Abstract
We conducted a comprehensive metabolic phenotyping of potato (Solanum tuberosum L. cv Desiree) tuber tissue that had been modified either by transgenesis or exposure to different environmental conditions using a recently developed gas chromatography-mass spectrometry profiling protocol. Applying this technique, we were able to identify and quantify the major constituent metabolites of the potato tuber within a single chromatographic run. The plant systems that we selected to profile were tuber discs incubated in varying concentrations of fructose, sucrose, and mannitol and transgenic plants impaired in their starch biosynthesis. The resultant profiles were then compared, first at the level of individual metabolites and then using the statistical tools hierarchical cluster analysis and principal component analysis. These tools allowed us to assign clusters to the individual plant systems and to determine relative distances between these clusters; furthermore, analyzing the loadings of these analyses enabled identification of the most important metabolites in the definition of these clusters. The metabolic profiles of the sugar-fed discs were dramatically different from the wild-type steady-state values. When these profiles were compared with one another and also with those we assessed in previous studies, however, we were able to evaluate potential phenocopies. These comparisons highlight the importance of such an approach in the functional and qualitative assessment of diverse systems to gain insights into important mediators of metabolism.
Recent years have seen rapid advances in the application of efficient tools to create and characterize genetic diversity both within plants and other biological systems. The tandem development of transgenic knockout populations, transposon insertions, chemical gene machines, and the genotyping of single nucleotide polymorphisms within large populations have paved the way to a far more substantial base of genetic diversity than imagined a few years ago (Aarts et al., 1993; Schaefer and Zryd, 1997; Strepp et al., 1998; Cho et al., 1999; Zu et al., 1999). That these developments have occurred in parallel with both the elucidation of complete genomes of several organisms and the rapid development of multiparallel technologies to describe properties of the biological systems (for review, see Celis et al., 2000) has provided the driving force behind many genomics initiatives. The most visible of these technologies is expression profiling (Lockhart et al., 1996; Ruan et al., 1998; Terryn et al., 1999; Aharoni et al., 2000; Richmond and Somerville, 2000); however, techniques for describing the protein (Shevchenko et al., 1996; Santoni et al., 1998; Chang et al., 2000) and metabolite complement (Duez et al., 1996; Matsumoto and Kuhara, 1996; Fiehn et al., 2000; Roessner et al., 2001) of the cell are now being widely developed.
Despite these recent advances, much research effort in the plant field is still focused on the phenotyping of the available genetic diversity on simple traits. In plants, the most common phenotypic screens are based on conditional lethality (for example, see Springer et al., 1995; Chekanova et al., 2000; Kampranis et al., 2000), fertility (for example, see Aarts et al., 1993; Lang et al., 1994), or an easily identifiable phenotype such as dwarfism or abnormal leaf development (for example, see Vollbrecht et al., 1991; Pepper et al., 1994; Bennett et al., 1996; Soppe et al., 1999; Hanzawa et al., 2000; Ramachandran et al., 2000) and mutants identified by biochemical phenotyping still represent a minority (for example, see Gibson et al., 1994; Dörmann et al., 1999). Although this approach has undoubtedly been a success in the identification of developmental mutants, and the consequent functional assignment of the respective genes, it is clear that many genes do not play a role in the determination of the visible phenotype of an organism. It has recently been estimated that up to 85% of genes present in yeast are not required for survival and only a few of these are altered in the chemical processes involved in energy production or growth (Cornish-Bowden and Cardenas, 2001).
It seems likely that plants will contain a similar proportion of “silent genes” that therefore would be overlooked in the type of screen described above. Countless studies in which plant enzyme activities have been altered by mutation or transgenesis without a resultant change in visible phenotype back this up. For this reason, we recently developed a method to allow phenotyping at the level of the metabolite capable of routinely identifying and quantifying the level of the major constituent metabolites within the potato (Solanum tuberosum L. cv Desiree) tuber (Roessner et al., 2000). In a first approach, we evaluated whether this protocol, in combination with bioinformatic techniques based on standard statistical methods, was capable of distinguishing systems that were genetically or environmentally modified (Roessner et al., 2001). Here, the metabolic phenotypes of a further three genotypes, this time altered in a different metabolic pathway—that of starch synthesis, and a further three environmentally altered potato tuber systems are discussed to demonstrate the general applicability of this approach. The resultant metabolic complements were then compared with each other and to those previously determined using hierarchical cluster analysis (HCA) and principal component analysis (PCA). The primary aims of this work were 2-fold; on the one hand, to perform a more detailed characterization of the perturbed tuber systems in the hope of gaining a fuller understanding of the interactions between their component pathways, proteins, and metabolites; and on the other hand, we wanted to determine the clustering patterns of the metabolic complements of tubers exhibiting genetic modifications in different metabolic pathways. In particular, we proposed to evaluate possible phenocopies by assessing the similarities and differences between these samples and samples that were metabolically perturbed by incubation of various concentrations of sugars.
RESULTS
Environmental Perturbation of Wild-Type Tuber Discs
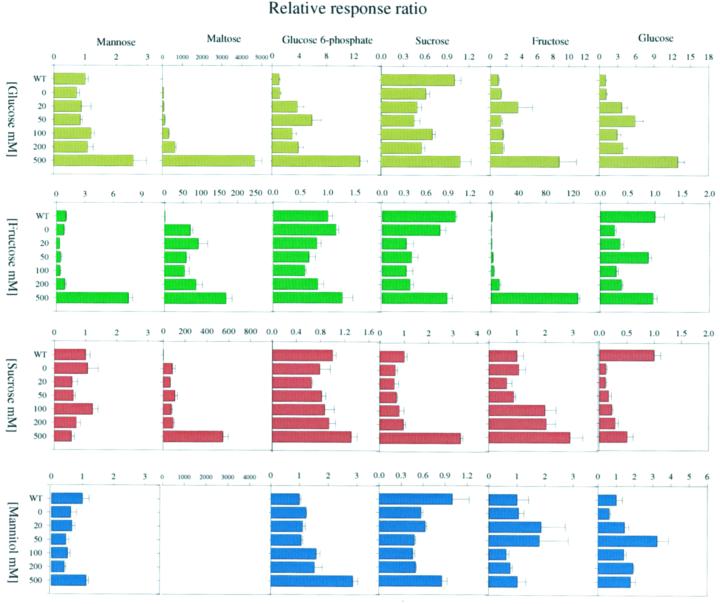
In our previous study, we characterized the metabolic changes following short-term incubation of potato tuber parenchyma tissue in various concentrations of Glc (Roessner et al., 2001). Here, we present additional data from parallel incubations of tuber tissue in varying concentrations of Suc, Fru, and mannitol. Discs were cut directly from developing tubers of healthy 10-week-old wild-type potato plants and incubated for 2 h in buffered medium {10 mm MES [2-(N-morpholino)-ethanesulfonic acid]-KOH, pH 6.5} containing 20, 50, 100, 200, and 500 mm of the appropriate sugar. The metabolite complement of these discs was then assessed using a recently established gas chromatography (GC)-mass spectrometry (MS) protocol (Roessner et al., 2001). In addition, discs that were incubated in buffer alone were analyzed to evaluate the number of changes that were due merely to this treatment. From the resultant profiles, it became apparent that significant changes occurred only in the discs incubated in 100, 200, or 500 mm of the sugars and only slight changes were observed in those incubated in 20 or 50 mm sugar. We always observed a striking increase in the cellular level of the fed sugar; however, there were other dramatic changes in metabolism, some of which were comparable with those reported in a similar, smaller experiment carried out by Geiger et al. (1998), but others are novel to this study. The most interesting of these changes are highlighted in Figure 1; in addition, the full data set can be viewed on our web page (http://www.mpimp-golm.mpg.de/willmitzer/ metabolic-profiling-e.html).
Figure 1.
Primary changes in metabolite levels following 2-h incubation in Glc, Fru, Suc, or mannitol. Metabolites were determined in discs from developing potato tubers of wild-type plants incubated in Glc (dark yellow), Fru (dark green), Suc (dark red), or mannitol (dark blue). Data are normalized to the mean response calculated for the wild-type steady-state levels of each measure batch. (To allow comparison between measure batches, individual wild-type values were normalized in the same way.) Values presented are the mean ± se of four independent determinants.
From the many changes determined following these incubations, the ones of particular interest were increases in the Fru and hexose-P pools with only a mild parallel increase in Suc following incubation in Glc (Roessner et al., 2001; data presented here for ease of comparison). Also, following this treatment, the levels of Man and ascorbate increased (Fig. 1), whereas the levels of amino acids did not. A similar pattern of changes was observed in tuber discs fed with varying concentrations of Fru. Changes observed on incubation in buffer alone were minor. The level of Glc (although lower than the wild-type steady-state level) tended to increase with increasing Fru concentration, whereas the level of Suc showed no clear trend under the different experimental conditions. Although similarities were also observed in the Man and maltose levels of discs incubated in either Fru or Glc, there was no clear trend in the level of Glc 6-phosphate and the total amino acid content actually decreased on incubation with higher concentrations of Fru. In contrast to the hexose-fed samples, Suc feeding led to very few changes in the metabolite profiles of the discs despite the fact that it resulted in significant increases in the levels of Glc and a marked (up to 3-fold) increase in the level of Fru. The notable exceptions to this statement are the 2-fold increase in ascorbate and total amino acids and also an increase in maltose in discs incubated in 500 mm Suc. Because there were very few changes in the metabolic profiles following incubations in mannitol, the majority of the above changes were most probably a direct consequence of the sugars themselves or metabolic products thereof rather than a general osmotic response. A notable exception to this was the pattern of fluctuation in Suc levels following incubation in various concentrations of Glc, Fru, and mannitol; although these fluctuations were relatively minor, they were very similar indicating some influence of osmotic factors on metabolism.
HCA and PCA of the Metabolic Complements of the Incubated Samples
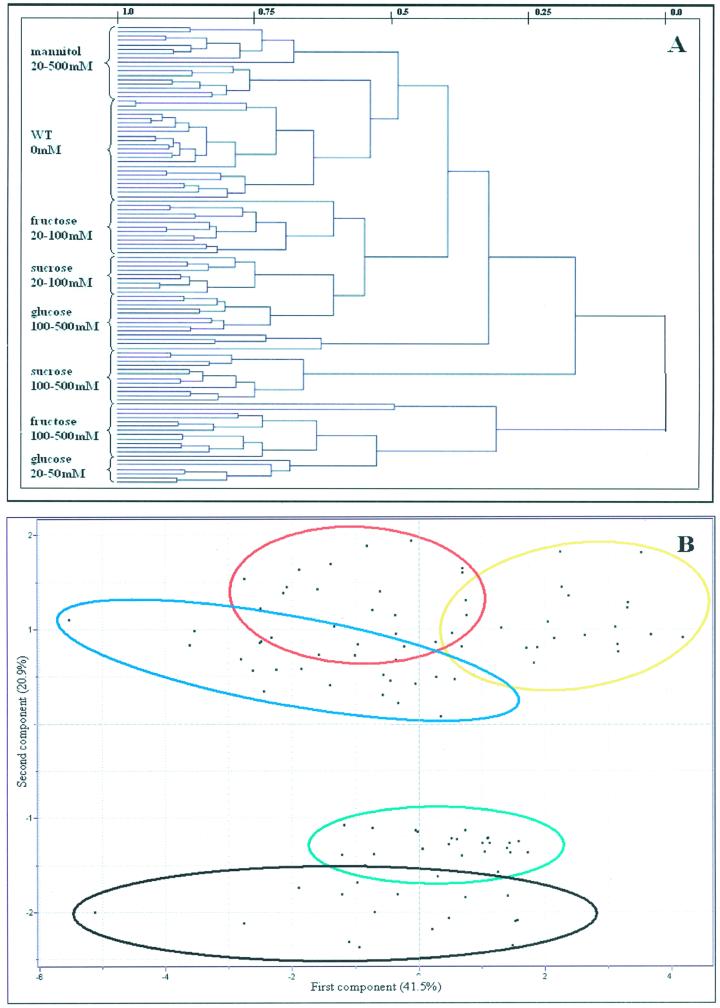
From the size of the data set obtained, it is clear that simple point-by-point analysis represents a daunting task. For this reason, we decided to also apply contemporary bioinformatic tools to our data set. Given that there is a fair degree of natural variation in metabolite levels (exemplified by the relative values of discs incubated in buffer alone; Fig. 1), we chose to plot all individual chromatograms, rather than the mean values presented earlier. A further advantage of taking this approach is that it means we are able to assess if individual discs incubated in the same sugar and/or concentration exhibited similar behavior with respect to their total metabolic profile. Applying cluster analysis to the full data set obtained following GC-MS analysis of the above samples revealed interesting results. HCA showed that the tuber discs incubated in buffer alone had the most similar metabolite complement to the steady-state wild-type levels (Fig. 2A), suggesting that the incubation alone had relatively minor implications for metabolism. Furthermore, all mannitol-fed samples formed a distinct cluster that was more similar to the samples incubated in buffer alone than that of any of the other fed samples. This finding provides further support to our earlier claim that the response of metabolism to the exogenous supply of sugars was largely not determined by osmotic effects. Other clusters that formed, with increasing distance from the wild-type steady state, are samples incubated in low concentrations of Fru, in low concentrations of Suc, and finally in high concentrations of Glc and Suc. It is surprising that samples incubated in high concentrations of Fru and low concentrations of Glc represent individual clusters. In summary, however, these data demonstrate the difficulties inherent in resolving a large number of similar samples by a hierarchical approach. Taking a second, complementary approach—that of PCA—similar trends were revealed, although results from the two approaches were not in absolute agreement. Using PCA, the samples incubated in buffer alone coclustered with the wild-type steady-state samples but once again the mannitol-fed samples form a discrete cluster not too distant from this cluster. Although discs incubated at a defined concentration of a sugar essentially coclustered, they did not form discrete clusters from the other incubations in the respective sugar. The Glc-, Fru-, and Suc-fed samples, however, were distinct from the wild-type and mannitol clusters and formed essentially discrete clusters (represented by the elipses in Fig. 2B), with only minor overlaps.
Figure 2.
A, Dendogram obtained following HCA of the metabolic profiles of the analyzed environmentally modified systems. Wherever possible, individual branches are grouped in brackets for ease of reading. B, PCA of the metabolite profiles of the analyzed environmentally modified systems. Samples representing wild-type tissue incubated in various concentrations of Glc (red circle), Fru (blue circle), Suc (yellow circle), and mannitol (green circle) are marked as described for ease of comparison. PCA vectors 1 and 2 were chosen for best visualization of differences between experimental treatments and include 62.4% of the information derived from metabolic variances.
Loadings, which define the most important components with respect to the clustering behaviors of these samples, revealed that maltose, Fru, and Glc were major components of these analyses. This is somewhat predictable because they exhibited the largest changes following treatment with Glc, Fru, Suc, and mannitol; however, Asn, Trp, Ala, Tyr, Lys, glycerol, and Arg also contributed significantly to the cluster formation (data not shown). Moreover, the PCA result was essentially the same when the fed compounds were excluded from the cluster analysis, demonstrating that the clusters were not formed exclusively on the increase in the introduced metabolite and thus allowing us confidence in interpreting the data with respect to general changes in metabolism (data not shown).
Metabolic Profiles of Transgenic Plants Impaired in Starch Synthesis
Our previous study also concentrated on the analysis of the metabolic complements of three different transgenics altered in their Suc mobilization (Suc phosphorylase expressers and plants engineered to have a highly increased invertase activity both in combination with, and independently of, glucokinase activity; Roessner et al., 2001). Here, we decided to extend this study to encompass three further transgenic potatoes, this time inhibited in starch synthesis (ADP glucose pyrophosphorylase [AGPase], Müller-Röber et al., 1992; plastidial PGM, Tauberger et al., 2000, Fernie et al., 2001b; and cytosolic phosphoglucomutase [PGM], Fernie et al., 2001c). These transgenics have been preliminarily characterized at the metabolic level where we previously postulated that they phenocopied one another. However, these previous studies were limited in the scope of metabolites determined; therefore, we decided to perform a more comprehensive analysis of the metabolite complement of these lines. In an initial experiment, the various transgenic plants were grown alongside one another under identical greenhouse conditions and samples were harvested from developing tubers. We chose the lines AGP-85 and AGP-93 (Müller-Röber et al., 1992); cPGM-29, cPGM-44, and c-PGM 66 (Fernie et al., 2001c); and pPGM-5, pPGM-9, and pPGM-60 (Tauberger et al., 2000; Fernie et al., 2001b) for this study because the primary metabolic changes in these lines are well documented and are characteristic of those found on expression of the respective transgenes.
We confirmed that these lines had similar changes in the introduced enzyme activity and in the major storage carbohydrate pools as previously reported and were as such suitable for further experimentation. Following this, we extracted six replicate samples from the same plants as used for the primary characterization and separated and characterized the detectable hydrophilic metabolite complement using GC-MS. Due to the large sample size of this experiment, containing eight transgenic lines and therefore a total of 48 independent transgenic tuber samples, we extracted a separate set of wild types per each set of transgenics, despite the fact that all plants were grown under identical conditions, to allow us independent references for each individual machine run. Results from this analysis are presented in Table I. The data set is comprised of 49 metabolites defined with respect to their chemical nature including sugars, sugar alcohols, amino acids, organic acids, and several miscellaneous compounds. The majority was found to alter within the transgenic lines. From perusal of the table, it becomes apparent that the plastidial PGM and AGPase lines exhibit similar changes in metabolite pool sizes, whereas patterns of change in the poolsizes of the cytosolic lines are less clear (Table I). AGPase and pPGM lines are characterized by dramatic reductions in many amino acids, most notably Ala, Arg, Asp, Lys, Phe, Ser, Trp, and Tyr and also in organic acids exemplified by decreases in isocitrate, oxalate, and shikimate in both transgenic plants. Furthermore, both systems had elevated levels of hexose-phosphates. Despite these similarities, marked differences also occurred between these transgenic lines—for example, only AGPase lines were characterized by large increases in mannitol and Man and only pPGM line 60 exhibited increases in Glc and malate. In contrast, the majority of the changes in metabolite levels observed in the cPGM plants are quite different to those observed in the transgenic lines studied here. There is no clear trend in the levels of many metabolites because many amino acids decrease and many phosphorylated intermediates increase in the lines cPGM-29 and cPGM-44, whereas the same amino acids increase in line cPGM-66, which is furthermore characterized by a decrease in the level of Glc-6-phosphate.
Table I.
Comparison of metabolic levels in wild-type developing potato tubers with those in tubers of transgenic potato plants
| WT | cPGM29 | cPGM44 | cPGM66 | WT | pPGM5 | pPGM9 | pPGM60 | WT | AGP-85 | AGP-93 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ala | 1.00 ± 0.07 | 0.57 ± 0.11 | 0.51 ± 0.11 | 1.48 ± 0.06 | 1.00 ± 0.13 | 0.26 ± 0.04 | 0.40 ± 0.07 | 0.22 ± 0.05 | 1.00 ± 0.19 | 0.32 ± 0.04 | 0.16 ± 0.03 |
| Arg | 1.00 ± 0.08 | 0.87 ± 0.18 | 0.74 ± 0.17 | 2.21 ± 0.14 | 1.00 ± 0.07 | 0.60 ± 0.05 | 0.88 ± 0.24 | 0.58 ± 0.13 | 1.00 ± 0.18 | 0.74 ± 0.05 | 0.47 ± 0.08 |
| Asn | 1.00 ± 0.18 | 0.87 ± 0.15 | 0.84 ± 0.17 | 0.65 ± 0.03 | 1.00 ± 0.15 | 0.57 ± 0.16 | 0.65 ± 0.19 | 0.27 ± 0.08 | 1.00 ± 0.24 | 0.61 ± 0.09 | 0.40 ± 0.12 |
| Asp | 1.00 ± 0.04 | 0.92 ± 0.06 | 0.88 ± 0.05 | 1.15 ± 0.05 | 1.00 ± 0.12 | 0.62 ± 0.09 | 0.53 ± 0.06 | 0.55 ± 0.10 | 1.00 ± 0.09 | 0.57 ± 0.06 | 0.41 ± 0.04 |
| β-Ala | 1.00 ± 0.13 | 0.66 ± 0.10 | 0.93 ± 0.15 | 0.99 ± 0.11 | 1.00 ± 0.16 | 0.42 ± 0.06 | 0.40 ± 0.05 | 0.29 ± 0.05 | 1.00 ± 0.18 | 0.58 ± 0.07 | 0.55 ± 0.15 |
| Cys | n.d. | n.d. | n.d. | n.d. | 1.00 ± 0.05 | 0.75 ± 0.08 | n.d. | n.d. | n.d. | n.d. | n.d. |
| GABA | 1.00 ± 0.04 | 0.84 ± 0.07 | 0.74 ± 0.09 | 0.41 ± 0.03 | 1.00 ± 0.13 | 0.65 ± 0.09 | 0.63 ± 0.06 | 0.83 ± 0.08 | 1.00 ± 0.13 | 0.82 ± 0.10 | 0.79 ± 0.18 |
| Glu | 1.00 ± 0.03 | 1.04 ± 0.06 | 0.95 ± 0.05 | 1.12 ± 0.04 | 1.00 ± 0.14 | 0.29 ± 0.11 | 0.54 ± 0.04 | 0.61 ± 0.12 | 1.00 ± 0.08 | 1.06 ± 0.11 | 0.75 ± 0.10 |
| Gln | 1.00 ± 0.22 | 1.02 ± 0.23 | 0.98 ± 0.20 | 1.20 ± 0.21 | 1.00 ± 0.11 | 0.54 ± 0.05 | 1.00 ± 0.38 | 0.51 ± 0.11 | 1.00 ± 0.09 | 0.84 ± 0.07 | 0.63 ± 0.19 |
| Gly | 1.00 ± 0.11 | 0.83 ± 0.13 | 0.64 ± 0.06 | 0.97 ± 0.08 | 1.00 ± 0.08 | 0.94 ± 0.12 | 0.63 ± 0.08 | 0.52 ± 0.09 | 1.00 ± 0.06 | 0.98 ± 0.09 | 0.85 ± 0.17 |
| His | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Homo-Ser | 1.00 ± 0.09 | 1.24 ± 0.24 | 1.27 ± 0.05 | 1.51 ± 0.17 | 1.00 ± 0.17 | 0.55 ± 0.04 | 1.11 ± 0.17 | 1.07 ± 0.17 | 1.00 ± 0.06 | 1.22 ± 0.09 | 1.11 ± 0.10 |
| Ile | 1.00 ± 0.08 | 0.81 ± 0.09 | 0.68 ± 0.12 | 1.66 ± 0.11 | 1.00 ± 0.31 | 0.30 ± 0.03 | 1.06 ± 0.26 | 0.65 ± 0.12 | 1.00 ± 0.14 | 1.02 ± 0.09 | 0.62 ± 0.08 |
| Leu | 1.00 ± 0.19 | 0.68 ± 0.15 | 0.65 ± 0.21 | 3.07 ± 0.50 | 1.00 ± 0.13 | 0.39 ± 0.06 | 1.17 ± 0.37 | 0.49 ± 0.13 | 1.00 ± 0.21 | 0.98 ± 0.15 | 0.49 ± 0.06 |
| Lys | 1.00 ± 0.08 | 0.60 ± 0.07 | 0.49 ± 0.14 | 2.53 ± 0.19 | 1.00 ± 0.11 | 0.67 ± 0.03 | 1.04 ± 0.44 | 0.41 ± 0.12 | 1.00 ± 0.22 | 0.52 ± 0.04 | 0.26 ± 0.03 |
| Met | 1.00 ± 0.04 | 0.76 ± 0.07 | 0.49 ± 0.12 | 1.11 ± 0.05 | 1.00 ± 0.13 | 0.56 ± 0.05 | 0.92 ± 0.18 | 0.67 ± 0.14 | 1.00 ± 0.14 | 0.72 ± 0.08 | 0.49 ± 0.10 |
| Orn | 1.00 ± 0.17 | 0.76 ± 0.14 | 0.87 ± 0.23 | 1.67 ± 0.20 | 1.00 ± 0.11 | 0.45 ± 0.10 | 1.10 ± 0.30 | 0.63 ± 0.12 | 1.00 ± 0.18 | 1.01 ± 0.15 | 0.77 ± 0.14 |
| 5-Oxy-Pro | 1.00 ± 0.14 | 0.74 ± 0.15 | 0.97 ± 0.05 | 0.99 ± 0.13 | 1.00 ± 0.13 | 0.35 ± 0.04 | 1.07 ± 0.37 | 0.46 ± 0.14 | 1.00 ± 0.11 | 0.82 ± 0.08 | 0.64 ± 0.17 |
| Phe | 1.00 ± 0.06 | 0.51 ± 0.08 | 0.36 ± 0.11 | 1.23 ± 0.05 | 1.00 ± 0.16 | 0.67 ± 0.23 | 0.83 ± 0.23 | 0.34 ± 0.08 | 1.00 ± 0.14 | 0.46 ± 0.09 | 0.18 ± 0.02 |
| Pro | 1.00 ± 0.09 | 1.33 ± 0.29 | 0.51 ± 0.12 | 2.41 ± 0.52 | 1.00 ± 0.12 | 0.53 ± 0.02 | 0.45 ± 0.09 | 0.38 ± 0.11 | 1.00 ± 0.05 | 1.24 ± 0.11 | 0.79 ± 0.14 |
| Ser | 1.00 ± 0.10 | 0.83 ± 0.15 | 0.57 ± 0.07 | 1.44 ± 0.08 | 1.00 ± 0.26 | 0.52 ± 0.02 | 0.62 ± 0.09 | 0.49 ± 0.10 | 1.00 ± 0.14 | 0.77 ± 0.08 | 0.55 ± 0.11 |
| Thr | 1.00 ± 0.08 | 0.82 ± 0.13 | 0.64 ± 0.08 | 1.22 ± 0.07 | 1.00 ± 0.11 | 0.18 ± 0.02 | 0.68 ± 0.12 | 0.56 ± 0.11 | 1.00 ± 0.14 | 1.04 ± 0.11 | 0.76 ± 0.15 |
| Trp | 1.00 ± 0.17 | 0.48 ± 0.14 | 0.22 ± 0.09 | 1.46 ± 0.27 | 1.00 ± 0.12 | 0.27 ± 0.03 | 1.40 ± 0.64 | 0.19 ± 0.06 | 1.00 ± 0.33 | 0.37 ± 0.08 | 0.14 ± 0.02 |
| Tyr | 1.00 ± 0.08 | 0.48 ± 0.11 | 0.28 ± 0.12 | 1.37 ± 0.07 | 1.00 ± 0.12 | 0.70 ± 0.05 | 1.23 ± 0.53 | 0.30 ± 0.10 | 1.00 ± 0.30 | 0.43 ± 0.07 | 0.08 ± 0.01 |
| Val | 1.00 ± 0.04 | 0.93 ± 0.05 | 0.96 ± 0.05 | 1.19 ± 0.02 | 1.00 ± 0.11 | 0.56 ± 0.09 | 1.00 ± 0.19 | 0.86 ± 0.16 | 1.00 ± 0.06 | 1.22 ± 0.07 | 0.92 ± 0.12 |
| α-Ketogluterate | 1.00 ± 0.07 | 1.89 ± 0.23 | 1.79 ± 0.36 | 0.75 ± 0.12 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Ascorbate | 1.00 ± 0.25 | 0.48 ± 0.15 | 0.53 ± 0.22 | 1.09 ± 0.37 | 1.00 ± 0.17 | 0.86 ± 0.07 | 0.67 ± 0.20 | 0.51 ± 0.10 | 1.00 ± 0.02 | 0.93 ± 0.08 | 0.88 ± 0.09 |
| Citrate | 1.00 ± 0.15 | 1.04 ± 0.15 | 1.09 ± 0.07 | 1.33 ± 0.03 | 1.00 ± 0.31 | 0.75 ± 0.05 | 0.74 ± 0.12 | 0.76 ± 0.08 | 1.00 ± 0.18 | 0.78 ± 0.03 | 0.44 ± 0.07 |
| Fumarate | 1.00 ± 0.19 | 2.22 ± 0.70 | 0.85 ± 0.11 | 0.57 ± 0.07 | 1.00 ± 0.14 | 0.80 ± 0.06 | 0.69 ± 0.05 | 0.84 ± 0.08 | 1.00 ± 0.08 | 1.10 ± 0.07 | 1.01 ± 0.05 |
| Glycerate | 1.00 ± 0.04 | 0.97 ± 0.20 | 0.82 ± 0.07 | 0.98 ± 0.05 | 1.00 ± 0.13 | 0.58 ± 0.04 | 0.82 ± 0.16 | 0.82 ± 0.08 | n.d. | n.d. | n.d. |
| Isocitrate | 1.00 ± 0.03 | 0.56 ± 0.04 | 0.39 ± 0.05 | 3.15 ± 1.80 | 1.00 ± 0.13 | 1.13 ± 0.14 | 0.72 ± 0.17 | 0.41 ± 0.09 | 1.00 ± 0.12 | 0.60 ± 0.07 | 0.52 ± 0.07 |
| Malate | 1.00 ± 0.04 | 2.14 ± 0.17 | 2.53 ± 0.36 | 1.33 ± 0.26 | 1.00 ± 0.11 | 0.58 ± 0.11 | 1.00 ± 0.15 | 1.88 ± 0.18 | 1.00 ± 0.13 | 1.26 ± 0.09 | 1.33 ± 0.13 |
| Oxalate | n.d. | n.d. | n.d. | n.d. | 1.00 ± 0.22 | 1.13 ± 0.11 | 0.23 ± 0.05 | 0.37 ± 0.10 | 1.00 ± 0.28 | 0.29 ± 0.02 | 0.26 ± 0.01 |
| Quinate | 1.00 ± 0.37 | 1.99 ± 0.49 | 1.89 ± 0.50 | 0.77 ± 0.16 | 1.00 ± 0.26 | 0.58 ± 0.04 | 0.63 ± 0.11 | 0.63 ± 0.17 | 1.00 ± 0.04 | 0.70 ± 0.07 | 0.75 ± 0.10 |
| Shikimate | 1.00 ± 0.06 | 0.89 ± 0.11 | 0.88 ± 0.04 | 0.61 ± 0.06 | 1.00 ± 0.17 | 0.79 ± 0.06 | 0.40 ± 0.07 | 0.47 ± 0.06 | 1.00 ± 0.09 | 0.41 ± 0.02 | 0.29 ± 0.02 |
| Succinate | 1.00 ± 0.17 | 0.93 ± 0.25 | 1.44 ± 0.36 | 1.09 ± 0.17 | 1.00 ± 0.18 | 0.44 ± 0.04 | 0.46 ± 0.06 | 0.37 ± 0.02 | 1.00 ± 0.25 | 0.35 ± 0.03 | 0.28 ± 0.02 |
| Threonate | 1.00 ± 0.13 | 0.64 ± 0.16 | 0.57 ± 0.06 | 0.52 ± 0.11 | 1.00 ± 0.29 | 0.42 ± 0.06 | n.d. | n.d. | n.d. | n.d. | n.d. |
| Fru | 1.00 ± 0.19 | 2.12 ± 0.66 | 23.35 ± 12.51 | 1.82 ± 0.32 | 1.00 ± 0.51 | 0.38 ± 0.04 | 0.35 ± 0.04 | 3.28 ± 2.14 | 1.00 ± 0.17 | 5.84 ± 3.37 | 10.52 ± 6.15 |
| Gal | 1.00 ± 0.07 | 1.39 ± 0.24 | 1.01 ± 0.16 | 0.94 ± 0.10 | 1.00 ± 0.37 | 1.15 ± 0.21 | 0.71 ± 0.07 | 1.40 ± 0.34 | n.d. | 192.50 ± 26.22 | 282.17 ± 41.34 |
| Glc | 1.00 ± 0.26 | 2.30 ± 0.59 | 4.46 ± 0.58 | 1.34 ± 0.30 | 1.00 ± 0.35 | 1.02 ± 0.31 | 1.79 ± 0.40 | 2.72 ± 0.37 | 1.00 ± 0.18 | 1.18 ± 0.21 | 1.18 ± 0.14 |
| Inositol | 1.00 ± 0.15 | 1.29 ± 0.31 | 0.88 ± 0.05 | 0.51 ± 0.02 | 1.00 ± 0.24 | 0.47 ± 0.04 | 0.52 ± 0.05 | 0.40 ± 0.06 | 1.00 ± 0.06 | 0.58 ± 0.03 | 0.88 ± 0.05 |
| Mannitol | 1.00 ± 0.04 | 1.12 ± 0.14 | 1.81 ± 0.37 | 0.43 ± 0.04 | 1.00 ± 0.08 | 0.68 ± 0.02 | 0.52 ± 0.04 | 1.45 ± 0.13 | 1.00 ± 0.12 | 1.97 ± 0.18 | 3.07 ± 0.52 |
| Man | 1.00 ± 0.05 | 1.43 ± 0.19 | 1.44 ± 0.18 | 1.06 ± 0.16 | 1.00 ± 0.36 | 1.30 ± 0.20 | 0.85 ± 0.13 | 1.81 ± 0.29 | 1.00 ± 0.09 | 1.99 ± 0.13 | 2.90 ± 0.32 |
| Suc | 1.00 ± 0.06 | 1.20 ± 0.17 | 0.92 ± 0.08 | 1.18 ± 0.17 | 1.00 ± 0.35 | 1.58 ± 0.15 | 0.87 ± 0.23 | 1.89 ± 0.28 | 1.00 ± 0.14 | 1.16 ± 0.14 | 1.22 ± 0.10 |
| Fru-6-P | 1.00 ± 0.07 | 1.33 ± 0.10 | 1.50 ± 0.09 | 0.97 ± 0.04 | 1.00 ± 0.24 | 1.64 ± 0.19 | 1.38 ± 0.121 | 1.79 ± 0.16 | 1.00 ± 0.05 | 1.42 ± 0.07 | 1.65 ± 0.07 |
| Glu-6-P | 1.00 ± 0.04 | 1.43 ± 0.10 | 1.55 ± 0.09 | 0.86 ± 0.02 | 1.00 ± 0.25 | 1.33 ± 0.13 | 1.10 ± 0.07 | 1.83 ± 0.11 | 1.00 ± 0.14 | 2.31 ± 0.12 | 2.82 ± 0.20 |
| 3-PGA | 1.00 ± 0.11 | 1.70 ± 0.12 | 2.04 ± 0.30 | 1.62 ± 0.42 | 1.00 ± 0.28 | 1.29 ± 0.15 | 1.30 ± 0.19 | 1.42 ± 0.31 | n.d. | n.d. | n.d. |
| 6-P-Gluconate | 1.00 ± 0.22 | 1.63 ± 0.40 | 0.86 ± 0.23 | 1.72 ± 0.53 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Phosphate | 1.00 ± 0.04 | 0.97 ± 0.03 | 0.92 ± 0.06 | 1.06 ± 0.02 | 1.00 ± 0.07 | 0.88 ± 0.07 | 0.80 ± 0.08 | 0.81 ± 0.06 | 1.00 ± 0.05 | 0.75 ± 0.06 | 0.57 ± 0.03 |
Data are normalized to the mean response calculated for the wild type of each measure batch. (To allow comparison between measure batches, individual wild-type values were normalized in the same way.) Values presented are the mean ± se of the mean of six independent determinants. Those that are significantly different from wild type are identified in bold type. n.d., Compounds that were not determined in a particular set of chromatograms; 3-PGA, 3-phosphoglyceric acid; GABA, 4-amino butyric acid.
HCA and PCA of the Metabolic Complement of Transgenic Potato Tubers
Next, we decided to compare the transgenics analyzed in this study with those studied in our previous study (Roessner et al., 2001). When taken together, data from these seven transgenic plants constituted 19 transgenic plant lines, each separate genotype with its own corresponding wild type and yielding approximately 8,000 data points. To gain a rapid way to analyzing this data set, data-mining tools were once again applied to the data. Using HCA and PCA, it was possible to determine tubers that exhibited similar changes in their metabolite profile as well as those that have distinct differences in the level of certain metabolites.
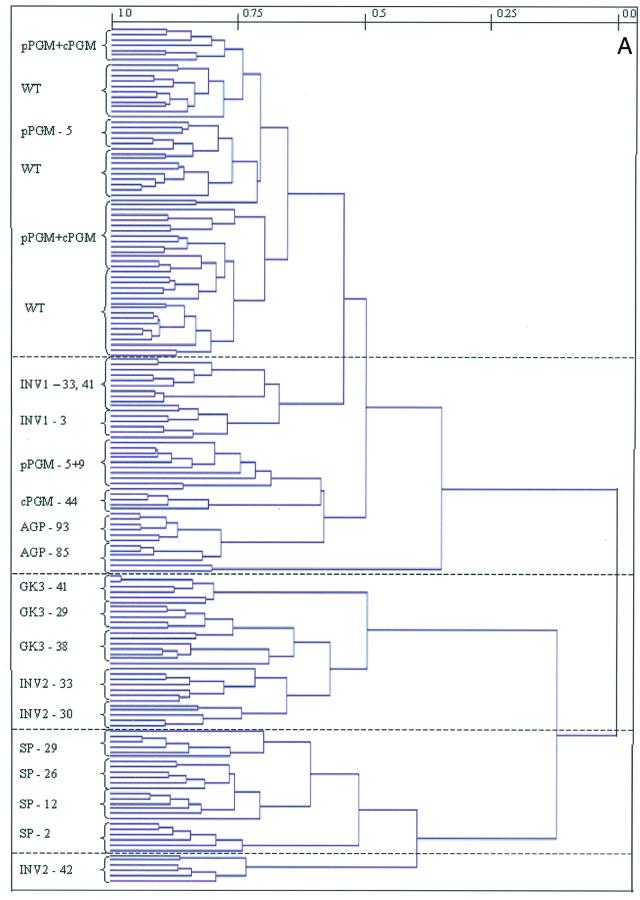
When HCA was applied to the data set obtained from the analysis of the transgenic potato tubers mentioned above (Fig. 3A), two large clusters could be observed. In the lower cluster, all samples were characterized by a highly increased Suc mobilization within the cytosol (INV2, GK3, and SP), whereas the upper cluster included wild-type tubers and those exhibiting alterations in the levels of starch synthetic enzymes. When the lower cluster is analyzed in detail, it is clear that the cluster formation is the same as that obtained from the application of HCA to only these transgenics (Roessner et al., 2001). Again, the tubers expressing the invertase in the cytosol and these in combination with the glucokinase were clustered together and the tubers expressing the Suc phosphorylase formed a distinct cluster. The only difference between the two analyses was that line INV2-42, which represents the weakest line of the INV2 lines, clustered differently. However, it is an inherent feature of this form of cluster analysis that a different cluster pattern is formed because a new hierarchy is established whenever a data set is expanded or contracted. In contrast to the lower cluster, the upper cluster can be divided into several subclusters. However, only the tubers expressing the invertase in the apoplast and those that are inhibited in the AGPase activity resolved independently. In contrast, the tubers reduced in the expression of the plastidial or cytosolic phosphoglucomutase essentially coclustered with wild-type tubers. Some tubers inhibited in the expression of either one or the other isoform of phosphoglucomutase did not fall into this subcluster but gave separate distinct clusters of their own.
Figure 3.
A, Dendogram obtained following HCA of the metabolic profiles of the genetically modified potato tubers. Wherever possible individual branches are grouped in brackets for ease of reading. B, PCA of the metabolite profiles of all analyzed genetically modified potato tubers. Samples representing wild-type, pPGM, and cPGM tubers (light-green circle), AGPase tubers (dark-green circle), apoplastic invertase expressing tubers (light-blue circle), INV2-30; INV2-33, and GK3 tubers (yellow circle), INV2-42 (red circle), and SP (dark-blue circle) are marked as described for ease of comparison. PCA vectors 1 and 2 were chosen for best visualization of differences between experimental treatments and include 70.6% of the information derived from metabolic variances.
When PCA was applied to the data set, similar trends to those described above were observed. Once again, the INV2, together with the GK3 lines and SP lines, constitutes single and independent clusters and the samples of the INV2-42 line clustered on its own (Fig. 3B). The other transgenic lines (INV1, αPGMI, αPGMII, and αAGP) clustered independently to the Suc mobilizers. Within this subcluster, only the INV1 lines and the αAGP lines were resolved into subclusters. Lines inhibited in the activity of either plastidial or cytosolic phosphoglucomutase clustered together with the wild-type tubers and within this cluster it was not possible to identify subclusters that represented individual transgenic plants. Loadings indicated that the metabolites that exhibited a large contribution to the cluster formation, i.e. PT00, maltose, Trp, 6-phosphogluconate, maltitol, and trehalose, represent novel compounds only detectable in certain transgenic tubers with respect to wild-type tubers. Other compounds that were detected in all analyzed tubers and contributed significantly to the clustering result were Glc, Gal, Suc, Man, Fru, inositol, α-ketogluterate, fumerate, and the phosphorylated intermediates 3-phosphoglyceric acid, Glc-, and Fru-6-phosphate (data not shown).
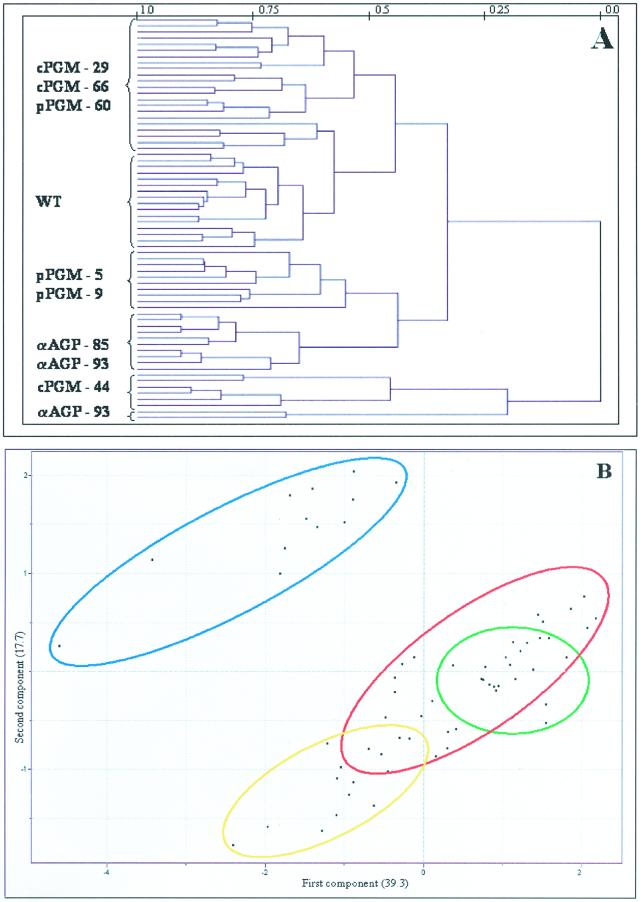
Resolution of Genotypes Impaired in Starch Synthesis
Analysis of the data obtained after metabolite profiling of all analyzed transgenic potato tubers revealed that the tubers of the INV2-, GK3-, and SP-lines that are altered in cytosolic Suc mobilization formed distinct clusters, suggesting that these tubers represent individual phenotypes despite bearing genes targeted to the same metabolite. However, the other transgenic lines formed one single large cluster, in which the samples of the INV1 and AGPase lines could nevertheless be distinguished, indicating that the metabolite complements of all these tubers are to some extent similar.
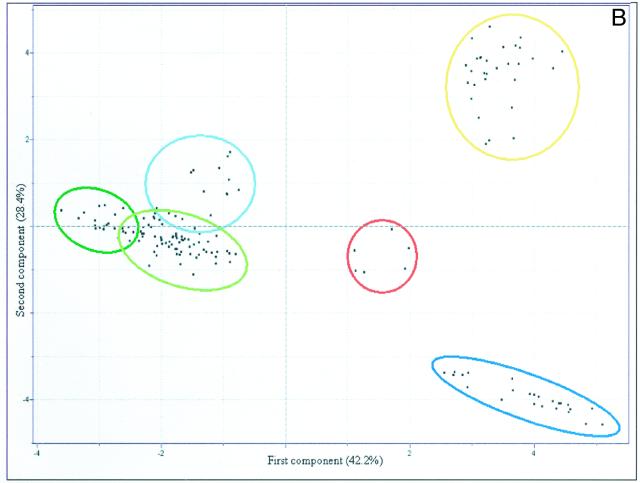
HCA was next applied to a subset of the data including only results obtained from profiling the AGPase, cPGM, and pPGM lines to simplify the comparison and thus allow discrimination of patterns that were obscured in graphs containing the full data set. Following this analysis, three major clusters could be resolved (Fig. 4A). The upper cluster contained two subclusters, the first included all wild-type samples, and the second mainly consisting of samples from the cPGM tubers (but also included samples of pPGM-60, exhibiting the weakest inhibition of the plastidial phopshoglucomutase; Tauberger et al., 2000; Fernie et al., 2001d). The lower major cluster could be further divided into two subclusters, one including the strongest lines of the pPGM plants and the other including both AGPase lines, revealing that these transgenic tubers have very similar metabolite complements. The final subcluster, samples of cPGM-44, joined the other samples at a great distance. This is not so surprising because previous studies established that this line exhibited the most severe reduction in cytosolic phosphoglucomutase activity and furthermore was characterized by the most dramatic phenotypic and metabolic changes (Fernie et al., 2001c). When PCA was applied exclusively to theses lines, however, a slightly different picture emerged (Fig. 4B). Using this method, it was possible to clearly identify the samples derived from plants inhibited in the activity of AGPase within one independent and distinct cluster, whereas the samples of the plants expressing reduced levels of either plastidial or cytosolic phosphoglucomutase clustered together with wild-type samples. However, the pPGM samples were in a distinct subcluster to that of the cPGM samples, which showed considerable overlap with wild-type samples. Loadings from this PCA revealed that Gal was the metabolite that contributed the most to the cluster formation with the amino acids Gln, Tyr, Trp, Ala, Phe, Leu, Lys, Pro, and Asn, the hexose monophosphates and Fru also having a large influence on the pattern formation (data not shown).
Figure 4.
A, Dendogram obtained following HCA of the metabolic profiles of the genetically modified potato systems impaired in starch synthesis. Wherever possible, individual branches are grouped in brackets for ease of reading. B, PCA of the metabolite profiles of the genetically modified potato systems impaired in starch synthesis. Samples representing wild type (green circle), pPGM (yellow circle), cPGM tubers (red circle), and AGPase tubers (blue circle) are marked as described for ease of comparison. PCA vectors 1 and 2 were chosen for best visualization of differences between experimental treatments and include 57.0% of the information derived from metabolic variances.
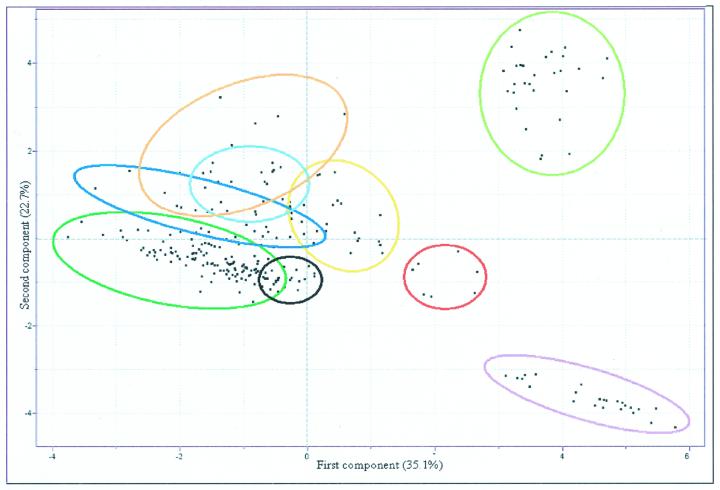
Comparison of Metabolic Complements of Genetically and Environmentally Modified Systems
The above examples detail independent analysis of genetic and environmentally modified samples. Following these analyses, it was decided to compare all modified systems within a single analysis. HCA analysis of the resultant combined data set resulted in a very complex dendogram that was nevertheless, as would be expected, very similar to the dendograms of independent analysis of genetic or environmentally manipulated potato tubers (Fig. 2, A and B). However, when PCA was applied to this data set, several interesting results could be observed more clearly (Fig. 5). Samples of the INV2-, GK3-, and SP-lines formed a clustering pattern that was similar to that observed previously (Fig. 3A; Roessner et al., 2001). In contrast, all other samples transgenically and environmentally manipulated systems assembled in one single cluster. The wild-type samples, samples incubated only in buffer, and samples of the AGPase and phosphoglucomutase transgenics could be identified in one large cluster (overlapping with the samples incubated in mannitol), suggesting that the metabolic complements of all these samples are very similar in comparison with those of the other samples analyzed. The loadings for this complete analysis were very similar to those obtained when PCA was carried out on only the genetically diverse systems, with the exception that malate played a far larger contribution to the pattern formation in this analysis (data not shown).
Figure 5.
PCA of the metabolite profiles of all analyzed environmentally and genetically modified systems. Samples representing wild type; tubers incubated in buffer alone pPGM, cPGM, AGPase tubers (dark-green circle); mannitol-fed tubers (black circle); Fru-fed tubers (dark-blue circle); Suc-fed tubers (yellow circle); Glc-fed tubers (light-red circle); apoplastic invertase-expressing tubers (light-blue circle); INV2-30, INV2-33, and GK3 tubers (light-green circle); INV2-42 (dark-red circle); and SP (lilac circle) are marked as described for ease of comparison. PCA vectors 1 and 2 were chosen for best visualization of differences between experimental treatments and include 57.8% of the information derived from metabolic variances.
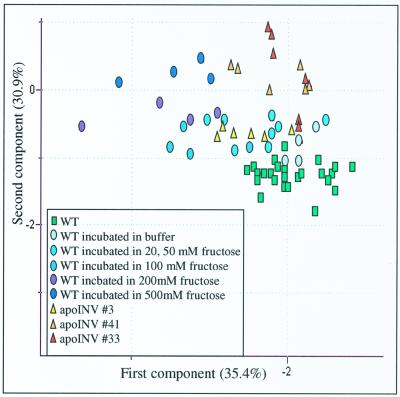
It is interesting that there was a clear separation to the samples incubated in Suc, Fru, and Glc; Glc completely and Fru partially overlapped with the samples of the INV1 tubers, thus allowing us to refine our previous statement that the elevation of extracellular Glc was able to phenocopy apoplastic expression of invertase because it is clear that elevation of extracellular Fru also phenocopied this manipulation. An in-depth comparison of changes in the chromatograms of the three modified systems confirmed that they were indeed very similar. Furthermore, when this region of the PCA is expanded and annotated, there is a clear trend of distance from the wild-type steady-state samples that can be observed both with increasing activity of the transgene and also with increasing external hexose concentrations (Fig. 6).
Figure 6.
PCA of the metabolic profiles of wild-type potato tubers, Fru-fed tuber discs, and apoplastic invertase-expressing tubers. PCA vectors 1 and 2 were chosen for best visualization of differences between experimental treatments and include 53.3% and 65.4% of the information derived from metabolic variances, respectively.
DISCUSSION
This study illustrates the potential of comprehensive metabolic analysis coupled to statistical methods of cluster analysis for phenotypic studies and thus by implication for functional genomics. We have previously used the techniques described in this paper to phenotype transgenic lines exhibiting enhanced Suc mobilization (Roessner et al., 2001). Here, we chose to extend this profiling to encompass lines impaired in starch synthesis either by inhibition of AGPase, plastidial PGM, or cytosolic PGM and to tuber discs incubated in various concentrations of Suc, Glc, Fru, or mannitol. All the transgenic lines have been fairly well characterized at the metabolic and morphological levels. The metabolic profiles of these systems revealed changes in the levels of very many metabolites with extracts from lines with reduced expression of AGPase and pPGM displaying very similar changes with respect to wild type. That these tubers displayed similar metabolic complements is in keeping with our previous suggestions that these transgenics phenocopy one another (Tauberger et al., 2000; Fernie et al., 2001c).
Furthermore, it is interesting that there were large changes in metabolites levels within the transgenic systems studied here, yet not all the plants displayed a visible phenotype. This is, however, not so surprising because results from many previous studies have indicated a high level of functional redundancy in plant systems, and furthermore, in many cases where visible phenotypes were observed, enzyme activity often needed to be reduced below a certain low threshold level. Examples of this include the miniature phenotype of maize (Zea mays) mutant min1 (Cheng et al., 1996) and the decreased potato tuber yield observed on the antisense repression of Suc synthase (Zrenner et al., 1995), which were only observed on the loss of 90% and 70% of the respective gene activities. The fact that we could discriminate these lines from the wild type on the basis of their metabolic complements therefore provides support for the importance of a broad metabolic screen in comprehensive phenotyping strategies.
The metabolic consequences of sugar feeding were relatively minor with incubation of tuber discs in low concentrations of sugar having very little effect on metabolism; however, incubations in high concentrations led to dramatic changes in the levels of most metabolites. That large effects on metabolism were observed following incubation in high levels of sugars is in agreement with many gene expression studies revealing large changes in transcript levels under such conditions (for example, see Godt et al., 1995; Roitsch et al., 1995). Furthermore, these data are generally in close agreement with the smaller metabolite data sets obtained following incubation of tuber discs (Geiger et al., 1998) and Chenopodium rubrum suspension cultures (Hatzfeld et al., 1990) in media containing high sugar concentrations. However, the fact that so many metabolic changes occur following these short-term feedings may have implications for many earlier studies on carbohydrate mediated gene expression (see Huang et al., 1993; Fu et al., 1995; Hesse and Willmitzer, 1996) because it follows that incubation of tissue in high concentrations of sugars results in dramatic changes in metabolites other than that supplied, thereby complicating the interpretation of changes in transcript level following such experiments. On a more positive note, several interesting results emerge from these feeding experiments, two of which are particularly striking. Following incubation of potato tuber discs in high concentrations of either Glc, Fru, Suc, and mannitol, there was a large increase in Trp, despite there being a tendential decrease in total amino acid content following the majority of these incubations. Because this specific increase in Trp was also observed on incubation in mannitol, it can be best interpreted as a stress response. Similar increases in Trp biosynthesis in response to stress have been characterized in many species, perhaps most thoroughly in Eschericia coli (Yanofsky and Horn, 1994); however, this effect has also been commonly observed in plant species (for example, see Liuz et al., 1995; Mobley et al., 1999; Pustovoitova et al., 2000). In addition, samples incubated in high levels of Suc exhibited increases in the level of every single amino acid. This finding is consistent with the results of our previous study in which we showed that all amino acids increased in tuber tissue of several lines expressing Suc-degrading proteins under a tuber-specific promoter but not in the leaves (Roessner et al., 2001). When taken together, these data give strong support for our theory that the potato tuber has the required machinery to synthesize amino acids de novo. Increased synthesis of many amino acids has previously been observed following incubation of tobacco leaf tissue in 25 mm Suc, where it correlated with an increased rate of nitrate assimilation and an increased oxogluterate synthesis (Morcuende et al., 1998). However, the exact reason for the increase in amino acid contents observed in this study following incubation in Suc but not in Glc, Fru, or mannitol remains mysterious especially when it is considered that our previous observations were made in transgenic tubers exhibiting very low levels of Suc.
In addition to analysis of the changes in individual metabolites, we analyzed the metabolic complements using the statistical tools HCA and PCA. We performed a number of analyses with respect to sugar-fed samples and genetically modified samples to identify phenocopies at the level of the metabolic complement. The two different statistical approaches gave slightly different results; however, this is to be expected because they effectively ask different questions. The primary purpose of HCA is to present data in a manner that emphasizes natural groupings, whereas PCA reduces dimensionality of the data and allows display of linear combinations of the original independent variables that account for maximal amounts of variation. However, the fact that similar results were obtained using both approaches shows that the conclusions are not inherently biased by the methodology used. The combination of these approaches gave many insights into the similarity of the systems studied.
We mentioned above that on individual analysis of the component metabolites in the transgenic systems that pPGM plants appeared to phenocopy the AGPase plants. This observation was also made from the results of HCA, being particularly apparent when only those samples genetically impaired in starch synthesis were taken into consideration. In this example, however, the results of the PCA were somewhat different. If only the most important components of the analysis were considered (i.e. the first component axis), AGPase and pPGM lines seemed fairly similar; however, they were not on consideration of further components. That said, when all analyzed transgenic systems were compared together it became clear that those impaired in starch synthesis are clearly distinct from those exhibiting increased Suc mobilization. This is in itself intriguing because earlier measurements documented that the lines expressing Suc-degrading proteins were also characterized by a reduced starch accumulation (Sonnewald et al., 1997; Trethewey et al., 1998, 2001). Therefore, these data may imply that the reduction of starch accumulation in these Suc-mobilizing lines is not a direct result of the inhibition of either isoform of phosphoglucomutase or of AGPase; however, further biochemical studies are required to assess if this interpretation is correct. The fact that the Suc mobilizing lines are more distinct from wild type than those inhibited in starch synthesis is also very interesting, although perhaps not so surprising because the lines enhanced in Suc mobilization have previously been demonstrated to exhibit marked differences in Suc mobilization and resynthesis and starch synthesis and glycolysis, whereas those impaired in starch synthesis appear to be altered only in starch synthesis and Suc resynthesis. However, these data do support theories of an essential role for the supply to and subsequent metabolism of Suc within heterotrophic plant tissues (Riesmeier et al., 1994; Gottwald et al., 2000).
A further insight gained using principal component analyses was that Fru feeding can, at least to a limited extent, phenocopy transgenic potato tubers expressing invertase in the apoplast. However, from both the PCA analysis and the analyzed chromatograms, it is clear that these two systems diverge at high concentrations of Fru and high activity of invertase and therefore these systems are not absolute phenocopies of one another. Despite this fact, these results mean that we have to modify our earlier conclusion to state that apoplastic invertase-expressing tubers can be phenocopied by exogenous application of hexoses and as such the putative sugar-sensing factor present in the apoplast (Lalonde et al., 1999; Fernie et al., 2000) may not be as specific for Glc as we had previously postulated.
Although phenocopying of genetic manipulation has also previously been achieved in plants on a number of other occasions (for example, see Tsuchimoto et al., 1993; Lehman et al., 1996; Cheng and Chourey, 1999; Yephremov et al., 1999; Adams et al., 2000; Beemster and Baskin, 2000), very few attempts have been made to distinguish metabolic phenocopies either within plants or any other species (Feifel et al., 1993; Zhou et al., 1998; Marx et al., 1999). The data obtained in the current study are particularly interesting when considered alongside those from a recent studies of the Minature1 mutant of maize also characterized by a deficiency of apoplastic invertase and exhibiting dramatically impaired growth with developing seeds typically exhibiting a loss of 70% to 80% of the normal seed weight (Cheng et al., 1996). Anatomical, biochemical, and histological data obtained from in vitro kernel development experiments revealed that the mutant phenotype remains irrespective of Suc or hexose supply in the culture medium (Cheng and Chourey, 1999). The authors convincingly argue that in this case, the invertase-mediated release of sugars, and not the exogenous supply of sugars, is critical for appropriate carbon partitioning and normal seed development in maize. Although our study is restricted to the levels of metabolites following incubation in sugar, and we have no idea whether the sugar supply is able to complement the increased rate of cell division previously observed in the apoplastic invertase tubers (Tauberger et al., 1999), it is clear here that the exogenous supply of sugar is able to phenocopy the metabolic complement of the apoplastic invertase tubers. It is surprising that the exogenous application of sugars is able to phenocopy a deficiency in invertase activity in potato tuber tissue but not in maize; however, it is likely that routes of sugar metabolism and/or mechanisms of sugar sensing differ between these species.
In summary, the examples chosen here show that phenotyping and consequently the identification of phenocopies can be rapidly carried out at a metabolic basis. Furthermore, although we showed that pPGM and AGPase lines phenocopy one another, at least to a limited extent, they are still distinguishable. This result gives valuable insight into the high resolving power of metabolic profiling because it constitutes evidence that this method is able to resolve two genotypes even when they differ only in the expression levels of consecutive enzyme activities within a simple linear pathway.
MATERIALS AND METHODS
Plant Materials
Potato (Solanum tuberosum L. cv Desiree) was obtained from Saatzucht Lange AG (Bad Schwartau, Germany). The generation and selection of the transgenic lines used here have been described previously by Müller-Röber et al. (1992), Sonnewald et al. (1997), Trethewey et al. (1998, 2001), Tauberger et al. (1999), and Fernie et al., (2001c). Plants were maintained in tissue culture with a 16-h-light, 8-h-dark regime on Murashige and Skoog medium (Murashige and Skoog, 1962) that contained 2% (w/v) Suc. In the greenhouse, plants from transgenic lines and wild-type controls were grown in parallel under the same light regime with a minimum of 250 μmol photons m−2 s−1 at 22°C. In this paper, the term developing tubers is used for tubers (over 10 g fresh weight) harvested from healthy 10-week-old plants and the term set of transgenics is used to describe plants expressing the same transgene. Samples were harvested by cutting discs into liquid nitrogen directly from tubers attached to the mother plant and were taken from six independent plants per line.
Chemicals
All chemicals and pure standard substances were purchased from either Sigma-Aldrich Chemie GmbH (Deisenhofen, Germany) or Merck KgaA (Darmstadt, Germany).
Confirmation of Preliminary Biochemical Characteristics of Transgenic Lines
Extraction and assaying of AGPase and phosphoglucomutase activities were carried out according to Fernie et al. (2001b). Carbohydrate levels were determined exactly as described by ap Rees and Morrell (1990).
Extraction, Derivatization, and Analysis of Potato Tuber Metabolites Using GC-MS Analysis
Potato tuber tissue (100 mg) was extracted and derivatized exactly as described by Roessner et al. (2000). Sample volumes of 1 μL were then injected with a split ratio of 25:1 using a hot needle technique. The GC-MS system was comprised of an AS 2000 autosampler, a GC 8000 gas chromatograph, and a Voyager quadrole mass spectrometer (ThermoQuest, Manchester, UK). GC was performed on a 30-m SPB-50 column with 0.25-μm film thickness (Supelco, Bellfonte, CA). The injection temperature was set at 230°C, the interface was set at 250°C, and the ion source was adjusted to 200°C. Helium was used as the carrier gas at a flow rate of 1 mL min−1. The analysis was performed using the temperature program described in Roessner et al. (2000). Mass spectra were recorded at 2 scan s−1 with an m/z 50 to 600 scanning range. Peaks were assigned and quantified and all data were normalized to the mean response calculated for the wild-type control of each measured batch; to allow comparison between the samples, individual wild-type values were normalized in the same way (Roessner et al., 2001). The recovery of small representative amounts of each metabolite through the extraction, derivatization, storage, and quantification procedures has been documented previously (Roessner et al., 2000).
Incubations of Potato Tuber Slices
Fru, Suc, and mannitol incubations were performed essentially as described by Geiger et al. (1998). Discs were cut directly from developing tubers from wild-type plants and washed three times in 10 mm MES-KOH (one disc was cut from a tuber of four independent plants per incubation). They were then placed in 100-mL flasks (eight discs per flask) containing 5 mL of incubation medium (10 mm MES- KOH, pH 6.5), supplemented with 0, 20, 50, 100, 200, and 500 mm of sugar and incubated with agitation (at 150 rpm) for 2 h, after which an aliquot of the incubation media was immediately taken and frozen in liquid N2 for subsequent analysis. Samples were then washed three times in 10 mm MES-KOH (pH 6.5) before they were dried and frozen in liquid N2 for subsequent analysis. Analysis of the tuber extracts was carried out as described above with the exception that the Glc level of the sample was quantified by calibration as described previously (Roessner et al., 2000; Fernie et al., 2001a).
Cluster Analysis
HCA and PCA were obtained using the informatic program Pirouette 2.6 (Infometrix, Woodinville, WA). HCA allows the presentation of complete linkage clusters results in a dendogram represented by a similarity factor between 1 and 0 with 1 being the most similar. All HCAs described in this paper use the Euclidean distance to calculate the matrix of all samples and are transformed into log10 to allow better comparison of large and small numbers. For PCA, a covariance matrix approach was taken in which the n-dimensional data set was transformed by log10 and then further into a second n-dimensional data set in which what was designated as the most important information of the original data set was stored in the first few dimensions. The results of these analyses were then presented as a two-dimensional graphical display of the data in which the presented ellipses cover points belonging to the defined biological population.
Statistical Analysis
Where two observations are described in the text as different, this means that they were determined to be statistically significantly different by performing Student's t tests using the algorithm incorporated into Microsoft Excel 7.0. (Microsoft Corp., Seattle).
Footnotes
LITERATURE CITED
- Aarts MGM, Dirkse WG, Stiekama WJ, Pereira A. Transposon tagging of a male sterility gene in Arabidopsis. Nature. 1993;36:715–717. doi: 10.1038/363715a0. [DOI] [PubMed] [Google Scholar]
- Adams S, Vinkenoog R, Spielman M, Dickinson HG, Scott RJ. Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development. 2000;127:2493–2502. doi: 10.1242/dev.127.11.2493. [DOI] [PubMed] [Google Scholar]
- Aharoni A, Keizer LCP, Bouwmeester HJ, Sun Z, Alvarez-Huerta M, Verhoeven HA, Blaas J, van Houwelingen AMML, De Vos RCH, van der Voet H. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell. 2000;12:647–661. doi: 10.1105/tpc.12.5.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees T, Morrell S. Carbohydrate metabolism in developing potatoes. Am Potato J. 1990;67:835–847. [Google Scholar]
- Beemster GTS, Baskin TI. Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 2000;124:1718–1727. doi: 10.1104/pp.124.4.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Milner PA, Walker AR, Schulz B, Feldmann KA. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science. 1996;273:948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- Celis JE, Kruhoffer M, Gromova I, Frederiksen C, Ostergaard M, Thykjaer T, Gromov P, Yu J, Palsdottir H, Magnusson N. Gene expression profiling: monitoring transcription and translation products using DNA microarrays and proteomics. FEBS Lett. 2000;480:2–16. doi: 10.1016/s0014-5793(00)01771-3. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Chourey PS. Genetic evidence that invertase-mediated release of hexoses is critical for approriate carbon partitioning and normal seed development in maize. Theor Appl Genet. 1999;98:485–495. [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS. The Minature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996;8:971–983. doi: 10.1105/tpc.8.6.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WWP, Huang L, Shen M, Webster C, Berlingame AL, Roberts JKM. Patterns of protein synthesis and tolerance of anoxia in root tips of maize seedlings acclimated to a low-oxygen environment, and identification of proteins by mass spectrometry. Plant Physiol. 2000;122:295–317. doi: 10.1104/pp.122.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem. 2000;275:33158–33166. doi: 10.1074/jbc.M005493200. [DOI] [PubMed] [Google Scholar]
- Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet. 1999;23:203–207. doi: 10.1038/13833. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A, Cardenas ML. Silent genes given voice. Nature. 2001;409:571–572. doi: 10.1038/35054646. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Balbo I, Benning C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 1999;284:2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- Duez P, Kumps A, Mardens Y. GC-MS profiling of urinary organic acids evaluated as a quantitative method. Clin Chem. 1996;42:1609–1615. [PubMed] [Google Scholar]
- Feifel N, Kucher K, Fuchs L, Jedrychowski M, Schmidt E, Antonin KH, Bieck PR, Gleiter CH. Role of cytochrome-P4502D6 in the metabolism of brofaromine: a new selective MAO-A inhibitor. Bone. 1993;25:119–122. doi: 10.1007/BF00315394. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Reismeier JW, Martiny A, Ramalingam S, Willmitzer L, Trethewey RN. Consequences of the expression of a bacterial glucokinase in potato tubers. Aust J Plant Physiol. 2000;27:827–833. [Google Scholar]
- Fernie AR, Roessner U, Geigenberger P. The sucrose analog palatinose leads to a stimulation of sucrose degradation and starch synthesis when supplied to discs of growing potato tubers (Solanum tuberosum) Plant Physiol. 2001a;125:1967–1977. doi: 10.1104/pp.125.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Roessner U, Trethewey RN, Willmitzer L. The contribution of plastidial phosphoglucomutase to the control of starch synthesis within the potato tuber. Planta. 2001b;213:418–426. doi: 10.1007/s004250100521. [DOI] [PubMed] [Google Scholar]
- Fernie AR, Tauberger E, Lytovchenko A, Roessner U, Willmitzer L, Trethewey RN (2001c) Antisense repression of cytosolic phosphoglucomutase in potato (Solanum tuberosum) results in a severe growth retardation, reduction in tuber number and altered carbon metabolism. Planta (in press) [DOI] [PubMed]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fu HY, Kim SY, Park WD. High-level tuber expression and sucrose inducibility of a potato Sus4 sucrose synthase gene require 5′and 3′ flanking sequences and the leader intron. Plant Cell. 1995;7:1387–1394. doi: 10.1105/tpc.7.9.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger M, Stitt M, Geigenberger P. Metabolism in slices from growing potato tubers responds differently to addition of glucose and sucrose. Planta. 1998;206:234–244. [Google Scholar]
- Gibson S, Falcone DL, Browse J, Somerville C. Use of transgenic plants and mutants to study the regulation and function of lipid-composition. Plant Cell Environ. 1994;17:627–637. [Google Scholar]
- Godt DE, Riegel A, Roitsch T. Regulation of sucrose synthase expression in Chenopodium rubrum: characterization of sugar induced expression in photoautotrophic suspension cultures and sink tissue specific expression in plants. J Plant Physiol. 1995;146:231–238. [Google Scholar]
- Gottwald JR, Krysan PJ, Young JC, Evert RF, Sussman MR. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc Natl Acad Sci USA. 2000;97:13979–13984. doi: 10.1073/pnas.250473797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y. ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 2000;19:4248–4256. doi: 10.1093/emboj/19.16.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld WD, Dancer J, Stitt M. Fructose-2, 6-bisphosphate, metabolites and “coarse” control of pyrophosphate:fructose-6-phosphate phosphotransferase during triose-phosphate cycling in heterotrophic cell suspension cultures of Chenopodium rubrum. Planta. 1990;180:205–211. doi: 10.1007/BF00193997. [DOI] [PubMed] [Google Scholar]
- Hesse H, Willmitzer L. Expression analysis of a sucrose synthase gene from sugar beet (Beta vulgaris L.) Plant Mol Biol. 1996;30:863–873. doi: 10.1007/BF00020799. [DOI] [PubMed] [Google Scholar]
- Huang N, Chandler J, Thomas BR, Koizumi N, Rodriguez RL. Metabolic regulation of alpha-amylase expression in transgenic cell cultures of rice (Oryza sativa L) Plant Mol Biol. 1993;23:737–747. doi: 10.1007/BF00021529. [DOI] [PubMed] [Google Scholar]
- Kampranis SC, Damianova R, Atallah M, Toby G, Kondi G, Tsichlis PN, Makris AM. A novel plant glutathione S-transferase/peroxidase suppresses bax lethality in yeast. J Biol Chem. 2000;275:29207–29216. doi: 10.1074/jbc.M002359200. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. The dual function of sugar carriers: transport and sugar sensing. Plant Cell. 1999;11:707–726. doi: 10.1105/tpc.11.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JD, Ray S, Ray A. SIN 1, a mutation affecting female fertility in Arabidopsis, interacts with MOD1, its recessive modifier. Genetics. 1994;137:1101–1110. doi: 10.1093/genetics/137.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Liuz Y, Silverstone AL, Wu YM, Shang FY. Formation of N-Malonyl-L-tryptophan in water-stressed tomato leaves. Phytochemistry. 1995;40:691–697. [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Marx SJ, Agarwal SK, Heppner C, Kim YS, Kester MB, Goldsmith PK, Skarulis MC, Spiegel AM, Burns AL, Debelenko LV. The gene for multiple endocrine neoplasia type 1: recent findings. Bone. 1999;25:119–122. doi: 10.1016/s8756-3282(99)00112-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto I, Kuhara T. A new chemical diagnostic method for inborn errors of metabolism by mass spectrometry: rapid, practical, and simultaneous urinary metabolite analysis. Mass Spectrom Rev. 1996;15:43–57. doi: 10.1002/(SICI)1098-2787(1996)15:1<43::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mobley EM, Kunkel BN, Keith B. Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene. 1999;240:115–123. doi: 10.1016/s0378-1119(99)00423-0. [DOI] [PubMed] [Google Scholar]
- Morcuende R, Krapp A, Hurry V, Stitt M. Sucrose feeding leads to increased rates of nitrate assimilation, increased rates of oxogluterate synthesis and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta. 1998;206:394–409. [Google Scholar]
- Müller-Röber B, Sonnewald U, Willmitzer L. Antisense inhibition of the ADPglucose pyrophosphorylase in transgenic potato leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J. 1992;11:1229–1238. doi: 10.1002/j.1460-2075.1992.tb05167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene-expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Pustovoitova TN, Bavrina TV, Zhdanova NE. Drought tolerance of transgenic tobacco plants carrying the iaaM and iaaH genes of auxin biosynthesis. Russ J Plant Physiol. 2000;47:380–385. [Google Scholar]
- Ramachandran S, Christensen HEM, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, Chua NH. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 2000;124:1637–1647. doi: 10.1104/pp.124.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond T, Somerville S. Chasing the dream: plant EST microarrays. Curr Opin Plant Biol. 2000;3:108–116. doi: 10.1016/s1369-5266(99)00049-7. [DOI] [PubMed] [Google Scholar]
- Roitsch T, Bittner M, Godt DE. Induction of apoplastic invertase of Chenopodium rubrum by D-glucose and a glucose analog and tissue-specific expression suggest a role in sink-source regulation. Plant Physiol. 1995;108:285–294. doi: 10.1104/pp.108.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR. Metabolic profiling and phenotyping of genetically and environmentally modified plant systems. Plant Cell. 2001;13:11–29. doi: 10.1105/tpc.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography: mass spectrometry. Plant J. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Santoni V, Rouquie D, Doumas P, Mansion M, Boutry M, Degand H, Dupree P, Packman L, Sherrier J, Prime T. Use of proteome strategy for tagging proteins present at the plasma membrane. Plant J. 1998;16:633–641. doi: 10.1046/j.1365-313x.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zryd JP. Efficient gene targeting in the moss Physcomitrella patens. Plant J. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorn O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: large scale identification of yeast proteins from two-dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnewald U, Hajiraezaei MR, Kossmann J, Heyer A, Trethewey RN, Willmitzer L. Expression of a yeast invertase in the apoplast of potato tubers increases tuber size. Nat Biotechnol. 1997;15:794–797. doi: 10.1038/nbt0897-794. [DOI] [PubMed] [Google Scholar]
- Soppe WJJ, Bentsink L, Koornneef M. The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidosis thaliana. Development. 1999;126:4763–4770. doi: 10.1242/dev.126.21.4763. [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. Gene trap tagging of PROFILERA, an essential MCM2–3-5-like gene in Arabidopsis. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R. Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancesteral tubulin. Proc Natl Acad Sci USA. 1998;95:4368–4376. doi: 10.1073/pnas.95.8.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauberger E, Fernie AR, Emmermann M, Renz A, Kossmann J, Willmitzer L, Trethewey RN. Antisense inhibition of plastidial phosphoglucomutase provides compelling evidence that potato tuber amyloplasts import carbon from the cytosol in the form of glucose-6-phosphate. Plant J. 2000;23:43–53. doi: 10.1046/j.1365-313x.2000.00783.x. [DOI] [PubMed] [Google Scholar]
- Tauberger E, Hoffmann-Benning S, Fleischer-Notter H, Willmitzer L, Fisahn J. Impact of invertase overexpression on cell size, starch granule formation and cell wall properties during tuber development in potatoes with modified carbon allocation patterns. J Exp Bot. 1999;50:477–486. [Google Scholar]
- Terryn N, Rouze P, Van Montagu M. Plant genomics. FEBS Lett. 1999;452:3–6. doi: 10.1016/s0014-5793(99)00591-8. [DOI] [PubMed] [Google Scholar]
- Trethewey RN, Fernie AR, Bachmann A, Fleischer-Notter H, Geigenberger P, Willmitzer L. Glucose-independent induction of glycolysis in heterotrophic plant tissue. Plant Cell Environ. 2001;24:357–365. [Google Scholar]
- Trethewey RN, Geigenberger P, Riedel K, Hajirezaei MR, Sonnewald U, Stitt M, Riesmeier JW, Willmitzer L. Combined expression of glucokinase and invertase in potato tubers leads to a dramatic reduction in starch accumulation and a stimulation of glycolysis. Plant J. 1998;15:109–118. doi: 10.1046/j.1365-313x.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S, van der Krol AR, Chua NH. Ectopic expression of PMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell. 1993;5:843–853. doi: 10.1105/tpc.5.8.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C, Horn V. Role of regulatory features of the trp operon of Eschericia coli in mediating a response to a nutritional shift. J Bacteriol. 1994;176:6245–6254. doi: 10.1128/jb.176.20.6245-6254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA. 1998;95:10294–10299. doi: 10.1073/pnas.95.17.10294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Salanoubat M, Willmitzer L, Sonnewald U. Evidence for a crucial role of sucrose synthase for sink strength using transgenic potato plants. Plant J. 1995;7:97–107. doi: 10.1046/j.1365-313x.1995.07010097.x. [DOI] [PubMed] [Google Scholar]
- Zu T, Peterson DJ, Tagliani L, St. Clair G, Baszczynski CL, Bowen B. Targetted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc Natl Acad Sci USA. 1999;96:8768–8773. doi: 10.1073/pnas.96.15.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]