Abstract
Salmonella enterica mutants defective in Dam methylase are strongly attenuated in virulence and release a large amount of proteins to the extracellular medium. The extent to which these two phenotypes are linked is unknown. Using a proteomic approach, we identified Sb6, Sb13, and Sb36 as proteins present in larger amounts in culture supernatants of an S. enterica serovar Typhimurium dam mutant than in those of the wild-type strain. These three proteins are encoded in the Salmonella prophage ST64B. Higher amounts of ST64B phage DNA and tailless viral capsids were also detected in supernatant extracts of the dam mutant, suggesting that Dam methylation negatively regulates the excision of ST64B. Reverse transcription-PCR analysis revealed that the expression of two ST64B genes encoding a putative antirepressor and a phage replication protein increases in the dam mutant. The SOS response also augments the excision of ST64B. Infection assays performed with phage-cured strains demonstrated that ST64B does not carry genes required for virulence in the mouse model. Evidence was also obtained discarding a relationship between the high excision of ST64B and the envelope instability or virulence attenuation phenotype. Taken together, these data indicate that ST64B excises at a high rate in dam mutants due to the loss of repression exerted by Dam on phage genes and induction of the SOS response characteristic of these mutants. The exacerbated excision of ST64B does not however contribute to the incapacity of dam mutants to cause disease.
Deoxyadenosine methyltransferase (Dam) is an enzyme conserved in diverse groups of proteobacteria that specifically methylates the deoxyadenosine residue of 5′-GATC-3′ sites in the DNA. This event is essential for postreplicative mismatch repair, DNA replication, chromosome segregation, transposition of insertion sequences, phage DNA packaging, and bacterial conjugation (42). An additional role assigned to Dam methylation is regulation of gene expression. Thus, the methylation status of GATC sites located in the upstream region of certain genes affects binding of regulatory proteins (34, 42). Regulation of gene expression by Dam methylation has been shown for the pyelonephritis-associated pili (pap) genes of uropathogenic Escherichia coli (35), the genes encoding Pef fimbria in Salmonella enterica serovar Typhimurium (48), and genes involved in the conjugal transfer of serovar Typhimurium virulence plasmid (9).
In 1999, two independent studies demonstrated that lack of Dam methylase causes strong attenuation of S. enterica virulence (26, 33) whereas mutations affecting DNA mismatch repair such as mutL have no effect on virulence (26). These observations indicated that virulence attenuation was not the result of a defect in the mismatch repair system and raised the possibility that Dam methylation could modulate expression of virulence genes. It was later demonstrated that the deficiency of Dam methylase causes envelope instability in S. enterica (53). This alteration was denoted by the presence of large amount of membrane vesicles in the supernatant of cultures of dam mutants (53). The release of membrane material to the extracellular medium correlated with a high protein content in extracts prepared from culture supernatants of dam mutants.
Release of envelope material occurs spontaneously in actively growing bacteria (5). However, in S. enterica, but not in Escherichia coli, the lack of Dam methylation results in decreased association with peptidoglycan of membrane proteins required for maintenance of envelope integrity, such as OmpA, TolB, peptidoglycan-associated lipoprotein (Pal), and Braun's murein lipoprotein (53). These observations supported the idea that Dam methylation could regulate the expression of some Salmonella-specific function(s) involved directly or indirectly in maintenance of envelope integrity.
One of the consequences of the envelope alteration displayed by S. enterica dam mutants is the impaired secretion of SipC, an effector protein encoded in Salmonella pathogenicity island 1 (SPI-1) that is required for efficient bacterial entry into nonphagocytic cells (25, 53). Another phenotype of dam mutants that is linked to the envelope instability is an increased sensitivity to antimicrobial peptides, bile salts, and hydrogen peroxide (32). Changes in the structure of the bacterial envelope may also lead to exacerbated exposure and/or release of antigenic bacterial products.
Vaccination tests with dam mutants of serovar Typhimurium have shown that these strains elicit a cross-protective immunity that extends to other Salmonella serovars (17, 32). This observation suggests that the antigen(s) or changes in bacterial physiology linked to the to dam mutation may be conserved among salmonellae. dam mutants have been shown to act as vaccine strains not only in murine models, but also in chickens and calves (16-18, 26). The important role of Dam methylation for virulence has also been shown in other bacterial pathogens as Haemophilus influenzae, Pasteurella multocida, Yersinia pseudotuberculosis, and Vibrio cholerae (12, 40, 64). However, a recent work claimed a modest effect of dam mutations in Shigella pathogenicity (38).
To define more precisely the primary basis of the virulence attenuation linked to dam mutations, we decided to perform a proteomic study to identify proteins released to the extracellular medium by S. enterica serovar Typhimurium dam mutants. Our rationale implied that the massive release of membrane material and proteins occurring in dam mutants could act as a factor negatively influencing the virulence capacity of these mutants. The study revealed that, unlike the parental wild-type strain, the mutants defective in Dam methylase release significant amounts of proteins of a recently described Salmonella bacteriophage named ST64B (23, 46, 62). This prophage was first identified in serovar Typhimurium epidemic strain DT64 and has a mosaic structure, carrying fragments of the virulence genes sspH2, sopE, and orgA inserted in the open reading frames of the tail genes (46). Although susceptible to induction, the ST64B prophage is defective in morphogenesis, producing tailless particles that are noninfectious (46). ST64B-related prophages have been found in other serovar Typhimurium strains, and their defective nature correlated with the presence of a frameshift mutation in the sb21 gene, which could prevent the correct assembly of the phage tail (23). Here, we show that although the ST64B prophage is not required for virulence in the mouse model, its excision rate is finely modulated by Dam methylation and the SOS response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Salmonella strains used in this work are listed in Table 1. Deletion of entire phages or specific phage genes was performed by the λ-Red-mediated recombination method described by Datsenko and Wanner (14). Primers used for the construction of these mutants are listed in Table 2. The deletion alleles were checked by general transduction with the P22 HT 105/1 int201 phage and by PCR analysis and sequencing. Strains were grown in commercial LB broth and LB agar (USB Corporation, Ohio) at 37°C and when required, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; cloramphenicol, 10 μg ml−1; kanamycin, 30 μg ml−1; and tetracycline, 10 μg ml−1.
TABLE 1.
Salmonella strains used in this study
| Strain | Strain | Relevant genotype | Source or referencea |
|---|---|---|---|
| S. enterica | |||
| Subspecies I serovar Typhimurium | SL1344 | hisG64 rpsL, mouse virulent | 37 |
| SV1610 | SL1344 dam-228::MudJ | 26 | |
| SV4204 | SL1344 dam-201::Tn10dTet | J. Casadesús | |
| MD1101 | SL1344 Δsb49::Kn | This study | |
| MD1102 | SL1344 Δsb50::Kn | This study | |
| MD1103 | SL1344 Δsb49sb50::Kn | This study | |
| MD1104 | SL1344 ST64B [Δ(int-pin)::Kn] | This study | |
| MD1202 | SL1344 ST64B [Δ(int-pin)::Kn] dam-201::Tn10dTet | This study | |
| MA6247 | SL1344 Gifsy-1[−] Gifsy-2[−] | 23 | |
| MD1203 | SL1344 Gifsy-1[−] Gifsy-2[−] dam-201::Tn10dTet | This study | |
| MA7551 | SL1344 Gifsy-1[−] Gifsy-2[−] ST64B [Δ(int-pin)::Kn] | 23 | |
| MD1201 | SL1344 Gifsy-1[−] Gifsy-2[−] ST64B [Δ(int-pin)::Kn] dam-201::Tn10dTet | This study | |
| MA7891 | SL1344 Gifsy-1[−] Gifsy-2[−] ST64B [Δ(int-pin)::Kn] SopEΦ [Δ(int-B)::lacZ-Cm] | This study | |
| MD0200 | SL1344 Gifsy-1[−] Gifsy-2[−] ST64B [Δ(int-pin)::Kn] SopEΦ [Δ(int-B)::lacZ-Cm] dam-201::Tn10dTet | This study | |
| MA8234 | SL1344 ST64B [Δ(int-xis)::Kn] | This study | |
| MD1221 | SL1344 ST64B [Δ(int-xis)::Kn] dam-201::Tn10dTet | This study | |
| MA8226 | SL1344 Δ[ilv-leuO]3308::sodC1 Gifsy-2[−] Gifsy-1[−]::pNFB14 (Ampr) SopEΦ [Δ(int-B)::lacZ-Cm] ST64B [Δ(int-pin)::Kn] | This study | |
| MD1222 | SL1344 Δ[ilv-leuO]3308::sodC1 Gifsy-2[−] Gifsy-1[−]::pNFB14 (Ampr) SopEΦ [Δ(int-B)::lacZ-Cm] ST64B [Δ(int-pin)::Kn] dam-201::Tn10dTet | This study | |
| LT2 | Wild type | Lab stock | |
| SV3000 | LT2 dam-201::Tn10dTet | 60 | |
| 14028s | Wild type, mouse virulent | 22 | |
| SV4392 | 14028s dam-201::Tn10dTet | 53 | |
| MD1105 | 14028s ST64B [Δ(int-pin)::Kn] | This study | |
| MD1214 | 14028s pEG108 (cea::lacZ, Kn) | This study | |
| MD1215 | 14028s lexA (Ind−) pEG108 (cea::lacZ, Kn) | This study | |
| SV4536 | 14028s Δdam230 | 51 | |
| MD1213 | 14028s Δdam230/pEG108 (cea::lacZ, Kn) | This study | |
| MD1210 | 14028s mutS121::Tn10 | This study | |
| MD1216 | 14028s mutS121::Tn10/pEG108 (cea::lacZ, Kn) | This study | |
| MD1217 | 14028s mutS121::Tn10 lexA (Ind−)/pEG108 (cea::lacZ, Kn) | This study | |
| MD1211 | 14028s Δdam230 mutS121::Tn10 | This study | |
| SV4854 | 14028s Δdam230 mutS121::Tn10/pEG108 (cea::lacZ, Kn) | 51 | |
| MD1212 | 14028s Δdam230 mutS121::Tn10 lexA (Ind−)/pEG108 (cea::lacZ, Kn) | This study | |
| Subspecies I serovar Typhi | 5866 | Clinical isolate | R. Rotger |
| Subspecies II | SARC-3 | SGSC | |
| Subspecies IIIa | SARC-5 | SGSC | |
| Subspecies IIIb | SARC-7 | SGSC | |
| Subspecies IV | SARC-9 | SGSC | |
| Subspecies VI | SARC-13 | SGSC | |
| Subspecies VII | SARC-15 | SGSC | |
| S. bongori subspecies V | SARC-11 | SGSC |
SGSC, Salmonella Genetic Stock Center; University of Calgary, Calgary, Canada.
TABLE 2.
Oligonucleotides used to generate deletions mutants
| Oligonucleotidea | Sequence (5′→3′)b | Prophage or gene deleted |
|---|---|---|
| sb49-3D | ATGACACCACGTCAACGCCGTCAACATCGTAACGCAATAGAAAAAGCCGCTGTGTAGGCTGGAGCTGCTTC | sb49 |
| sb49-4R | TCAAGATATTTTTTCATTTTGCTTTCTCATTGCATCCCTGAATGCGAGAAAATTCCGGGGATCCGTCGACC | sb49 |
| sb50-1D | ATGCTTGGCAAAGCTGCGCCGCGATGGCAGCGACGAAAAGCAAGTCAATTAGTGTAGGCTGGAGCTGCTTC | sb50 |
| sb50-2R | ACATGCCAGCAGAAGCTGCGCAATTTCTCCGCTCGCTGAAACTGCCAGTCAATTCCGGGGATCCGTCGAC | sb50 |
| pp291 | AGAATATCGCGCAGTACCAGTATGTTTCCGCCGTTCATCACAGATCATCGAGCTCTCCCG | ST64B |
| pp293 | ATACCTGAAAGCTGCTTCTGGTATGTGCTGCATGACCGCATGTAGGCTGGAGCTGCTTCG | ST64B |
| pp339 | ACTGTACTTCTGCTTGTCTTTTGCCGTTCCCTCATAGTCTCATATGAATATCCTCCTTAG | SopEΦ |
| pp340 | TTAACTCCCTTCCGGTTAGCCGATAACAGAATCCAGTACATGTAGGCTGGAGCTGCTTCG | SopEΦ |
Preparation of protein extracts for two-dimensional gel electrophoresis.
Bacteria were first grown overnight in 2 ml LB medium without shaking at 37°C. This culture was used to inoculate 30 ml of LB medium (dilution 1:30) and growth was continued without shaking at 37°C for another overnight period. Bacteria were then spun down by centrifugation at 10,000 × g, 10 min, 4°C, and supernatant was filtered in a Millipore 0.22-μm filter. Extracellular proteins were precipitated in 6% trichloroacetic acid (15 min, 4°C), washed twice in acetone, and solubilized in rehydration sample buffer containing BioLytes 3/10, 8 M urea, 4% cholamidopropyldimethylammoniopropanesulfonate (CHAPS), and 10 mM dithiothreitol, 0.2% (wt/vol) (Bio-Rad Laboratories, California). Protein concentration was determined with the RC-DC protein assay (Bio-Rad Laboratories, California) according to the manufacturer's instructions.
For two-dimensional polyacrylamide gel electrophoresis (PAGE), samples containing ∼150 μg of protein were diluted in rehydration sample buffer BioLytes 4/7 and focused in the first dimension with 11-cm-length pH range 4 to 7 ReadyStrips IPG using the PROTEAN isoelectric focusing cell (Bio-Rad Laboratories, California). The second dimension was performed on 8% acrylamide gels using the sodium dodecyl sulfate-Tris-Tricine system (56) in the Mini Protean 3 apparatus (Bio-Rad). Proteins were silver stained using the PlusOne staining kit (Amersham Biosciences, Sweden).
Peptide mass fingerprinting.
Protein spots that appeared differentially in extracts prepared from cultures of dam mutants were excised from the gel. Gel slices were washed in water and acetonitrile, dried by vacuum centrifugation and resuspended in digestion buffer containing trypsin (10 ng ml−1 sequencing grade; Promega, Madison, WI). Incubation was for 40 min on ice. After discarding the supernatant, 20 to 40 μl of digestion buffer was added and the digestion was continued for 18 h at 37°C. The released tryptic peptides were analyzed by mass spectrometry using matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Briefly, 0.5-μl aliquots of the digestion solution were manually deposited onto a stainless steel MALDI probe and allowed to dry at room temperature. Then 0.5 μl of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in 33% aqueous acetonitrile and 0.1% trifluoroacetic acid) was added and the samples were allowed to dry at room temperature.
Samples were measured on a Bruker Reflex III MALDI-TOF mass spectrometer (Bruker-Franzen Analytic GmbH, Bremen, Germany) equipped with the SCOUT source in the positive ion reflector mode by using delayed extraction and AutoXecute acquisition software. The ion acceleration voltage was 20 kV. The equipment was first externally calibrated by employing protonated mass signals from a peptide mixture covering the 1,000- to 4,000-m/z range, and thereafter, every spectrum was internally calibrated using signals arising from trypsin autoproteolysis. The measured tryptic peptide masses were transferred automatically through the mass spectrometry BioTools program as inputs to search automatically the nonredundant NCBI database using Mascot software (Matrix Science).
PCR amplification.
PCR amplifications were performed on either bacteria collected from colonies or purified DNA using Taq DNA polymerase according to standard protocols. The presence of ST64B prophage was assessed using primers sb49-1D (5′-CCA GAG TGC GTG GGT TAA AT-3′) and sb49-2R (5′-ACC CGG ATC GTC GAC AAA TA-3′). The excised phage DNA form was detected using primers sb27-1D (5′-ACA CGG GCT GTA CTG GAT TC-3′) and sb28-2R (5′-CTT GAC TGC ACT TTC CAC G-3′). The frameshift mutation in the sb21 tail gene was confirmed by PCR and sequencing with the primers pp439 (5′-TGC CGG TTA TTG CTG ATG-3′) and pp440 (5′-CGG CAA AAT ATG GTC ACG-3′) (23). The PCR control reactions for DNA content were made with the primers fimY-1 (5′-GAG TTA CTG AAC CAA CAG CT-3′) and fimY-2 (5′-GCC GGT AAA CTA CAC GAT GA-3′) (fimY gene) (65) or, alternatively, YF2D (5′-TAG CTG CTT TGC TGG CC-3′) and YF5R (5′-CGG GCA ACA GTT TAC GC-3′) (igaA gene) (11).
The presence of other Salmonella prophages was tested with the following primers: Gifsy-1, xis gene, pp63 (5′-GGA GTG TGG TCG GTG TAC G-3′) and pp64 (5′-CTT GGC GAC GAA CTG CAC G-3′); Gifsy-2, sodC gene, pp3 (5′-CCT CCG CCA TCC CCG TGA CC-3′) and pp4 (5′-ACG TGC ACC GCC ACC ACC C-3′); and SopEΦ, sopE gene, pp17 (5′-CAT CAA TCA GAT GGA CAT AGC-3′) and pp18 (5′-CTG GCC GGA GAA ACC AGC TC-3′).
To assess the presence of phage DNA in culture supernatants and unless otherwise indicated, bacteria were grown in the same conditions as for preparation of extracellular protein extracts (see above). After filtration, the culture supernatant was treated twice with phenol-chloroform and DNA was precipitated with ice-cold isopropanol. The DNA-containing pellet was resuspended in an appropriate volume of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
RNA purification and RT-PCRs.
Total RNA was purified from bacteria grown to exponential phase to an optical density at 600 nm (OD600) of 0.2 using the Total RNA isolation system (Promega, Madison, WI) as described (20). The reverse transcription (RT)-PCR was performed on 1 μg of RNA using the High-Capacity cDNA archive kit (Applied Biosystems, Foster City, CA) following the manufacturer's instructions. The relative expression levels of the following genes were assessed: sb41 [primers 41A (5′-TTA TCT ATC TGC GCA AGG GC-3′) and 41B (5′-CAG GTT GAG CGA GGG TTG-3′)]; sb42 [primers 42A (5′-CAT ATT GCT GAC CAG TGC GA-3′) and 42B (5′-GCT TTC CAG GCT TTG TAG G-3′)]; ompA [primers omp1 (5′-TGT AAG CGT CAG AAC CGA TAC G-3′) and omp2 (5′GAG CAA CCT GGA TCC GAA AG-3′)]; and igaA (primers YF2D and YF5R listed above), giving amplicons for sb41 (406 bp), sb42 (442 bp), ompA (73 bp), and igaA (125 bp).
Competition index and median lethal dose experiments in mice.
Female BALB/c mice (7 to 8 weeks old) were used for competition index assays. Mice were inoculated intraperitoneally with a 1:1 mixture of two strains comprising a total of ∼105 CFU. After 48 to 72 h, mice were sacrificed and the competition index values for spleen and liver were determined as described by Beuzón and Holden (4). The 50% lethal dose assays were performed as described (15).
Transmission electronic microscopy.
Supernatants from bacterial cultures were prepared in the same conditions as for two-dimensional PAGE analysis. Once filtered in a Millipore 0.22-μm filter, the supernatant was centrifuged 20 min and 4°C at 300,000 × g. The pellet was resuspended in an appropriate volume of TE buffer to obtain a 200-fold concentration relative to the initial volume of culture supernatant. Samples were then analyzed by transmission electron microscopy as previously described (52).
RESULTS
Identification of ST64B phage proteins in culture supernatants of dam mutants.
The deficiency in Dam methylase in S. enterica serovar Typhimurium causes envelope alterations manifested by the accumulation of membrane particulate material in the extracellular medium (53). We hypothesized that this condition could favor the release of specific proteins to the extracellular medium by the dam mutant. This possibility was assessed by a two-dimensional PAGE proteomic analysis.
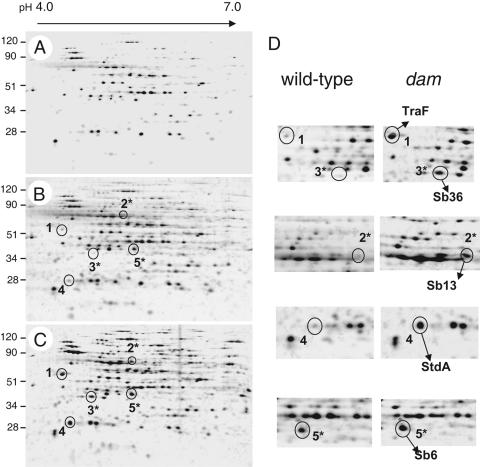
As previously reported (26), and despite the fact that the cultures reached the same optical density, the extracellular protein content of the sample of the dam mutant was ∼3-fold higher than that of the sample obtained from cultures of the wild-type strain SL1344. Thus, when samples were normalized to an equivalent bacterial culture volume (10 ml), protein spots in the two-dimensional gels were in general more intense in the sample of the dam mutant (Fig. 1A and C). Comparison of these two samples revealed that five proteins were enriched in the culture supernatant of the dam mutant (Fig. 1C, spots 1 to 5). These differences were also evident when comparing the protein pattern of the dam mutant with that obtained from twice the amount of culture volume of the wild-type strain (20 ml) (Fig. 1B and C). Peptide mass-fingerprinting analysis identified these proteins as: TraF (spot 1); Sb13 (spot 2); Sb36 (spot 3); StdA (spot 4); and Sb6 (spot 5) (Table 3).
FIG. 1.
Extracellular proteins present in culture supernatants of serovar Typhimurium strains SL1344 (wild-type, dam+) and SV1610 (dam). Two-dimensional electrophoresis was applied to resolve extracellular proteins present in supernatants of the following strains and culture volumes: (A) wild-type, 10 ml; (B) wild-type, 20 ml; and (C) dam mutant, 10 ml. Bacteria were grown overnight in LB medium in static conditions at 37°C. Proteins that appeared more prominent in the samples of the dam mutant are highlighted (spots 1 to 5). Spots labeled with an asterisk (2*, 3*, and 5*) correspond to ST64B phage proteins. (D) Enlargement of the gel areas in which differences in the protein pattern were detected. Shown are the five proteins identified by peptide-mass fingerprinting. The size of the molecular weight markers is indicated in kilodaltons.
TABLE 3.
Proteins identified in the supernatant fraction of cultures of S. enterica serovar Typhimurium dam mutantsa
| Spot no. | Size (kDa) | pI | Protein (Swiss Prot accesion no.) | Putative function | Theoretical sizeb (kDa) | Theoretical pIb | No. of matching peptides/total peptides analyzed in spot | Protein sequence coverage (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 40 | 4.5 | TraF (Q9R2H5) | Pilus assembly protein | 43 | 5.05 | 6/13 | 16 |
| 2 | 60 | 5.5 | Sb13 (Q8HAC5) | Tail sheath protein, phage ST64B | 53 | 5.31 | 7/18 | 19 |
| 3 | 32 | 5.0 | Sb36 (Q8HAA2) | Hypothetical protein, phage ST64B | 30 | 5.00 | 9/24 | 32 |
| 4 | 25 | 4.8 | StdA (Q8ZM89) | Putative fimbria-like protein | 27 | 6.81 | 3/6 | 22 |
| 5 | 40 | 5.6 | Sb6 (Q8HAD2) | Major capsid protein precursor, phage ST64B | 44 | 5.41 | 12/32 | 34 |
Spot numbers refer to proteins indicated in Fig. 1. Sizes and isectric points were estimated from the relative positions of the protein spots.
Theoretical values of the precursor unprocessed form.
TraF has been reported as a protease required for mobilization of conjugative plasmids and donor-specific bacteriophages (27). TraF is encoded in the pSLT virulence plasmid of S. enterica serovar Typhimurium strain LT2 (45). StdA is the putative major subunit encoded in the std fimbrial operon (39). The remaining three abundant proteins in the culture supernatant of the dam mutant, Sb6, Sb13, and Sb36, are all encoded by genes annotated in the genome of serovar Typhimurium bacteriophage ST64B (accession no. AY055382). This ST64B sequence was obtained from phage particles isolated upon treatment of the epidemic serovar Typhimurium DT64 strain with mitomycin C (46). In agreement with a recent study (23), our proteomic analysis confirmed the presence of the ST64B prophage in serovar Typhimurium strain SL1344. Furthermore, these data demonstrate that the absence of Dam methylase results in the accumulation of ST64B phage proteins in the extracellular medium.
ST64B prophage is carried only by some strains of serovar Typhimurium.
The genome of strain SL1344 is currently under gap closure status in the Wellcome Trust-Sanger Institute (http://www.sanger.ac.uk/Projects/Salmonella/). Using the BLAST service provided at the Sanger Institute, the genome of SL1344 was scanned for the presence of the 40,149-base-pair genome of ST64B annotated in the databases, which encompasses a total of 56 open reading frames (ORFs). The ST64B prophage integrates within the tRNA-encoding gene serU of SL1344 (23). The serU integration site is also conserved in serovar Typhimurium strain DT64 (46). Our in silico analysis also denoted that none of the ST64B sequences are present in the completely sequenced genomes of serovar Typhi strain CT18 (accession no. AL513382); serovar Typhi strain Ty2 (accession no. AE014613); serovar Paratyphi A strain ATCC9150 (accession no. CP000026); serovar Choleraesuis strain SC-B67 (accession no. AE017220); or Salmonella bongori (http://www.sanger.ac.uk/Projects/Salmonella/).
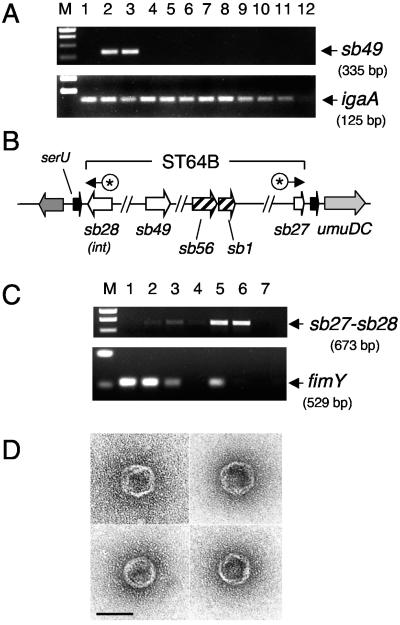
The distribution of the ST64B phage in the genus Salmonella was assessed by PCR using primers specific to the ST64B phage gene sb49. The assay was made on strains of the Salmonella Reference Collection C (SAR-C), representative of the diverse S. enterica subspecies and the species S. bongori. The experiment showed that the sb49 gene is present in serovar Typhimurium strains SL1344 and 14028s, but absent in the rest of the Salmonella serovars, subspecies, and species analyzed (Fig. 2A). Thus, like other phages previously identified in serovar Typhimurium (8, 24, 28), ST64B distribution is restricted to specific strains.
FIG. 2.
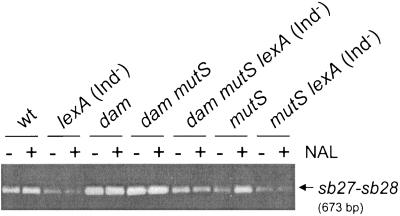
ST64B prophage presents a narrow distribution in the Salmonella genus and is induced at a high rate in dam mutants. (A) PCR-mediated amplification of products specific to the genes sb49 (ST64B phage) and igaA (present in all salmonellae). The assay was performed on DNA collected from colonies of the following Salmonella strains (lane 1) LT2; (lane 2) SL1344; (lane 3) 14028s; (lane 4) 5866; (lane 5) SARC3; (lane 6) SARC5; (lane 7) SARC7; (lane 8) SARC9; (lane 9) SARC11; (lane 10) SARC13; (lane 11) SARC15; and (lane 12) negative control (no DNA). Two of the three subspecies I serovar Typhimurium strains tested, SL1344 and 14028s, carry the ST64B prophage. See Table 1 for complete strain descriptions (B) Schematic representation of the ST64B phage genome integrated in the bacterial chromosome. Indicated with asterisks (*) are the relative positions of primers designed on sb27 and sb28 to amplify the excised form of phage DNA. (C) PCRs performed with sb27-sb28 primers in DNA template isolated from (lane 1) a colony of wild-type strain serovar Typhimurium SL1344; (lane 2) a colony of serovar Typhimurium SV1610 (dam); (lane 3) supernatant of the SL1344 culture; (lane 4) filtered supernatant of the SL1344 culture; (lane 5) supernatant of the SV1610 (dam) culture; (lane 6) filtered supernatant of the SV1610 (dam) culture; (lane 7) negative control with no DNA. As a control for the presence of chromosomal DNA, the same samples were used for PCR amplification with primers specific for the bacterial gene fimY (see Materials and Methods). Lane M, size markers. (D) ST64B phage particles seen in extracts prepared from the culture supernatant of the dam mutant. Bar, 50 nm.
Detection of ST64B viral particles and phage DNA in the culture supernatant of dam mutants.
The release of ST64B phage proteins to the extracellular medium by dam mutants suggested that the lack of Dam methylation could result in enhanced expression of ST64B genes. It was also possible that this alteration could be accompanied by an increase in the excision rate of this prophage. To test these hypotheses, a PCR assay was designed with primers complementary to sequences flanking the ST64B attP site (genes sb27 and sb28) (44). The primers were oriented in such a way that a PCR product is obtained only when the phage DNA has excised from the chromosome (Fig. 2B).
The presence of this excised form of phage DNA was assessed by PCR directly on bacteria (colony taken from plate) and nonfiltered and filtered culture supernatants (Fig. 2C). The amount of sb27-sb28 PCR product was prominent in samples prepared from colonies and nonfiltered culture supernatants of the dam mutant (Fig. 2C, top panel, lanes 5 and 6). Importantly, an sb27-sb28 PCR product of equal intensity was obtained from the filtered supernatants, in which we were unable to amplify a control bacterial gene, fimY (Fig. 2C, lower panel, lane 6). These results confirmed that the template of the sb27-sb28 PCR product detected in our assay corresponded to excised phage DNA and not to prophage DNA present in the extracellular medium as a result of bacterial lysis. Interestingly, we were able to detect the excised DNA form in culture supernatants of the wild-type strain SL1344 (Fig. 2C, top panel, lane 3), suggesting that the ST64B prophage may also excise spontaneously in this strain, albeit to a much lower extent than in the dam mutant. Taken together, these data demonstrated that release of ST64B proteins by dam mutants correlates with a higher excision rate of the ST64B prophage in these mutants.
Excision of the ST64B prophage is accompanied by formation of phage particles that remain defective for infectivity.
It has recently been shown that the ST64B prophage carried by serovar Typhimurium strain SL1344 harbors a frameshift mutation that causes a premature stop codon of the tail gene sb21 (23). This defect may prevent the correct expression of tail genes and hence the assembly of the tail that is required for phage infectivity. Unlike strains SL1344 and 14028s, the epidemic strain DT104 does not carry this sb21 frameshift mutation, which may explain the higher number of infective phage particles released by this strain (23). Other ST64B tail genes present a mosaic structure, carrying fragments with homology to virulence genes (46). This particular arrangement in the tail gene region has also been proposed as a defect impairing proper expression of tail genes.
To determine whether the ST64B phage particles released by dam mutants remain defective, culture supernatants of the dam mutant were used to infect recipient ST64B-cured strains in SL1344 and 14028s genetic backgrounds. Despite numerous attempts, no plaques of lysis were observed (data not shown). We also confirmed by PCR and sequencing the presence of the sb21 frameshift mutation in ST64B phage DNA collected from filtered culture supernatants of the dam mutant (data not shown). These data argued that, despite the increase in ST64B phage particles noted in the culture supernatant of dam mutants, the phages released by these mutants remain defective for infection. As a further evidence, transmission electron microscopy analysis revealed that the ST64B phage particles present in culture supernatant of dam mutants were tailless viral particles of icosahedrical shape and ∼45 nm in width (Fig. 2D). These phage particles were not seen in supernatants of the wild-type strain, probably due to the very low spontaneous induction rate of ST64B occurring in this strain (Fig. 2C). Altogether, these observations indicate that although the induction rate of the ST64B prophage is increased in dam mutants, the viral particles generated remain defective for infection of susceptible strains.
Excision rate of other Salmonella prophages in mutants defective in Dam methylase.
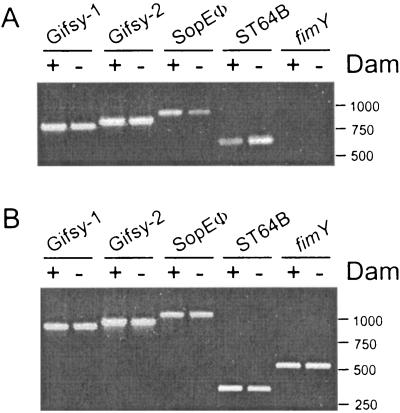
To test whether the lack of Dam methylation could also have consequences for the excision rate of other Salmonella prophages, the relative amount of DNA specific to the phages Gifsy-1, Gifsy-2, and SopEΦ was monitored by PCR in extracts prepared from culture supernatants of the wild-type dam+ strain SL1344 and its isogenic dam mutant SV1610 (Fig. 3A). Control PCRs were also performed on bacteria grown on plates (Fig. 3B). Unlike the case with ST64B, similar amounts of PCR product specific to the Gifsy-1 and Gifsy-2 phages were obtained from the culture supernatant extracts of strains carrying dam+ or dam alleles (Fig. 3A). On the other hand the level of SopEΦ-specific product was lower in the culture supernatant extract of the dam mutant compared to that of the wild-type strain (Fig. 3A). Importantly, no PCR product specific for a bacterial gene (fimY) was obtained in any of the culture supernatant extracts, confirming that the PCR products observed in these samples were amplified from DNA of phages present in the extracellular medium. As expected, the reactions performed on bacteria grown on plates did not show any difference in the amount of the respective PCR products. Taken together, these data demonstrate that among the prophages tested, ST64B is the only prophage whose excision is negatively modulated by Dam methylation.
FIG. 3.
Effect of the dam mutation on the excision rate of different Salmonella prophages. Shown is the PCR-mediated amplification of products specific to genes of the following prophages: Gifsy-1 (xis gene, product size 785 bp); Gifsy-2 (sodC1 gene, product size 856 bp); SopEΦ (panel A, sopE gene, product size 1,042 bp); and ST64B (sb27-sb28 product of size 673 bp; and panel B, sb49 gene product of size 335 bp). As a control, a product specific for a bacterial gene fimY (product size 529 bp), was included for comparison. (A) Samples prepared from culture supernatant; (B) samples prepared from bacteria grown on plates. The strains used were SL1344 (wild-type, dam+) and SV1610 (dam). The assay was repeated two times with identical results.
Dam methylation regulates the expression of genes present in the ST64B prophage.
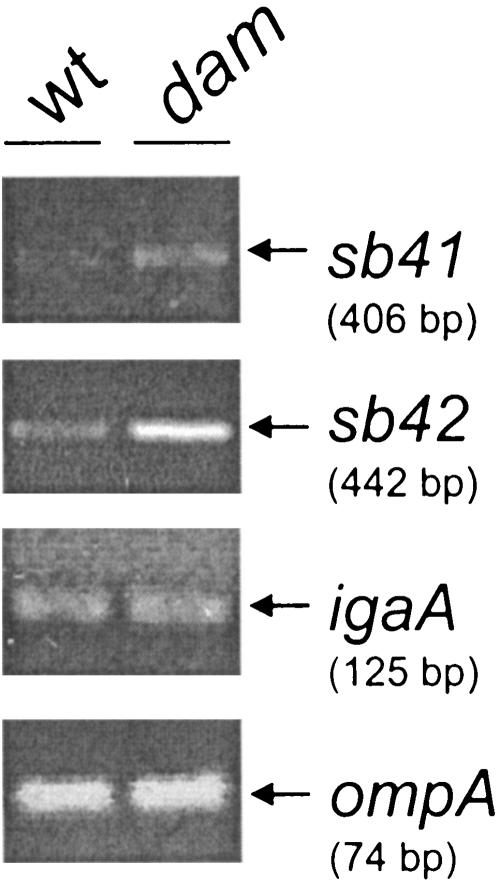
One of the biological roles of Dam methylation is to regulate gene expression. We considered the possibility that the increase in the excision of ST64B occurring in dam mutants could result from changes in the expression of ST64B genes promoting the induction of the prophage. Upon in silico analysis of the ST64B genome sequence, we noticed that the regulatory region of the putative operon encompassing the sb40-sb41-sb42-sb43-sb44-sb45 genes is highly enriched in GATC sites. In fact, three GATC sites exist in a stretch of 169 nucleotides located within the 298-nucleotide sb39-sb40 intergenic region. The annotation found in the databases assigns to the genes of this operon diverse functions, including a hypothetical phage antirepressor (Sb41), a replication protein (Sb42), a putative Dam methylase (Sb44), a putative crossover junction endodeoxyribounclease (Sb45), and proteins of unknown function (Sb40 and Sb43). sb41 and sb42 are clearly linked to functions involved in phage induction, so their expression level was assessed by RT-PCR. This assay revealed that the expression of sb41 and sb42 is higher in the dam mutant than in the wild-type strain (Fig. 4), indicating that the excision of ST64B is regulated negatively by Dam methylation at the level of gene expression.
FIG. 4.
Increased expression of the ST64B genes sb41 and sb42 in dam mutants. Total RNA was extracted from SL1344 (wild type) and SV1610 (dam) bacteria grown to exponential phase in LB medium at 37°C (final OD600 of ∼0.3) and further subjected to an RT-PCR with primers specific to the ST64B genes sb41 and sb42. These two genes encode functions related to phage induction. The expression of two bacterial genes, ompA and igaA, was also monitored for comparison. The size of each amplicon is indicated. The assay was repeated three times with similar results.
SOS response contributes to excision of the ST64B phage.
A deficiency of Dam methylase has been shown to cause induction of the SOS response in both Escherichia coli and S. enterica (49, 60). SOS induction has also been implicated in the stimulation of phage gene expression and the induction of the λ phage and other lambdoid phages (63). The ST64B prophage was catalogued as a member of the λ phage family based in its genome architecture (46). Thus, we analyzed whether the control of the excision rate of ST64B by Dam methylation depended on the induction of SOS.
A series of isogenic strains having a lexA (Ind−) mutation was constructed in both dam+ and dam genetic backgrounds. The presence of the lexA (Ind−) mutation in these strains was monitored with a plasmid-borne lacZ transcriptional fusion to the cea gene, which is activated in response to SOS induction (51). Since the combination of lexA (Ind−) and dam mutations is lethal (57), this particular construction was made in a mutS background. The experiment also included the challenge of all strains of the series with nalidixic acid, a quinolone antibiotic that induces SOS. Bacteria were grown in the absence and presence of the antibiotic for 4 h and culture supernatant extracts were processed for detection by PCR of the excised form of the ST64B phage DNA (sb27-sb28 amplicon, see Fig. 2C).
The excision of ST64B diminished in strains harboring the lexA (Ind−) mutation, regardless of whether they carried a dam+ or dam allele (Fig. 5). Interestingly, the excision of ST64B detected in the dam mutS lexA (Ind−) mutant was lower than that observed in either dam or dam mutS mutants (Fig. 5). This result showed that the SOS response might favor the induction of ST64B independently of the regulation exerted by Dam. The induction of SOS mediated by nalidixic acid led to a clear increase of the excision of ST64B in strains having a dam+ background (Fig. 5). Taken together, these data demonstrate that Dam methylation and the SOS response control the excision rate of the ST64B prophage.
FIG. 5.
SOS response augments the excision rate of the ST64B prophage. A series of isogenic strains defective for Dam methylase or induction of the SOS response were used to monitor the excision rate of ST64B. Bacteria were collected in logarithmic exponential phase after 4 h of growth in LB medium (final OD600 of ∼0.3). A culture in parallel of each of the strains was also incubated for the same period of time in the presence of 1 μg ml−1 nalidixic acid (NAL). No changes in the final OD600 were registered in untreated versus treated cultures. Shown is the PCR product specific to the excised form of the ST64B genome (sb27-sb28 amplicon) detected in samples of culture supernatants. This series of isogenic strains was constructed in the 14028s genetic background (see Table 1 for details). The assay was repeated three times with similar results.
ST64B prophage is not required for virulence in the murine typhoid model.
Prophages carried by many bacterial pathogens contain genes encoding virulence factors (reviewed in references 7 and 8). In the case of Salmonella, factors that alter antigenicity, effector proteins involved in invasion, and enzymes required for intracellular survival are known to be encoded by prophages (7, 8). Mmolawa et al. tentatively assigned a putative function of ST64B in virulence based in the presence in this phage of fragments with partial homology to sequences of the virulence genes sspH2, sopE, and orgA (46). Upon analysis of the ST64B ORFs, we realized that the sb49 and sb50 genes display no homology to sequences available in the databases and that they have a 48% and 45% G+C content, respectively, slightly lower than the G+C percentage of the ST64 genome, 51%, or, the average of the bacterial genome, 52%.
These features led us to first investigate the requirement for the sb49 and sb50 gene products in serovar Typhimurium virulence, so we constructed SL1344 isogenic strains defective for each of these genes. In parallel, a strain lacking the entire ST64B prophage was used. All these mutants behaved like the virulent wild-type strain in invasion and intracellular proliferation and survival assays performed in cultured macrophage, epithelial, and fibroblast cell lines (data not shown). When tested in the BALB/c mouse infection model, the Δsb49, Δsb50, Δsb49sb50, and Δ[ST64B] mutants were also able to compete with the wild-type virulent strain for colonization of target organs as liver or spleen (data not shown). Median lethal dose experiments showed that the Δ[ST64B] mutant penetrates the intestinal epithelium efficiently and produces systemic disease when administered by the oral route (data not shown). These observations demonstrate that none of the genes encoded in ST64B is required for serovar Typhimurium pathogenesis in the murine typhoid model.
Increased excision of the ST64B prophage does not contribute to the phenotypes of envelope instability and virulence attenuation of dam mutants.
Our proteomic analysis indicated that a large amount of ST64B proteins is released to the extracellular medium in dam mutants, an alteration that could be potentially related to envelope instability. This assumption takes into account the fact that some phage-encoded functions are known to have lytic activity against bacterial envelope components as the peptidoglycan (21). Thus, we tested whether expression of functions encoded in an S. enterica serovar Typhimurium prophage (ST64B, Fels-2, Gifsy-1, Gifsy-2, or SopEΦ) could contribute to the envelope instability displayed by dam mutants.
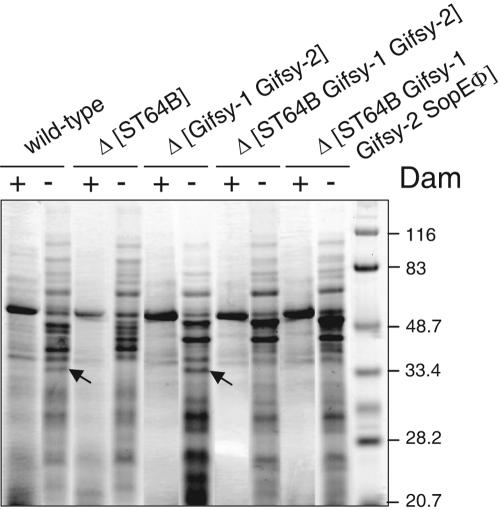
The envelope stability of wild-type strain SL1344, which does not contain the Fels-2 prophage, was compared to that of isogenic strains carrying diverse repertories of phages (cured for ST64B and/or phages Gifsy-1, Gifsy-2, and SopEΦ, strains MD1104, MA6247, MA7551, and MA7891; see Table 1). A second pair of isogenic strains carrying the dam mutation in addition were used in parallel (strains MD1202, MD1203, MD1201, and MD0200; see Table 1). The analysis of the extracellular protein extracts showed that all the strains constructed in a dam background release a large amount of proteins to the medium, irrespective of the number of phages deleted (Fig. 6). These observations discarded the possibility that a function(s) encoded by ST64B or any of the other Salmonella prophages carried by SL1344 (Gifsy-1, Gifsy-2, and SopEΦ) contributes to the cell envelope instability of the dam mutants.
FIG. 6.
Increased excision of ST64B does not contribute to the phenotype of envelope instability of dam mutants. Protein extracts were prepared from culture supernatants of the following isogenic strains: SL1344, wild type; SV1610, dam; MD1104, Δ[ST64B]; MD1202, Δ[ST64B] dam; MA6247, Δ[Gifsy-1 Gifsy-2]; MD1203, Δ[Gifsy-1 Gifsy-2] dam; MA7551, Δ[ST64B Gifsy-1 Gifsy-2]; MD1201, Δ[ST64B Gifsy-1 Gifsy-2] dam; MA7891, Δ[ST64B Gifsy-1 Gifsy-2 SopEΦ]; and MD0200, Δ[ST64B Gifsy-1 Gifsy-2 SopEΦ] dam. Bacteria were grown overnight in LB medium at 37°C in static conditions. The amount loaded per lane corresponded to the same number of bacteria (2 × 109). The sizes of the prestained molecular weight markers are indicated in kilodaltons. Arrows highlight a prominent protein that is only observed in dam mutants carrying the ST64B prophage. The assay was repeated three times with similar results.
These phage-cured strains were also used in competition experiments in BALB/c mice. When combined with the dam mutant, the isogenic mutant lacking in addition the four prophages (ST64B, Gifsy-1, Gifsy-2, and SopEΦ) displayed a higher level of attenuation of virulence (competition indexes of 0.059 and 0.027 in liver and spleen, respectively) (Table 4). This phenotype could be caused by the absence of virulence genes located in these prophages, namely sodC1 and gtgE in Gifsy-2 (24, 36). To verify this, a new dam mutant was constructed lacking the four prophages (ST64B, Gifsy-1, Gifsy-2, and SopEΦ) but carrying sodC1 and gtgE integrated in the chromosome. This strain displayed the same level of virulence attenuation as the dam mutant (Table 4), confirming that the excision of these prophages does not alter the capacity of the bacteria to cause disease. Further evidence supporting this conclusion was obtained in the case of ST64B when testing a derivative dam mutant lacking only the ST64B gene encoding the putative excisionase (xis). As expected, no sb27-sb28 PCR product was detected in culture supernatants of this specific mutant (data not shown). The competition experiments demonstrated the dam ST64B Δxis bacteria display the same level of attenuation as the dam mutant (Table 4).
TABLE 4.
Virulence properties in BALB/c mice of serovar Typhimurium dam mutants cured of prophages or with a defect in excision
| Strain 1a | Strain 2a | Competition indexb
|
|
|---|---|---|---|
| Liver | Spleen | ||
| dam | Wild type | 0.0012 ± 0.001 | 0.0009 ± 0.0002 |
| dam Δ[Gifsy-1, Gifsy-2, SopEΦ, ST64B] | dam | 0.059 ± 0.013 | 0.027 ± 0.0003 |
| dam Δ[Gifsy-1, Gifsy-2, SopEΦ, ST64B] sodC1+gtgE+ | dam | 0.97 ± 0.15 | 1.09 ± 0.54 |
| dam [ST64B Δxis] | dam | 1.79 ± 0.48 | 1.17 ± 0.46 |
All these strains are isogenic to wild-type strain SL1344 (see Table 1 for details).
Values were calculated upon intraperitoneal challenge of three BALB/C mice per mixed infection. Organ homogenates were prepared at 48 to 72 hours postinfection.
DISCUSSION
This work was initiated with a proteomic analysis designed to identify proteins released to the extracellular medium specifically by dam mutants. The aim of this approach was to obtain insights on the basis of the envelope instability displayed by these mutants. The study provided evidence for a role of Dam methylation in negatively modulating the level of the TraF protein, which controls the mobilization of conjugative plasmids. This finding agrees with the inhibition of the transfer of the F plasmid and the virulence plasmid pSLT by Dam methylation uncovered by genetic approaches (9, 61).
We also identified StdA, the major subunit of Std fimbriae, as another protein overproduced by dam mutants. The synthesis of Std fimbriae has been shown to be silenced by serovar Typhimurium during static growth in LB medium whereas it is up-regulated upon infection of bovine ligated loops (39). Interestingly, two GATC sites are present just upstream of stdA, the first gene of the stdABC fimbrial operon, suggesting that, like the pap and pef operons (35, 48), the methylation state of these GATC sites may play a relevant role in regulation. We have recently obtained data with bacteria grown in laboratory conditions confirming that the regulation of Dam methylation exerted on the stdABC operon takes place primarily at the level of gene expression (10). Why Dam methylation seems to be specifically regulating the expression of fimbrial operons, which are fragments of DNA acquired recently during the differentiation of distinct genera, species, and serovars (19), is an open question that deserves further investigation.
As in the case of the fimbrial operons, this study has provided evidence for regulation mediated by Dam on genes with a very narrow distribution among bacteria as it is the prophage ST64B, present exclusively in a few strains of S. enterica serovar Typhimurium (23). This case therefore constitutes an example of regulation by Dam methylation of DNA fragments acquired very recently, even after the diversification of serovar Typhimurium.
ST64B has been shown to be a defective prophage (46). Our infectivity assays revealed that the ST64B viral particles visualized in the culture supernatant of dam mutants remain noninfectious, supporting the idea that the tail genes of ST64B might not be properly expressed (23, 46). Furthermore, the infection assays performed in mice demonstrated that the lack of ST64B has no effect on virulence, discounting the suggestion made by others that this prophage could carry virulence genes (46). The possibility of a potential role of ST64B in colonization of other hosts by Salmonella spp. cannot be formally discarded. Such a contribution of prophage-encoded genes to the colonization of specific hosts has recently been shown for serovar Typhimurium genes located in the Gifsy-1 and Gifsy-2 prophages, which are required for virulence in calves but dispensable in chicken colonization (47).
The comparative analysis of the excision rate of Salmonella prophages in the dam mutant revealed that ST64B is the only prophage that apparently increase its excision rate in comparison with the other prophages tested (Gifsy-1, Gifsy-2, and SopEΦ). Interestingly, the excision of SopEΦ, which like ST64B is carried exclusively by certain serovar Typhimurium strains, also seems to be controlled by Dam methylation, although in this case the regulation is positive. Altogether, these results were fully compatible with the proteomic analysis, designed to identify exclusively those proteins present in higher amounts in the extracts of the dam mutant, as it was the case for the ST64B phage proteins. It is hence possible that proteins from the other phages were present in the two-dimensional gels but were not selected for mass spectrometry analysis due to their relatively equivalent (Gifsy-1 and Gifsy-2) or probably lower amounts (SopEΦ) in the samples of the dam mutant.
The changes registered at the level of ST64B proteins were consistent with the up-regulation of ST64B genes located in the sb40-sb45 operon, supposedly involved in phage induction. That was the case for sb41, encoding a putative antirepressor protein, and sb42, encoding a phage replication protein. The alteration in the expression of these genes fits with the enrichment of GATC sites observed in the regulatory region of the sb40-sb45 operon operon (three sites in a 169-nucleotide stretch mapping in the sb39-sb40 intergenic region). The regulation of the sb41 and sb42 genes by Dam methylation constitutes another example of regulation of a phage gene expression by Dam as the previously reported for the mom gene of bacteriophage Mu. mom, which encodes a protein linked to DNA modification, is regulated by Dam methylation, although in this case its expression decreases in dam mutants (29). A factor modulating mom expression is the host OxyR regulatory protein, which has preference for binding to the hemimethylated form of the mom promoter (31). Given that a common trait in the mode of regulation of gene expression by Dam is to modulate the binding of regulatory proteins to the promoter region, it is tempting to assume that a similar mechanism may apply for the case of the sb40-sb45 ST64B operon. Further work is undoubtedly required to sustain this hypothesis.
The sb40-sb45 operon contains a gene, sb44, encoding a putative Dam methylase (46). The presence of a gene encoding a Dam methylase is a feature shared by many phages of bacteria and even archaea (1-3, 13, 30, 41, 43, 50, 54). The exact function of these phage Dam methylases remains unknown in most cases. An exception is that of the P1-encoded Dam methylase, which methylates the pac site to be recognized by endonucleases involved in the phage packaging process (58). It is also known that some bacteriophage genomes contain a remarkably small number of GATC sites (6, 44, 59), raising the possibility that phage-encoded Dam methylases could exert their function on the host bacterial genome to modulate, for example, integration and/or excision of the phage. This postulate seems however unlikely in the cases of the P1 and ST64B phages, which contain a normal number of GATC sites (43). Nonetheless, the probable production of Sb44 in dam mutants should be investigated further to determine whether this phenomenon has consequences in the bacterial physiology or the induction rate of the ST64B prophage.
Previous studies have shown that the SOS response contributes to the excision of lysogenic phages as lambda and other lambdoid phages in E. coli, the CTXΦ phage in Vibrio cholerae, and P22 in S. enterica (63). These observations led us to investigate whether the enhanced excision of ST64B observed in dam mutants could be merely a consequence of the moderate induction of SOS reported for these mutants in both E. coli and S. enterica (49, 60). Our results suggest that SOS and Dam methylation modulate the excision of the ST64B prophage. The SOS response also seems to be responsible for the basal excision of ST64B in wild-type bacteria since it is effectively abrogated by a lexA (Ind−) mutation. This observation suggests that, in wild-type bacteria, SOS counteracts the negative regulation exerted by Dam methylation on the expression of ST64B genes. The fact that the excision of ST64B diminishes in dam mutants carrying the lexA (Ind−) mutation may simply indicate that the induction of SOS occurring in dam mutants exacerbates the excision resulting from the loss of repression of the sb40-sb45 operon.
We also sought to determine whether the enhanced excision of ST64B could constitute per se an alteration contributing to the phenotypes of envelope instability or virulence attenuation. The experiments performed with an ST64B excisionase-defective mutant and phage-cured strains demonstrated that no relationship exists between these events. This conclusion is further supported by other indirect evidence, such as the envelope instability displayed by dam mutants of serovar Typhimurium strain LT2 (53), which does not carry the ST64B prophage. The data obtained in parallel with strains cured of the other prophages analyzed, Gifsy-1, Gifsy-2, and SopEΦ, also discarded the possibility that a function expressed in any of these prophages could elicit damage to the cell envelope.
In summary, this study reports a novel phenotype for S. enterica serovar Typhimurium dam mutants consisting of exacerbated excision of the defective ST64B prophage. A future challenge will be to uncover the identity of the regulatory proteins and the underlying mechanisms that control the rate at which this prophage excises.
Acknowledgments
We thank L. Bossi and J. Casadesús for helpful discussions and communicating unpublished data. We also thank R. Rotger for sending strains.
This work was supported by a grant from the Spanish Ministry of Education and Science (BIO2004-03455-C02-01). A. Alonso was supported by a postdoctoral fellowship from the Comunidad de Madrid and M. G. Pucciarelli is an Investigator of the “Ramón y Cajal” program from the Spanish Ministry of Education and Science.
REFERENCES
- 1.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, H. P., U. Ziese, and W. Zillig. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409-416. [DOI] [PubMed] [Google Scholar]
- 3.Baranyi, U., R. Klein, W. Lubitz, D. H. Kruger, and A. Witte. 2000. The archaeal halophilic virus-encoded Dam-like methyltransferase M. phiCh1-I methylates adenine residues and complements dam mutants in the low salt environment of Escherichia coli. Mol. Microbiol. 35:1168-1179. [DOI] [PubMed] [Google Scholar]
- 4.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 5.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaisdell, B. E., A. M. Campbell, and S. Karlin. 1996. Similarities and dissimilarities of phage genomes. Proc. Natl. Acad. Sci. USA 93:5854-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., and H. Brussow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 8.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camacho, E. M., and J. Casadesús. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 10.Camacho, E. M., A. Serna, M. G. Pucciarelli, F. García-del Portillo, and J. Casadesús. Regulation of the Salmonella enterica fimbrial operon stdABC by DNA denine methylation. Unpublished data.
- 11.Cano, D. A., G. Domínguez-Bernal, A. Tierrez, F. García-Del Portillo, and J. Casadesus. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., D. B. Paulsen, D. W. Scruggs, M. M. Banes, B. Y. Reeks, and M. L. Lawrence. 2003. Alteration of DNA adenine methylase (Dam) activity in Pasteurella multocida causes increased spontaneous mutation frequency and attenuation in mice. Microbiology 149:2283-2290. [DOI] [PubMed] [Google Scholar]
- 13.Citron, M., M. Velleman, and H. Schuster. 1989. Three additional operators, Op21, Op68, and Op88, of bacteriophage P1. Evidence for control of the P1 Dam methylase by Op68. J. Biol. Chem. 264:3611-3617. [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domínguez-Bernal, G., M. G. Pucciarelli, F. Ramos-Morales, M. García-Quintanilla, D. A. Cano, J. Casadesús, and F. García-del Portillo. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53:1437-1449. [DOI] [PubMed] [Google Scholar]
- 16.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine 21:3249-3258. [DOI] [PubMed] [Google Scholar]
- 17.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2001. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect. Immun. 69:7950-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants prevent colonization of newly hatched chickens by homologous and heterologous serovars. Int. J. Food Microbiol. 80:153-159. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 21.Fastrez, J. 1996. Phage lysozymes. EXS (Basel) 75:35-64. [DOI] [PubMed] [Google Scholar]
- 22.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Figueroa-Bossi, N., and L. Bossi. 2004. Resuscitation of a defective prophage in Salmonella cocultures. J. Bacteriol. 186:4038-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-272. [DOI] [PubMed] [Google Scholar]
- 25.Galán, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 26.García-del Portillo, F., M. G. Pucciarelli, and J. Casadesús. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. USA 96:11578-11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haase, J., and E. Lanka. 1997. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J. Bacteriol. 179:5728-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardt, W. D., H. Urlaub, and J. E. Galán. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hattman, S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84:367-388. [DOI] [PubMed] [Google Scholar]
- 30.Hattman, S., and E. G. Malygin. 2004. Bacteriophage T2Dam and T4Dam DNA-[N6-adenine]-methyltransferases. Prog. Nucleic Acid Res. Mol. Biol. 77:67-126. [DOI] [PubMed] [Google Scholar]
- 31.Hattman, S., and W. Sun. 1997. Escherichia coli OxyR modulation of bacteriophage Mu mom expression in dam+ cells can be attributed to its ability to bind hemimethylated Pmom promoter DNA. Nucleic Acids Res. 25:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heithoff, D. M., E. Y. Enioutina, R. A. Daynes, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. Salmonella DNA adenine methylase mutants confer cross-protective immunity. Infect. Immun. 69:6725-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 1999. An essential role for DNA adenine methylation in bacterial virulence. Science 284:967-970. [DOI] [PubMed] [Google Scholar]
- 34.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 99:16470-16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernday, A. D., B. A. Braaten, and D. A. Low. 2003. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell 12:947-957. [DOI] [PubMed] [Google Scholar]
- 36.Ho, T. D., N. Figueroa-Bossi, M. Wang, S. Uzzau, L. Bossi, and J. M. Slauch. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 38.Honma, Y., R. E. Fernandez, and A. T. Maurelli. 2004. A DNA adenine methylase mutant of Shigella flexneri shows no significant attenuation of virulence. Microbiology 150:1073-1078. [DOI] [PubMed] [Google Scholar]
- 39.Humphries, A. D., M. Raffatellu, S. Winter, E. H. Weening, R. A. Kingsley, R. Droleskey, S. Zhang, J. Figueiredo, S. Khare, J. Nunes, L. G. Adams, R. M. Tsolis, and A. J. Baumler. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 40.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapfhammer, D., J. Blass, S. Evers, and J. Reidl. 2002. Vibrio cholerae phage K139: complete genome sequence and comparative genomics of related phages. J. Bacteriol. 184:6592-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobner-Olesen, A., O. Skovgaard, and M. G. Marinus. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154-160. [DOI] [PubMed] [Google Scholar]
- 43.Lobocka, M. B., D. J. Rose, G. Plunkett 3rd, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClelland, M. 1984. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J. Mol. Evol. 21:317-322. [DOI] [PubMed] [Google Scholar]
- 45.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 46.Mmolawa, P. T., H. Schmieger, and M. W. Heuzenroeder. 2003. Bacteriophage ST64B, a genetic mosaic of genes from diverse sources isolated from Salmonella enterica serovar Typhimurium DT64. J. Bacteriol. 185:6481-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson, B., and D. Low. 2000. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol. Microbiol. 35:728-742. [DOI] [PubMed] [Google Scholar]
- 49.Peterson, K. R., K. F. Wertman, D. W. Mount, and M. G. Marinus. 1985. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol. Gen. Genet. 201:14-19. [DOI] [PubMed] [Google Scholar]
- 50.Piekarowicz, A., and J. Bujnicki. 1999. Cloning of the Dam methyltransferase gene from Haemophilus influenzae bacteriophage HP1. Acta Microbiol. Pol. 48:123-129. [PubMed] [Google Scholar]
- 51.Prieto, A. I., F. Ramos-Morales, and J. Casadesus. 2004.Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pucciarelli, M. G., and F. García-Del Portillo. 2003. Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48:573-585. [DOI] [PubMed] [Google Scholar]
- 53.Pucciarelli, M. G., A. I. Prieto, J. Casadesús, and F. García-del Portillo. 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148:1171-1182. [DOI] [PubMed] [Google Scholar]
- 54.Radlinska, M., and J. M. Bujnicki. 2001. Cloning of enterohemorrhagic Escherichia coli phage VT-2 Dam methyltransferase. Acta Microbiol. Pol. 50:161-167. [PubMed] [Google Scholar]
- 55.Robertson, G. T., A. Reisenauer, R. Wright, R. B. Jensen, A. Jensen, L. Shapiro, and R. M. Roop, 2nd. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 57.Stambuk, S., and M. Radman. 1998. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics 150:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sternberg, N., and J. Coulby. 1990. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc. Natl. Acad. Sci. USA 87:8070-8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Styriak, I., P. Pristas, and P. Javorsky. 2000. Lack of GATC sites in the genome of Streptococcus bovis bacteriophage F4. Res. Microbiol. 151:285-289. [DOI] [PubMed] [Google Scholar]
- 60.Torreblanca, J., and J. Casadesús. 1996. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 144:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torreblanca, J., S. Marques, and J. Casadesús. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 152:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tucker, C. P., and M. W. Heuzenroeder. 2004. ST64B is a defective bacteriophage in Salmonella enterica serovar Typhimurium DT64 that encodes a functional immunity region capable of mediating phage-type conversion. Int. J. Med. Microbiol. 294:59-63. [DOI] [PubMed] [Google Scholar]
- 63.Waldor, M. K., and D. I. Friedman. 2005. Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8:459-465. [DOI] [PubMed] [Google Scholar]
- 64.Watson, M. E., Jr., J. Jarisch, and A. L. Smith. 2004. Inactivation of deoxyadenosine methyltransferase (dam) attenuates Haemophilus influenzae virulence. Mol. Microbiol. 53:651-664. [DOI] [PubMed] [Google Scholar]
- 65.Yeh, K. S., T. H. Chen, C. W. Liao, C. S. Chang, and H. C. Lo. 2002. PCR amplification of the Salmonella typhimurium fimY gene sequence to detect the Salmonella species. Int. J. Food Microbiol. 78:227-234. [DOI] [PubMed] [Google Scholar]