. Primary neurogenesis was not rescued when RNA for SHARP(2843–3127) lacking the RBP-binding site was coinjected with Notch-1ΔE RNA (97/129 = 75%, compared with 119/162 = 73%), which showed disrupted primary neurogenesis upon injection with Notch-1ΔE. Overexpression of SHARP alone produced more primary neurons and a broader zone of lateral primary neurons in the injected sides (Figure 7D–F shows several examples of this phenotype). This effect is dose dependent, since more embryos showed the neurogenic phenotype with increasing SHARP RNA concentration (65/151 = 50% with 1.2 ng SHARP RNA; and 71/107 = 67% with 2.4 ng SHARP RNA). Taken together, these results suggest that SHARP interferes with Notch signalling during primary neurogenesis in Xenopus.
Fig. 7. SHARP rescues Notch-1-dependent inhibition of primary neurogenesis in X.laevis, and produces a mild neurogenic phenotype when overexpressed alone. Embryos were injected with 100 pg GFP expression plasmid alone (A) or together with 100 pg mNotch-1ΔE RNA (B) and 2.4 ng full-length SHARP RNA (C) in one cell at the 2-cell stage. A range of examples for the phenotype produced by SHARP alone (D–F) is shown. Whole mount in situ hybridization for N-tubulin shows primary neurons. The injected sides are marked with an asterisk.
Discussion
One central event in Notch signalling is the conversion of the DNA-binding protein RBP-Jκ/CBF-1 from a repressor to an activator of transcription. Interaction with the intracellular domain of Notch changes the properties of RBP-Jκ, probably by displacing a complex of interacting proteins that repress transcription, and by recruitment of other proteins that activate. Chromatin modifications, in particular acetylation and deacetylation of histones, may play an important role in this switch of transcriptional regulation. In a former study, we identified p300, a common coactivator mediating transcriptional activation by Notch through supporting the recruitment of histone acetyl transferase (HAT) activity to Notch responsive promoters (Oswald et al., 2001). Others have shown that P/CAF and GCN5 physically interact with Notch-IC (Kurooka and Honjo, 2000). Notch-IC may also recruit transcriptional coactivators specific for Notch signalling. The gene Mastermind has been identified in multiple genetic screens for modifiers of Notch phenotypes in Drosophila. The human homologue of Mastermind, MAML1, functions as a transcriptional coactivator by directly interacting with the ankyrin repeat domain of Notch (Wu et al., 2000) and by recruiting p300/CBP (Fryer et al., 2002). In the absence of Notch, RBP-Jκ/CBF-1 might exert its repressive function by interacting with corepressor complexes recruiting HDAC activity to chromatin (Kao et al., 1998; Hsieh et al., 1999).
Here we report SHARP to be a novel RBP-Jκ/CBF-1-interacting protein. Functional analyses revealed that SHARP acts as a corepressor, and sequence analysis revealed the presence of four putative RNA recognition motifs (RRMs) and five putative nuclear localization signals. We mapped the interaction domain of SHARP with RBP-Jκ to amino acids 2803–2817. SHARP belongs to the Spen (split ends) family of proteins, which contains RRMs in their N-terminal part and a highly conserved C-terminal domain called the SPOC domain. The SPOC domain has been shown to be essential for Spen function in Drosophila (Chen and Rebay, 2000; Kuang et al., 2000). Spen-like proteins have been identified in Caenorhabditis elegans, Drosophila, mouse and human. Spen proteins are detectable as early as the cellular blastoderm in Drosophila and are ubiquitously nuclear during early development (Kuang et al., 2000). In our study, expression of SHARP overlapped with expression of Notch-1; however, SHARP was also expressed in other tissues, suggesting that SHARP may have other functions in addition to modulation of Notch signalling during embryogenesis.
While this work was in progress, Shi and coworkers described SHARP as an SMRT/HDAC-1-associated repressor protein, which was isolated in a yeast two-hybrid screen using the C-terminus of the corepressor SMRT as a bait (Shi et al., 2001). In their study, SHARP was implicated in nuclear receptor signalling, and was shown to associate with components of the NuRD complex including HDAC1, HDAC2, MTA2, MBD3 and RbAp48 through a C-terminal repression domain. Moreover, SHARP was shown to interact with the RNA coactivator SRA via the RRMs located in the N-terminal part of the protein. An interaction domain for nuclear receptors was identified in the SHARP protein based on sequence homology to other nuclear receptor binding proteins (Shi et al., 2001). The mouse homologue of SHARP, Mint, was identified as a binding partner of Msx2, a homeodomain repressor protein involved in osteoblast differentiation (Newberry et al., 1999). These studies suggest that Spen-like proteins might be involved in various regulatory pathways.
We show here that SHARP reduced transactivation mediated by RBP–VP16. This reduction was sensitive to TSA, indicating that in vivo SHARP may act as a repressor. This is consistent with the ability of SHARP to interact with HDACs and components of the NuRD complex. SHARP also repressed the activity of a luciferase construct derived from the human HES–1 promoter. In this case, repression was dependent on functional RBP-binding sites. Two models of RBP-Jκ-mediated repression have been postulated. RBP-Jκ has been shown to interact with the transcriptional coactivators TFIIA and dTAFII110, a subunit of TFIID. The domain of dTAFII110 targeted by RBP-Jκ is the same domain that interacts with TFIIA, but is different from the domain that interacts with SP1. Repression can be thwarted when stable transcription pre-initiation complexes are formed before RBP-Jκ addition, suggesting that RBP-Jκ interaction with TFIIA and TFIID perturbs optimal interactions between these cofactors (Olave et al., 1998). In contrast to the specific targeting of the basal transcription machinery by RBP-Jκ, others have favoured the model of transcriptional repression via chromatin remodelling by recruiting HDAC corepressor complexes to RBP-Jκ (Kao et al., 1998; Hsieh et al., 1999; Zhou et al., 2000a,b). RBP-Jκ has been shown to interact with a corepressor complex containing SMRT and the histone deacetylase HDAC-1 (Kao et al., 1998). In addition, CBF1-interacting corepressor (CIR) was isolated in a two-hybrid screen using RBP-Jκ/CBF-1 as a bait. CIR acted as a repressor in transient transfection assays, and bound to histone deacetylase and SAP30 (Hsieh et al., 1999).
The Ski-interacting protein (SKIP) was also shown to interact with RBP-Jκ/CBF-1. SKIP was originally identified as an interacting partner of the avian retroviral oncogene, v-Ski, whose cellular homologue, c-Ski, has been recognized as a component of the HDAC corepressor complex that associates with the Mad and thyroid hormone receptor complexes. SKIP interacted with SMRT and CIR in the corepressor complex (Zhou et al., 2000a,b). Interestingly, SKIP not only associated with RBP-Jκ and SMRT, but also interacted with the fourth ankyrin repeat of Notch-IC. A mutant form of Notch-IC with an AA to EF amino acid substitution in the fourth ankyrin repeat not only failed to fully transactivate a Notch-dependent reporter construct, but also failed to interact with SKIP (Zhou et al., 2000b). Therefore, SKIP is postulated to be a component of both the corepressor complex and the RBP-Jκ–Notch complex. We show that after overexpression of human SKIP in HEK-293 cells, endogenous SHARP protein colocalized with SKIP. In cotransfection assays, SKIP facilitated the repressive effect of SHARP, suggesting that SHARP and SKIP are components of a corepressor complex recruited by RBP-Jκ–CBF1.
Does SHARP play a role in Notch signalling? An important function of Notch signalling is to limit the number of neurons developing from neural precursor cells via lateral inhibition. Overexpression of a dominant active form of Notch results in a reduction of primary neurons in Xenopus embryogenesis (Chitnis et al., 1995). Conversely, inhibition of Notch signalling results in the production of more primary neurons in the neural plate. We have shown that coinjection of SHARP mRNA with Notch-1ΔE rescued the formation of primary neurons normally blocked by excessive Notch signalling. Additionally, overexpression of SHARP alone induced a neurogenic phenotype. Taken together, the data suggest that SHARP might be a novel molecular component of the switch from repression to transactivation of target genes during Notch signalling.
Our protein–protein interaction (Figure 2B) and DNA-binding studies (Figure 4B) suggest that RBP interacts either with SHARP or with Notch-IC, supporting the idea that formation of SHARP-containing or Notch-containing RBP complexes could be exclusive. We present a model in which different DNA-bound RBP complexes mediate transcriptional repression versus transactivation. Gel filtration experiments carried out recently (Jeffries et al., 2002), identified a 1.5 MDa Notch-IC ‘enhancer complex’, indicating that the activator (and probably also repressor complex discussed here) contains many as yet not identified component proteins. However, SHARP may act as a molecular switch for the conversion of the activator into the repressor in the absence of activated Notch. In this model, SHARP would function as an adapter to link RBP with the HDAC chromatin remodelling machinery. This is supported by the previously reported interaction between SHARP and SMRT (Shi et al., 2001), the colocalization of SHARP and RBP (Figure 5C) as well as SHARP and SKIP (Figure 5D), and the fact that SHARP function was sensitive to an HDAC inhibitor (Figure 6A) in our experiments. SHARP was originally identified as a corepressor involved in steroid receptor signalling. The results reported here support an additional role for SHARP as a corepressor in Notch signalling. In addition, we suggest that SHARP may play a general role as a molecular switch to create repression complexes in Notch-independent signalling pathways.
Materials and methods
Yeast two-hybrid screen
Yeast two-hybrid assays were carried out with the MATCHMAKER system (Clontech) according to the manufacturer’s instructions. A human embryonic liver two-hybrid library (Stratagene) and pGBT9-RBP2N were cotransformed into yeast strain HF7c. Approximately 5 × 106 transformants were screened. Selection was performed on yeast minimal medium Leu-Trp-His plates. Colonies were picked 6 days after transformation and confirmed by β-galactosidase assays. Plasmids were isolated from yeast, and retransformed together with pGBT9-RBP2N. Retransformed positive clones were subjected to sequencing. The full-length SHARP cDNA sequence was made by using expression sequence tags and an RT–PCR approach, and verified by sequencing.
Plasmids
The human cDNA for RBP2N was cloned into pBluescriptKS (Stratagene) resulting in pBlueRBP2N. To create pBlueRBP2N-ATG the start ATG of the RBP2N cDNA was deleted by PCR, and the PCR product was cloned into pBluescriptKS. The bait vector for two-hybrid screening was constructed by cloning RBP2N-ATG into pGBT9 (Clontech). PCR cloning approaches into pcDNA-3 (Invitrogen) were used to generate all human SHARP expression constructs. The luciferase reporter plasmids pGa981/6 and Hes-1-LUC as well as the expression plasmids pCMV-RBP-VP16 and pcDNA-3-mNotch-1-IC were described previously (Oswald et al., 2001). The mutated HES-1-specific reporter construct HES-1mut-LUC was made using an in vitro mutagenesis system (Stratagene), as specified by the manufacturer. The SKIP cDNA was amplified from a human placenta cDNA library, and inserted into pGEM™-T (Promega). The start ATG was deleted by PCR and cloned into pcDNA-3–FLAG-1 to generate the expression plasmid for N-terminally Flag-tagged SKIP. The cDNA for murine Notch-1 cloned into pcDNA-3 (Invitrogen) resulting in pcDNA-3-mNotch-1. To generate the dominant active form of mNotch-1, an EcoRI/NotI fragment, corresponding to the extracellular part of mNotch-1 was replaced by a PCR product resulting in pcDNA-3-mNotch-1ΔE. Primer sequences and cloning details for all plasmids are available on request.
Cell lines and extracts
The cell lines HEK-293 (ATCC CRL 1573) MiaPaCa-2, PANC-1 and HeLa (ATCC CCL 2) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco), and SUP-T1 cells were grown in RPMI 1640 medium (Gibco) supplemented with 2 mM l-glutamine. All cell lines were maintained with 10% fetal calf serum at 37°C under 5% CO2. Whole-cell lysates for immunoprecipitation and western blotting were prepared as described previously (Oswald et al., 2001). Protein concentrations were determined using the Bradford method (Bio-Rad).
Cell-free synthesized proteins
Proteins were translated in vitro in the presence of [35S]methionine using a reticulocyte lysate-coupled transcription/translation system according to the manufacturer’s instructions (Promega). The quality of translation and labelling was monitored on SDS–PAGE.
Pull-down assay
Purification of bacterially expressed GST–RBP2N and GST–Notch-1-IC proteins and in vitro interaction assays were performed as described previously (Oswald et al., 1998).
Electrophoretic mobility shift assays (EMSAs)
Nuclear extracts were prepared as described previously (Oswald et al., 1998). Approximately 5–10 µg of reticulocyte lysate or nuclear extracts from SUP-T1 cells were used for EMSAs in a binding buffer consisting of 10 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 0.5 mM dithiothreitol (DTT) and 4% glycerol. For the binding reaction, 2 µg poly(dI-dC) (Pharmacia) and ∼0.5 ng of 32P-labelled oligonucleotides were added. The sequence of the double-stranded oligonucleotide FO233 (5′-CCTGGAACTATTTTCCCACGGTGCCCTTCCGCCCATTTTCCCACGAGTCG-3′) corresponds to the two RBP-Jκ-binding sites within the EBV TP-1 promoter. For antibody perturbation experiments, 0.5 µg of the K0043 antibody directed against RBP-Jκ, 0.4 µg of the H-131 antibody against the N-terminus of human Notch-1 (Santa Cruz), 0.4 µg of the C20 antibody against the C-terminus of human Notch-1 (Santa Cruz) or 5 µg of an antibody directed against the Flag epitope (M5, Sigma) was added to the reaction mixture. The reaction products were separated on 5% Tris–glycine–EDTA (TGE) polyacrylamide gels at room temperature.
DNA transfection and luciferase assay
A total of 106 HEK-293 cells were transfected in 90 mm culture dishes with 5–10 µg of plasmid DNA expressing human SHARP proteins using calcium phosphate coprecipitation as described previously (Oswald et al., 2001). Proteins were prepared 24 h after transfection, and the extracts were assayed for protein expression and subcellular localization. HeLa cells were transfected (2 × 105) in 35 mm culture dishes with 2 µg of reporter plasmid DNA together with various amounts of expression plasmid using the Fugene transfection reagent (Roche). Luciferase activity was determined from at least four independent transfections with 20 µl of cleared lysate in a LB 9501 luminometer (Berthold) using the luciferase assay system from Promega. All transfections were normalized using total cellular protein concentrations (Bradford assay; Bio-Rad).
Immunofluorescence assay
HEK-293 cells were cultured on glass cover slips in a 25-well plate (Bibby Sterilin Ltd) at a density of 105 cells per cm2. After 16 h, cells were transfected with 1 µg of SHARP construct using the calcium phosphate coprecipitation method. Cells were rinsed in PBS 24 h after transfection, fixed in 100% methanol on ice for 2 min and permeabilized with 0.1% Triton X-100. Non-specific immunostaining was blocked by incubating the cells in 3% BSA in PBS with 0.1% TWEEN-20. SHARP was detected using an anti-Flag antibody (Sigma) as well as rabbit antisera (α-SHARP.1, α-SHARP.2) directed against SHARP. Staining was performed using an FITC-(fluorescein-iso-thiocyanate) conjugated goat anti-mouse IgG (Sigma), or a Cy™3-conjugated donkey anti-rabbit IgG (Dianova) secondary antibody. After washing and mounting, the cells were analysed using a TCS 4D Leica confocal microscope.
Western blotting
Western blotting was performed as previously described (Oswald et al., 2001). To analyse coimmunoprecipitated RBP2N-GFP proteins, membranes were incubated with the primary antibody directed against GFP protein (Roche). As secondary antibody a 1:7000 dilution of peroxidase-conjugated sheep anti-mouse IgG (Sigma) was used. Hyperacetylated histones were detected using 1:1000 diluted polyclonal rabbit antisera against acetylated H3 and H4 proteins (Upstate), followed by 1:2500 diluted peroxidase-conjugated goat anti-rabbit antibody (Upstate).
Coimmunoprecipitation
Immunoprecipitation (IP) was carried out using cell extracts from HEK-293 cells 24 h after cotransfection with various pcDNA3-SHARP constructs together with pcDNA3-RBP2N–GFP. Cells were lysed in 900 µl CHAPS lysis buffer consisting of 10 mM 3-[(cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM Tris–HCl pH 7.9, 150 mM LiCl, 2 mM EDTA, 5 mM NaF, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 1 mM DTT, 0.5 mM phenylmethylsulfonyl flouride (PMSF) and incubated on ice for 40 min. The lysate was cleared at 80 000 g for 30 min. Extracts were incubated with 40 µl of an agarose-conjugated anti-Flag antibody (M2, Sigma) at 4°C overnight. Beads were washed three times with CHAPS lysis buffer containing 500 mM LiCl. After a further wash with CHAPS lysis buffer containing 150 mM LiCl, the beads were resuspended in SDS–polyacrylamide gel loading buffer and used for western blotting.
Injection of mRNA into X.laevis embryos and whole–mount in situ hybridization
Microinjections were performed with in vitro fertilized X.laevis embryos (Nasco), dejellied in 2% cysteine hydrochloride in 0.1× Modified Barth’s Solution High-Salt MBSH; 10 mM HEPES pH 7.4, 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.82 mM MgSO4, 0.41 mM CaCl2, 0.66 mM KNO3) and staged according to Nieuwkoop and Faber (1975).
Capped mRNA of mNotch-1ΔE and full-length SHARP were transcribed in vitro for injection using the mMESSAGEmMACHINE kit (Ambion), then purified on RNeasy columns (Qiagen). The GFPpCS2 expression plasmid (100 pg), mNotch-1ΔE mRNA (100 pg) and SHARP mRNA (2.4 ng) were injected in a volume of 10 nl into one cell of two-cell stage embryos. The embryos were cultured in 0.1× MBSH at 18°C until stage 14–15, sorted into groups injected either on the left or right side based on GFP fluorescence, then fixed in MEMFA (0.1 M MOPS pH 7.2, 2 mM EGTA, 1 mM MgSO4, 4% paraformaldehyde) overnight at 4°C. Whole mount in situ hybridization was performed using a digoxigenin-labelled antisense RNA probe for N-tubulin as described previously (Harland, 1991; Chitnis et al., 1995). Stained embryos were post-fixed overnight in MEMFA and depigmented with 10% H2O2 in methanol. The uninjected side was used as control for normal primary neurogenesis.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We wish to thank U.Wegenka for critically reading the manuscript. We also thank T.Honjo for providing the RBP-Jκ-specific antibody K0043. For excellent technical assistance, we thank E.Rüber, R.Rittelmann and S.Schirmer. This study was supported by the Deutsche Forschungs gemeinschaft, Sonderforschungsbereich 497, C4.
References
- Beatus P. and Lendahl,U. (1998) Notch and neurogenesis. J. Neurosci. Res., 54, 125–136. [DOI] [PubMed] [Google Scholar]
- Brou C. et al. (2000) A novel proteolytic cleavage involved in Notch signalling: the role of the disintegrin-metalloprotease TACE. Mol. Cell, 5, 207–216. [DOI] [PubMed] [Google Scholar]
- Chen F. and Rebay,I. (2000) Split ends, a new component of the Drosophila EGF receptor pathway, regulates development of midline glial cells. Curr. Biol., 10, 943–946. [DOI] [PubMed] [Google Scholar]
- Chitnis A., Henrique,D., Lewis,J., Ish-Horowicz,D. and Kintner,C. (1995) Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature, 375, 761–766. [DOI] [PubMed] [Google Scholar]
- De Strooper B. et al. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature, 398, 518–522. [DOI] [PubMed] [Google Scholar]
- Dou S., Zeng,X., Cortes,P., Erdjument-Bromage,H., Tempst,P., Honjo,T. and Vales,L.D. (1994) The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol. Cell. Biol., 14, 3310–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S.E., St-Pierre,B. and Leow,C.C. (1998) Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr. Top. Microbiol. Immunol., 228, 273–324. [DOI] [PubMed] [Google Scholar]
- Fryer C.J., Lamar,E., Turbachova,I., Kintner,C. and Jones,K.A. (2002) Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev., 16, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R.M. (1991) In situ hybridisation: an improved whole-mount method for Xenopus embryos. Methods Cell Biol., 36, 685–695. [DOI] [PubMed] [Google Scholar]
- Hsieh J.J., Zhou,S., Chen,L., Young,D.B. and Hayward,S.D. (1999) CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou,C., Logeat,F., Schroeter,E.H., Kopan,R. and Israel,A. (1995) Signalling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- Jarriault S., Le Bail,O., Hirsinger,E., Pourquie,O., Logeat,F., Strong,C.F., Brou,C., Seidah,N.G. and Israel,A. (1998) Delta-1 activation of notch-1 signalling results in HES-1 transactivation. Mol. Cell. Biol., 18, 7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries S., Robbins,D.J. and Cabobianco,A.J. (2002) Characterization of a high-molecular-weight Notch complex in the nucleus of NotchIC-transformed RKE cells and in a human leukemia cell line. Mol. Cell. Biol., 22, 3927–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.Y., Ordentlich,P., Koyano,N., Tang,Z., Downes,M., Kintner,C.R., Evans,R.M. and Kadesch,T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang B., Wu,S.C., Shin,Y., Luo,L. and Kolodziej,P. (2000) Split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development, 127, 1517–1529. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Tani,S., Tamura,K., Minoguchi,S., Kurooka,H. and Honjo,T. (1999) Delta-induced Notch signalling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem., 274, 7238–7244. [DOI] [PubMed] [Google Scholar]
- Kurooka H. and Honjo,T. (2000) Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem., 275, 17211–17220. [DOI] [PubMed] [Google Scholar]
- Newberry E.P., Latifi,T. and Towler,D.A. (1999) The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry, 38, 10678–10690. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P.D. and Faber,J. (1975) Normal Table of Xenopus laevis (Daudin), 2nd edn. Elsevier/North-Holland, Amsterdam.
- Olave I., Reinberg,D. and Vales,L.D. (1998) The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev., 12, 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F., Liptay,S., Adler,G. and Schmid,R.M. (1998) NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol. Cell. Biol., 18, 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F., Täuber,B., Dobner,T., Bourteele,S., Kostezka,U., Adler,G., Liptay,S. and Schmid,R.M. (2001) p300 acts as a transcriptional coactivator for mammalian notch-1. Mol. Cell. Biol., 21, 7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- Shi Y., Downes,M., Xie,W., Kao,H.Y., Ordentlich,P., Tsai,C.C., Hon,M. and Evans,R.M. (2001) Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev., 15, 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. and Adachi,A. (1998) Nuclear access and action of notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- Struhl G. and Greenwald,I. (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature, 398, 522–555. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y., Furukawa,T., Tun,T., Han,H. and Honjo,T. (1998) LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol., 18, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Aster,J.C., Blacklow,S.C., Lake,R., Artavanis-Tsakonas,S. and Griffin,J.D. (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nature Genet., 26, 484–489. [DOI] [PubMed] [Google Scholar]
- Zhou S. and Hayward,S.D. (2001) Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol. Cell. Biol., 21, 6222–6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Fujimuro, M., Hsieh,J.J.-D., Chen,L., Hayward,S.D. (2000a) A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol., 74, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Fujimuro,M., Hsieh,J.J., Chen,L., Miyamoto,A., Weinmaster,G. and Hayward,S.D. (2000b) SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol., 20, 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]