Abstract
A major response of eukaryotic cells to the presence of unfolded proteins in the lumen of the endoplasmic reticulum (ER) is to activate genes that encode ER-located molecular chaperones, such as the binding protein. This response, called the unfolded protein response, requires the transduction of a signal from the ER to the nucleus. In yeast (Saccharomyces cerevisiae) and mammalian cells, an ER-located transmembrane receptor protein kinase/ribonuclease called Ire1, with a sensor domain in the lumen of the ER, is the first component of this pathway. Here, we report the cloning and derived amino acid sequences of AtIre1-1 and AtIre1-2, two Arabidopsis homologs of Ire1. The two proteins are located in the perinuclear ER (based on heterologous expression of fusions with green fluorescent protein). The expression patterns of the two genes (using β-glucuronidase fusions) are nearly nonoverlapping. We also demonstrate functional complementation of the sensor domains of the two proteins in yeast and show that the Ire1-2 protein is capable of autotransphosphorylation. These and other findings are discussed in relation to the involvement of these genes in unfolded protein response signaling in plants.
The endoplasmic reticulum (ER), which in plant cells is composed of numerous distinct morphological domains with specific functions (Staehelin, 1997; Zheng and Staehelin, 2001), is a three-dimensional network of membranous tubules and sheets that extends throughout the cytoplasm. At the periphery of the cell, it underlies the plasma membrane, and around the nucleus it links up with the nuclear envelope. The role of the rough ER in the biosynthesis, modification, folding, and export of secreted, vacuolar, and membrane proteins of plant cells has been the subject of numerous studies (for review, see Vitale and Denecke, 1999; Vitale and Gallili, 2001). About one-third of all cellular proteins are thought to be synthesized on the rough ER. Correct folding of newly synthesized proteins in the lumen of the ER is a prerequisite for their transport to other cellular destinations. To promote polypeptide folding and subunit assembly, the ER lumen contains molecular chaperones, disulfide exchange proteins, and a system to synthesize and attach Asn-linked glycans to nascent polypeptides. Malfolded and unassembled proteins are subject to a quality control process that retains them in the ER to restore them to normal conformation or that eliminates them via a degradation after transport to the vacuole, or in the cytosol, after retrotranslocation across the ER membrane (for review, see Bonifacino and Weissman, 1998; Mori, 2000).
When stress causes protein folding to be slowed, the presence of unfolded proteins in the ER triggers several cellular responses. One such response, called the unfolded protein response (UPR), results in the enhanced expression of a large number of genes. As part of the UPR, the expression of genes encoding ER chaperones such as the binding protein (BiP) and protein disulfide isomerase (PDI) is enhanced. Genome-wide expression studies show that the UPR affects not only ER chaperone genes, but multiple functions of the ER and the rest of the secretory pathway as well (Travers et al., 2000). The second cellular response to unfolded proteins, so far only described in mammalian cells, consists of a transient attenuation in the rate of protein synthesis so that less protein enters the ER (see Brostrom and Brostrom, 1998; Harding et al., 2000). Together, these two responses minimize the presence of unfolded proteins in the ER.
In yeast (Saccharomyces cerevisiae) and mammalian cells where the UPR has been studied in some detail, signaling from the ER to the nucleus is mediated by the transmembrane protein kinase/ribonuclease Ire1. This protein has a sensor domain in the lumen of the ER that is thought to sense the presence of unfolded proteins. BiP itself has been shown to be the ligand for this sensor domain (Bertolotti et al., 2000; Okamura et al., 2000). In yeast, the nuclease domain initiates the splicing of the mRNA that encodes a transcription factor (Hac1p) that is ultimately responsible for the transcriptional activation of the UPR target genes. Yeast has only one Ire1 gene, but humans and mice have at least two genes for this protein (for review, see Kaufman, 1999; Silverman and Williams, 1999). Given the complex nature of the UPR, the two mammalian IRE1 proteins may affect different downstream processes. Both proteins appear to signal the induction of chaperones as shown by their expression in cultured cells (Tirasophon et al., 1998; Wang et al., 1998). Iwawaki et al. (2001) recently reported that human IRE1β induces translational repression through 28S rRNA cleavage in response to ER stress. The attenuation of protein synthesis is also signaled via the protein kinase dsRNA-dependent (PKR)-like ER kinase (PERK) in mammalian cells. This kinase has a related lumenal sensor domain (Harding et al., 1999), but lacks a ribonuclease domain. In mammals, under unstressed condition, BiP represses UPR signaling through the association with the lumenal regions of PERK and IRE1, but the accumulation of mis-folded proteins in the ER relieves this repression by bringing about the release of BiP from the lumenal domains of PERK and IRE1 (Harding et al., 2000). Similar regulation is also observed in yeast (Okamura et al., 2000).
A variety of stress conditions, including treatment with tunicamycin, inhibit the proper folding and oligomerization of proteins in the lumen of the ER (Pelham, 1989) and tunicamycin is routinely used as a way to induce the UPR. Treatment of plant cells or tissues with tunicamycin induces BiP and other ER chaperones (Fontes et al., 1991; Shorrosh and Dixon, 1992). D'Amico et al. (1992) showed that treatment of bean (Phaesoleus vulgaris) cotyledons with tunicamycin not only induces BiP, but that BiP can be co-immunoprecipitated with the newly synthesized (unglycosylated and presumably malfolded) polypeptides present in the ER. More recent evidence suggests that BiP and calreticulin form an abundant complex in the ER (Crofts et al., 1998). In tobacco (Nicotiana tabacum) and soybean (Glycine max), BiP is encoded by a small gene family (Denecke et al., 1991; Kalinski et al., 1995), whereas in Arabidopsis there are only two BiP genes (Koizumi, 1996). These early investigations set the stage for all later work on the role of chaperones in protein folding, protein assembly, and quality control in the ER of plant cells (for review, see Vitale and Denecke, 1999).
With respect to the UPR itself, there is almost no information regarding plant cells. The floury-2 endosperm mutant of maize (Zea mays) presents an interesting case of ER stress in plants. This mutant produces an aberrant zein storage protein with a defective signal peptide-processing site (Coleman et al., 1995) and the seeds have dramatically increased levels of BiP and other chaperones (Boston et al., 1991; Li and Larkins, 1996; Coleman et al., 1997; Gillikin et al., 1997). This system was recently used to show that the enzymes of lipid metabolism associated with the ER are up-regulated (Shank et al., 2001). Tunicamycin, which inhibits the synthesis of dolichol-linked glycans (Takatsuki et al., 1975), has been most widely used to induce the UPR in plant cells. Overexpression of the gene encoding the enzyme that is inhibited by tunicamycin makes plants less sensitive to this antibiotic and obviates the UPR (Koizumi et al., 1999).
The role of BiP in ER stress was explored by Leborgne-Castel et al. (1999) who created lines of tobacco that overexpress BiP. They found that overexpression from a transgene caused down-regulation of the endogenous BiP genes and greatly reduced the UPR. They also observed that tunicamycin down regulated the level of α-amylase, a secreted protein, compared with a cytosolic protein. Co-expression of BiP restored amylase synthesis, suggesting that there was insufficient BiP to take care of the large increase in the demand for folding required by the overexpressed α-amylase. Because this particular amylase is not a glycosylated protein, the ER stress resulting from tunicamycin treatment could be signaling through the PERK pathway. However, an exhaustive search of the Arabidopsis database did not reveal any PERK homologs.
Here, we report the presence of two Ire1 homologs in the ER of Arabidopsis cells. The derived amino acid sequences have the three domains characteristic of Ire1 proteins found in other organisms: a lumenal sensing domain, a protein kinase domain, and a ribonuclease domain. The two proteins share 41% amino acid identity. Introduction into Δire1 yeast cells of chimeric genes encoding the lumenal domains of either of the Arabidopsis Ire1 proteins and the other two domains of yeast Ire1 results in yeast strains that respond to tunicamycin by enhancing the expression of an introduced UPR element (UPRE)-lacZ reporter construct. Analysis of plants transformed with AtIre1 promoter-β-glucuronidase (GUS) fusions shows that the two genes have nearly non-overlapping patterns of expression.
RESULTS
Accumulation of Chaperone mRNAs Is Induced by Tunicamycin
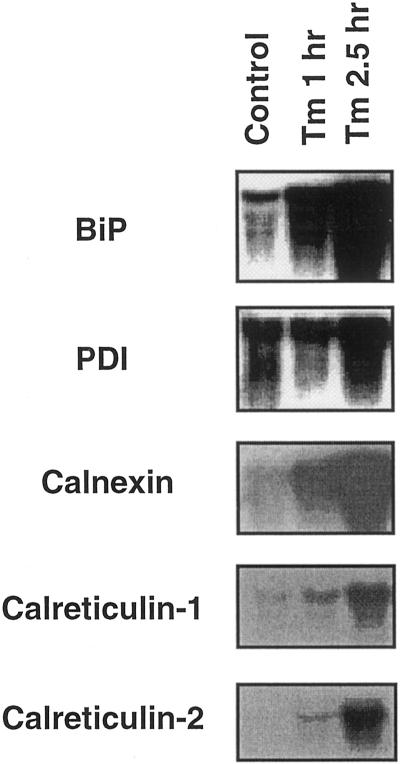
Tunicamycin (5 μg mL−1) treatment of immersed leaf segments causes rapid changes in ER chaperone mRNAs (Fig. 1). mRNAs of five chaperones (BiP, PDI, calnexin, calreticulin-1, and calreticulin-2) increased rapidly upon tunicamycin treatment. BiP mRNA was also induced by two chemical stresses: azetidine-2-carboxylate and dithiothreitol (results not shown). These results confirm and extend earlier work with plant tissues, demonstrating the existence of the UPR in plants.
Figure 1.
Cooperative induction of mRNA of the ER chaperones in Arabidopsis by tunicamycin. Total RNA was extracted from Arabidopsis seedlings after addition of tunicamycin (5 μg mL−1) to the culture medium. Incubation periods after treatment were 0 h (Control), 1 h (Tm 1 h), and 2.5 h (Tm 2.5 h). The RNA blot was probed with the cDNAs of BiP, PDI, calnexin, calreticulin-1, and calreticulin-2.
Cloning of the Arabidopsis Ire1 Homologs
Ire1p of yeast is a large transmembrane protein (1,115 amino acids) with a three-domain structure: A large sensor domain is followed by a protein kinase domain and a nuclease domain.
The sensor domain and the kinase domain are separated by a transmembrane helix. Using the amino acid sequence of yeast Ire1p, we conducted a search for homologs of this protein in the Arabidopsis database. We found two genomic sequences that, when translated, showed considerable sequence identity with Ire1 proteins of other organisms. We designated these genes AtIre1-1 and AtIre1-2. AtIre1-1 appeared to be a complete coding frame in a long genomic sequence (accession no. AB016884), but AtIre1-2 was only a partial sequence, at the end of a BAC clone (accession no. B28768) when we found it. We amplified a full-length cDNA of AtIre1-1 by reverse transcriptase (RT)-PCR. For AtIre1-2, we first amplified a portion of the cDNA according to the known sequence and then this fragment was used as a probe to screen a cDNA library of Arabidopsis. The cDNA we obtained was almost full length and in the meantime a complete genomic sequence for AtIre1-2 was released (accession no. AC007584). This allowed us to isolate a full-length cDNA for AtIre1-2 by RT-PCR using primers derived from the genomic sequence. The sequences of the AtIre1-1 and AtIre1-2 cDNAs that we obtained did not match the predicted coding sequences found in the database because the predictions of some of the intron splicing sites were incorrect. In the case of AtIre1-1, the first intron was not detected by computer analysis. For AtIre1-2, the C-terminal portion of the protein, encoding the kinase and RNase domains, was predicted as a protein. The correct nucleotide sequences of AtIre1-1 and AtIre1-2 have been deposited in the database (accession nos. AB049936 and AB049937, respectively). The derived amino acid sequences of the two proteins are shown in Figure 2.
Figure 2.
Alignment of Ire1 homologs from various organisms. Two Arabidopsis homologs (AtIre1-1 and AtIre1-2; this study), yeast Ire1p (Ire1p, accession no. Z11701), human Ire1α (hIre1α, accession no. AF059198), and mouse Ire1β (mIre1β, accession no. AF071777) were aligned using ClustalW software. Amino acid residues conserved in three out of five sequences are boxed. The ATP binding motif (VAVKR) and the Ser/Thr protein kinase motif (DLKPQN) are underlined.
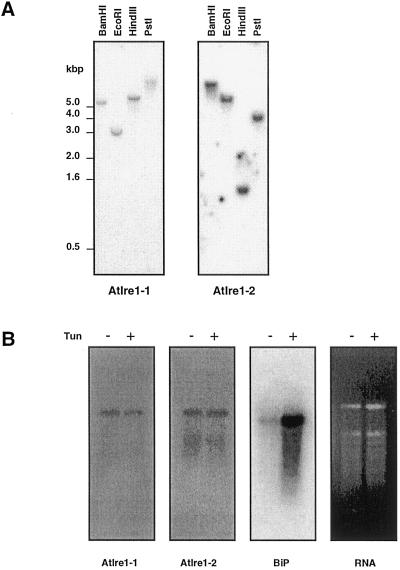
A comparison of the cDNA and genomic nucleotide sequences showed that AtIre1-1 and AtIre1-2 have five and six introns, respectively. The splice sites are conserved between the two genes except that the last intron of AtIre1-2 is missing from AtIre1-1. A genomic Southern analysis was carried out using gene-specific probes for each gene (Fig. 3A). A single band in each restriction digest indicated that AtIre1-1 and AtIre1-2 exist as a single copy in the genome of Arabidopsis.
Figure 3.
A, Genomic Southern analysis of AtIre1-1 and AtIre1-2. Genomic DNA of Arabidopsis digested with two restriction enzymes was fractionated by agarose gel, blotted, and probed with a fragment of either AtIre1-1 or AtIre1-2 cDNA. B, mRNA abundance analysis of AtIre1-1, AtIre1-2, and BiP. Poly(A+) RNA was isolated from Arabidopsis seedlings with no treatment or treated with tunicamycin (5 μg mL−1 for 4 h). Two micrograms of RNA sample from each treatment was used for northern blotting. Probes used were same as in A for AtIre1-1 and AtIre1-2. BiP probe was the same as in Figure 1.
Characteristics of the Amino Acid Sequences
AtIre1-1 and AtIre1-2 encode open reading frames of 881 and 841 amino acids, respectively. Although the polypeptides are smaller than those of human (977 amino acids) and yeast (1,115 amino acids), the derived Arabidopsis amino acid sequences revealed the structural features of yeast and mammalian Ire1 proteins. Namely, they have short hydrophobic regions at the N-terminal end and near the middle of the proteins, representing the likely signal peptides and transmembrane domains. Following the putative transmembrane domain, they contain kinase and RNase domains. These two C-terminal domains have significant sequence identity with the homologous domains of other Ire1 proteins (30%–40%). The kinase domain has a critical Lys residue in the conserved VAVKR domain and the signature sequence of Ser/Thr kinases (DLKPEN) is represented as DLKPQN (underlined in Fig. 2). The N-terminal regions representing the likely sensor domains are much less conserved among the different homologs (<10%). Comparing AtIre1-1 with AtIre1-2, the C-terminal domains also have a higher sequence identity (65%) than the N-terminal regions (28%).
Expression of the AtIre Genes
To detect transcripts of AtIre1-1 and AtIre1-2, a northern-blot analysis was done. Using total RNA, we were never able to detect any signals. Therefore, we isolated poly(A+) RNA for a northern blot, and in this way we detected weak signals for AtIre1-1 and AtIre1-2 (Fig. 3B). A search in the Arabidopsis expressed sequence tag (EST) database did not produce any ESTs for AtIre1-1 or AtIre1-2, but in August 2000, a single EST of AtIre1-2 was reported in developing seeds and added to the database with our annotation. Together, these results indicate that expression levels of the AtIre1 genes are very low in Arabidopsis. Unlike BiP, which is highly induced by tunicamycin, the levels of the AtIre transcripts were not changed by tunicamycin treatment.
AtIre1-2 Has Protein Kinase Activity
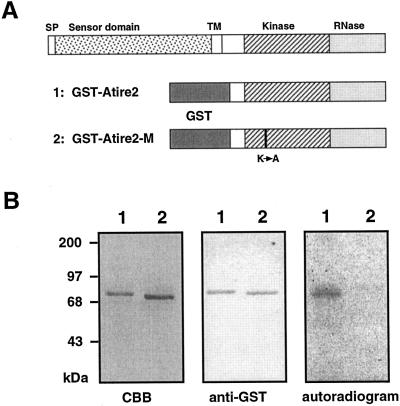
Ire1p of yeast and its mammalian homologs have autophosphorylation activity in vitro. To determine if AtIre1 has protein kinase activity, we expressed in Escherichia coli a fusion polypeptide consisting of glutathione S-transferase (GST) and the C-terminal half of AtIre1-2 containing the protein kinase and ribonuclease domains. A second construct carried a K442A mutation in which Lys-442 was mutated to Ala (see Fig. 4). This mutation inactivates the enzymatic activity of other protein kinases. We purified the recombinant protein on a glutathione affinity column and obtained a single band of 73 to 75 kD on a Coomassie Brilliant Blue-stained gel. This band reacted with an anti-GST serum and its molecular size was consistent with that of the fusion protein. Incubation with 32P-labeled ATP resulted in the labeling of the same polypeptide as was identified by the GST antiserum. The GST fusion polypeptide was not labeled with 32P in the K442A mutant. We suggest on the basis of this experiment that AtIre1-2 is a trans-phosphorylating protein kinase similar to other receptor kinases.
Figure 4.
A and B, Autophosphorylation activity of the C-terminal domain of AtIre1-2. A, Domain structure of Ire1 (top) and of the GST fusions showing the location of the K to A mutation. B, The fusion protein of GST with the C-terminal domain of AtIre1-2 was expressed in E. coli and affinity purified on glutathione-Sepharose. Purified protein incubated with γ-32P ATP was subjected to SDS-PAGE. The gel was Coomassie stained (CBB) and exposed to x-ray film (autoradiogram). The same fraction was used for an immunoblot analysis with anti-GST antibody (anti-GST). In A and B, lane 1 has the wild-type GST-AtIre1-2 fusion and lane 2 has the mutant GST-AtIre1-2 fusion. There is no 32P labeling of the fusion polypeptide of the mutant (lane 2 in the autoradiogram).
Subcellular Localization of AtIre1-Green Fluorescent Protein (GFP) Fusions
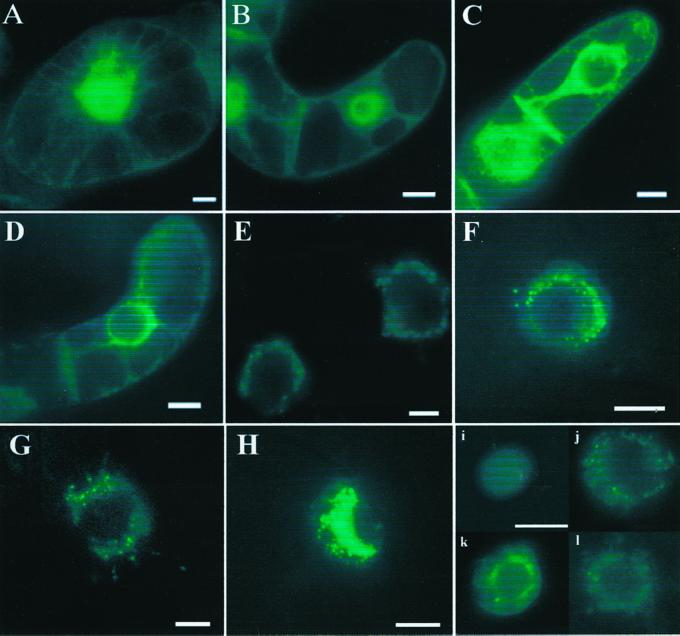
To determine the subcellular location of AtIre1, we made translational fusions with the GFP and expressed these constructs in cultured tobacco cells (BY-2 cells). As controls, we used GFP by itself and a construct in which GFP has a signal peptide and a carboxyterminal ER retention motif (Lys-Asp-Glu-Leu or KDEL). The expression of all constructs was driven by the cauliflower mosaic virus (CaMV) 35S promoter. The cells were examined by fluorescence microscopy. In cells expressing GFP alone (Fig. 5, A and B), the entire cytoplasm but not the vacuoles is diffusely labeled and the nucleus appears bright green, indicating that GFP enters the nucleus readily. Figure 5, C and D, show the results with the GFP-KDEL construct. Fluorescence is seen in the perinuclear zone, in cytoplasmic strands, and in the newly formed cross walls (possibly associated with plasmodesmata), but not in the nucleoplasm. In cells transformed with the AtIre1-1-GFP construct (Fig. 5, E and F) the fluorescence is confined to the perinuclear area. Figure 5G shows that the location of AtIre1-2-GFP is similar to that of AtIre1-1-GFP. Figure 5H is an image of a cell transformed with the mutant K442A AtIre1-2-GFP construct. The parallel construct of AtIre1-1-GFP (K487A) showed the same localization (data not shown). This mutation produces a nonfunctional protein, and the transformed cells invariably show brighter fluorescence. Treatment of cells with brefeldin A (10 μg mL−1 for 90 min), which causes redistribution of Golgi proteins (Staehelin and Driouich, 1997), did not change the distribution of the AtIre1 proteins (data not shown). A similar treatment with brefeldin A caused the redistribution of a GFP fusion with the Golgi marker enzyme α-mannosidase I (data not shown). These results rule out the Golgi as the location of the AtIre1 proteins.
Figure 5.
Localization of AtIre1-1-GFP and AtIre1-2-GFP fusion proteins by fluorescence microscopy. Epifluorescence images of tobacco BY-2 cells (A–H), and purified nuclei from tobacco BY-2 cells (i–l). A, B, and i, BY-2 cells express normal GFP. Note green fluorescence in the nucleoplasm. C, D, and j, BY-2 cells expressing GFP-KDEL. Note absence of fluorescence in the nucleoplasm. E, F, and k, BY-2 cells expressing AtIre1-1-GFP. G and l, Cells expressing AtIre1-2-GFP. H, BY-2 cell expressing the AtIre1-2(K442A)-GFP. Bars = 0.01 mm.
Figure 5, i through l, are of isolated nuclei from cells expressing GFP alone (Fig. 5i), GFP-KDEL (Fig. 5j), the AtIre1-1-GFP construct (Fig. 5k), and the AtIre1-2-GFP construct (Fig. 5l). The nucleoplasm is labeled in Figure 5i, indicating that GFP itself stays in the nucleus during nuclear isolation (NI). Together, these data support the interpretation that AtIre1-1 and AtIre1-2 are located in the nuclear envelope and/or the ER that is in close proximity to the nucleus.
The N-Terminal Regions of AtIre1-1 and AtIre1-2 Function as Sensors
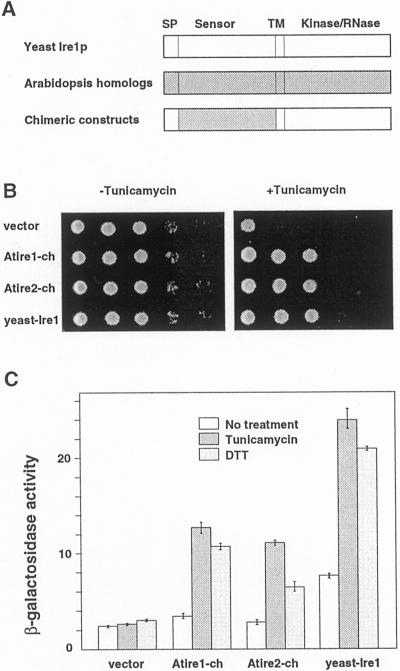
Although the amino acid sequences of the N-terminal region of AtIre1-1 and AtIre1-2 are very dissimilar from that of yeast Ire1p, we investigated whether these regions can function as sensors of stress in yeast cells. To test this possibility, we made chimeric genes of the N-terminal domains of AtIre1-1 and AtIre1-2, and the C-terminal domains of yeast Ire1p. The N-terminal domain of yeast Ire1p, except the signal peptide, was replaced with the N-terminal domain of either AtIre1-1 or AtIre1-2 (see Fig. 6A). These constructs were introduced into a yeast Δire1 mutant, using a single-copy (ARS and CEN) plasmid vector pRS313 (Sikorski et al., 1989). As a positive control, pRS313-carrying yeast IRE1 was used. The various yeast transformants were grown on synthetic dextrose plates without or with tunicamycin (0.2 μg mL−1). Growth of the yeast Δire1 mutant containing the empty vector was severely inhibited by tunicamycin. This inhibition of growth was rescued by transformation with yeast IRE1. Growth inhibition was also rescued with the two chimeric constructs Atire1-ch and Atire2-ch (Fig. 6B). To confirm that the UPR was induced when the yeast cells were subjected to ER stress, we measured the expression of β-galactosidase driven by a 22-bp UPRE of the yeast BiP gene (KAR2) promoter. In the yeast Δire1 mutant transformed with empty vector only, the induction of β-galactosidase was not observed upon treatment with either tunicamycin or dithiothreitol. Yeast Δire1 cells complemented with yeast IRE1 showed the expected response to these stresses. In yeast Δire1 cells transformed with the chimeric constructs, the chemical stresses also induced the accumulation of the enzyme (Fig. 5C). These results indicate that the N-terminal domains of the Arabidopsis Ire1-1 and Ire1-2 proteins can act as ER stress sensors in yeast.
Figure 6.
A through C, Complementation of yeast ire1 deletion mutant with chimeric constructs of yeast Ire1p and Arabidopsis Ire1 homologs. A, Schematic view of the chimeric constructs used for yeast complementation. Chimeric constructs consisted of the sensor domains of AtIre1-1 or AtIre1-2, and other parts (signal peptide and C-terminal half) of yeast Ire1p. B, Growth of Δire1 strains of yeast complemented with chimeric constructs in the absence or presence of tunicamycin. Δire1 containing vector only (vector), chimeric constructs for AtIre1-1 (Atire1-ch) and AtIre1-2 (Atire2-ch), and yeast IRE1 were grown on synthetic dextrose plates without tunicamycin (−Tunicamycin) and with tunicamycin (+Tunicamycin, final concentration at 0.2 μg mL−1). Approximately 1 × 105 cells were spotted on the left column of each plate. Series of one-tenth dilution of cells were spotted on the right side. Yeast cells were grown for 3 d at 30°C. C, Activity of β-galactosidase of yeast cells containing each construct. Cells were incubated at 30°C with 2 μg mL−1 tunicamycin for 4 h, or 1 mm dithiothreitol for 2 h, and their β-galactosidase activity was measured.
AtIre1-1 and AtIre1-2 Have Nonoverlapping Expression Patterns
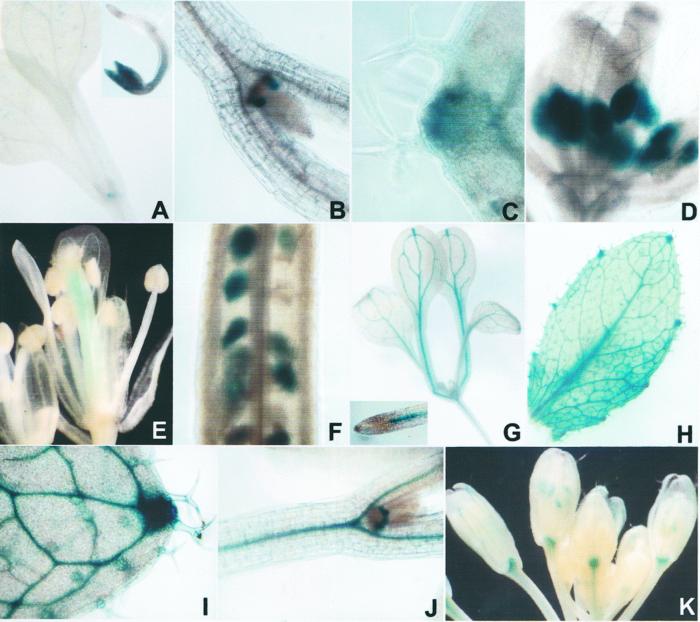
We made AtIre1 promoter::GUS fusions for both genes and examined their expression in Arabidopsis. GUS activity was found to be very low, in agreement with the finding that there were very few accessions of Ire-1 genes in the EST databases. The 1,996-bp promoter of AtIre1-1 drives GUS expression in the apical meristem (Fig. 7, A and B), at leaf margins where vascular bundles end (Fig. 7C) in the anthers before pollen is formed (Fig. 7D), and in the ovules at a very early stage of development (Fig. 7, E and F). There is no expression in more mature embryos. AtIre1-1 is also strongly expressed in the cotyledons immediately after germination (inset in the upper right hand corner of Fig. 7A) but not later on (Fig. 7A). Thus, the expression of AtIre1-1 in the plant is extremely restricted to certain tissues at specific developmental times. The 1,994-bp promoter of AtIre1-2 drives expression of GUS in the vascular bundles of young plants (Fig. 7G), leaves (Fig. 7, G and H), in roots (inset in Fig. 7G), seedlings (Fig. 7J), and in the receptacles of flowers and vascular bundles of the petals (Fig. 7K). This gene appears to be more widely expressed than AtIre1-1, but it is clearly not generally expressed throughout the plant.
Figure 7.
A through K, Histochemical staining of GUS of Arabidopsis plants transformed with AtIre1-promoter::GUS fusions. A through F, AtIre1-1 and G-K: AtIre1-2. A and B, Seedlings with fully expanded cotyledons (note staining of the meristems); A inset, 48-h-old seedling with stained cotyledons; C, leaf margin; D, flower with immature anthers; E, flower after pollination, only the gynecium is stained and anthers are not longer stained; F. ovules; G, seedling; G inset, root tip; H and I, rosette leaf; J, seedling; K, flowers.
DISCUSSION
In this report, we show that the genes of five different ER chaperones are induced during the UPR in Arabidopsis and we present the molecular characteristics of two ER-located transmembrane protein kinases, AtIre1-1 and AtIre1-2, whose homologs are involved in UPR signaling in other organisms; the two AtIre1 homologs have nearly nonoverlapping expression patterns. One of the two proteins is shown to function as a protein kinase (the other one was not tested) and the sensor domains of the proteins function in sensing unfolded proteins in yeast. It remains to be demonstrated that the two proteins are involved in the UPR of plant cells.
The UPR in Plant Cells
In the most recent major review on the functions of the ER in plant cells (Vitale and Denecke, 1999), no mention is made of the UPR, although ER quality control is discussed in detail. This omission is simply a reflection of the fact that there have been no systematic studies of the UPR in plants, although it was shown some years ago that tunicamycin increases the level of BiP mRNA and protein. In Figure 1, we show that 5 different ER-located chaperones are induced by tunicamycin. Plants, we conclude, behave similarly to other eukaryotes with respect to this important cellular process (Kaufman, 1999; Mori, 2000). This is not surprising, because the UPR is thought to control many essential aspects of ER function, not only those that relate to protein import, processing, and export (Travers et al., 2000). Furthermore, unfolded proteins are constantly being delivered to the ER of all cells, and these proteins must be either folded and assembled, or exported for degradation. Tunicamycin prevents protein folding by inhibiting the synthesis of high-Man glycans in the ER. In a similar manner, dithiothreitol can prevent protein folding by inhibiting disulfide bond formation, and this chemical has been shown to increase the levels of mRNA for two chaperones, BiP and PDI (Denecke et al., 1995). Nelson et al. (1997) did not find up-regulation of calreticulin mRNA after heat shock, but it is not clear to what extent the treatment interfered with protein folding. A related type of ER stress in maize endosperm, caused by the presence of an excessive amount of zein polypeptides with uncleaved signal peptides, has been shown to be accompanied by the up-regulation of calnexin, BiP, and PDI (Fontes et al., 1991; Li and Larkins, 1996; Wrobel, 1996). It is more than likely that the zein polypeptides are in an unfolded state in the ER of these cells.
The Arabidopsis Ire1 Proteins
Homologs of yeast IRE1 have been identified in several other eukaryotic species including Drosophila melanogaster, Cenorhabditis elegans, human, and mouse. Alignment of the derived amino acid sequences shows that the most conserved features of the proteins are the kinase and ribonuclease domains. The two Arabidopsis Ire1 proteins are somewhat smaller than the yeast protein but contain the same three domains, with greater sequence identity in the two enzymatic domains compared with the putative sensor domain. Each protein species is encoded by a unique gene. In mice and humans, two IRE-1 proteins, each encoded by a different gene, have so far been identified. The gene encoding AtIre1-1 is located on chromosome 5 (Bac K16H17), whereas the gene encoding AtIre1-2 is on chromosome 2 (Bac mjb20). The translation product from this gene is incorrectly annotated in the database as the putative protein comprises only the carboxyterminal half of AtIre1-2 and does not include the transmembrane and sensor domains.
AtIre1-GFP Fusion Proteins Are Located in the Perinuclear ER
The results presented in Figure 5 are consistent with a localization of the AtIre1-GFP fusion proteins in the perinuclear ER. The fluorescence obtained with the AtIre1-GFP constructs was much less than that obtained with GFP alone or GFP-KDEL. This lower fluorescence may be the result of the property of the Ire1 to regulate its own expression at the level of mRNA (Tirasophon et al., 1998). Localization of Ire1 in the ER of yeast cells was first postulated by Cox et al. (1993) and by Mori et al. (1993) on the basis of the molecular properties of the Ire1 proteins: a signal peptide, a transmembrane domain, and high-Man glycans. Light microscopy analysis of COS1 cells transfected with epitope-tagged mouse Ire1 shows colocalization with the ER marker ribophorin 2 (Wang et al., 1998). The clearer confocal images in the study of Tirasophon et al. (1998) show a perinuclear localization for human Ire1 transfected into COS1 cells, very much like the images shown in Figure 5. Brefeldin A at 5 mg mL−1 did not alter the distribution of fusion proteins in the cells, although it caused, as expected, a redistribution of the soybean Golgi marker α-mannosidase I (data not shown). We conclude that AtIre1 is present in the nuclear envelope and in the ER immediately adjacent to the nucleus. Deletion of the ribonuclease domains (data not shown) and a single amino acid mutation in the kinase domain that prevented kinase activity did not alter the observed subcellular distribution of the fusion protein. So far, all studies have been done with overexpressed Ire1 proteins and we cannot rule out the possibility that endogenous Ire1 is confined to the nuclear envelope itself. Upon induction of the UPR in mammalian cells, human Ire1 is proteolytically cleaved, and fragments containing the kinase and nuclease domains accumulate in the nucleus (Niwa et al., 1999). It may be relevant in this respect that both AtIre1 proteins have putative nuclear localization signals at the N-terminal end of this combined enzymatic domain (KKKKSKK in AtIre1-1 and KKKKNRK in AtIre1-2).
AtIre1-2 Is an Autophosphorylating Protein Kinase
Ire1 proteins are transmembrane receptor protein kinases that oligomerize when BiP is detached from the receptor domain (Bertolotti et al., 2000; Okamura et al., 2000). Oligomerization then leads to trans-autophosphorylation of the cytoplasmic effector domains (Shamu and Walter, 1996; Bertolotti et al., 2000). BiP thus serves as an inhibitory ligand for these receptor kinases, which mechanistically are similar to other transmembrane protein kinase receptors. AtIre1-1 and ArIre1-2 have a DLKPQN motif that closely resembles the consensus motif for Ser/Thr kinases (DLKPEN; Lindberg et al., 1992). Incubation of the purified GST fusion protein with 32P-ATP resulted in the phosphorylation of the fusion protein. Lys-442 is contained in a motif (VAVKR) conserved in all protein kinases that contacts the α- and β-phosphates of ATP and is essential for catalysis. Mutation of this Lys residue (K442A) resulted in the inactivation of the protein kinase. This result confirms that the phosphorylation of the AtIre1-2-GST fusion protein was caused by the intrinsic kinase activity of the protein and not due to a contaminating kinase. Using a yeast two-hybrid system, Welihinda and Kaufman (1996) demonstrated that yeast Ire1p forms oligomers and that oligomer formation is a prerequisite for activation of the kinase. Whereas we do not show that oligomer formation is required for kinase activity, GST is known to form dimers in nature, and the phosphorylation results described here with the chimeric protein parallel those observed with yeast Ire1p. It is therefore likely that AtIre1 proteins also form dimers in vivo.
Sensing Unfolded Proteins
The AtIre1 amino acid sequences we describe here have the typical tripartite structure of other eukaryotic Ire1 proteins. The N-terminal lumenal domain is thought to be the sensor domain of the protein. To demonstrate this function, several studies have relied on the expression of a chimeric gene in an Δire1 strain of yeast that harbors a single copy of the lacZ gene under the control of the UPR element of the yeast BiP gene (KAR2). Accumulation of β-galactosidase after treatment with tunicamycin is then a convenient way to measure the UPR. Liu et al. (2000) showed that the N-terminal domains of human, mouse, and C. elegans Ire1 linked to the transmembrane and C-terminal portions of yeast IRE1 could drive β-galactosidase expression in a Δire1 yeast strain treated with tunicamycin. Our results (Fig. 5) similarly show that the sensor domains of AtIre1-1 and AtIre1-2 in chimeric constructs with the C-terminal portion of yeast IRE1, induce β-galactosidase when the complemented Δire1 strain is treated with tunicamycin. A similar approach was used by Bertolotti et al. (2000) to show the functional equivalence of the lumenal domains of the mammalian Ire1p and PERK transmembrane protein kinases.
But how does Ire1 sense the presence of unfolded proteins in the ER? Indirect observations suggest that unfolded proteins are not sensed directly, but indirectly through the level of free BiP. For example, cells that overexpress Kar2p/BiP respond less well to ER stress (Dorner et al., 1992; Kohno et al., 1993). In mammalian cells, BiP was found to be associated with both IRE1 and PERK (Bertolotti et al., 2000), the two proteins involved in sensing ER stress. Recent experiments show that in yeast, in the absence of stress, BiP/Kar2p binds to the lumenal domain of Ire1p and keeps this protein in an inactive unphosphorylated state. Stress results in the release of BiP/Kar2p from Ire1p and activation of Ire1p and the UPR pathway (Okamura et al., 2000). In tobacco, overexpression of BiP also alleviates ER stress (Leborgne-Castel et al., 1999); therefore, a similar regulation mechanism might be conserved in plants.
What Is the Function of the Two AtIre1 Genes?
Like humans, Arabidopsis has two putative AtIre1 genes, although yeast has only one. The yeast UPR is a linear pathway, but the mammalian UPR is more complex with diversity of downstream signals (Urano et al., 2000a). When overexpressed, either mammalian gene (human Ire1α or mouse Ire1β) can activate the BiP promoter, as does overexpression of the yeast gene in yeast, suggesting that this portion of the role of Ire1 is conserved between yeast and mammals (Tirasophon et al., 1998; Wang et al., 1998). We were unable to overexpress the AtIre1 genes in Arabidopsis. That is, when plants transformed with a CaMV35S::AtIre1-1 or AtIre1-2 were tested they were found to have normal levels of AtIre1 transcripts. BiP genes were not overexpressed in those plants. We postulate that the gene product down-regulates the expression of the introduced genes. The two genes are differentially expressed in Arabidopsis. In mammals also, Ire1α (Tirasophon et al., 1998) is ubiquitously expressed, whereas Ire1β is expressed only in gastrointestinal epithelial cells (Urano et al., 2000b).
In this study, we dealt primarily with that aspect of the UPR that concerns the induction of chaperones. However, the UPR has two components: up-regulation of chaperone genes and attenuation of translation. In mammalian cells, the attenuation of protein synthesis in response to ER stress in mediated by the protein PERK, another ER stress sensor protein (Harding et al., 2000). An exhaustive search of the Arabidopsis database did not reveal any PERK homologs. Iwawaki et al. (2001) recently found that in human cells IRE1β signals the attenuation of protein synthesis during ER stress. Whether in plants the Ire1 proteins signal an attenuation of protein synthesis remains to be examined. It is also possible that in the course of evolution these ER-located protein kinases assumed an entirely different function, unrelated to the UPR.
MATERIALS AND METHODS
Plant Material
Sterile seeds of Arabidopsis (ecotype Columbia) were germinated in one-half-strength Murashige and Skoog medium containing 1% (w/v) Suc and further cultured in the light/dark cycle of 16 h of light and 8 h of darkness with gentle shaking. Two-week-old plantlets were treated with tunicamycin, which was added to the culture medium at final concentration of 5 μg mL−1. Non-sterile seeds growing in soil for 4 weeks were used for stable transformations.
The tobacco (Nicotiana tabacum) cell line BY-2 was used in stable transformation assays for heterologous expression of chimeric AtIre1-GFP fusion proteins. The BY-2 cells were maintained and subcultured by 1:30 dilution in growth medium (Murashige and Skoog salts medium at pH 5.7 supplemented with 3% [w/v] Suc, 0.1 g L−1 myoinositol, 1 mg L−1 thiamine, 0.18 g L−1 KH2PO4, 2 μm 2,4-dichlorophenoxyacetic acid, and suitable antibiotic) at 24°C in darkness and shaken at 113 rpm.
RNA Preparation and Northern Analysis
For northern analysis, total RNA was isolated (Koizumi et al., 1999), fractionated by agarose gel electrophoresis, and transferred to a nylon membrane. The membrane was probed with 32P-labeled cDNAs of BiP (Koizumi, 1996), PDI, calnexin (Huang et al., 1993), and calreticulin-1 and -2 (Nelson et al., 1997). cDNAs of calnexin and PDI were amplified by RT-PCR with primers CN-1 (ATGAGACAACGGCAACTATTTTCC) and CN-2 (AAGACAAAAATTTCTCAAACTTGG), and primer PDI-1 (CTCGTGAAGCTGAGGGTATTG) and PDI-2 (AAGATTGGAGCAAGCTTTGG), respectively. Calreticulin cDNAs was a generous gift of Dr. Donald E. Nelson (University of Arizona, Tucson). Hybridization was carried out as described previously (Koizumi et al., 1999). For detection of Ire1 homologs, poly(A+) RNA was prepared using PolyATtract mRNA isolation system (Promega Corporation, Madison, WI) according to the manufacturer's instructions, and subjected to northern analysis.
Isolation of Promoters and cDNAs Encoding Ire1 Homologs
To isolate the full coding region of AtIre1-1 cDNA, RT-PCR was carried out with primer 1-a (AAAGCGATGAGAGGATCTGC) and primer 1-b (GAAGAAAAGAATCCTAGAATACAGTGG). A specific PCR product, approximately 3 kb in length, was cloned into the plasmid vector pCR 2.1-TOPO (Invitrogen, Carlsbad, CA). For AtIre1-2, RT-PCR was carried out using primer 2-a (CGTTTGTTAAACCCACACCC) and 2-b (TGAACTTGAATTTCCGGAGG) to amplify an approximately 0.4-kb fragment. This fragment was used as a probe to screen a cDNA library constructed in HybirZAP-2.1 (Stratagene, La Jolla, CA), and nearly full-length cDNA was isolated. Finally, primer 2-c (ATGCCGCCGAGATGTCCTTTCC) and 2-b were used to amplify a full-length cDNA encoding AtIre1-2. This PCR product was also cloned into pCR 2.1-TOPO vector. Nucleotide sequences of both cDNAs were determined by DNA sequencing using synthetic primers.
AtIre1 promoters were amplified by PCR using forward primers at positions −1,996 bp for AtIre1-1 and −1,994 bp for AtIre1-2 and reverse primers at the initiation codon of both genes. PCR products were cloned into the plasmid pCR2.1-TOPO (Invitrogen). DNA sequences were confirmed. To make transcriptional fusions with GUS gene, the 35S promoter of pBl121 was substituted by either AtiIre1-1 promoter (vector called pbinAtIre1-1p::GUS) or AtIre1-2 promoter (vector called pbinAtIre1-2p::GUS).
To make the fusions with GFP, AtIre1-1, AtIre1-1(K487A), AtIre1-2, and AtIre1-2(K442A) cDNAs were translationally fused to the 5′ end of GFP by recombinant PCR. The GFP gene was a gift of Dr. Yasuo Niwa (University of Shizuoka, Japan; Chiu et al., 1996). Amplified transgenes and control of the GFP gene alone were introduced into the pCR2.1-TOPO. All constructs were confirmed by DNA sequencing.
For stable expression in BY2 cells, the SpeI/XhoI restriction fragment from the pCR2.1 constructs carrying the GFP gene by itself, the AtIre1-1::GFP and the AtIre1-1(K487A)::GFP transgenes, and the AvrII/XhoI restriction fragments from the AtIre1-2::GFP and the AtIre1-2(K442A)::GFP transgenes were inserted in the plant expression vector pIG121-Hm (a gift from Kenzo Nakamura, Nagoya University, Japan) by substitution of the XbaI/XhoI fragment. This expression vector contains the CaMV 35S promoter and the nos 3′-polyadenylation tail in an expression cassette that permits two selection options: hygromycin and kanamycin. The GFP-KDEL gene containing plasmid, pER-GFP, was provided by Dr. Dolors Ludevid (Consejo Superior de Investigaciones Cientificas, Barcelona), and the BY-2 cells expressing the soybean (Glycine max) α-1-2 mannosidase I-GFP Golgi marker was provided by Dr. Andrew Staehelin (University of Colorado, Boulder).
Genomic Southern Analysis
A part of AtIre1-1 encoding C-terminal portion amplified with primer 1-c (CGAGAGCACAAGATGTTATGC) and primer 1-b was used as a probe to detect AtIre1-1. For AtIre1-2, the PCR fragment obtained with primer 2-a and 2-b was used as well. Genomic DNA of Arabidopsis was prepared as described previously (Koizumi et al., 1999), digested with appropriate restriction enzymes, and subjected to Southern hybridization using the probes mentioned above.
Transformation Procedures
To generate transgenic BY-2 cells expressing the GFP fusions, all the GFP constructs were transferred to Agrobacterium tumefaciens AGL-O following the method described by Lazo et al. (1991). Absorbance (0.9) of an overnight A. tumefaciens culture was used to transform 5 mL of a 4-d-old BY-2 cell culture previously diluted once in fresh growth medium. After 2 d of incubation at 26°C in darkness, BY-2 cells were washed five times with growth medium containing 100 mg L−1 kanamycin and 500 mg L−1 carbenicillin. Cells were plated in presence of kanamycin and carbenicillin at the same concentrations and incubated in darkness at 26°C. Calli resistant to kanamycin formed after 3 weeks of selection and were then moved to fresh plates monthly. Callus cells were resuspended in fresh growth medium before examination with an epifluorescence microscope.
Transgenic Arabidopsis plants expressing GUS protein under the regulation of either of the AtIre1 promoters were produced by infiltration (Clough and Bent, 1998) in 200 mL of A. tumefaciens AGL-O containing the binary vector pbinAtIre1-1p::GUS or pbinAtIre1-2p::GUS. Seeds from infiltrated plants were collected and the seedlings selected on kanamycin.
Isolation of Nuclei
BY-2 protoplasts were prepared from 2-week-old calli. BY-2 cells were incubated with growth medium plus 5.41% (w/v) betaine, 1% (w/v) cellulase, 0.5% (w/v) macerozyme, and 0.1% (w/v) bovine serum albumin Fraction V for 5 h at 26°C in darkness. Protoplasts were sedimented at 50 g during 4 min and washed twice with growth medium with betaine. The pellet was resuspended in NI buffer supplemented with the following protease inhibitors: 0.4 mm phenylmethylsulfonyl fluoride, 5 μg mL−1 aprotinin, 5 μg mL−1 pepstatin A, and 5 μg mL−1 leupeptin. The NI buffer contains 10 mm MES [2-(N-morpholino)-ethanesulfonic acid], 0.2 m Suc, 10 mm NaCl, 10 mm KCl, 2.5 mm EDTA, 2.5 mm dithiothreitol, 0.1 mm Spermine, and 0.5 mm Spermidine. Protoplasts were disrupted by six passages through a 21-gauge needle on ice. The released nuclei were concentrated on a Percoll step gradient prepared in NI buffer and containing layers of 50% (w/v) Percoll, 7% (v/v) Percoll, and sample. The gradient was centrifuged for 10 min at 100g at 4°C. Purified nuclei at the 50% to 7% (v/v) Percoll interface were washed three times by a one-tenth dilution with NI buffer plus 20% (v/v) glycerol. Nuclei were resuspended in fresh NI buffer/glycerol before examination in the epifluorescence microscope.
In Vitro Kinase Assay
A part of AtIre1-2, amplified with primer 2-d (GAATTCAAAAAGTTTTCGTCGAGGGG) and primer 2-e (CTCGAGTTAGATGATGTCCCATTTGAAG), was subcloned in pGEX-2T (Amersham-Pharmacia Biotech, Uppsala) with EcoRI and XhoI, yielding a plasmid harboring a fusion protein of GST and the C terminal portion of AtIre1-2 (GST-AtIre2). At the same time, a mutant construct in which Lys-442 was converted to Ala (GST-AtIre2-M) was also made. These plasmids were introduced in Escherichia coli (BL21) and the fusion proteins were induced by addition of IPTG. The fusion proteins were purified with glutathione-Sepharose beads as described previously (Ikeda et al., 1999). Approximately 50 ng of purified proteins were incubated with 0.1 μCi of [32P]ATP in kinase buffer containing 20 mm sodium phosphate (pH 8.0) and 10 mm MgCl2 in a final volume of 20 μL at 37°C for 10 min. The reaction product was fractionated by SDS-PAGE and subjected to autoradiography.
Complementation of Yeast (Saccharomyces cerevisiae) ire1 Mutants
The Δire1 yeast strain KMY1515 (MATα ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 Δire1::TRP1 lys2-801::LYS2-UPRE- CYC1-lacZ; Okamura et al., 2000) was transformed with the following plasmids. pRS313, a yeast centromeric plasmid vector (Sikorski and Hieter, 1989), and pR313-IRE1 (Okamura et al., 2000), which contains the yeast IRE1 gene, were used as a negative and a positive control, respectively. To make chimeric constructs, an EcoRI site was introduced after the nucleotide sequence encoding the signal peptide (after Arg-31) and a BamHI site was introduced before the sequence for the transmembrane domain (before Glu-521) of pR313-IRE1. The nucleotide sequences encoding the sensor domain of AtIre1-1 or AtIre1-2 were amplified with primer 1-d (GAATTCGGATCTGAAATCTCCAAGTCC) and 1-e (GGATCCGCTAGCAAAGCCTGCCTGTTTCG) or primer 2-f (GAATTCGGCGGCGCTGCCGACGTAG) and 2-g (GGATCCTCCAAACAAATATGTATATTTCTGC) and replaced with that of yeast Ire1 using EcoRI and BamHI sites obtaining Atire1-ch or Atire2-ch. KMY1515 cells transformed with either of these plasmids were incubated in synthetic dextrose medium (Kaiser et al., 1994) supplemented with uracil (100 mg L−1) and His (50 mg L−1) at 30°C with or without tunicamycin. Cells were also used for β-galactosidase activity assays according to Kaiser et al. (1994).
Histochemical Staining
Whole transgenic seedlings or organs were treated for 20 min in cold 90% (v/v) acetone, then washed once with GUS buffer (25 mm sodium phosphate buffer, pH 7, 0.5 mm KFe(CN)2, 0.5 mm KFe(CN)3, and 10 mm EDTA), and incubated overnight with 2 mm 5-bromo-4-chloro-3-indolyl β-d-glucuronide in GUS buffer at 37°C in the dark. The GUS staining solution was removed, and the tissues were dehydrated by increasing the ethanol concentrations gradually from 70% (v/v) to absolute ethanol. Samples where visualized in the light microscope.
Footnotes
This work has been supported by a grant from the Department of Energy (Office of Energy Biosciences, grant no. DE–FG03–86ER13497) to M.J.C., a fellowship from the Ministry of Science and Technology of Spain to I.M.M., and grants from the Research for the Future Program (JSPS–RFTF00L01604) of the Japan Society for the Promotion of Science.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010636.
LITERATURE CITED
- Bertolotti A, Zhang YH, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boston RS, Fontes EBP, Shank BB, Wrobel RL. Increased expression of the maize immunoglobulin binding protein homolog B-70 in 3 zein regulatory mutants. Plant Cell. 1991;3:497–505. doi: 10.1105/tpc.3.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brostrom CO, Brostrom MA. Regulation of translational initiation during cellular responses to stress. Prog Nucleic Acids Res Mol Biol. 1998;58:79–125. doi: 10.1016/s0079-6603(08)60034-3. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coleman CE, Clore AM, Ranch JP, Higgins R, Lopes MA, Larkins BA. Expression of a mutant alpha-zein creates the floury2 phenotype in transgenic maize. Proc Natl Acad Sci USA. 1997;94:7094–7097. doi: 10.1073/pnas.94.13.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CE, Lopes MA, Gillikin JW, Boston RS, Larkins BA. A defective signal peptide in the maize high-lysine mutant floury-2. Proc Natl Acad Sci USA. 1995;92:6828–6831. doi: 10.1073/pnas.92.15.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding ER resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Crofts AJ, Leborgne-Castel N, Pesca M, Vitale A, Denecke J. BiP and calreticulin form an abundant complex that is independent of endoplasmic reticulum stress. Plant Cell. 1998;10:813–823. doi: 10.1105/tpc.10.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amico L, Valsasina B, Daminati MG, Fabbrini MS, Nitti G, Bollini R, Ceriotti A, Vitale A. Bean homologs of the mammalian glucose-related proteins: induction by tunicamycin and interaction with newly synthesized seed storage proteins in the endoplasmic reticulum. Plant J. 1992;2:443–455. doi: 10.1111/j.1365-313x.1992.00443.x. [DOI] [PubMed] [Google Scholar]
- Denecke J, Carlsson LE, Vidal S, Höglund AS, Ek B, van Zeijl MJ, Sinjorgo KM, Palva ET. The tobacco homolog of mammalian calreticulin is present in protein complexes in vivo. Plant Cell. 1995;3:391–406. doi: 10.1105/tpc.7.4.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denecke J, Goldman MHS, Demolder J, Seurinck J, Botterman J. The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–1035. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner AJ, Wasley LC, Kaufman RJ. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EBP, Shank BB, Wrobel RL, Moose SP, O'Brian GR, Wurtzel ET, Boston RS. Characterization of an immunoglobulin binding protein homolog in maize floury-2 endosperm mutant. Plant Cell. 1991;3:483–496. doi: 10.1105/tpc.3.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillikin JW, Zhang F, Coleman CE, Bass HW, Larkins BA, Boston RS. A defective signal peptide tethers the floury-2 zein to the endoplasmic reticulum membrane. Plant Physiol. 1997;114:345–352. doi: 10.1104/pp.114.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Huang L, Franklin AE, Hoffman NE. Primary structure and characterization of an Arabidopsis thaliana calnexin-like protein. J Biol Chem. 1993;268:6560–6566. [PubMed] [Google Scholar]
- Ikeda Y, Koizumi N, Kusano T, Sano H. Sucrose and cytokinin modulation of WPK4, a gene encoding a SNF1-related protein kinase from wheat. Plant Physiol. 1999;121:813–820. doi: 10.1104/pp.121.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K. A novel translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nature Cell Biol. 2001;3:158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM. Binding-protein expression is subjected to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- Kaufman R. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Develop. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N. Isolation and responses to stress of a gene that encodes a luminal binding protein in Arabidopsis thaliana. Plant Cell Physiol. 1996;37:862–865. doi: 10.1093/oxfordjournals.pcp.a029023. [DOI] [PubMed] [Google Scholar]
- Koizumi N, Ujino T, Sano H, Chrispeels MJ. Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol. 1999;121:353–362. doi: 10.1104/pp.121.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N, Jelitto-Van Dooren EPWM, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell. 1999;11:459–469. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CP, Larkins BA. Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol Biol. 1996;30:873–882. doi: 10.1007/BF00020800. [DOI] [PubMed] [Google Scholar]
- Lindberg RA, Quinn AM, Hunter T. Dual-specificity protein kinases: will any hydroxyl do? Trends Biochem Sci. 1992;17:114–119. doi: 10.1016/0968-0004(92)90248-8. [DOI] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress-response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething M-J, Sambrook J. A transmembrane protein with cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Nelson DE, Glaunsinger B, Bohnert HJ. Abundant accumulation of the calcium-binding molecular chaperone calreticulin in specific floral tissues of Arabidopsis thaliana. Plant Physiol. 1997;114:29–37. doi: 10.1104/pp.114.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Sidrauski C, Kaufman RJ, Walter P. A role for presenilin-1 in nuclear accumulation of Ire-1 fragments and induction of the mammalian unfolded protein response. Cell. 1999;99:691–702. doi: 10.1016/s0092-8674(00)81667-0. [DOI] [PubMed] [Google Scholar]
- Okamura K, Kimata Y, Higashio H, Tsuru A, Kohno K. Dissociation of Kar2p/BiP from an endoplasmic reticulum sensory molecule, Ire1p, triggers unfolded protein response in yeast. Biochem Biophys Res Commun. 2000;279:445–450. doi: 10.1006/bbrc.2000.3987. [DOI] [PubMed] [Google Scholar]
- Pelham HRB. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- Shank KJ, Su P, Brglez I, Boss WF, Dewey RE, Boston RS. Induction of lipid meatabolic enzymes during the endoplasmic reticulum stress response in plants. Plant Physiol. 2001;126:267–277. doi: 10.1104/pp.126.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorrosh BS, Dixon RA. Molecular characterization and expression of the alfalfa protein with sequence similarity to mammalian ERP72, a glucose regulated endoplasmic reticulum protein containing active site sequences of protein disulphide isomerase. Plant J. 1992;2:51–58. doi: 10.1046/j.1365-313x.1992.t01-50-00999.x. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman RH, Williams BRG. Translational control perks up. Nature. 1999;397:208–209. doi: 10.1038/16586. [DOI] [PubMed] [Google Scholar]
- Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Driouich A. Brefeldin A effects in plants: are different Golgi responses caused by different sites of action? Plant Physiology. 1997;114:401–403. doi: 10.1104/pp.114.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki A, Kohno K, Tamura G. Inhibition of biosynthesis of polyisoprenol sugars in chick embryo microsomes by tunicamycin. Agric Biol Chem. 1975;39:2089–2091. [Google Scholar]
- Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Urano F, Bertolotti A, Ron D. IRE1 and efferent signaling from the endoplasmic reticulum. J Cell Sci. 2000a;113:3697–3702. doi: 10.1242/jcs.113.21.3697. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000b;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vitale A, Denecke J. The endoplasmic reticulum: gateway of the secretory pathway. Plant Cell. 1999;11:615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Gallili G. The endomembrane system and the problem of protein sorting. Plant Physiol. 2001;125:115–118. doi: 10.1104/pp.125.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-Z, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae: oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- Wrobel RL. Expression of molecular chaperones in endoplasmic reticulum of maize endosperm. PhD Thesis. Raleigh: North Carolina State University; 1996. [Google Scholar]
- Zheng HQ, Staehelin LA. Nodal endoplasmic reticulum, a specialized from of ER found in gravity sensing root tip columella cells. Plant Physiol. 2001;125:252–265. doi: 10.1104/pp.125.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]