Abstract
Sequencing of the Arabidopsis genome has led to the identification of thousands of new putative genes based on the predicted proteins they encode. Genes encoding tRNAs, ribosomal RNAs, and small nucleolar RNAs have also been annotated; however, a potentially important class of genes has largely escaped previous annotation efforts. These genes correspond to RNAs that lack significant open reading frames and encode RNA as their final product. Accumulating evidence indicates that such “non-coding RNAs” (ncRNAs) can play critical roles in a wide range of cellular processes, including chromosomal silencing, transcriptional regulation, developmental control, and responses to stress. Approximately 15 putative Arabidopsis ncRNAs have been reported in the literature or have been annotated. Although several have homologs in other plant species, all appear to be plant specific, with the exception of signal recognition particle RNA. Conversely, none of the ncRNAs reported from yeast or animal systems have homologs in Arabidopsis or other plants. To identify additional genes that are likely to encode ncRNAs, we used computational tools to filter protein-coding genes from genes corresponding to 20,000 expressed sequence tag clones. Using this strategy, we identified 19 clones with characteristics of ncRNAs, nine putative peptide-coding RNAs with open reading frames smaller than 100 amino acids, and 11 that could not be differentiated between the two categories. Again, none of these clones had homologs outside the plant kingdom, suggesting that most Arabidopsis ncRNAs are likely plant specific. These data indicate that ncRNAs represent a significant and underdeveloped aspect of Arabidopsis genomics that deserves further study.
The recent completion of the sequencing of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000) represents an important advance in our knowledge of plant biology and also an important contribution to the understanding of general genomic organization and evolution. Through analysis of the sequence, more than 25,000 putative genes encoding proteins and structural RNA species have been identified. The annotation of the genome was based on the prediction of genes by a combination of algorithms specifically optimized with parameters based on known Arabidopsis genes and by analyzing similarities to known proteins and expressed sequence tags (ESTs). The sensitivity and selectivity of the gene prediction software used by the Arabidopsis Genome Initiative has been comprehensively assessed and shown to be highly efficient to predict protein-coding genes (Pavy et al., 1999; Arabidopsis Genome Initiative, 2000).
With the exception of structural RNA genes, the main criterion for gene prediction from the genome sequence has been the presence of open reading frames (ORFs). GeneMark.hmm (Lukashin and Borodovsky, 1998) was deemed to be the most efficient tool to predict Arabidopsis genes, based on the evaluation of several gene prediction programs (Pavy et al., 1999). However, one default parameter used by GeneMark.hmm to predict a gene is the presence of an ORF of at least 100 amino acids (aa; J. Besemer, personal communication). This parameter has been used routinely since the annotation of the first eukaryotic genome sequenced, Saccharomyces cerevisiae (Goffeau et al., 1996). To generate the Arabidopsis annotation, other programs were also used in addition to GeneMark.hmm. Additional criteria, such as homology to known proteins or genes from other species, splice site prediction, and the presence of polyadenylation sites and TATA boxes, were also applied (Arabidopsis Genome Initiative, 2000; for a detailed description of annotation strategies, see The Institute of Genomic Research Web site, http://www.tigr.org/tdb/edb2/ath1/htmls/annotation.html; and the Munich Information Center for Protein Sequences Web site, http://mips.gsf.de/proj/thal/proj/proj_overview.html). However, these annotation strategies still depend on the presence of a significant ORF to identify putative genes.
Some genes encode RNAs, rather than proteins, as their final products. tRNA, rRNA, and the small nuclear RNAs and nucleolar RNAs have been studied extensively, and are relatively straightforward to identify by homology searches or with specialized algorithms (e.g. Lowe and Eddy, 1997, 1999). It has become apparent recently that in addition to these structural RNAs, other non-coding RNAs (ncRNAs) exist that lack protein-coding capacity and exert their action mainly or exclusively at the RNA level (Eddy, 1999; Caprara and Nilsen, 2000; Erdmann et al., 2000; 2001). Analyses of the properties and functions of ncRNAs have indicated that they can act as gene regulators, as part of biotic and abiotic stress signals, or as part of RNA-protein complexes with various enzymatic and structural activities. A number of ncRNAs are processed in an mRNA-like manner; consequently, they undergo splicing and have poly(A+) tails and, presumably, caps (for review, see Erdmann et al., 2000). Current strategies for genome annotation, although efficient in the identification of protein-coding genes, rarely detect these ncRNA genes due to the lack of a significant ORF.
The presence of ncRNAs has been described in several systems. For example, in prokaryotic and eukaryotic systems, RNase P RNA catalytically processes the 5′ end of tRNA (Altman and Kirsebom, 1999), and signal recognition particle (SRP) RNA is involved in protein transport across the endoplasmic reticulum (ER; Walter et al., 2000). In most eukaryotes, telomerase RNA serves as the template for the reverse transcriptase that synthesizes telomeric DNA (Blackburn, 2000). mei RNA helps regulate the initiation of meiosis in Saccharomyces pombe (Watanabe and Yamamoto, 1994). In Caenorhabditis elegans, ncRNAs are intimately involved in larval development. Two small ncRNAs, lin-4 (22 nucleotides [nts]; Lee et al., 1993) and let-7 (21nt; Reinhart et al., 2000) are required for the transition from the first to the second larval stage and from late larval to adult cell fates, respectively (for review, see Moss, 2000). Both of these transcripts are natural antisense RNAs complementary to the 3′-untranslated regions (UTRs) of different sets of protein-coding mRNAs. Data indicate that both lin-4 and let-7 RNAs negatively regulate the translation of their target mRNAs by binding to these sequences.
Dosage compensation is one of the most intensely studied processes involving ncRNAs. In mammalian systems, the Xist RNA is critical for X inactivation in females. The RNA physically coats the inactivated X chromosome (Barr body) and is probably involved in changes in chromatin architecture (Willard and Salz, 1997; Panning and Jaenisch, 1998). It is interesting that Xist is apparently regulated by another ncRNA, an antisense RNA called Tsix (Lee et al., 1999). Drosophila melanogaster achieves dosage compensation by a different mechanism, doubling the expression from the single X chromosome in males. This process involves two male-specific ncRNAs, roX1 and roX2 (Amrein and Axel, 1997; Meller et al., 1997), that are essential for the formation of a regulatory complex on specific chromosomal domains (Akhtar et al., 2000).
Only a few ncRNAs from plants have been reported. One of the first transcripts identified as a ncRNA in plants was CR20 (Teramoto et al., 1996), a gene from cucumber (Cucumis sativus) that is repressed by cytokinins and by stress or developmental conditions (Teramoto et al., 1995). This gene is part of a family of ncRNAs with members in several plant species (Taylor and Green, 1995; Teramoto et al., 1996; van Hoof et al., 1997). GUT15 (gene with unstable transcript 15) is another characterized member of this family. This transcript was first identified as one of the most unstable transcripts in tobacco (Nicotiana tabacum) cell cultures (Taylor and Green, 1995). The fact that transcripts of this family are hormonally regulated and have unstable transcripts suggest that they may play a role in regulatory processes, although their true functions are unknown. Another interesting family of ncRNAs present in plants is typified by Mt4 in Medicago truncatula (Burleigh and Harrison, 1998) and TPSI1 in tomato (Lycopersicon esculentum; Liu et al., 1997). As with the GUT15/CR20 family, these genes are regulated by biotic (cytokinins) and abiotic (phosphate starvation) signals. Several short non-conserved ORFs are present in the Mt4/TPSI1 family, and all of the transcripts show regions of absolute identity at the nt level (Martín et al., 2000). The high degree of nt sequence conservation and low level of ORF conservation suggest that the final product of these genes is RNA and not protein. It is interesting that both the GUT15/CR20 and Mt4/TPSI1 families appear to be plant specific, and reports of plant homologs of animal ncRNAs are virtually absent from the literature. The analysis of additional ncRNA candidates should indicate whether kingdom specificity is a common feature of ncRNAs.
Data supporting the existence of additional ncRNAs in Arabidopsis have emerged recently. One putative ncRNA described in Arabidopsis was identified through the generation of Arabidopsis mutants by transformation with an activation tag construct. A mutant recovered from this population, jaw, displayed altered growth and leaf shape (Weigel et al., 2000). In this line, the insertion occurred in a region of DNA where no evident ORFs were present. A 2-kb genomic probe adjacent to the insertion site detected a transcript of approximately 0.5 kb that was up-regulated in the mutant, suggesting that JAW encodes an ncRNA. Other ncRNA candidates have been identified by searching for cDNAs corresponding to long contiguous sequences (Terryn et al., 1998; Kato et al., 1999). Although these initial efforts to identify and characterize individual ncRNAs have been encouraging, genome annotations have not incorporated systematic searches for the identification of ncRNAs on a global scale.
In this work, we initiated a systematic sequence analysis for ncRNAs in plants as a first step toward evaluating their significance. We examined Arabidopsis for the presence of ncRNAs found in other kingdoms and collected and reanalyzed potential Arabidopsis ncRNAs reported previously. A key aspect of our study was to screen genomic sequences corresponding to 20,000 Arabidopsis ESTs for those that exhibit characteristics of ncRNAs. We also detected peptide-coding genes that have ORFs too small to be detected with general annotation protocols. Our results indicate that there is a significant number of ncRNAs in plants, the vast majority of which appear to be plant-specific. The use of ESTs present in the Arabidopsis Functional Genomics Consortium (AFGC) microarrays for this initial analysis allowed us to survey existing data on regulation of gene expression for these putative ncRNAs.
RESULTS
Most Known or Annotated ncRNAs in Arabidopsis Lack Homologs in Animals
Table I compiles the list of known or putative ncRNAs that were deduced from published information at the onset of this analysis, including several discussed in the introduction. AtGUT15 and AtCR20-1 comprise two members of a family of genes that lack a long ORF (Teramoto et al., 1996; van Hoof et al., 1997). Members of this family are present in several plant species. At4 (Burleigh and Harrison, 1999) and AtIPS1 (Martín et al., 2000) are Arabidopsis orthologs of two proposed ncRNAs that are induced during phosphate starvation in M. truncatula and tomato, respectively (Liu et al., 1997; Burleigh and Harrison, 1998). Three ESTs located adjacent to AtCR20-1 correspond to regions of the genome annotated as containing no ORF. EST contig analysis indicates that they could be part of the AtCR20-1 transcript and that what was reported as AtCR20-1 is a partial clone of 758 nt, whereas the full-length cDNA is approximately 1.5 kb. Although the sequence of the JAW RNA is not known, it has been characterized as a ncRNA based on its expression in the presence of, and linkage to, an activation tag (Weigel et al., 2000). The other ncRNA candidates were implicated as ncRNAs from analyses of cDNAs corresponding to contiguous regions of Arabidopsis sequences (Terryn et al., 1998; Kato et al., 1999). Two are potential antisense-coding genes. A few may be chimeric clones or correspond to protein-coding regions based on further scrutiny (see comments in Table I).
Table I.
ESTs or transcripts previously suggested to be non-coding RNAs in Arabidopsis
| Name | Accession No. | Presence in Other Species | Annotation | Comments | Regulation or Phenotype |
|---|---|---|---|---|---|
| AtGUT15a | U84973 | Several plant species | AtGUT15, unknown protein | Alternate splicing, lacks long conserved ORF | Unstable mRNA |
| AtCR20-1b | D79218 | Several plant species | AtCR20-1, non-coding RNA | Homolog of AtGUT15 | ↓ By cytokinins |
| At4c | AF055372 | M. truncatula | Putative protein | No conserved ORFs | ↑ By Pi-starvation |
| AtlPS1d | AF236376 | Tomato | – | At4 homolog (TPSI1/Mt4 family) | ↓ By cytokinins |
| ↑ By Pi-starvation | |||||
| JAWe | – | Glycine max? | – | 2 kb of genomic probe detected 0.5-kb mRNA | Altered growth and shape in overexpressor |
| 179K9T7f | H37319 | – | Contains no ORF | Adjacent to AtCR20 | – |
| 248G6T7f | W43209 | – | Contains no ORF | Adjacent to AtCR20 | – |
| E6G11T7f | AA042352 | – | Contains no ORF | Adjacent to AtCR20 | – |
| ATH132404g | AJ132404 | – | Antisense transcript, AKL kinase-like gene | – | – |
| ZCF83h | – | – | – | RNA antisense of ZCW32 (AB028232) | Poly(A+) found |
| ZCF120h | AB028200 | – | – | – | – |
| ZCF112h | AB028193 | – | – | Homologous sequence found in Arabidopsis | – |
| ZF2h | AB028197 | – | – | – | – |
| RXF6h | AB008026 | – | – | Chimeric RNA? | – |
| RXW18h | AB008024 | Brassica rapa | Acyl carrier-like protein | Chimeric RNA? | – |
| ZCF44h | AB028227 | – | Unknown protein | Predicted introns are present in the cDNA | – |
| ZCF58h | AB028192 | G. max? | Repeat region, rpt family = “AT rich” | Has ORF, truncated RNA? | – |
vanHoff et al. (1997).
Annotated by Munich Information Center for Protein Sequences.
Because several of the cellular processes that involve ncRNAs in non-plant systems also occur in plants, we used the program BLAST (Altschul et al., 1990) to search for plant homologs of known ncRNAs from mammals and other animals, yeast, and bacteria. An important resource used to conduct this search was the ncRNA database containing reported ncRNAs from various systems (Erdmann et al., 2001). We used the RNAs present in this database, along with several more from the literature, as queries for BLAST analysis. It was surprising that of 33 different ncRNAs from bacteria, fungi, and animals (for a complete list, see http://www.prl.msu.edu/PLANTncRNAs/page2.html), none were found in Arabidopsis. This collection of ncRNAs comprises transcripts involved in a variety of processes, including imprinting, chromosomal silencing, transcriptional regulation, and responses to biotic and abiotic stress (Erdmann et al., 2000). To extend our analysis, we searched for homologs of these ncRNAs in other plant species. Negative results were obtained in every case. The only known ncRNA that is present in animals and for which a homolog has been found in plants is SRP RNA, the RNA component of the signal recognition particle that directs ribosomes translating secretory and membrane proteins to the ER (Gorodkin et al., 2001). SRP RNA has been conserved throughout evolution and is found in all organisms studied so far (Walter et al., 2000).
In Silico Search of New ncRNAs
As mentioned, several ncRNAs have mRNA-like modifications, such as polyadenylation and splicing. Based on these observations, we predicted that some ncRNAs should be present in EST collections because these collections are derived from cDNA libraries enriched for poly(A+) RNA. This idea was supported by the fact that ESTs corresponding to several previously known ncRNAs (such as AtGUT15, AtCR20-1, and At4) are present in the PRL2 EST collection (Newman et al., 1994). In accordance, we performed a computational analysis to identify new potential ncRNAs from a combination of two EST collections (Newman et al., 1994; White et al., 2000) that are being used by the AFGC for the microarray project (see http://afgc.stanford.edu). These sets, comprising a total of about 20,000 ESTs, were chosen because they have largely been “cleaned” of redundant sequences. Further, because these sequences are represented on the AFGC microarrays, we could obtain important information about the regulation of accumulation of putative ncRNA transcripts. Finally, the use of ESTs to begin our systematic search for ncRNAs provided us with the certainty that any sequence identified as a ncRNA would be transcribed.
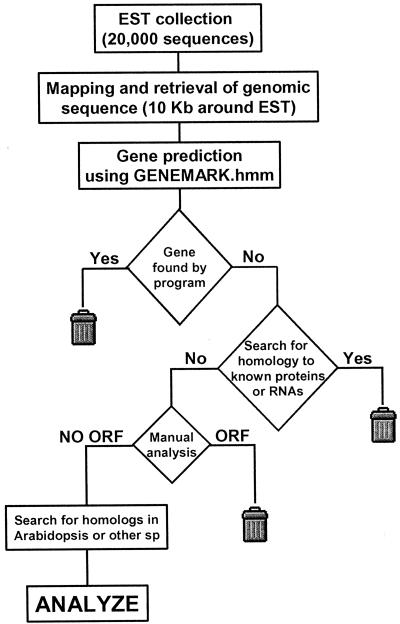
A flow chart of the computational screening is represented in Figure 1. The first step of the search involved the mapping of each individual EST sequence to the genome and the retrieval of 10 kb of genomic sequence around the EST. Next, these genomic sequences were used as queries in the gene prediction program GeneMark.hmm (Lukashin and Borodovsky, 1998), which has been evaluated as the most accurate program to predict Arabidopsis genes (Pavy et al., 1999). This program has been trained to predict genes from Arabidopsis sequences and, as mentioned, has a default cutoff for an ORF of at least 100 aa. Because they would code for proteins of more than 100 aa, sequences that yielded positive predictions that included the EST sequence were discarded. In this step, more than 18,000 sequences were identified as protein-coding genes and therefore were discarded. ESTs corresponding to genomic sequences that were not predicted to contain genes were then analyzed using BLAST to find any similarity to known proteins. We set a low arbitrary cutoff to avoid losing bona fide ncRNAs that could share some sequence homology with protein-coding genes and because we planned to analyze individual genes further to discard protein-coding genes (see below). Thus, any EST with a BLAST match with an E score lower than 10−5 was discarded. Then, we searched for homologs of the remaining ESTs (284) in other Arabidopsis and non-Arabidopsis EST collections. Subsequently, these 284 ESTs were individually analyzed using a combination of six-frame translation and the FGENE gene prediction program (Salamov and Solovyev, 2000) to discard any protein-coding sequence that had escaped the initial screening. Because the EST sequences are not entirely accurate, the genomic sequence corresponding to each EST was retrieved, and the EST sequence was corrected to match the genome sequence before the analysis. Finally, the genomic annotations that correlated with the EST position were retrieved, and those ESTs that were predicted as part of a gene or that were obvious UTRs (determined after analysis of EST contigs) were also discarded. Table II shows a summary of the output of each of the steps of this strategy.
Figure 1.
Schematic representation of the computational methods and individual inspection used to identify ESTs with characteristics of potential ncRNAs or with ORFs smaller than 100 aa.
Table II.
Output of the computational screening of the ACFG EST collection and further analysis
| Computational Analysis of the Initial EST Collection | Manual Individual Analysis of the 284 Remaining ESTs | ||
|---|---|---|---|
| Starting genomic sequences corresponding to EST collection | ∼20,000 | Annotated as protein by genome project | 178 |
| Genes found with GENEMARK and discarded | ∼18,000 | Putative protein or exon-like | 16 |
| Remaining | ∼2,000 | UTR | 34 |
| Homology to known proteins (10−5 cutoff)—dis carded | ∼1,700 | Transposon? | 1 |
| Final output | 284 | Discarded (low quality sequence, no genomic sequence, etc.) | 16 |
| Remaining candidates | 39 | ||
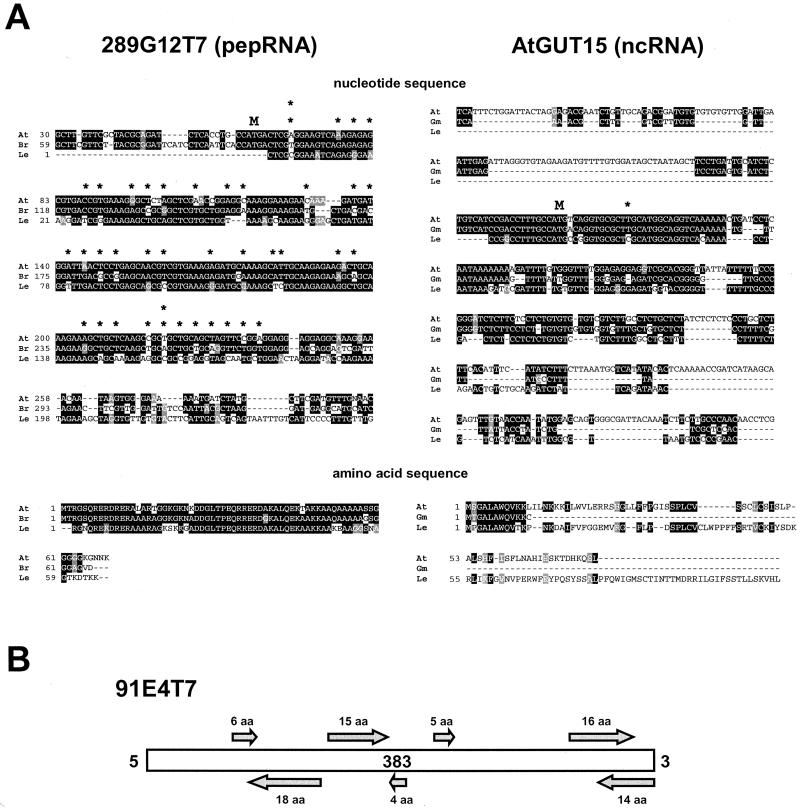
The 39 sequences remaining after the filtering strategy outlined above corresponded not only to putative ncRNAs but also to peptide-coding RNAs (pepRNAs) with ORFs smaller than 100 aa. These sequences were then classified as putative ncRNAs or pepRNAs based on several criteria. A peptide-coding gene would be under selective pressure to preserve the aa sequence. Therefore, neutral nt changes in the third position of codons could more readily accumulate, and small deletions or insertions would be favored only when comprising multiples of three bases to preserve the reading frame. In contrast, if the gene corresponds to a ncRNA, the nt sequence itself would be subject to selection pressure, and base changes in the third position of the “codon” would not be favored over changes in either of the other two positions. In addition, insertions or deletions in factors of three are not favored over those of other sizes because there is no ORF to be preserved. It was possible to examine these characteristics in those cases where we found homologous ESTs in other species or within Arabidopsis. An example of this analysis is presented in Figure 2A. In this example, comparison of a putative pepRNA (289G12T7, left) from three different plant species revealed changes in almost every third-base position. However, the conservation of the longest potential ORF was high. As expected, small deletions and insertions were present in this gene, but all conserved the reading frame. In contrast, an ncRNA (AtGUT15, right) showed high conservation at the nt level (only one third-position base change). Although the conservation at the aa level was high for an 11-aa segment of a putative ORF coincidental with the region of high homology at the nt level, the potential ORFs from the three different homologs had a significant variation in length, from 12 to 111 aa, mainly due to the fact that insertions or deletions did not conserve the reading frame in this case. It is interesting that the original GUT15 gene was described as a peptide-coding gene because both the tobacco and Arabidopsis genes have putative ORFs of 78 and 75 aa, respectively (van Hoof et al., 1997). However, the identification of homologs in other species and the analysis described here allowed us to classify AtGUT15 as a putative ncRNA.
Figure 2.
Criteria used to classify individual ESTs as putative ncRNAs or pepRNAs. A, Criterion used for ESTs with homologs in other species. Left, Comparison of a putative peptide-coding Arabidopsis (At) EST (289G12T7) with homologs from B. rapa (Br) and tomato (Le) at the nt (top) and aa (bottom) levels. Right, Same comparison for an EST corresponding to an ncRNA (AtGut15) with homologs from G. max (Gm) and tomato. Black and gray boxes indicate identity and similarity, respectively. Asterisk, Indicates third-position base changes that do not produce changes in aa of the corresponding ORF. M, Start codon of the longest conserved ORF. B, Criterion used for ESTs without homologs in other species. In this case, six-frame translation was used to predict ORFs. ESTs with no significant ORF, such as EST 91E4T7 depicted in this figure, were classified as ncRNAs. Arrows indicate length and strand of putative ORFs.
In some cases, homologs in Arabidopsis or other species were not found, but the potential ORFs present in the transcript lacked an AUG start codon or were very short. Transcripts in this category were also considered to be putative ncRNAs. An example of this type of transcript is shown in Figure 2B. Based on the criteria presented above, the 39 transcripts obtained as the final output (Table III) were classified into putative ncRNAs, putative pepRNAs, or uncategorized (i.e. could be either ncRNA or pepRNA genes). The uncategorized group included ESTs that did not have homologs in other species and that had the potential to code for small peptides. As mentioned, several of the known ncRNAs have potential ORFs that are not conserved between homologs. As a consequence, although the uncategorized ESTs have the potential to code for small peptides, their classification is not possible until homologous sequences are found in other species.
Table III.
Final list of putative ncRNAs and peptide-coding RNAs (pepRNAs) obtained after the computational screening of a set of Arabidopsis ESTs
| EST | Accession No. | Longest ORFa | Homologs in Arabidopsis or Other Speciesb |
|---|---|---|---|
| aa | |||
| ncRNAs | |||
| 117K6XP | AA395209 | 38 | At |
| 120I16T7 | R87017 | 43 | At |
| 121B21T7 | T43554 | 20 | – |
| 132C18T7 | T45772 | 22 | – |
| 135G1XP | AA394537 | 24 | – |
| 135I17XP | AA394545 | 43 | Bn |
| 166E1T7 | R30601 | 19 | – |
| 178K15T7 | H36788 | 75 | Gm, Le, St, Nt, Mt, Lj, etc. |
| 186C13T7 | H37736 | 23 | At |
| 198O8T7 | AA712521 | 58 | Ha, Cs, Gm |
| 207P16T7 | N37235 | 36 | – |
| 39E2T7 | T13766 | 7 | – |
| 46E11T7 | T14074 | 22 | – |
| 600034440R1 | BE525895 | 50 | – |
| 600034527R1 | BE525960 | 16 | – |
| 600036624R1 | BE527687 | 20 | – |
| 600037943R1 | BE528770 | 20 | – |
| 600039726R1 | BE530252 | – | – |
| 91E4T7 | T20691 | 18 | – |
| pepRNAs | |||
| 118D15T7 | T43362 | 22 | Br, Zm, Pb, Hv, Le, etc. |
| 140K5XP | AA404821 | 64 | Zm, Os |
| 158O18T7 | R30241 | 72 | Gm, Mt, Pt, Le, Ta, etc. |
| 172I7T7 | H36461 | 56 | At |
| 179K3XP | AA651289 | 69 | Ga, Gm |
| 195G23T7 | AA712735 | 29 | Le |
| 206H8T7 | AA597669 | 84 | Ds, Gm, Mt, Os |
| 248N24T7 | AA597974 | 23 | Ga, Mc, Pb, Le, St, etc. |
| 289G12T7 | AA650884 | 40 | Le, Pt, Lj, Gm, Bc |
| Uncategorized | |||
| 155P4T7 | T88108 | 50 | – |
| 167H14T7 | R64808 | 25 | – |
| 170D20T7 | AA720338 | 58 | – |
| 186A14T7 | H37727 | 98 | – |
| 219K22T7 | N38534 | 74 | – |
| 226E4T7 | N65318 | 39 | – |
| 33D12T7 | T13664 | 76 | – |
| 600034309R1 | BE525791 | 70 | – |
| 600034421R1 | BE525881 | 47 | – |
| 91P14T7 | T21475 | 59 | – |
| 91P19T7 | T21480 | 37 | – |
The reported ORF corresponds to the longest ORF that starts with ATG in each case.
At, Arabidopsis; Bn, Brassica napus; Gm, G. max; Le, tomato; St, Solanum tuberosum; Nt, tobacco; Mt, M. truncatula; Lj, Lotus japonicus; Ha, hybrid aspen (Populus tremula × Populus tremuloides); Cs, cucumber; Br, B. rapa; Zm, Zea mays; Pb, Populus balsamifera; Hv, Hordeum vulgare; Os, Oryza sativa; Pt, Pinus taeda; Ta, Triticum aestivum; Ga, Gossypium arboreum; Ds, Descurainia sophia; Mc, Mesembryanthemum crystallinum; Bc, Brassica campestris.
Characterization of Putative ncRNAs Using Public DNA Microarray Data
The availability of expression profiles for genes that were represented in the EST collections used as the starting material for our screen allowed us to analyze changes in transcript accumulation in response to a variety of stimuli. This was carried out through a search of the AFGC microarray data in the Stanford Microarray Database available on the Web (http://genome-www4.stanford.edu/c/s.dll/ewing/queryCloneList.pl). Only data that showed similar ratios in duplicate experiments and with ratios higher than 2 or lower than 0.5 were used for this analysis. A ratio of 2 (or 0.5) is normally used as the cutoff for significant differences in gene expression detectable by microarray analyses (for example, see DeRisi et al., 1997), although for experiments with multiple replicates, smaller changes are also statistically valid (Wildsmith and Elcock, 2001). Table IV shows a summary of the most prominent characteristics of five putative ncRNAs and five putative pepRNAs. It is evident from this analysis that certain putative ncRNAs (and pepRNAs) are highly regulated, showing tissue specificity and responsiveness to biotic and abiotic signals. In other cases, no significant changes were found (for example, see EST 248N24T7 in Table IV), indicating that some may have constitutive roles. Although Table IV only shows data for selected examples, data for all the ESTs are available in our database (see below).
Table IV.
Analysis of transcript accumulation for representative putative ncRNAs and pepRNAs using public data from AFGC
| EST | Category | Level of Expressiona | Tissue Specificity | Response to Stimuli or Other Characteristics |
|---|---|---|---|---|
| 121B21T7 | ncRNA | Low | Leaves | Repressed by potassium nitrate |
| 178K15T7 | ncRNA | Low/medium | Low in leaves | Unstable transcript; light-repressed |
| 186C13T7 | ncRNA | Low | Roots, cell specific in flowers | Dark-repressed |
| 91E4T7 | ncRNA | Low | Leaves | Circadian |
| 120I16T7 | ncRNA | Low/medium | Roots and leaves | Circadian |
| 289G12T7 | pepRNA | Medium | Leaves | Decreased during light-harvesting complex disassembly; unstable |
| 118D15T7 | pepRNA | Medium | All | Unstable transcript |
| 158O18T7 | pepRNA | Low | n.d.b | Unstable |
| 206H8T7 | pepRNA | Low | Leaves | Decreased during light-harvesting complex disassembly. |
| 248N24T7 | pepRNA | High | All | Constitutive |
Level of expression was estimated as a function of the average intensity of signal in microarray experiments. Low, Less than 5,000 units; medium, between 5,000 and 15,000 units; high, more than 15,000 units of intensity.
n.d., No consistent data available.
Creation of a Plant ncRNA Database
To facilitate the incorporation of our data into the new body of resources generated by the increasing availability of data from genome-wide analyses, we created a public database of plant ncRNAs and pepRNAs that is available on a Web site (http://www. prl.msu.edu/PLANTncRNAs/). This database is designed to be a source to find known or putative ncRNAs present in Arabidopsis. The site consists of a section with descriptions of previously reported or annotated ncRNAs of Arabidopsis, the full list of animal, bacterial, and fungal ncRNAs that were used to search for homologs in Arabidopsis, and a list with the results of the in silico search for ncRNAs described above. A form that allows researchers to submit new ncRNAs to the database is also included.
For each EST found in our in silico search, we included a link to its GenBank record to facilitate the sequence retrieval. There is also a link to the AFGC site, where the transcript profile for each EST can be found and analyzed further. The current AFGC microarrays include ESTs primarily from the PRL2 library (Newman et al., 1994). ESTs from a developing seed library (White et al., 2000) will be included in the next generation of AFGC microarrays (E. Wisman, personal communication). Some information regarding the transcript profile of these ESTs can be found currently in the Arabidopsis Developing Seed Microarrays Web site (Girke et al., 2000).
To date, Stanford Microarray Database contains data from more than 160 AFGC microarrays from a variety of experiments that examine genotype differences, developmental processes and responses to biotic and abiotic signals. Moreover, this number should greatly increase in the coming year, making this resource a powerful tool to begin an analysis of potential roles for each of these genes.
DISCUSSION
ncRNAs are an emerging class of transcripts with intriguing characteristics and important roles in cellular physiology. The present work is a first step toward the systematic identification and study of this type of RNA in plants. Analysis of only a fraction of the existing ESTs yielded nearly 40 new putative ncRNAs and pepRNAs that have escaped previous annotation efforts. In addition, further scrutiny of known genes showed that, in some cases, probable ncRNAs had been annotated as peptides, whereas others that had been described as putative ncRNAs are most likely peptide-coding genes. None of the putative ncRNAs identified in this study had homologs outside the plant kingdom. This work illustrates that ncRNA genes are an underdeveloped area of plant genomics. The identification and characterization of poorly studied classes of genes is essential if we are to elucidate the function of all the genes of Arabidopsis (Chory et al., 2000).
How Many ncRNAs Are Estimated to Exist in Arabidopsis?
At this early stage in the analysis, we do not have definitive information about the number of ncRNAs present in any organism. Yeast is the only system where systematic searches for nonannotated transcripts, including putative ncRNAs, have been performed. In yeast, Serial Analysis of Gene Expression (SAGE) identified 170 tags that did not correspond to predicted ORF regions in the genome (Velculescu et al., 1997). A SAGE analysis will not detect all RNAs in an organism, and not all SAGE tags outside ORFs are new transcripts (some could be long UTRs, for example). Nevertheless, if we estimate that there are approximately 170 ncRNAs or pepRNAs out of approximately 6,000 genes in yeast, then this would indicate that 2% to 3% of yeast genes fall into this category. The search for RNAs transcribed from large gaps between predicted genes in yeast also argues for many hidden ncRNAs (Olivas et al., 1997). This strategy identified several new genes, including one that encodes a small nucleolar RNA and 16 that represent unique transcripts ranging in size from 161 to 1,200 nt, with the larger gaps between predicted ORFs giving rise to the larger transcripts. By extension, many small RNAs may be hidden within gaps smaller than 2 kb.
The number of ncRNAs in Arabidopsis cannot be precisely predicted as yet, but even conservative estimates indicate it is substantial. One way to estimate this number is to extrapolate from a study involving exhaustive cloning of cDNAs corresponding to a relatively long contiguous sequence. It was reported recently that seven cDNAs that could correspond to ncRNAs or pepRNAs were cloned out of the 50 cDNAs found in a 300-kb region of chromosome 1 (Kato et al., 1999). Several of these were more accurately categorized in our study (e.g. as chimeric RNAs or truncated RNAs; see Table I), with the availability of the complete genome sequence. However, even if there are only one or two ncRNA or pepRNA genes in this region, the estimated percentages (2%–4%) would be as high as in yeast. Consistent with this estimate was the finding that one gene in a 40-kb contig was a natural antisense transcript (Terryn et al., 1998; Terryn and Rouzé, 2000).
In our in silico search for ncRNAs in the AFGC EST collection, we identified 39 putative ncRNAs and pepRNAs represented in a population that is arguably biased against ncRNAs and that represents only about 40% of the protein-coding genes. Like most cDNA libraries, those giving rise to these EST clones were size selected to avoid small cDNAs, so the population that is expected to contain many ncRNAs and pepRNAs would be lost. ncRNAs that lack a poly(A+) tail would also not be represented in the EST collection we screened. Finally, our screening strategy was designed to avoid false positives and consequently it could filter out ESTs that represent bona fide ncRNAs. A clear example of this is the fact that antisense RNAs are lost in our screening because they have high degrees of homology to protein-coding genes and would be discarded by our automated selection process. However, the existence of this type of ncRNA in plants is well documented (Terryn and Rouzé, 2000). These arguments indicate that the number of putative ncRNAs we identified from Arabidopsis is likely a vast understimate. Thus, although it is not possible to indicate how many ncRNAs the Arabidopsis genome encodes without further experiments, current indications suggest there are a sizable number.
The manual analysis of the selected sequences allowed the classification of most of them as putative ncRNAs or putative pepRNAs. As indicated, the lack of homologs from other species prevented the assignment of some of these ESTs to one of these categories. Although our classification has been as precise as possible with the available data, the nature of this classification is expected to be dynamic; i.e. as more data become available, some ESTs could be moved from one category to another or discarded as part of protein-coding genes. A definitive classification will require extremely detailed mutagenesis experiments designed to identify directly whether a specific gene acts as an RNA or a peptide. Even with this type of data, it is sometimes difficult to address this question completely. For example, ENOD40, a gene that is induced during the early stages of nodule formation in Medicago sativa was first described as an ncRNA due to a lack of significant ORFs (Crespi et al., 1994). Later, detailed mutagenesis experiments showed that translation of two small peptides and the RNA itself are important for ENOD40 function during the nodulation process (van de Sande et al., 1996; Sousa et al., 2001).
pepRNAs, such as the one shown in Figure 2A, were an interesting by-product of our analysis that will also contribute to the complete annotation of the Arabidopsis genome. Small peptides have been recently described as a novel type of signaling molecule in plants. Key examples are systemin, involved with the wound response, CL3 implicated in flower development, the S-pollen peptides that function in sporophytic self-incompatibility, phytosulfokines that are associated with cell proliferation, and others (for review, see Schopfer and Nasrallah, 2000; Ryan and Pearce, 2001). Thus, similar to the situation for ncRNAs, it is expected that small peptides will be found to have important roles in a variety of processes.
It is important to note that although all the pepRNAs we identified have homologs in other species, only a few ncRNAs do. This feature is, in part, a consequence of the screening strategy utilized here, and could also be explained by biological characteristics of the transcripts analyzed. For our analysis, it is necessary that pepRNAs have homologs to be classified as such. A transcript with the potential to encode a small peptide but for which no homologs have been identified and therefore cannot be analyzed as shown in Figure 2 is designated as uncategorized. From the biological perspective, several ncRNAs are unstable transcripts or low-abundance transcripts, which could make them more difficult to find in smaller EST collections from other plants. As a consequence, we do not expect that ncRNAs will be reclassified as pepRNA when more homologs are found, although this could be the case for the uncategorized RNAs. In addition, because ESTs are partial sequences, the classification of each EST can change due to further analysis of individual clones (for example, after obtaining the sequence of the entire clone).
Most ncRNAs Are Predicted to Be Plant Specific
During our analysis and classification of ESTs, a striking observation was made. None of the putative ncRNAs discovered in this study were found to have homologs outside of the plant kingdom. The same is true for the previously described plant ncRNAs (data not shown). Furthermore, our search for Arabidopsis genes homologous to animal, bacterial, or fungal ncRNAs did not produce any significant matches. In the Arabidopsis genome, plant-specific protein coding genes are a significant component (Arabidopsis Genome Initiative, 2000), so the presence of many plant-specific ncRNA genes is also expected. In many cases, this could be due to ncRNAs being involved in kingdom-specific processes, as concluded from a study on the evolution of C. elegans let-7, which is involved in late embryo development (Pasquinelli et al., 2000). Homologs of this small ncRNA are found in a range of higher animal species but not in jellyfish, sponges, yeast, Escherichia coli, or Arabidopsis. Another small ncRNA from nematodes, lin-4 RNA, is restricted to the Caenorhabditae genus (Pasquinelli et al., 2000). In some cases, the observed kingdom specificity may not be explained by kingdom-specific functions but instead by distantly related ncRNAs that have evolved so extensively to accommodate changes in their targets that they are no longer recognized as homologs. ncRNAs that participate in similar processes in different kingdoms such as gene dosage or stress responses might be related in this way.
On the other hand, some ncRNAs with essential or “housekeeping” functions are expected to be conserved among kingdoms, as is the case for SRP RNA, the RNA component of the signal recognition particle that directs ribosomes to the ER. The reaction catalyzed by SRP must have been of great importance early in evolutions because phylogenetic evidence clearly points to an ancient function for SRP, one that perhaps reaches as far back as the hypothetical “RNA world” (Walter et al., 2000). Similar conservation is expected for other ncRNAs such as telomerase RNA or RNase P RNA. Although these RNAs have not been found in plants so far, the fact that other components of the telomerase and RNase P complexes have been identified (Arends and Schon, 1997; Fitzgerald et al., 1999) suggests that these RNAs also exist in plants.
Future Prospects
One advantage of beginning this analysis with the EST collections used to generate the AFGC microarrays is that expression data are available for virtually all the putative ncRNAs and pepRNAs identified in this study and will continue to accumulate. At this stage, such data can help us begin to hypothesize roles for some ncRNAs and pepRNAs, which could then be prioritized for functional analyses. For example, the putative ncRNA corresponding to EST 178K15T7 is unstable and light repressed, as indicated in Table IV. It will be interesting to examine whether over- or underexpression of this gene will lead to phenotypes consistent with a light-regulatory function. Some previously described ncRNAs, such as CR20, Mt4/TPSI1, and JAW, also have interesting regulatory features with potential functional significance. As more ncRNAs are incorporated into arrays and gene chips, and more gene expression profiling data accumulate, clustering and other analysis tools should help reveal networks of ncRNAs and the genes they regulate. A putative function for ncRNAs is gene regulation, probably through sequence-specific mechanisms. A quick homology search for segments of these putative ncRNAs with the entire genome showed that in fact some of them have small regions with sequence homology to other parts of the genome. For example, AtGUT15 and AtCR20-1 share a region of high homology. These genes are located in chromosomes 2 and 4, respectively. The comparison of their sequence with the entire genome showed that part of the highly conserved region is also present in chromosome 1 (data not shown), suggesting that this region of chromosome 1 could be a target sequence for the putative regulatory activity of this gene family. These exciting data should provide the foundation for a more detailed functional analysis of ncRNAs that should have a high potential impact.
The work presented here emphasizes the importance of future studies to identify a complete collection of ncRNAs from plants. Promising approaches might include the creation of cDNA libraries specific for small RNAs, the analysis of larger EST collections, and the development of new algorithms for comparative genome analysis or the identification of antisense ncRNAs. Implementation of these approaches should assure that the part of the Arabidopsis genome devoted to the production of ncRNAs will not remain underdeveloped for long.
MATERIALS AND METHODS
Computational Analysis
Arabidopsis EST sequences from the PRL2 library (Newman et al., 1994), seed-specific EST library (White et al., 2000), and Arabidopsis bacteria artificial chromosome (BAC) sequences were obtained from the National Center for Biotechnology Information (NCBI) using the batch ENTREZ program. ESTs were compared with the BAC sequences using the program blastall, also obtained from NCBI, which can take a multiple FASTA-formatted file of sequences as query (Altschul et al., 1997). The output of the blastall program was parsed with a PERL program to extract the coordinates of the BAC sequence corresponding to the EST. Another PERL program was used to extract 5,000 bp on each side of the sequence corresponding to the EST. This genomic sequence was analyzed by the gene-finding program GeneMark.hmm (Lukashin and Borodovsky, 1998), and a PERL program was used to evaluate whether an EST overlapped any predicted transcribed regions. This program produced a file containing the accession numbers of the EST and corresponding BAC coordinates. All the ESTs that did not overlap with a predicted transcribed region were retrieved and compared with the nonredundant protein database of NCBI using BLASTX. Any EST that produced an E score = 10−5 or smaller was discarded. The remaining ESTs were compared with the sequences in the dbEST database using BLASTN to obtain homologous ESTs from Arabidopsis and other species to be used in the manual analysis of each individual EST that remained after the different filtering steps. Default parameters were used in all the sequence analysis programs.
Manual Analysis
The final set of ESTs was evaluated by a combination of computational analysis and individual inspection. Genomic sequences corresponding to 2,500 bp on each side of every individual EST were analyzed using FGENE (Salamov and Solovyev, 2000; http://dot.imgen.bcm.tmc.edu:9331/gene-finder/gf.html) prediction to detect potential genes that could have been missed by GeneMark.hmm. The genomic annotation of the region where each EST was mapped was also retrieved. In any case where the gene prediction or annotation overlapped with an EST, such an EST was discarded. Then, each EST sequence was corrected based on the corresponding genomic data because EST sequences generally contain mistakes. These corrected sequences were analyzed using BLASTN, BLASTX, and TBLASTX to look for homology to known or predicted genes or proteins. Finally, six-frame translation and analysis of EST contigs were used to search for potential ORFs and to evaluate the possibility that a given EST would correspond to UTRs of predicted protein-coding genes.
Note Added in Proof
Due to the availability of additional sequence information, some of the ESTs in Table III have been reclassified. Please see http://www.prl.msu.edu/PLANTncRNAs/for an update classification.
ACKNOWLEDGMENTS
We thank Drs. Mike Thomashow and James Kastenmayer (Michigan State University) for critical reading of the manuscript and Nicole LeBrasseur (Michigan State University) for critical reading of the manuscript and editorial assistance.
Footnotes
This work was supported by the National Science Foundation (grant no. DBN9872638 to P.J.G.) and by the U.S. Department of Energy (grant no. DE–FG02–91ER20021 to P.J.G.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010501.
LITERATURE CITED
- Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- Altman S, Kirsebom L. Ribonuclease P. In: Gesteland RF, Cech T-R, Atkins JF, editors. The RNA World. Ed 2. Plainview, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 351–380. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Arends S, Schon A. Partial purification and characterization of nuclear ribonuclease P from wheat. Eur J Biochem. 1997;244:635–645. doi: 10.1111/j.1432-1033.1997.t01-1-00635.x. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. The end of the (DNA) line. Nat Struct Biol. 2000;7:847–850. doi: 10.1038/79594. [DOI] [PubMed] [Google Scholar]
- Burleigh SM, Harrison MJ. Characterization of the Mt4 gene from Medicago truncatula. Gene. 1998;216:47–53. doi: 10.1016/s0378-1119(98)00326-6. [DOI] [PubMed] [Google Scholar]
- Burleigh SM, Harrison MJ. The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol. 1999;119:241–248. doi: 10.1104/pp.119.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7:831–833. doi: 10.1038/82816. [DOI] [PubMed] [Google Scholar]
- Chory J, Ecker RJ, Briggs S, Caboche M, Coruzzi GM, Cook D, Dangl J, Grant S, Guerinot ML, Henikoff S. National Science Foundation-sponsored workshop report: “the 2010 project” functional genomics and the virtual plant: a blueprint for understanding how plants are built and how to improve them. Plant Physiol. 2000;123:423–426. doi: 10.1104/pp.123.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994;13:5099–5112. doi: 10.1002/j.1460-2075.1994.tb06839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Erdmann VA, Barciszewska MZ, Szymanski M, Hochberg A, de Groot N, Barciszewski J. The non-coding RNAs as riboregulators. Nucleic Acids Res. 2001;29:189–193. doi: 10.1093/nar/29.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann VA, Szymanski M, Hochberg A, de Groot N, Barciszewski J. Non-coding, mRNAs-like RNAs database Y2K. Nucleic Acids Res. 2000;28:197–200. doi: 10.1093/nar/28.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 2000;124:1570–1581. doi: 10.1104/pp.124.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Knudsen B, Zwieb C, Samuelsson T. SRPDB (Signal Recognition Particle Database) Nucleic Acids Res. 2001;29:169–170. doi: 10.1093/nar/29.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Suzuki M, Kuwahara A, Ooe H, Higano-Inaba K, Komeda Y. Isolation and analysis of cDNA within a 300 kb Arabidopsis thaliana genomic region located around the 100 map unit of chromosome 1. Gene. 1999;239:309–316. doi: 10.1016/s0378-1119(99)00403-5. [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation center. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Liu C, Muchhal US, Raghothama KG. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol Biol. 1997;33:867–874. doi: 10.1023/a:1005729309569. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000;24:559–567. doi: 10.1046/j.1365-313x.2000.00893.x. [DOI] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Moss EG. Non-coding RNAs: lightning strikes twice. Curr Biol. 2000;10:R436–R439. doi: 10.1016/s0960-9822(00)00528-5. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas WM, Muhlrad D, Parker R. Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res. 1997;25:4619–4625. doi: 10.1093/nar/25.22.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pavy N, Rombauts S, Dehais P, Mathe C, Ramana DV, Leroy P, Rouzé P. Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics. 1999;15:887–899. doi: 10.1093/bioinformatics/15.11.887. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ryan CA, Pearce G. Polypeptide hormones. Plant Physiol. 2001;125:65–68. doi: 10.1104/pp.125.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer CR, Nasrallah JB. Self-incompatibility: prospects for a novel putative peptide-signaling molecule. Plant Physiol. 2000;124:935–940. doi: 10.1104/pp.124.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C, Johansson C, Charon C, Manyani H, Sautter C, Kondorosi A, Crespi M. Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol. 2001;21:354–366. doi: 10.1128/MCB.21.1.354-366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CB, Green PJ. Identification and characterization of genes with unstable transcripts (GUTs) in tobacco. Plant Mol Biol. 1995;28:27–38. doi: 10.1007/BF00042035. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Toyama T, Takeba G, Tsuji H. Changes in expression of two cytokinin-repressed genes, CR9 and CR20, in relation to aging, greening and wounding in cucumber. Planta. 1995;196:387–395. [Google Scholar]
- Teramoto H, Toyama T, Takeba G, Tsuji H. Noncoding RNA for CR20, a cytokinin-repressed gene of cucumber. Plant Mol Biol. 1996;32:797–808. doi: 10.1007/BF00020478. [DOI] [PubMed] [Google Scholar]
- Terryn N, Gielen J, De Keyser A, Van Den Daele H, Ardiles W, Neyt P, De Clercq R, Coppieters J, Dehais P, Villarroel R. Sequence analysis of a 40-kb Arabidopsis thaliana genomic region located at the top of chromosome 1. Gene. 1998;215:11–17. doi: 10.1016/s0378-1119(98)00286-8. [DOI] [PubMed] [Google Scholar]
- Terryn N, Rouzé P. The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci. 2000;5:394–396. doi: 10.1016/s1360-1385(00)01696-4. [DOI] [PubMed] [Google Scholar]
- van de Sande K, Pawlowski K, Czaja I, Wieneke U, Schell J, Schmidt J, Walden R, Matvienko M, Wellink J, van Kammen A. Modification of phytohormone response by a peptide encoded by ENOD40 of legumes and a nonlegume. Science. 1996;273:370–373. doi: 10.1126/science.273.5273.370. [DOI] [PubMed] [Google Scholar]
- van Hoof A, Kastenmayer JP, Taylor CB, Green PJ. GUT15 cDNAs from tobacco (accession no. U84972) and Arabidopsis (accession no. U84973) correspond to transcripts with unusual metabolism and a short conserved ORF. Plant Physiol. 1997;113:1004. [Google Scholar]
- Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Hieter P, Vogelstein B, Kinzler KW. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- Walter P, Keenan R, Schmitz U. Perspectives: structural biology: SRP: where the RNA and membrane worlds meet. Science. 2000;287:1212–1213. doi: 10.1126/science.287.5456.1212. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–498. doi: 10.1016/0092-8674(94)90426-x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Todd J, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C. A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol. 2000;124:1582–1594. doi: 10.1104/pp.124.4.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildsmith SE, Elcock FJ. Microarrays under the microscope. J Clin Pathol. 2001;54:8–16. doi: 10.1136/mp.54.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Salz HK. Remodelling chromatin with RNA. Nature. 1997;386:228–229. doi: 10.1038/386228a0. [DOI] [PubMed] [Google Scholar]