Abstract
The Drosophila Dscam gene encodes 38,016 different proteins, due to alternative splicing of 95 of its 115 exons, that function in axon guidance and innate immunity. The alternative exons are organized into four clusters, and the exons within each cluster are spliced in a mutually exclusive manner. Here we describe an evolutionarily conserved RNA secondary structure we call the Inclusion Stem (iStem) that is required for efficient inclusion of all 12 variable exons in the exon 4 cluster. Although the iStem governs inclusion or exclusion of the entire exon 4 cluster, it does not play a significant role in determining which variable exon is selected. Thus, the iStem is a novel type of regulatory element that simultaneously controls the splicing of multiple alternative exons.
Alternative splicing is a common means of enhancing proteome diversity in higher eukaryotes. Although most genes encode transcripts that can be alternatively spliced to generate a relatively modest number of isoforms, there are several examples of genes that can potentially encode thousands of alternatively spliced mRNAs (1, 9). For example, mammals possess three neurexin genes, which encode proteins that function as cell adhesion molecules during synaptogenesis and intercellular signaling, that can potentially generate 2,250 different mRNAs (23, 28). The paralytic gene, which encodes the major voltage-gated action potential sodium channel in Drosophila species, can potentially generate 1,536 different mRNAs via alternative splicing (18, 29). The most dramatic example, however, is the Drosophila Dscam gene, which can potentially generate over 38,000 different mRNA isoforms (25).
Dscam encodes an axon guidance receptor that is required for nervous system development and axon pathfinding (25, 30). For example, olfactory receptor neurons that lack Dscam fail to form synapses with the correct glomeruli (15). More recently, Dscam has been shown to be expressed in immune-competent cells of the fly and to function in innate immunity (31). DSCAM contains an extracellular domain composed of 10 immunoglobulin (Ig) domains and 4 fibronectin type III domains, which are connected to a transmembrane domain and a cytoplasmic tail (25). Dscam contains 95 alternatively spliced exons that are organized into four clusters. The exon 4, 6, and 9 clusters contain 12, 48, and 33 alternative exons, respectively, each of which encodes different versions of three of the Ig domains. The exon 17 cluster contains two alternative exons that encode different transmembrane domains. Importantly, the exons within each cluster are alternatively spliced ina mutually exclusive manner. As a result, it is theoretically possible that 38,016 different mRNA isoforms, each of which would encode a distinct protein isoform, can be synthesized. Furthermore, because different classes of neurons express distinct sets of many different Dscam isoforms (20, 33), each DSCAM isoform engages in isoform-specific homotypic interactions (32), and the Dscam genes of distantly related insects all encode tens of thousands of isoforms (11), Dscam appears to encode a set of molecular tags that regulate neuronal connectivity. Thus, alternative splicing of Dscam plays an extremely important role in determining the specificity of neural wiring.
To understand the function of specific DSCAM isoforms it is necessary to determine their temporal and spatial expression patterns. Although the existence of all 38,016 potential isoforms has not been proven, 122 individual Dscam cDNA clones, of which 117 are unique, have been reported (15, 25). Moreover, all but 1 of the 95 alternative exons have been identified in at least one cDNA (3, 15, 20, 25). Thus, even if not all of the 38,016 potential isoforms are made, Dscam clearly generates a tremendously diverse repertoire of mRNAs and proteins. A few recent studies have examined where and when specific isoforms are expressed. Alternative splicing of the exon 4 cluster has been shown to be regulated throughout development (3, 20). The most dramatic example of this is exon 4.2, which is included at very low levels early in embryogenesis but increases throughout the remainder of development (3). More recently, microarray analysis has revealed that splicing of the exon 6 cluster and, in particular, the exon 9 cluster is also developmentally regulated (20). It is also clear that Dscam splicing is regulated in a tissue- and cell-specific manner (3, 20). For instance, isolated R3/R4 photoreceptors express a repertoire of isoforms distinct from that expressed by R7 photoreceptors (20). In addition, single-cell reverse transcription-PCR (RT-PCR) studies indicate that on the order of 50 Dscam isoforms are expressed in individual hemocytes and neurons (20). Interestingly, the splicing of each cluster appears to be controlled independently—the choice of an exon in one cluster does not affect exon choice in the other clusters (20). Despite the abundant evidence that Dscam alternative splicing is regulated, very little is known about the mechanisms involved in controlling Dscam alternative splicing.
Here we describe an intronic RNA secondary structure we refer to as the Inclusion Stem (iStem) that is required for efficient exon 4 inclusion. The iStem is located in the intron between exon 3 and the first exon 4 variant. Disruption of the iStem leads to the synthesis of Dscam transcripts that lack an exon 4 variant. Interestingly, although the iStem plays a major role in determining whether or not an exon 4 variant is included in the mRNA, it does not significantly affect which exon is included. The iStem is a novel type of regulatory element that simultaneously controls whether or not any of several exons are included in the mRNA.
MATERIALS AND METHODS
Exon 4 minigene.
The exon 4 minigene (pDscamWT) was constructed by cloning the complete exon 4 region of the Dscam gene beginning at the 5′ end of exon 3 and continuing into the intron downstream of exon 5. The exon 4 region was PCR amplified from 300 ng of D. melanogaster genomic DNA by use of Dmexon3US and DSCAMex5Rev primers. The PCR was carried out for 1 min at 94°C, 1 min at 55°C, and 15 min at 72°C for 35 cycles using 2.5 units of LA Taq (TaKaRa, Berkeley, CA). The PCR product was cloned into the vector pMT/V5-His-TOPO (Invitrogen, San Diego, CA) as described by the manufacturer.
Deletion mutants.
Deletion mutants were generated by PCR essentially as follows. Phosphorylated oligonucleotides flanking the region to be deleted were used for PCR with pDscamWT as a template. The reactions were carried out for 45 s at 94°C, 45 s at 55°C, and 9 min at 68°C for 28 cycles using 2.5 units of LA Taq (TaKaRa, Berkeley, CA). Next, 1.25 units of Pfu polymerase (Stratagene, La Jolla, CA) was added to the reaction mixtures, and the incubation continued at 72°C for 30 min to generate blunt ends. The PCR products were then cleaned up by phenol-chloroform extraction and ethanol precipitation. The PCR products were then self-ligated with T4 DNA ligase (Invitrogen, San Diego, CA) by incubation at room temperature overnight. The original template DNA was then destroyed by digestion with DpnI (New England Biolabs, Beverly, MA) at 37°C for 2 h. The reaction mixture was extracted once with phenol-chloroform, precipitated with ethanol, and resuspended in water, and the DNA was transformed into XL-1 Blue Escherichia coli.
Cell culture.
Drosophila S2 cells were grown at 27°C in a shaking suspension culture in Drosophila SFM (Invitrogen) supplemented with penicillin-streptomycin and glutamine. Prior to transfection, 1 × 106 cells were seeded into each well of a six-well culture dish and grown overnight at 27°C. Plasmid DNAs were transfected with Cellfectin reagent (Invitrogen) as described by the manufacturer. At 24 h posttransfection, transcription was induced by the addition of 500 μM copper sulfate and the cells were incubated at 27°C for 24 h.
RNA isolation from cells.
Total RNA was isolated by lysing the cells directly in the culture dish by adding 1 ml of TRIzol reagent (Invitrogen, San Diego) and passing the cell lysate through a pipette several times. The cell lysate was then transferred to a 1.5-ml centrifuge tube. A total of 100 μl of chloroform was added to the cell lysate, vortexed, and incubated on ice for 5 min. The samples were then centrifuged at 12,000 × g for 15 min at 4°C. The aqueous layer was removed to a new tube, 500 μl of isopropanol was added, and the samples were centrifuged at 12,000 × g at 4°C to precipitate the RNA. The supernatant was removed, and the pellets were resuspended in water.
RT-PCR.
Reverse transcription reactions were carried out at 42°C for 1 h, and the reaction mixtures contained 20 U RNase inhibitor (Invitrogen, San Diego, Calif.), 200 U Superscript II (Invitrogen), 5 μg total RNA, 500 ng random hexamers, and buffers provided by the manufacturer. A 3-μl amount of the RT reaction mixture was used as a template for PCR using a 32P-end-labeled primer that anneals upstream of exon 3 in the vector (MT forward) and a second unlabeled primer that anneals in exon 5 (Dmexon5ds). PCR was carried out for 30 s at 94°C, 15 s at 55°C, and 1 min at 72°C for 35 cycles using Taq DNA polymerase (Invitrogen).
SSCP.
To detect PCR products containing each of the exon 4 variants, we used single-strand conformation polymorphism (SSCP)-multidetection enhancement gels essentially as described previously (3). The SSCP gels contained 0.6× Tris-borate-EDTA buffer and 25% multidetection enhancement gel solution (BioWhittaker Molecular Applications, Walkersville, MD). A 10-μl amount of stop solution was added to 2 μl of each PCR, the mixtures were heated to 95°C for 2 min and placed on ice for 5 min, and 2 μl of each sample was loaded onto the gel. Each SSCP gel was run for 24 h at a maximum of 8 W and a constant temperature of 25°C. The gel was dried and exposed to film with an intensifying screen at −80°C overnight. Quantitation was performed using a Cyclone PhosphorImager system (Perkin Elmer).
Denaturing polyacrylamide gels.
To detect inclusion of an exon 4 variant, we used denaturing polyacrylamide gel electrophoresis. A 5-μl amount of stop solution (95% formamide, 0.25% bromophenol blue, 0.25% xylene cyanol) was added to 5 μl of each PCR and heated for 2 min. A total of 5 μl of the reaction was loaded on a 5% (19:1) 7 M urea gels and visualized by autoradiography. Quantitation was performed using a Cyclone PhosphorImager system (Perkin Elmer).
RNA isolation from flies.
Total RNA was isolated using LiCl-urea. Flies (0.2 to 0.5 g in weight) at various stages of development were homogenized in 3 ml of 3 M LiCl-6 M urea solution, and the homogenate was stored at −20°C overnight. The homogenate was removed and centrifuged at 13,000 rpm for 5 min. The pellet was resuspended in 300 μl of 1× TE (10 mM Tris [pH 8.0], 1 mM EDTA)-0.1% sodium dodecyl sulfate. The solution was extracted twice with phenol-chloroform, and the RNA was precipitated by adding 7.5 μl of 4 M NaCl and 750 μl of 100% ethanol.
Evolutionary analysis.
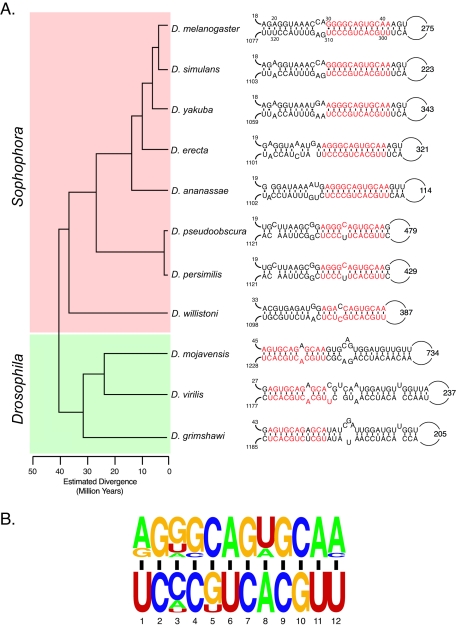
The sequences of the Drosophila Dscam genes have been previously reported (10, 11, 25). The pictogram (see Fig. 5) was generated using WebLogo (http://weblogo.berkeley.edu/logo.cgi) (6).
FIG. 5.
The iStem is conserved in other Drosophila species. (A) Phylogenetic tree of the Drosophila species analyzed. The Sophophora and Drosophila subgroups are highlighted in red and green, respectively. The structure of the iStem in each species is shown on the right. The distances from exon 3 to the 5′ boundary of the iStem and the distances from the 3′ boundary of the iStem to exon 4.1 are indicated for each structure. In addition, the size of the loop is indicated. The sequences that make up the core of the iStem are highlighted in red. (B) Pictogram representation of the sequences of the iStem core from all 11 Drosophila species.
RESULTS AND DISCUSSION
An intronic element required for exon 4 inclusion.
To begin characterizing the RNA sequence elements involved in Dscam exon 4 alternative splicing, we generated an exon 4 minigene. A portion of the Dscam gene beginning in exon 3 and ending in the intron downstream of exon 5 was cloned into a Drosophila expression vector containing the inducible metallothionein promoter to generate pDscamWT (Fig. 1A). This vector was transiently transfected into Drosophila S2 cells, and transcription was induced by the addition of CuSO4. Splicing was then analyzed by RT-PCR using a primer in exon 5 and a minigene-specific primer that anneals upstream of exon 3. Analysis of these PCR products on denaturing polyacrylamide gels revealed that the majority of the transcripts contain one of the exon 4 variants (Fig. 1B, lane 2). However, approximately 15% of the transcripts lacked an exon 4 variant and instead contained exon 3 spliced directly to exon 5. We analyzed the profile of exon 4 variants that are utilized both by sequencing cloned RT-PCR products and by resolving the RT-PCR products on a single-strand conformational polymorphism gel, which separates the molecules based on conformation rather than size (3). The majority of the transcripts from pDscamWT contain exons 4.12, 4.1, and 4.11, although exons 4.8, 4.10, 4.6, and 4.4 were also detectibly utilized (see Fig. 3A, lane 1). Thus, in S2 cells, transcripts derived from the minigene are spliced in a mutually exclusive manner and multiple exon 4 variants are selected. This system is therefore well suited for identifying and analyzing cis-acting sequences involved in Dscam exon 4 splicing.
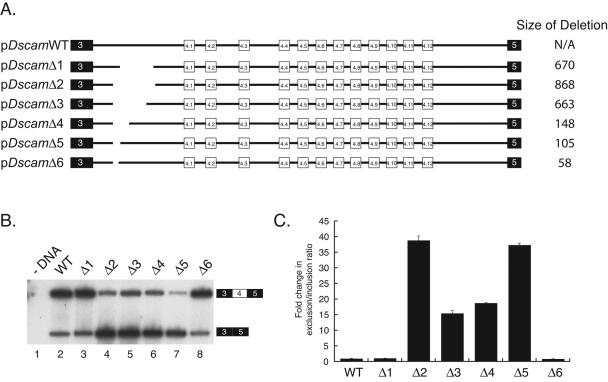
FIG. 1.
Identification of an element required for exon 4 inclusion. (A) Overview of the minigene constructs used to identify the iStem. The sizes of the deletions are indicated on the right. The intron between exons 3 and 4.1 is approximately 1.5 kb. (B) Analysis of exon 4 inclusion. Each minigene depicted in panel A was transiently transfected into Drosophila S2 cells and induced with CuSO4, and after 24 h, total RNA was harvested. The RNA was then used as a template in RT-PCRs using a reverse primer that anneals to exon 5 and a radiolabeled minigene-specific forward primer that anneals upstream of exon 3. The reaction products were then resolved on a denaturing polyacrylamide gel. WT, wild type. (C) Quantitation of the data in panel B. The gel in panel B was quantitated using a phosphorimager. The data are depicted as the severalfold change in the exon 4 exclusion/inclusion ratio in comparison to the wild-type minigene results. The error bars throughout this study were calculated from the average of at least three independent experiments.
FIG. 3.

Role of the iStem in exon 4 selection and exon 4 skipping in flies. (A) RNA was isolated from Drosophila S2 cells that were transiently transfected with the plasmids indicated. The RNA was used as a RT-PCR template, and the reactions were resolved by SSCP gel electrophoresis to analyze the frequency of exon 4 utilization. The identities of each band are indicated. WT, wild type. (B) Exon 4 skipping occurs in transcripts derived from the endogenous Dscam gene in the fly. RNA was isolated from D. melanogaster embryos, larvae, and adults. RT-PCR was performed using primers in exons 3 and 5, and the products were resolved on an agarose gel stained with ethidium bromide. The identities of the bands are indicated and were confirmed by cloning and sequencing.
We next began to identify RNA sequences required for various aspects of exon 4 splicing by generating deletions throughout the entire minigene and testing their effects on splicing in transfection experiments. Several of these deletions had profound effects on exon 4 splicing (J. M. Kreahling and B. R. Graveley, unpublished results). Here, we focus on an interesting set of deletions that removed portions of the 1,412-nucleotide (nt) intron between exons 3 and 4.1. Deletion of a 670-nt fragment encompassing nt 422 to 1090 of the intron (pDscamΔ1) had no effect on splicing of the minigene (Fig. 1B, lane 3). However, extending the 5′ boundary of this deletion to position 224 of the intron (pDscamΔ2) resulted in a dramatic increase in exon 4 skipping compared to pDscamWT results (Fig. 1B, lane 4; Fig. 1C). Additional deletions that progressively decrease the 3′ boundary of the deletion defined a 105-nt region (pDscamΔ5) that, when deleted, also resulted in a significant increase in exon 4 skipping (Fig. 1B, lane 7; Fig. 1C). In contrast, a slightly smaller 58-nt deletion encompassing nucleotides 224 to 280 (pDscamΔ6) displayed no more exon 4 skipping than the wild-type construct (Fig. 1B, lane 8; Fig. 1C). Thus, an element located between nt 280 and 422 of this intron (defined by the deletion boundaries in pDscamΔ1 and pDscamΔ6) is required for efficient exon 4 inclusion.
A stem-loop structure involved in exon 4 inclusion.
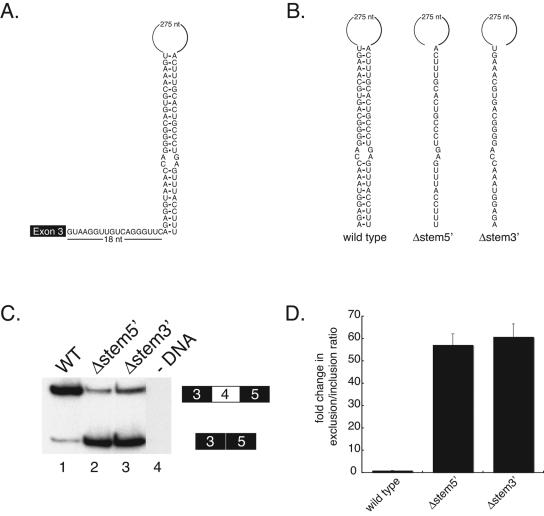
The sequence of the 105-nt segment deleted in pDscamΔ5 contains a pyrimidine-rich region and resembles a 3′ splice site. Although we have found that both U2 snRNP and U2AF can bind to this element in nuclear extracts, mutations that disrupt the binding of these splicing factors have little if any effect on exon 4 skipping (data not shown). This led us to hypothesize that this element may not function as a protein binding site in vivo. Detailed sequence analysis revealed that a 27-nt segment of this element could potentially base pair with a sequence located 18 nt downstream of exon 3 (Fig. 2A), forming a structure consisting of a 27-bp stem containing a 2-nt internal bulge and a 275-nt loop. We hypothesized that if the sequence element we initially identified functions to promote exon 4 inclusion by forming this stem-loop structure, disrupting this structure should affect the efficiency of exon 4 inclusion. We began testing this by disrupting the stem with deletions that removed either the 5′ or 3′ half of the stem (Fig. 2B). Indeed, we observed a dramatic increase in exon 4 skipping—deleting the 27 nt that make up either the 5′ or 3′ half of the stem resulted in ∼55-fold or ∼60-fold increases in the ratio of exon 4 exclusion/inclusion (Fig. 2C, lanes 2 and 3), respectively. This suggests that disrupting the formation of this RNA secondary structure has a significant effect on exon 4 splicing and that this structure is important for efficiently including an exon 4 variant. Hereafter, this RNA structural element will be referred to as the iStem.
FIG. 2.
Disruption of a putative RNA secondary structure results in exon 4 skipping. (A) Diagram of the potential RNA secondary structure formed by base pairing of sequences 18 nt downstream of exon 3 with a sequence in the region identified in Fig. 1. (B) Diagram of the locations of deletions made within the stem of the putative RNA secondary structure element. WT, wild type. (C) Polyacrylamide gel of RT-PCR products from RNA isolated from Drosophila S2 cells transiently transfected with the deletions constructs shown in panel B. Analysis of the data reveals that deletions made in the stem result in increased exon 4 skipping. (D) Quantitation of the data in panel C.
The iStem primarily controls exon inclusion but not exon selection.
Although the iStem has a profound effect on whether or not any of the exon 4 variants are included, it was not clear from the denaturing gel results whether the iStem plays a role in determining which of the exons are selected in those mRNAs that still contain an exon 4 variant. We therefore examined the impact of mutations of the iStem on exon 4 selection by resolving the RT-PCR products on SSCP gels (Fig. 3A). We find that disrupting the iStem has little impact on which of the exons is selected in those transcripts that contain an exon 4 variant (Fig. 3A). For instance, the frequency at which each exon 4 variant is selected is roughly equivalent between pDscamWT (Fig. 3A, lane 1) and pDscamΔstem3′ (Fig. 3A, lane 2), although the fraction of transcripts containing any exon 4 variant is significantly lower in pDscamΔstem3′. These results suggest that the iStem primarily functions to govern inclusion of the entire exon 4 cluster and that it does not play a role in exon 4 selection.
Exon 4 skipping occurs in flies.
Previous studies have not reported Dscam transcripts that lack an exon 4 variant (3, 15, 20, 25). However, our finding that the iStem is required for exon 4 inclusion led us to consider the possibility that Dscam exon 4 skipping might actually occur and be part of the normal gene expression program. Dscam transcripts that lack an exon 4 variant could still be translated, because exon 4 exclusion would not result in a frameshift. Due to the fact that the exon 4 variants encode sequences located in the second extracellular Ig domain, DSCAM isoforms synthesized from mRNAs lacking an exon 4 variant would be predicted to have protein interaction properties distinct from those of isoforms synthesized from exon 4-containing mRNAs. Based on previous results demonstrating that DSCAM isoforms exhibit isoform-specific homophilic interactions and that the identity of Ig domain 2 is important for the specificity of these interactions (32), it is likely that removal of this Ig domain would significantly affect the homophilic binding of DSCAM. Thus, it is entirely possible that exon 4 skipping could be biologically relevant.
To determine whether exon 4 skipping occurs in Drosophila species, we performed RT-PCR on RNA isolated from D. melanogaster embryos, larvae, and adults by use of primers in exons 3 and 5. Although the majority of transcripts that were amplified contained an exon 4 variant, a small fraction of the transcripts did indeed lack an exon 4 variant altogether (Fig. 3B). The identity of the exon-skipping PCR product was confirmed by cloning and sequencing (data not shown). This demonstrates that in the fly, exon 4 skipping occurs at a low but detectable frequency. The fact that exon 4 skipping occurs in the fly, coupled with the identification of a conserved RNA structural element that is required for efficient exon 4 inclusion, raises the possibility that exon 4 skipping is biologically relevant and perhaps controllable and that the iStem is the target of this regulation.
Sequences in the loop are not required for exon 4 inclusion.
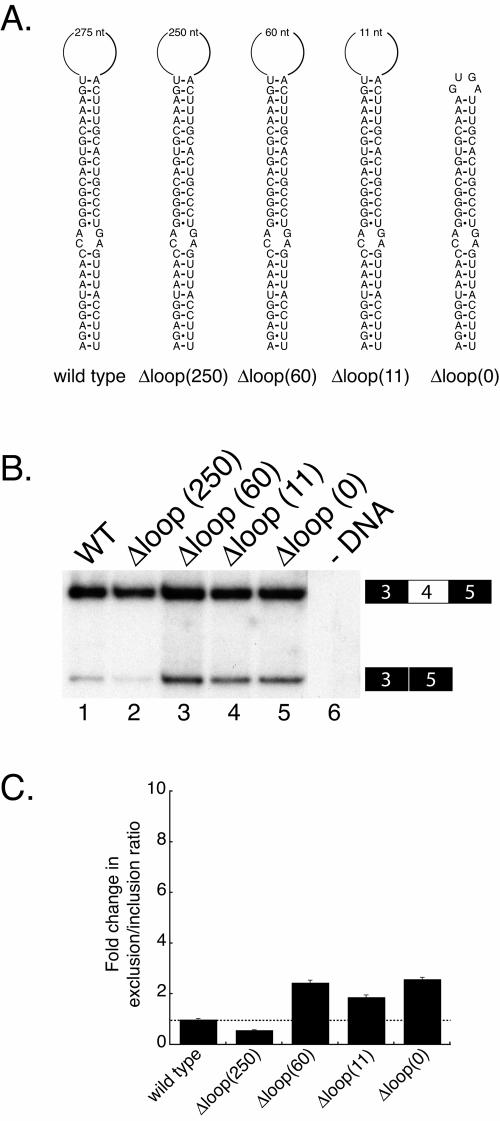
We next wanted to further characterize the iStem and sought to determine whether sequences in the 275-nt loop are required for exon 4 inclusion. To test this we made a series of deletions in the loop (Fig. 4A). These experiments revealed that decreasing the size of the loop from 275 nt to 250, 60, or 11 nt (Fig. 4B) does not result in a significant increase in exon 4 skipping; even completely deleting the loop, Δloop (0), had essentially no effect on the efficiency of exon 4 inclusion (Fig. 4B, lane 5). These data indicate that sequences within the loop are not required for efficient exon 4 inclusion and that the sequences involved in formation of the stem are the only sequences required for the function of the iStem.
FIG. 4.
Sequences within the loop of the iStem do not affect exon 4 inclusion. (A) Diagram of the deletions made within the loop of the iStem in the context of the pDscam minigene. (B) Polyacrylamide gel of RT-PCR products from RNA isolated from Drosophila S2 cells transiently transfected with the deletions constructs. (C) Quantitation of the data in panel B.
The iStem is evolutionarily conserved.
Genomic sequence is currently available for 11 members of the Drosophila subgenus (D. melanogaster, D. simulans, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. mojavensis, D. virilis, and D. grimshawi) that last shared a common ancestor at least 40 million years ago (22, 24). This allowed us to examine whether the iStem is conserved in other Drosophila species. We found evidence that the iStem is present in all 11 Drosophila species analyzed (Fig. 5A). In each species, the 5′ portion of the iStem is located an average of 24 nt downstream of the 5′ splice site of exon 3 and an average of 1,118 nt upstream of exon 4.1. In contrast, the size of the loop is quite variable, ranging from 114 nt in D. ananassae to 734 nt in D.mojavensis, which is consistent with our finding that deleting the loop has little effect on the function of the iStem.
The iStem in each species has a common core of 12 bp, which are highlighted in red in Fig. 5A and shown as a pictogram in Fig. 5B. Within this core there are two positions that show clear examples of nucleotide changes that maintain the overall structure. The first base pair of the core is either an A-U or G-U in all 11 species. The third base pair shows clear evidence of covariation and is a G-C, U-A, or A-U base pair. In addition, 5 of the 11 species contain a single-base bulge of C-U (D. pseudoobscura and D. persimilis), C-C (D. willistoni), or A-A (D. mojavensis and D. virilis) in the center of the core. However, it should be noted that all of these noncanonical nucleotide pairs have been shown to occur in other RNA structures (19).
The iStem is roughly twice the size of the core common among Drosophila species. However, the extent and location of the additional base pairs differ in members of the Sophophora subgroup (D. melanogaster, D. simulans, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, and D. willistoni) and the Drosophila subgroup (D. mojavensis, D. virilis, and D. grimshawi) (Fig. 5A). Whereas the iStem extends to the left of the core (proximal to the 5′ splice site) in members of the Sophophora subgroup, in the Drosophila subgroup the iStem extends to the right of the core (Fig. 5A). The sequences of the iStem extension are similar in all species of the Sophophora subgroup, with the exception of D. willistoni, which is the most distantly related member. Likewise, the sequences of the iStem extension are similar among the members of the Drosophila subgroup. Within the extensions specific to each group, there are several examples of covariation that maintain the overall integrity of the structures, although some nucleotide changes result in the introduction of internal bulges. The fact that the iStem has been conserved over millions of years suggests that it is functionally relevant to the splicing of the exon 4 cluster.
Compensatory mutations restore stem-loop function.
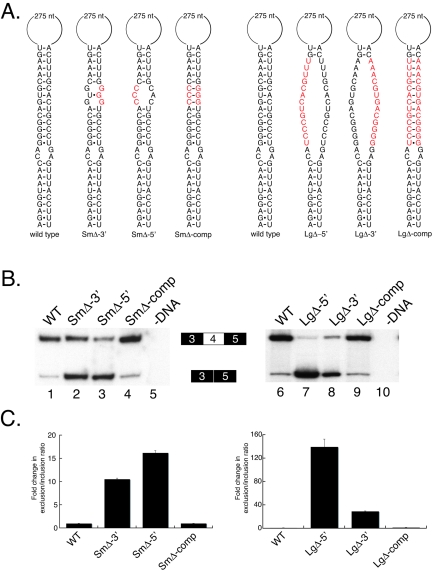
To obtain further evidence that the formation of the iStem is required for exon 4 inclusion, we tested a number of mutations and compensatory mutations in the core of the predicted stem (Fig. 6). We first generated two constructs that contained three point mutations in either the 3′ or 5′ sides of the core (SmΔ-3′ and SmΔ-5′) (Fig. 6A). These mutations result in ∼10-fold and ∼15-fold increases in the exon 4 exclusion/inclusion ratio, respectively (Fig. 6B, lanes 2 and 3). Importantly, combining these two mutations in the same construct, which should restore the formation of the RNA secondary structure, completely restores the efficiency of exon 4 inclusion to wild-type levels (Fig. 6B, lane 4).
FIG. 6.
Compensatory mutations restore the function of the iStem. (A) Diagram of mutations made in the core of the iStem. WT, wild type. (B) RT-PCR analysis of RNA isolated from S2 cells transiently transfected with the minigene constructs shown in panel A. RT-PCR products were resolved on a denaturing polyacrylamide gel. (C) Quantitation of the data in panel B.
We next generated constructs with more-severe mutations. Specifically, constructs were generated in which the sequence of one strand of the core was replaced with the sequence of the other strand (LgΔ-5′ and LgΔ-3′) (Fig. 6A). Either of these substitutions results in a significant (∼160-fold or ∼30-fold) change in the exon 4 exclusion/inclusion ratio (Fig. 6B, lanes 7 and 8). Furthermore, combining these two mutations in the same construct, which again should restore the ability of these sequences to form the RNA secondary structure, completely restores the efficiency of exon 4 inclusion to wild-type levels (Fig. 6B, lane 9). These results, together with the evolutionary conservation described above, provide strong evidence that these sequence elements engage in the formation of a long-range RNA secondary structure that is required for efficient exon 4 inclusion.
The iStem is a novel splicing element.
There are very few genes in any organism that contain clusters of more than two nonterminal, mutually exclusive alternative exons. One gene that shows splicing similar to the type of splicing that occurs for Dscam is the Drosophila Myosin heavy chain (Mhc) gene. Mhc contains multiple tandemly arranged exons—the exon 7 cluster contains four variants, the exon 9 cluster contains three variants, and the exon 11 cluster contains five variants (8). The five exons in the exon 11 cluster (exons 11a to 11e) are spliced in a mutually exclusive manner, and only one exon is typically utilized in individual muscles. For instance, exon 11e is the only variant included in the indirect flight muscle (IFM), whereas exon 11b is the only variant included in the jump muscle tergal depressor of the trochanter (13). An RNA element, called the conserved intronic element 3 (CIE3), located in the intron between the last exon 11 variant and exon 12 has been shown to be required for inclusion of exon 11e in the IFM (33, 34). When the CIE3 is deleted, exon 11e is skipped in the IFM and exon 10 is spliced directly to exon 12 (26, 27). From these results, the CIE3 appears functionally similar to the Dscam iStem. However, the Mhc CIE3 acts specifically on exon 11e because even when the CIE3 is deleted, the splicing of the four other exon 11 variants occurs normally in other muscles (26, 27). These results indicate that the CIE3 functions to specify which exon 11 variant is included. In contrast, the iStem is not involved in specifying the exon to be included but rather functions to control whether or not any of the exon 4 variants are included.
There are a number of examples of secondary structure elements in alternative splicing (2). The Saccharomyces cerevisiae rp51B pre-mRNA contains an intronic secondary structure that serves to bring the branchpoint into proximity of the 5′ splice site and therefore enhance spliceosome assembly (4). The yeast Actin gene contains secondary structure elements that function to alter the distance between splicing signals that enhances the efficiency of splicing of a distal 3′ splice site by sequestering the proximal 3′ splice site (2, 7). Moreover, stem-loop structures play an important role in the inclusion of an internal exon in one of the rare yeast genes that contain two introns (14). There are other examples, such as the Drosophila Adh pre-mRNA, where secondary structures function as negative elements by sequestering splice site sequences from the splicing machinery (2, 5). In the chicken β-tropomyosin pre-mRNA, a secondary structure has been proposed to form that functions to sequester exon 6B so that it is excluded from the mRNA (16). These few examples serve to illustrate that RNA secondary structure plays an important role in alternative splicing.
More recently, RNA structure has been found to play a key role in the mutually exclusive splicing of the exon 6 cluster in Dscam (10). In this case, a sequence upstream of each exon 6 variant known as the selector sequence base pairs with an additional sequence element called the docking site that is located downstream of constitutive exon 5. These interactions serve to juxtapose the exon 6 variant that is to be included with exon 5. Because only one selector sequence at a time can interact with the docking site these competing RNA secondary structures appear to be at the heart of the mechanism that ensures that only one exon 6 variant is included.
However, our data indicate that the iStem functions in a manner different from previously identified secondary structural elements that participate in pre-mRNA splicing. First, the iStem does not appear to function by simply bringing the exon 4 variants closer to exon 3. This interpretation is supported by the observation that exon skipping occurs in pDscamΔ2, which deletes 868 nt encompassing part of the iStem and therefore decreases the distance between exon 3 and exon 4.1 to a greater extent than would the iStem alone (329 nt) (Fig. 1B, lane 4). Second, the iStem does not appear to function to bring an intronic splicing regulatory element closer to exon 3. This is supported by the fact that deleting sequences downstream of the iStem does not have a significant effect on exon 4 skipping. Third, the iStem does not appear to sequester any regulatory elements, because deleting sequences within the loop does not have an effect on exon 4 inclusion. Finally, although at first glance the iStem may appear analogous to the selector sequence-docking site interactions identified in the exon 6 cluster (10), by virtue of their location and the fact that each appears to be involved in controlling the number of exons that are included, they are in fact quite distinct. The elements in the exon 6 cluster, in particular the docking site, are much more conserved than the iStem (10). Moreover, elements analogous to the selector sequences that would base pair with the upstream portion of the iStem are not present at other locations in the exon 4 cluster. Thus, the iStem appears to be a novel type of RNA structural element involved in Dscam splicing.
Model for iStem action.
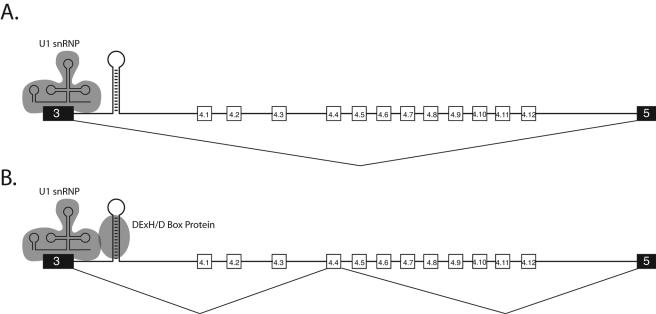
Due to the evolutionarily conserved proximity of the iStem to exon 3 and the fact that the iStem affects the inclusion of all 12 exon 4 variants equally, it seems most likely that the iStem acts on the 5′ splice site of exon 3 (Fig. 7). One possibility is that the iStem promotes the assembly of a specific protein complex at the 5′ splice site of exon 3 that confers upon exon 3 the ability to splice to one of the exon 4 variants. The iStem could do this by serving as a binding site for a splicing regulator or splicing regulatory complex.
FIG. 7.
Speculative model for iStem function. Due to the fact that the location of the iStem is conserved and always located just downstream of the 5′ splice site of exon 3 and that disrupting the function of the iStem interferes with the splicing of all 12 exon 4 variants, it is most likely that the iStem modulates the activity of the 5′ splice site of exon 3. One possibility is that in the absence of the iStem or when a protein is not bound to the iStem, the complex containing U1 snRNP that assembles on the 5′ splice site is incapable of splicing to one of the exon 4 variants. As a result, exon 3 is spliced directly to exon 5 (A). In contrast, when a protein binds to the iStem, it may form a complex with U1 snRNP at the 5′ splice site of exon 3, and this complex is then capable of splicing to one of the exon 4 variants (B). As explained in the text, one type of protein that could recognize the iStem is a DExH/D-box protein.
What types of regulators could recognize the iStem? If a protein or complex interacts with the iStem, it would need to do so in a sequence-independent manner. An RNA interference screen was recently conducted to identify proteins that regulate Dscam alternative splicing (21). Although none of the Drosophila double-stranded RNA binding proteins tested had an impact on the splicing of exon 4, depletion of several DExH/D-box proteins resulted in an increase in exon 4 skipping (21). One of these DExH/D-box proteins identified in the screen is Rm62, the Drosophila homolog of the human p68 helicase. Interestingly, p68 helicase has previously been shown to modulate the binding of U1 snRNP to 5′ splice sites (17) and functions as an alternative splicing regulator (12). Thus, it is possible that the iStem serves as a binding site for a DExH/D-box protein (such as Rm62) that interacts with U1 snRNP bound to the 5′ splice site of exon 3, resulting in a complex that is competent to splice to one of the exon 4 variants. In the absence of the iStem, or the DExH/D-box protein, the complex would not assemble and the 5′ splice site of exon 3 and, as a result, the exon 4 variants would be skipped. Testing this model will require the development of an in vitro splicing system for Dscam; such a system is currently unavailable.
Acknowledgments
We are grateful to members of our laboratory, Klemens Hertel, Kristen Lynch, and Robert Reenan, for discussions and comments on the manuscript.
This work was supported by a National Institutes of Health grant (GM 067842) to B.R.G.
REFERENCES
- 1.Black, D. L. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103:367-370. [DOI] [PubMed] [Google Scholar]
- 2.Buratti, E., and F. E. Baralle. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 24:10505-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celotto, A. M., and B. R. Graveley. 2001. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics 159:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charpentier, B., and M. Rosbash. 1996. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA 2:509-522. [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., and W. Stephan. 2003. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc. Natl. Acad. Sci. USA 100:11499-11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshler, J. O., and J. J. Rossi. 1991. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 5:1252-1263. [DOI] [PubMed] [Google Scholar]
- 8.George, E. L., M. B. Ober, and C. P. Emerson, Jr. 1989. Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol. Cell. Biol. 9:2957-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100-107. [DOI] [PubMed] [Google Scholar]
- 10.Graveley, B. R. 2005. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 123:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graveley, B. R., A. Kaur, D. Gunning, S. L. Zipursky, L. Rowen, and J. C. Clemens. 2004. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. RNA 10:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guil, S., R. Gattoni, M. Carrascal, J. Abián, J. Stévenin, and M. Bach-Elias. 2003. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol. Cell. Biol. 23:2927-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastings, G. A., and C. P. Emerson, Jr. 1991. Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J. Cell Biol. 114:263-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howe, K., and M. J. Ares. 1997. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. USA 94:12467-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummel, T., M. L. Vasconcelos, J. C. Clemens, Y. Fishilevich, L. B. Vosshall, and S. L. Zipursky. 2003. Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 37:221-231. [DOI] [PubMed] [Google Scholar]
- 16.Libri, D., A. Piseri, and M. Y. Fiszman. 1991. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science 252:1842-1845. [DOI] [PubMed] [Google Scholar]
- 17.Liu, Z. R. 2002. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 22:5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loughney, K., R. Kreber, and B. Ganetzky. 1989. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell 58:1143-1154. [DOI] [PubMed] [Google Scholar]
- 19.Nagaswamy, U., M. Larios-Sanz, J. Hury, S. Collins, Z. Zhang, Q. Zhao, and G. E. Fox. 2002. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res. 30:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neves, G., J. Zucker, M. Daly, and A. Chess. 2004. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat. Genet. 36:240-246. [DOI] [PubMed] [Google Scholar]
- 21.Park, J. W., K. Parisky, A. M. Celotto, R. A. Reenan, and B. R. Graveley. 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA 101:15974-15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell, J. R. 1997. Progress and prospects in evolutionary biology: the Drosophila model. Oxford University Press, New York, N.Y.
- 23.Rowen, L., J. Young, B. Birditt, A. Kaur, A. Madan, D. L. Philipps, S. Qin, P. Minx, R. K. Wilson, L. Hood, and B. R. Graveley. 2002. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics 79:587-597. [DOI] [PubMed] [Google Scholar]
- 24.Russo, C. A., N. Takezaki, and M. Nei. 1995. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 12:391-404. [DOI] [PubMed] [Google Scholar]
- 25.Schmucker, D., J. C. Clemens, H. Shu, C. A. Worby, J. Xiao, M. Muda, J. E. Dixon, and S. L. Zipursky. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101:671-684. [DOI] [PubMed] [Google Scholar]
- 26.Standiford, D. M., M. B. Davis, W. Sun, and C. P. Emerson, Jr. 1997. Splice-junction elements and intronic sequences regulate alternative splicing of the Drosophila myosin heavy chain gene transcript. Genetics 147:725-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Standiford, D. M., W. T. Sun, M. B. Davis, and C. P. Emerson, Jr. 2001. Positive and negative intronic regulatory elements control muscle-specific alternative exon splicing of Drosophila myosin heavy chain transcripts. Genetics 157:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabuchi, K., and T. C. Sudhof. 2002. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics 79:849-859. [DOI] [PubMed] [Google Scholar]
- 29.Thackeray, J. R., and B. Ganetzky. 1995. Conserved alternative splicing patterns and splicing signals in the Drosophila sodium channel gene para. Genetics 141:203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J., C. T. Zugates, I. H. Liang, C. H. Lee, and T. Lee. 2002. Drosophila Dscam is required for divergent segregation of sister branches and suppresses ectopic bifurcation of axons. Neuron 33:559-571. [DOI] [PubMed] [Google Scholar]
- 31.Watson, F., R. Puttmann-Holgado, F. Thomas, D. Lamar, M. Hughes, M. Kondo, V. Rebel, and D. Schmucker. 2005. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309:1874-1878. [DOI] [PubMed] [Google Scholar]
- 32.Wojtowicz, W. M., J. J. Flanagan, S. S. Millard, S. L. Zipursky, and J. C. Clemens. 2004. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell 118:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan, X. L., J. C. Clemens, G. Neves, D. Hattori, J. J. Flanagan, T. Hummel, M. L. Vasconcelos, A. Chess, and S. L. Zipursky. 2004. Analysis of Dscam diversity in regulating axon guidance in Drosophila mushroom bodies. Neuron 43:673-686. [DOI] [PubMed] [Google Scholar]