Abstract
Elicitation or peroxide stimulation of grape (Vitis vinifera L. cv Touriga) vine callus cultures results in the rapid and selective in situ insolubilization of an abundant and ionically bound cell wall protein-denominated GvP1. Surface-enhanced laser desorption/ionization/time of flight-mass spectrometry analysis, the amino acid composition, and the N-terminal sequence of purified GvP1 identified it as an 89.9-kD extensin. Analysis of cell walls following the in situ insolubilization of GvP1 indicates large and specific increases in the major amino acids of GvP1 as compared with the amino acids present in salt-eluted cell walls. We calculate that following deposition, covalently bound GvP1 contributes up to 4% to 5% of the cell wall dry weight. The deposition of GvP1 in situ requires peroxide and endogenous peroxidase activity. Isoelectric focusing of saline eluates of callus revealed only a few basic peroxidases that were all isolated or purified to electrophoretic homogeneity. In vitro and in situ assays of extensin cross-linking activity using GvP1 and peroxidases showed that a 40-kD peroxidase cross-linked GvP1 within minutes, whereas other grapevine peroxidases had no significant activity with GvP1. Internal peptide sequences indicated this extensin peroxidase (EP) is a member of the class III peroxidases. We conclude that we have identified and purified an EP from grapevine callus that is responsible for the catalysis of GvP1 deposition in situ during elicitation. Our results suggest that GvP1 and this EP play an important combined role in grapevine cell wall defense.
Hyp-rich glycoproteins (HRGPs) are structural proteins that are abundant in the cell walls of higher plants (Showalter, 1993). Of this group, extensin, a family of HRGPs, has been studied the most. The deposition of extensin in cell walls is considered important in a wide range of plant physiological processes, including plant defense. It is thought that cross-linked extensin can increase the mechanical strength of cell walls through its intercalation and ionic interaction with other polymer systems (Cooper et al., 1987; Showalter, 1993). Evidence has been presented more recently for a direct covalent link between extensin and pectin (Qi et al., 1995). An increase in extensin deposition therefore could cause the overall level of cell wall cross-linking to rise dramatically, thereby increasing the cell wall rigidity and resistance to pathogen ingress.
The in muro deposition of HRGPs can be triggered by various environmental factors (Showalter, 1993) and has been repeatedly associated with cell wall toughening and plant disease resistance (Esqerrè-Tugaye et al., 1979; Baldwin et al., 1992; Brisson et al., 1994). The oxidative cross-linking of HRGPs can be rapidly initiated on perception of stress or elicitors without de novo protein synthesis (Bradley et al., 1992). Such rapid reactions therefore could rigidify cell walls as part of the plants primary defense and thus impede pathogen ingress until transcription-dependant defenses can be expressed (Brisson et al., 1994). In addition, there is overwhelming evidence supporting a major role for extensin deposition in the later, transcription-dependent response to pathogens, elicitors, and wounding (Showalter, 1993). Molecular studies have demonstrated repeatedly increased expression of extensin mRNA in defensive responses (Memelink et al., 1993; Shirsat et al., 1996).
Peroxidase and peroxide are believed to be required for the catalysis of extensin deposition (Cooper and Varner, 1984). In the tomato system, at least two peroxidases have been demonstrated to have a high capacity for the oxidative cross-linking of extensin, whereas other tomato peroxidases (Brownleader et al., 1995) and several others derived from different species (Schnabelrauch et al., 1996) did not. A basic peroxidase with a similarly high capacity recently was purified from lupin, whereas an acidic form presented negligible activity against extensin (Jackson et al., 1999). These results confirm an earlier suggestion by Everdeen et al. (1988) that extensin deposition in muro is mediated by specific class III peroxidases that can be referred to as extensin peroxidases (EPs). It follows that the deposition of extensin in cell wall defensive reactions requires the coordinated expression of extensin and EP at the cell wall of cells exposed to or undergoing an oxidative burst.
However, with very few exceptions (Bradley et al., 1992; Wojtaszec et al., 1997; Otte and Barz, 2000), the specific HRGPs involved in cell wall modification during plant defensive reactions have not been identified. In addition, the peroxidases responsible for catalysis of these defensive reactions in muro have been almost completely overlooked.
European cultures of grape (Vitis vinifera L. cv Touriga) vine are particularly susceptible to infection by Plasmopara viticola and Uncinula necator, the causal agents of downy and powdery mildew diseases. These diseases are difficult to control and can have a severe effect on grape production and quality. To date, studies of grapevine defensive responses to pathogen ingress have indicated the importance of fungicidal phytoalexins (Hain et al., 1993) and the deposition of phenolic compounds in the cell wall (Busam et al., 1997). In this report, we describe our study of extracellular proteins during elicitation of grapevine callus cultures. We present evidence that the rapid deposition of an abundant, 89.9-kD extensin (denominated GvP1) in response to elicitation is catalyzed specifically by a 40-kD EP. We discuss these results in light of the role of both this extensin and EP in grapevine defense.
RESULTS
Elicitation of Cell Wall Protein Insolubilization in Grapevine Callus
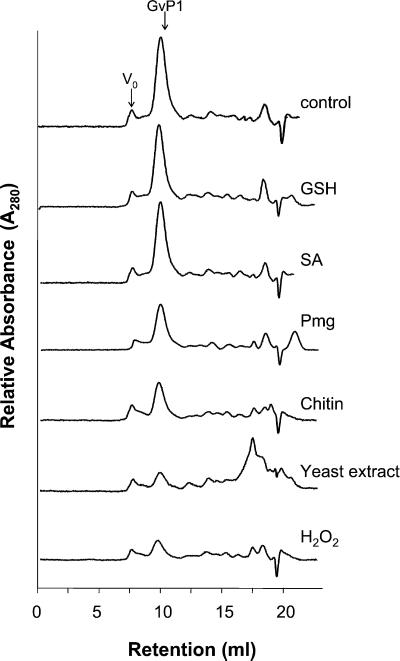
Saline eluates of intact, water-washed grapevine callus cultures provided a convenient and specific source of cell wall proteins. Superose-12 gel filtration chromatography of these eluates revealed that a single protein peak, GvP1, constituted a large proportion of the A280 absorbing material (protein) in these extracts (Fig. 1, trace control). A substantial proportion of GvP1 was insolubilized after exposing grapevine callus cultures for 15 min to either Pmg elicitor (a preparation of β-glucan elicitors from mycelial walls of Phytophthora megasperma Dresch f. sp. glycinea Kuan and Erwin (Hahn et al., 1992), chitin, yeast extract, or H2O2 (corresponding traces labeled on right), whereas the solubility of other cell wall proteins remained unaffected. GSH or salicylic acid had no effect on the solubility of GvP1, even when the time of incubation was increased to 1 h.
Figure 1.
The effect of different elicitors on the content of salt-soluble cell wall protein in grapevine callus. Superose-12 gel filtration chromatography was used to separate cell wall proteins. The traces were obtained from KCl extracts of 35 mg (fresh weight) of untreated callus (control) or those incubated for 60 min with 1 mm glutathione (GSH), 1 mm salicylic acid (SA), or for 15 min with 60 μg mL−1 (Glc equivalents) of Phytophthora megasperma Dresch f. sp. glycinea fungal elicitor (Pmg), 10 mg mL−1 chitin (chitin), 20 mg mL−1 yeast extract (yeast extract), and 100 μm hydrogen peroxide (H2O2).
Purification and Characterization of GvP1
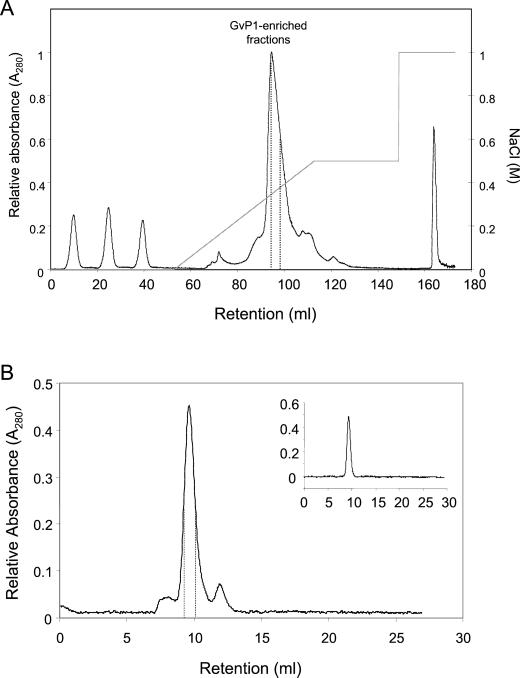
The insolubilization of GvP1 appears to constitute part of a rapid, cell wall response to elicitors; therefore, we sought to purify this protein for further characterization. Saline eluate obtained from 200 g of callus was subject to SP-Sepharose chromatography with a gradient of 0 to 0.5 m NaCl (Fig. 2A). Analysis of the resulting fractions by Superose-12 chromatography revealed that GvP1 eluted between 0.3 and 0.37 m NaCl. These fractions were pooled and further fractionated by Superose-12 chromatography (Fig. 2B). An aliquot analyzed for purity by Superose-12 chromatography (inset in Fig. 2B) demonstrated that GvP1 prepared in this way elutes as a single, Gaussian peak.
Figure 2.
Purification of GvP1 extensin and grapevine callus peroxidases. A, SP-Sepharose column chromatography of KCl eluates from intact cells. Fractions enriched in GvP1 are delimited by dashed lines. The three peaks eluted between 0 and 40 mL represent the nonbound material of three separate injections. B, Superose-12 fractionation of SP-Sepharose enriched GvP1 extensin. The fractions chosen are delineated by dashed lines. The inset chromatograph represents an analysis of the chosen, pooled fractions by Superose-12 chromatography.
The amino acid composition of GvP1is highly enriched in Hyp, and rich in Lys, Tyr, Ser, His, Pro, and Val (Table I). Together, these amino acids contribute 93 mol % of the amino acids of GvP1. Similar amino acid compositions are seen in extensins (Cassab, 1998; Table I). The N terminus of GvP1 was also sequenced (Table II). Residues 1 through 6 showed weak homology to the N terminus of other extensins. However, a BLAST search revealed that residues 6 through 20 were homologous to internal, repeated sequences of many extensins. These residues contains two Ser(Hyp)4 motifs, separated by a (Tyr)3Lys or (Tyr)3His motif. Both types of motifs are considered to be characteristic of extensin structure (Kieliszewski and Lamport, 1994).
Table I.
Amino acid composition of GvP1 extensin
| Amino Acid | Composition
|
||
|---|---|---|---|
| Tomato extensina | Potato extensinb | Grapevine GVP1 | |
| mol % | |||
| Hyp* | 34.3 | 30.0 | 46.1 |
| Lys* | 10.0 | 11.5 | 12.2 |
| Tyr* | 7.5 | 5.6 | 8.6 |
| Ser* | 8.0 | 2.0 | 8.2 |
| His* | 5.5 | 8.8 | 8.1 |
| Pro* | 10.8 | 20.0 | 5.9 |
| Val* | 9.6 | 10.1 | 3.6 |
| Ala | 3.5 | 1.2 | 2.7 |
| Phe | NDc | 0.2 | 2.2 |
| Ile | 0.4 | 0.6 | 1.0 |
| Met | NDc | NDc | 0.9 |
| Gly | 0.8 | 0.5 | 0.3 |
| Asp/Asn | 1.0 | 1.3 | 0.3 |
| Thr | 5.3 | 6.2 | 0.1 |
| Arg | 0.1 | 0.5 | 0.1 |
| Glu/Gln | 1.1 | 1.3 | NDc |
| Cys | NDc | NDc | NDc |
| Trp | NDc | NDc | NDc |
The compositions of tomato and potato extensins are shown for comparison. The major amino acids of GvP1 are indicated by an asterisk.
ND, Not detected.
Table II.
A comparison of the N-terminal sequence of grapevine extensin GvP1 and repeated sequences of other extensins
| Extensin Source/Accession No. | N-Terminal Amino Acid Sequences of GvP1 and of Repeated Sequences of Other Extensins | No. of Repeats |
|---|---|---|
| GvP1 | N Y H Y S Hy Hy Hy Hy Y Y Y K S Hy Hy Hy Hy Hy | – |
| Phaseolus vulgaris (AAA33765) | S Hy Hy Hy Hy Y Y Y K S Hy Hy Hy Hy | 9 |
| Brassica napus (227927) | S Hy Hy Hy Hy Y Y Y H S Hy Hy Hy Hy | 13 |
| Glycine max (T06782) | S Hy Hy Hy Hy Y Y Y H S Hy Hy Hy Hy | 10 |
Hydroxyproline residues are indicated as Hy.
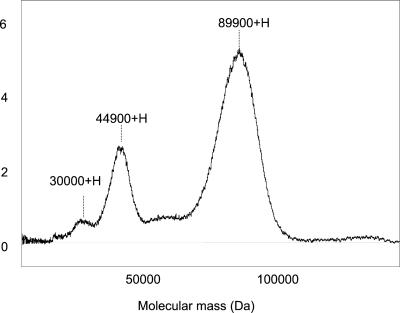
Surface-enhancedlaserdesorption/ionization (SELDI)-mass spectrometry (MS) demonstrated that GvP1 was homodisperse, with a molecular mass of 89.9 kD. This was confirmed by the presence of double and triple charged species at their appropriate m/z (Fig. 3). The total yield of amino acids from known quantities of GvP1 indicated that this extensin has a polypeptide of 410 to 420 amino acids in length (about 48.7 kD). Similar estimations could be made from the low coefficient of absorbance observed for GvP1 (ε280 = 0.542 mg mL−1 water at 24°C) and the Tyr content (8.4 mol %), to which the A280 of GvP1 can be attributed. These observations suggest that the polypeptide contributes only about 50% of the molecular mass of GvP1. Similar observations have been made for other extensins that tend to be highly glycosylated (Brownleader et al., 1996; Dey et al., 1997).
Figure 3.
SELDI-MS of GvP1. Labeled in the figure (left to right) are the m/z of the triple-, double-, and single-ionized species of GvP1.
These results indicate that the insolubilization of an extensin constitutes a rapid, cell wall response to stimulation by peroxide or elicitation in grapevine cultures.
Quantitative Effects of GvP1 Insolubilization on the Covalent Cell Wall Composition
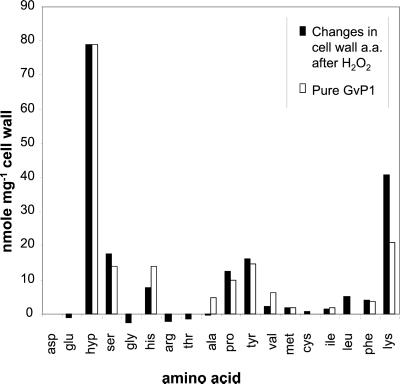
Next, we wished to quantify the impact of extensin deposition on the composition of the covalently bound cell wall structure. Using Gaussian analysis of Superose-12 chromatograms of salt eluates (see “Materials and Methods”), it was possible to quantify the level of salt-soluble GvP1 in callus cell walls before and after peroxide stimulation. These analyses indicated that peroxide caused >80% of monomeric GvP1 to be deposited in 15 min, resulting in a loss of 48 μg of monomeric GvP1 mg−1 cell wall (dry weight). It was important to demonstrate that this loss of salt-soluble GvP1 was due to the deposition of GvP1 at the cell wall matrix. Figure 4 demonstrates that overall, the changes in cell wall amino acid composition following H2O2 stimulation both qualitatively and quantitatively reflect the amino acid composition of pure GvP1. An exception to this was Lys, which, relative to other GvP1 amino acids, increased further than could be expected through the deposition of GvP1 alone. However, our chromatograms of cell wall hydrolysates indicated that Lys elutes together with unidentified compounds (results not shown), which made Lys quantification unreliable. The changes in the other major amino acids, however, confirm that GvP1 is deposited at the cell wall matrix and that the deposition of GvP1 accounts for the majority, if not all, of the changes in cell wall amino acid content.
Figure 4.
Peroxide-induced changes in cell wall amino acid composition. The changes (black bars) were calculated by subtracting the amino acid content of untreated cell walls from the amino acid content of cell walls treated with H2O2. The composition of GvP1 extensin (white bars) is presented for comparison.
Based on the increases in the major GvP1 amino acids in the covalent cell wall structure (Fig. 4), a deposition of 44 μg GvP1 mg−1 (DW) of covalently bound cell wall structure was estimated. Therefore, it seems reasonable to suggest that elicited GvP1 deposition can increase the dry weight of the covalently bound cell wall structure by 4% to 5%.
Inhibition of Peroxidase and Elicitor-Induced Deposition of GvP1
To determine if GvP1 deposition in response to elicitors is related to an oxidative burst, we have tested the effect of diphenyleneiodonium (DPI), an inhibitor of elicitor-induced peroxide production in some plant systems. DPI did not inhibit GvP1 deposition in response to exogenous H2O2. The deposition of GvP1 in response to chitin was completely inhibited after a brief incubation of callus cultures with 20 μm DPI (Table III). Pmg-induced deposition of GvP1 was similarly inhibited. Almost complete inhibition was obtained at 5 μm, whereas 1 μm only partially inhibited deposition.
Table III.
The effect of DPI, ascorbate (Asc.), or azide (NaN3) on chitin- or Pmg-elicited deposition of GvP1
| Treatment | Residual GvP1 |
|---|---|
| % | |
| H2O2 | 30.4 ± 4.6 |
| H2O2+ 20 μm DPI | 22.4 ± 3.9 |
| Pmg | 25.9 ± 4.1 |
| Pmg + 1 μm DPI | 47.9 ± 6.5 |
| Pmg + 5 μm DPI | 95.5 ± 7.7 |
| Pmg + 20 μm DPI | 83.1 ± 8.4 |
| Chitin | 30.9 ± 3.2 |
| Chitin + 20 μm DPI | 96.3 ± 6.8 |
| Chitin + 5 mm Asc. | 100 ± 12.4 |
| Chitin + 10 mm NaN3 | 74.1 ± 5.7 |
Superose-12 chromatography was utilized to separate cell wall proteins. Residual GvP1 was calculated from GvP1 peak height and is expressed as a percentage of untreated callus. The values represent means (±sd) of at least three independent experiments.
The treatment of cultures with azide or ascorbate also markedly reduced the level of GVP1 deposition induced by chitin (Table III), suggesting that GVP1 deposition is catalyzed by peroxidase. After washing cells in 1 m KCl, no residual peroxidase activity was detected, indicating that extensin deposition was catalyzed by salt-soluble, extracellular peroxidases.
Purification of Extracellular Peroxidases
An analysis of the peroxidases in saline eluates of callus by nonequilibrated isoelectric focusing (NEIEF) revealed that these contained only basic peroxidases; a minor, neutral-basic peroxidase and a group of highly basic peroxidases (Fig. 5B, lane cw).
Figure 5.
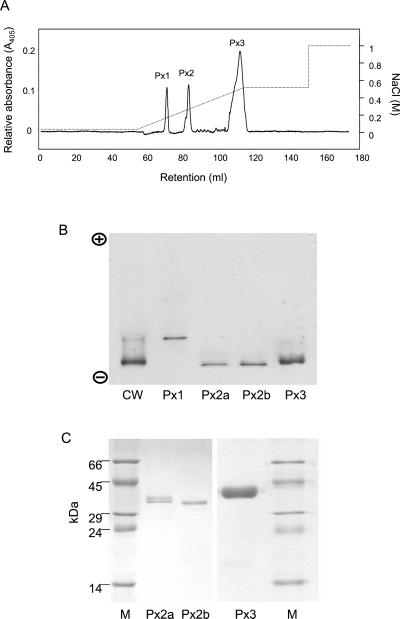
Fractionation of grapevine peroxidases from KCl eluates of grapevine cells by SP-Sepharose chromatography. The three distinct peroxidase pools (405 nm of absorbing material) are indicated as Px1, Px2, and Px3. The NaCl gradient is indicated as a dashed line. B, NEIEF; C, SDS-PAGE of grapevine peroxidases. In NEIEF, a saline eluate of cell walls (cw) was included for comparison. The isolated or purified peroxidases (Px1–3) are labeled at the bottom of each figure. M, Mr markers (labeled to the left of the figure).
SP-Sepharose chromatography of these saline eluates resolved three distinct peaks of peroxidase activity (Px 1–3; Fig. 5A). No peroxidase activity was detected in nonbinding fractions. Px 1 presented a Reinheitzal (Rz; ratio of A405/A275) value of 0.7 and corresponded to the least alkaline peroxidase in NEIEF gels of cell wall eluates (Fig. 5B, lane Px1). Px 2 was further fractionated by Mono-S chromatography into two pools: a minor pool (Px2a) of Rz 2.2 containing two proteins of aproximately 37 kD (Fig. 5C, lane Px2a), and a major pool (Px2b) of Rz 3.0 containing a single protein of 36 kD (lane Px2b). Both peroxidases migrated together in NEIEF and apparently contribute to the most alkaline peroxidase band of cell wall eluates (Fig. 5B, lanes Px2a,b). Px 3 contained the largest pool of peroxidase activity and 405 nm of absorbing material (Fig. 5A). This peroxidase pool was further fractionated by Mono-S followed by a Phenyl-Sepharose 6FF hydrophobic interaction column, to yield a single protein of 40 kD (Fig. 5C) with an Rz of 3.2. This peroxidase also appears to contribute to the most alkaline peroxidase band in cell wall eluates, although it migrates at a slightly slower rate than Px2a,b (Fig. 5B).
These results indicate that grapevine callus cultures have a very simplistic peroxidase polymorphism, and that we have isolated or purified all the major peroxidases from this material.
In Vitro and in Situ Assays of GvP1 Cross-Linking by Grapevine Peroxidases
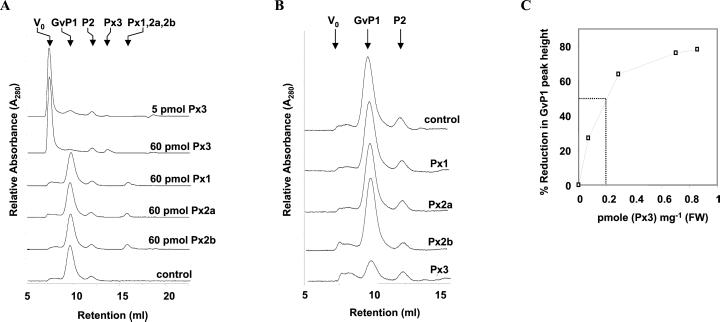
The in vitro activity of these purified or isolated peroxidase preparations was tested against GVP1 extensin in the presence of 100 μm H2O2. Figure 6A demonstrates that the addition of 5 pmol of Px3 resulted in near 100% cross-linking of 60 μg of GVP1, whereas no significant activity could be detected with the other peroxidases, even when 60 pmol of these peroxidases was utilized.
Figure 6.
Identification of grapevine peroxidases with high capacity for the cross-linking of GvP1 extensin. A, In vitro assay of extensin cross-linking by grapevine peroxidases. The different peroxidases and their quantities (pmole) assayed are indicated to the right of the figure. The elution volumes for the different protein species are indicated at the top. B, In situ assay for GvP1 extensin deposition by grapevine peroxidases. The different peroxidases assayed are indicated to the right of the figure. In all cases, 0.7 pmol of peroxidase mg−1 (fresh weight) cells were utilized, and KCl extracts from 35 mg (fresh weight) cells was analyzed. C, The relation of extensin deposition with the abundance of Px3 in cell walls.
The in vitro activities of extracted peroxidases can differ markedly from their activities in situ (Goldberg et al., 1983). We wished to determine if the differences observed in the in vitro GvP1 cross-linking activity of grapevine peroxidases was also evident in situ. Therefore, we have developed an assay that involves the removal of ionically bound constituents from intact grapevine cells and, following this step, the reconstitution of their cell walls with GvP1 and individual peroxidases (see “Materials and Methods”). The incubation of cells with 2.3 μg of GvP1 per mg (fresh weight) of cells (trace “control”) results in the binding of salt-soluble GvP1 in quantities similar to that present in untreated cells (1.7 μg per mg [fresh weight], or 60 μg per mg cell wall [dry weight]). Other cells were prepared with, in addition to GvP1, 0.7 pmol of either peroxidase per mg (fresh weight) of cells. Peroxide treatment of these cell preparations demonstrated that peroxidases 1, 2a, or 2b were unable to catalyze the deposition of GvP1, whereas Px3 catalyzed the deposition of the majority of GvP1 over 10 min (Fig. 6B). The relation of the quantity of GvP1 deposited with the abundance of Px3 is depicted in Figure 6C, and indicates that half-maximal GvP1 deposition over 10 min could be catalyzed with about 0.2 pmol Px3 mg−1 (fresh weight) cells.
These results clearly justify the classification of Px3 as an EP. Because we apparently have excluded a role for all other major extracellular peroxidases in the deposition of grapevine extensin, these results also strongly indicate that Px3 is uniquely responsible for the catalysis of GvP1 deposition in situ.
SDS-PAGE of Px3 revealed that this peroxidase had been purified to electrophoretic homogeneity (Fig. 5, lane Px3). The UV/Vis spectra of this 40-kD peroxidase demonstrated a Soret peak at 405 nm, with α- and β-absorption bands at 498 and 636 nm, respectively (data not shown). This spectra is very similar to that presented by the other grapevine peroxidases, and is typical of the high-spin spectra of class III peroxidases.
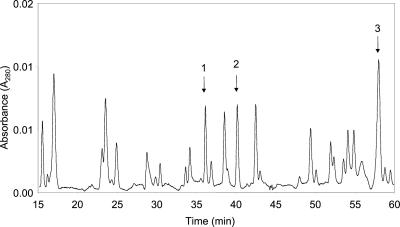
The polypeptide of Px3 was not accessible to N-terminal sequencing by Edman degradation, indicating that, like other class III peroxidases, it is blocked by derivatization of the N-terminal residue (Welinder, 1979). Therefore, Px3 was subject to trypsin digestion and product peptides separated by reverse-phase C18 column chromatography (Fig. 7). Peptides 1 through 3 (indicated in figure) were chosen for sequencing. Their sequences are shown in Table III. Peptide 1 yielded a sequence of nine amino acids that demonstrated high homology to the catalytic site of class III peroxidases, confirming that this EP is a class III peroxidase. Peptide 2 yielded a sequence of eight amino acids, which we have tentatively mapped to a semiconserved sequence located within helix J of known peroxidase structures. Peptide 3 had no significant homology with peroxidase sequences.
Figure 7.
Trypsin digest of Px3. Peptides 1 through 3 (indicated in the figure) were chosen for sequencing.
DISCUSSION
HRGPs have been purified from a variety of different sources (Brownleader and Dey, 1993; Dey et al., 1997). However, very few studies have demonstrated the deposition of specific HRGPs as part of the plant defense. Several studies have described increased expression of specific extensin genes in response to elicitation, plant-pathogen interaction, wounding, or other stress (Showalter et al., 1991; Kawalleck et al., 1995; Parmentier et al., 1995; Shirsat et al., 1996; Hirsinger et al., 1997). However, it is unclear from such studies whether the extensin genes expressed during plant defense are involved in modifying the covalent cell wall structure because extensin deposition was not shown.
We have demonstrated that our grapevine callus cultures present high amounts of an 89.9-kD protein (GvP1) at the cell wall. The amino acid composition and N-terminal sequence of GvP1 clearly identified this protein as a member of the extensin family of HRGPs. In addition, the polypeptide of GvP1 contributes about 50% of its native molecular mass, similar to other extensins that tend to be highly glycosylated, carbohydrate moieties accounting for 30% to 60% of their molecular mass (Brownleader et al., 1996; Dey et al., 1997).
Treatment of callus cultures with several elicitor preparations induced up to 80% of GvP1 to be insolubilized in 15 min. The time required for this insolubilization strongly suggests its independence from de novo transcription. These grapevine cells appear to require only an increase in apoplastic peroxide to initiate GvP1 insolubilization because the incubation of cells with 100 μm peroxide caused the loss of soluble GvP1 over the same time scale as elicitor treatments.
DPI has been reported to inhibit the oxidative burst in plants (Levine et al., 1994; Cazalé et al., 1998; Papadakis and Roubelakis-Angelakis, 1999). We find that 5 μm DPI effectively inhibited the deposition of GvP1 in response to Pmg, and partial inhibition was observed with 1 μm. This inhibitory effect appears to be due to the supression of H2O2 formation because DPI had no effect on GvP1 deposition in response to the addition of exogenous H2O2. Similarly low concentrations (0.1–10 μm) of DPI effectively inhibit the elicited formation of H2O2 from membrane-bound plant NADH-oxidases (Levine et al., 1994). Substantially higher concentrations (50–240 μm) of DPI are required to inhibit H2O2 production from NADH oxidase-peroxidase (Bolwell et al., 1998; Frahry and Schopfer, 1998), which can be important for the development of the oxidative burst in some plant species (Bolwell et al., 1995). The sensitivity of grapevine callus to DPI therefore suggests that the elicited deposition of GvP1 is driven by an oxidative burst from membrane-bound NADH oxidase.
Treatment of grapevine callus with salicylic acid or GSH did not induce GvP1 insolubilization, in contrast with results obtained in cell cultures of P. vulgaris and G. max, where the addition of either compound caused an H2O2-dependent deposition of structural proteins (Bradley et al., 1992). However, different plant species can place emphasis on different mechanisms for the production of active oxygen species in response to elicitation (Bolwell et al., 1998; Papadakis and Roubelakis-Angelakis, 1999). Our results indicate that increases in either GSH or salicylic acid are insufficient for the propagation of an oxidative burst in grapevine callus cultures. Nevertheless, that several other elicitor preparations can cause the insolubilization of GvP1 indicates that GvP1 insolubilization could be employed as a rapid cell wall defensive response to a broad spectrum of pathogens.
Evidence has been obtained to support a direct role for GvP1 in the modification of cell walls in response to elicitation. Estimates of GvP1 abundance indicated that this protein contributed 60 μg of extensin mg−1 cell wall (dry weight). Under normal culture conditions, secreted GvP1 appears to accumulate predominantly in its salt-soluble, monomeric form because salt-washed cell walls of control cultures have a relatively minor content of covalently bound Hyp. However, treatment of callus with peroxide or one of several elicitors causes the rapid and selective deposition of GvP1, as indicated by both the loss of salt-soluble GvP1, and a concomitant increase in the major amino acids of GvP1 in the covalent cell wall structure.
The deposition of GvP1 occurred quantitatively with the loss of 48 μg of GvP1 monomer mg−1 (dry weight) cell wall. Similar calculations could be made from the increases in major GvP1 amino acids in the cell wall, which indicated a deposition of 44 μg of GvP1 mg−1 (dry weight) cell wall. These calculations indicate that elicitation of extensin deposition can cause a remarkable 4% to 5% increase in the covalent cell wall dry weight.
The increase in cell wall cross-linking brought about by extensin deposition has been associated with cell wall toughening and increased resistance to fungal lytic enzymes (Esqerrè-Tugaye et al., 1979; Baldwin et al., 1992; Brisson et al., 1994), through the formation of an extensin network that may, in addition, be coupled to cell wall pectins (Qi et al., 1995). It is conceivable that the substantial deposition of extensin observed in grapevine callus cultures can increase cell wall resistance in a similar manner.
The deposition of extensin is thought to be catalyzed by class III peroxidases (Cooper and Varner, 1984). This appears to be true in grapevine callus cultures, where the elicitor-induced deposition of GvP1 extensin was inhibited by the peroxidase inhibitors azide and ascorbate. The extracellular peroxidase activity in grapevine callus apparently can be attributed to only four basic and ionically bound peroxidases, and all four were isolated or purified. Both in vitro and in situ tests of the activity of each peroxidase with GvP1 demonstrated that only Px3 had a high capacity for the oxidative cross-linking of this extensin. This confirms earlier in vitro studies that indicated extensin cross-linking activity is restricted to particular peroxidases (Brownleader et al., 1996; Schnabelrauch et al., 1996; Magliano and Casal, 1998; Jackson et al., 1999) that can be referred to as EPs (Everdeen et al., 1988).
The minor cell wall content of other peroxidases and their negligible capacity to cross-link extensin indicates that they are incapable of making any significant contribution to extensin cross-linking in situ. In contrast, the abundance of Px3 in cell walls and its high capacity for extensin cross-linking strongly indicates that the observed in situ deposition of GvP1 extensin during elicitation is catalyzed specifically by this peroxidase. To our knowledge, this work is one of only very few examples where the function of a particular class III peroxidase has been clearly associated with specific events in cell wall modification. Following the nomenclature of Everdeen et al. (1988), we refer to Px3 as GvEP (grapevine extensin peroxidase).
GvEP has been purified to electrophoretic homogeneity. This peroxidase presents a UV/Vis spectra qualitatively identical to those of other grapevine peroxidases (non-EPs) and class III peroxidases in general, indicating that this EP belongs to this class. Confirming this, two of the internal peptide sequences obtained from GvEP could be mapped to conserved sequences in helixes B and J of the class III peroxidase structure (Schuller et al., 1996; Gajhede et al., 1997; Mirza et al., 1999).
To date, the purification of only three other EPs has been reported (Brownleader et al., 1995; Schnabelrauch et al., 1996; Jackson et al., 1999) and none of these unusual peroxidases have been sequenced. Obtaining further peptide sequences of GvEP leading to the isolation of its cDNA clone therefore would be informative.
These results confirm that the deposition of extensin in cell wall defensive reactions requires the co-expression of extensin and specific peroxidases (an EP) at the cell wall of cells undergoing an oxidative burst. The quantitative purifications of both GvP1 and GvEP now allow us to probe their relationship to cell wall defensive responses in the undoubtedly more complex situation in planta.
MATERIALS AND METHODS
Growth Conditions for Grape (Vitis vinifera L. cv Touriga) Vine Callus
Grapevine callus was induced from leaf explants (0.5 cm2) of grapevine cv Touriga, placed on Murashige and Skoog-based tissue culture medium, containing (per liter) 4.4 g Murashige and Skoog basal salt mixture (Sigma, St. Louis), 20 g of Suc, 100 mg of casein hydrolysate, 5 g of polyvinylpyrrolidone 40T, 10 mg of 2,4-dichloro-phenoxyacetic acid, 2 mg of kinetin, and 2 g of Gelright (Sigma). The cultures were kept at 24°C in the dark. Callus was transferred after 4 weeks to fresh medium of the same composition except that hormonal supplements were 0.5 mg L−1 2,4-dichloro-phenoxyacetic acid and 0.215 mg L−1 kinetin, and were subcultured thereafter every 2 weeks.
Assay of GvP1 Deposition
Grapevine callus (approximately 1.5 g fresh weight) was vacuum infiltrated in 15 mL of sodium phosphate buffer (20 mm, pH 6.8) containing either 100 μm H2O2, 20 mg mL−1 of yeast extract, 10 mg mL−1 of chitin, 1 mm GSH, 1 mm salicylic acid, or 60 μg mL−1 of Glc equivalents of a crude preparation of the β-glucan elicitor from mycelial walls of Phytophthora megasperma Dresch f. sp. glycinea Kuan and Erwin (Hahn et al., 1992). For inhibition of elicited GvP1 insolubilization, callus tissues were infiltrated with sodium phosphate buffer containing 10 mg mL−1 of chitin and either 10 mm sodium azide, 5 mm ascorbate, or 1–20 μm DPI. As a control, callus tissues were infiltrated with sodium phosphate buffer alone. Unless otherwise stated, callus tissues were allowed to stand for 15 min at 24°C before washing with 30 mL of distilled water. Cell surface protein was eluted by gentle swirling in 3 mL of 1 m KCl for 2 min and the eluate collected by vacuum-assisted filtration through No. 1 filter paper (Whatman, Clifton, NJ). The eluate was clarified by centrifugation at 10,000g for 5 min. One hundred microliters of sample eluate was then loaded on to a Superose-12 gel-filtration FPLC column (HR 10/30, Amersham-Pharmacia Biotech, Uppsala) equilibrated with degassed 0.1 m sodium acetate buffer (pH 5.0), and run at a flow rate of 0.5 mL min−1. The eluate was monitored at 280 nm using a 2100 UV monitor (LKB, Bromma, Sweden) linked to a personal computer via a data acquisition card with analog to digital conversion (PC-512, National Instruments, Madrid). Control cultures presented high levels of GvP1 as a distinct peak eluting at 9.8 mL. Where treatments had caused GvP1 deposition, this peak was absent or substantially reduced.
Quantification of Extensin in Cell Wall Extracts
The relation of GvP1 peak height to micrograms GvP1 injected onto a Superose-12 column (conditions described above) was determined from known quantities (5–60 μg) of pure GvP1. To determine the quantity of GvP1 in cell wall eluates, their Superose-12 chromatograms were subject to Gaussian analysis. The contribution of proximal components to the GvP1 peak was subtracted, and the adjusted GvP1 peak height was used to quantify GvP1. The results were expressed as micrograms GvP1 mg−1 (fresh weight) of cells or as micrograms GvP1 mg−1 (dry weight) cell wall, as estimated from cells dried for 16 h at 80°C.
Purification of GVP1 and Grapevine Callus Peroxidases
Grapevine callus (200 g) was washed extensively with distilled water, then gently agitated in 500 mL of 1 m KCl in 20 mm sodium acetate (pH 4.5) to elute ionically bound cell surface proteins. The eluate was then collected by vacuum-assisted ultrafiltration through a 0.45-μm filter (Sartorius), equilibrated in 20 mm sodium acetate (pH 4.5), and concentrated to 20 mL by pressure-assisted filtration through a 10-kD cutoff membrane (Diaflow, Amicon, Beverly, MA). The concentrate was loaded onto a 1.5- × 20-cm SP-Sepharose column (Amersham-Pharmacia) equilibrated in 20 mm sodium acetate (pH 4.5), and washed with the same buffer at 2 mL min−1 until all nonbinding, A280-absorbing material had been removed. Bound proteins were then eluted within a 0 to 0.5 m NaCl gradient over 60 min at a flow rate of 2 mL min−1. Fractions containing GvP1 were identified by Superose-12 chromatography (conditions described above), where this extensin presents a distinct peak at 9.8 mL. SP-Sepharose-enriched fractions of GvP1 thus obtained were used in all in vitro and in situ extensin cross-linking assays. Fractions containing peroxidase were identified by their A405 and reaction with guaiacol. Further purification of these proteins is described below.
Purification of GvP1
Fractions enriched in GvP1 (0.35–0.41 m NaCl) were pooled and concentrated to 1 mL by centrifuge-assisted ultrafiltration (10-kD cutoff; Vivascience, Stonehouse, UK). Aliquots of 200 μL were loaded onto Superose-12 and fractions eluting between 9.2 and 10.2 mL were collected. These fractions were pooled, equilibrated in distilled water and freeze dried by Speed-Vac (Savant Instruments, Holbrook, NY).
Purification of Peroxidases
SP-Sepharose chromatography resolved cell wall peroxidases into three distinct peaks (Px1–3). Px1 was eluted between 0.14 and 0.17 m NaCl, Px2 between 0.27 and 0.28 m NaCl, and Px3 between 0.46 and 0.49 m NaCl. Px1 was concentrated and equilibrated in 20 mm sodium acetate buffer (pH 4.5), but otherwise was not processed further. Px2 was concentrated and equilibrated in 20 mm sodium acetate buffer (pH 4.5) and loaded onto a 1-mL Mono-S column (Pharmacia) equilibrated in the same buffer. After passing 10 mL of 0.15 m NaCl in equilibration buffer at 1 mL min−1, residual proteins were eluted within a 0.15 to 0.35 m NaCl gradient over 45 min at 1 mL min−1. Two peaks of peroxidase activity were detected in fractions eluting at 0.25 to 0.275 m NaCl (Px2a) and at 0.28 to 0.29 m NaCl (Px2b). Px3 was concentrated and equilibrated in 20 mm sodium acetate buffer (pH 4.5) and loaded onto a 1-mL Mono-S column (Pharmacia) equilibrated in the same buffer. After passing 10 mL of 0.25 m NaCl in equilibration buffer, residual proteins were eluted within a 0.25 to 0.5 m NaCl gradient over 45 min at 1 mL min−1. Peroxidase containing fractions (eluting at 0.47 m NaCl) were pooled, equilibrated in 1 m (NH4)2SO4 containing 50 mm K2HPO4 (pH 7.0), and loaded onto a 1-mL Phenyl Sepharose 6FF hydrophobic interaction column (Pharmacia) equilibrated with the same. The column was washed in equilibration buffer at 0.75 mL min−1 and all peroxidase activity collected in the nonbinding fraction.
Extensin Cross-Linking Assay
SP-Sepharose enriched GvP1 extensin (35 μg of GvP1) was incubated with 0.1 m sodium phosphate (pH 6.8) containing 100 μm H2O2. The reaction was initiated with the addition of selected peroxidase in a total volume of 50 μL. After 30 min in the dark at room temperature, the mixture was injected on to a Superose-12 gel-filtration column equilibrated with 0.1 m sodium acetate (pH 5.0). The eluate (0.5 mL min−1) was monitored at 280 nm. The extensin monomer eluted at 9.8 mL whereas cross-linked extensin eluted at the void volume (7.0 mL).
Assay of in Situ Deposition of GvP1 by Individual Grapevine Peroxidases
Grapevine callus was gently teased apart into small clumps (≤0.5 cm in diameter) and washed extensively in 20 mm sodium acetate buffer (pH 4.5) containing 1 m KCl by vacuum-assisted Buchner filtration. The washed cells were then resuspended in 20 mm sodium acetate buffer (pH 4.5) containing 2.3 μg of SP-Sepharose-enriched GvP1 and 0.7 pmol of either Px1, Px2 or Px3 per mg (fresh weight) of callus, and left to incubate at room temperature for 5 min with occasional agitation. The cells were then washed extensively in 20 mm sodium acetate buffer (pH 4.5) to remove unbound proteins. The cells were then collected by centrifugation at 3,500g and resuspended in sodium phosphate buffer (15 mm, pH 6.8) containing 100 μm H2O2, and incubated in the dark at 24°C for 10 min. The deposition of GvP1 was then assayed by Superose-12 chromatography as described above.
Electrophoresis
NEIEF utilized the LKB 2117 Multiphor II system, and 0.5-mm-thick polyacrylamide gels (5% T and 3% C), containing 5% (v/v) 3.5 to 10.0 ampholites. Gels were run at 4°C with the constant power of 0.15 W cm−1 width of gel and with a limiting voltage at 420 V for 1 h. All zymograms were developed using guaiacol as substrate as before (Jackson and Ricardo, 1994). SDS-PAGE was performed as described by Laemmli (1970), using 12% T and 3% C 0.75-mm polyacrylamide gels run in the Mini-Protean II electrophoresis apparatus (Bio-Rad, Hercules, CA) at 50 mA per gel and were stained with Coomassie blue R-250. The molecular mass (kilodalton) markers utilized were α-lactalbumin (14.2), trypsinogen (24.0), carbonic anhydrase (29.0), ovalbumin (45), and bovine serum albumin (66).
SELDI-MS
Mass spectra were obtained using a SELDI-mass spectrometer. GvP1 was spotted on to a reverse-phase (H4) protein array chip surface (C16 carbon backbone) and allowed to air dry. Samples were washed twice with 4 μL of 10% (v/v) acetonitrile and 0.7 μL of sinapinic acid matrix was added just before samples were allowed to dry. The matrix was a saturated solution in 50% (v/v) acetonitrile: water containing 0.5% (w/v) trifluoroacetic acid. The data from 50 laser shots were acquired and averaged to produce the spectrum. Insulin (Mr 5,733) was used as the external calibrant.
Amino Acid Analysis of GvP1
Pure GvP1 was acid hydrolyzed in 6 m HCl containing 1 mg mL−1 of phenol at 150°C for 1 h under nitrogen. Nor-Leu was added as an internal calibrant. The acid hydrolysates were derivatized with phenylisothiocyanate using a Waters pico-tag system and four samples corresponding to different quantities of GvP1 were analyzed on a Pico-Tag amino acid analysis system (Waters, Milford, MA) as per the manufacturer's instructions.
Effect of GvP1 Deposition on the Amino Acid Composition of Covalently Bound Cell Wall Protein
Grapevine callus (1.5 g) was vacuum infiltrated with 3 mL of 100 μm H2O2 for 15 min at 24°C to cause GvP1 deposition. Callus infiltrated with 3 mL of water under the same conditions was used as the untreated control. The callus was then washed in 20 mL of 1 m KCl to remove ionically bound cell wall protein and then ground in a pestle and mortar with liquid nitrogen. The homogenate was then washed by centrifugation at 4,500g for 5 min in 1% (w/v) Triton X-100, three times with 1 m KCl, and three times with distilled water. The washed cell wall pellet was freeze dried and acid hydrolyzed in 6 m HCl containing 1 mg mL−1 of phenol at 110°C for 16 h under nitrogen. The hydrolysate was clarified by ultrafiltration through a 0.2-μm filter (Millipore, Bedford, MA) and the amino acid composition determined under identical conditions used for amino acid composition analyses of GvP1.
Amino Acid Sequencing of GvP1 and GvEP
Urea (25 μL, 8 m) and 0.4 ammonium bicarbonate (pH 8.0) were added to 20 μL of GvP1 or Px3, vortexed, and centrifuged at 12,000 rpm. Tris (carboxy) ethylphosphine hydrochloride (5 μL, 45 mm; Pierce, Chester, UK) was then incubated with the denatured protein for 5 min at 37°C. The reaction mixture was cooled to room temperature and then 5 μL of 100 mm iodoacetamide was added in the dark for 20 min to alkylate Cys to carboxyamido methyl-Cys.
Tryptic fragments of Px3 were obtained by adding and 5 μL of sequencing grade and N-tosyl-l-phenylalanine chloromethyl ketone-treated porcine trypsin (Promega Corp., Southampton, UK) in 0.1 m ammonium bicarbonate and 60 μL of water to the Px3 incubation mixture (1:50 [w/v] trypsin:sample protein) for 18 h at 37°C. A control experiment containing all the above ingredients, with the exception of sample protein, and incubated under identical conditions was undertaken to ensure that the tryptic fragments were not derived from autolysis of trypsin. Tryptic fragments of Px3 were separated using a Symmetry 300, 5-μm, 150- × 2.1-mm C18 reverse-phase column. Tryptic fragments of Px3, and undigested GvP1, were sequenced using a Procise protein sequencer (model 491; Applied Biosystem, Warrington, UK) in the gas phase mode from Biobrene-treated glass fiber discs.
ACKNOWLEDGMENTS
We thank Lee Lomas (Ciphergen, Surrey, UK) for performing SELDI-mass spectrometric analyses and Micheal Hahn (University of Georgia, Athens) for the fungal elicitor.
Footnotes
The research was supported by the Fundação de Ciência e Tecnologia (PRAXIS/XXI; grants to P.A.P.J., C.S.P., and A.F.; project no. PRAXIS/2/2.1/BIO/1146/95).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010192.
LITERATURE CITED
- Baldwin T, Coen E, Dickinson H. The Ptl 1 gene expressed in the transmitting tissue of Antirrhinumencodes an extensin-like protein. Plant J. 1992;2:733–739. doi: 10.1046/j.1365-313x.1992.t01-14-00999.x. [DOI] [PubMed] [Google Scholar]
- Bolwell G, Davies D, Gerrish G, Auh C-K, Murphy T. Comparative biochemistry of the oxidative burstproduced by rose and french bean cells reveals two distinct mechanisms. Plant Physiol. 1998;116:1379–1385. doi: 10.1104/pp.116.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolwell GP, Butt VS, Davies DR, Zimmerlin A. The origin of the oxidative burst in plants. Free Rad Res. 1995;23:517–532. doi: 10.3109/10715769509065273. [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownleader M, Byron O, Rowe A, Trevan M, Welham K, Dey P. Investigations into the molecular size and shape of tomato extensin. Biochem J. 1996;320:577–583. doi: 10.1042/bj3200577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownleader M, Dey P. Purification of extensin from cell wall of tomato (hybrid of Lycopersicon esculentum and L. peruvianum) cells in suspension culture. Planta. 1993;191:457–469. doi: 10.1007/BF00195747. [DOI] [PubMed] [Google Scholar]
- Brownleader MD, Ahmed N, Trevan M, Chaplin M, Dey PM. Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 1995;109:1115–1123. doi: 10.1104/pp.109.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busam G, Junhanns KT, Kneusel RE, Kassemeyer H, Matern U. Characterization and expression of a 3-O-methyltransferase proposed for the induced resistance response of Vitis viniferaL. Plant Physiol. 1997;115:1039–1048. doi: 10.1104/pp.115.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- Cazalé A, Rouet-Mayer M, Barbier-Brygoo H, Mathieu Y, Laurière C. Oxidative burst and hypoosmotic stress in tobacco cell suspensions. Plant Physiol. 1998;116:659–669. doi: 10.1104/pp.116.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JB, Chen JA, Van Holst G, Varner JE. Hydroxyproline-rich glycoproteins of plant cell walls. Trends Biol Sci. 1987;12:24–27. [Google Scholar]
- Cooper JB, Varner JE. Crosslinking of soluble extensin in isolated cell walls. Plant Physiol. 1984;76:414–417. doi: 10.1104/pp.76.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Brownleader M, Pantelides A, Trevan M, Smith J, Saddler G. Extensin from suspension-cultured potato cells: a hydroxyproline-rich glycoprotein, devoid of agglutinin activity. Planta. 1997;202:179–187. doi: 10.1007/s004250050117. [DOI] [PubMed] [Google Scholar]
- Esqerrè-Tugaye M, Laffite C, Mazare D, Toppan A, A T. Cell surfaces in plant-microorganism interactions: evidence for the accumulation of hydroxyproline-rich glycoproteins in the cell wall of diseased plants as a defense mechanism. Plant Physiol. 1979;64:320–326. doi: 10.1104/pp.64.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everdeen DS, Kiefer S, Willard JJ, Muldoon EP, Dey PM, Li X, A LDT. Enzymic cross-linkage of monomeric extensin precursors in vitro. Plant Physiol. 1988;87:616–621. doi: 10.1104/pp.87.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahry G, Schopfer P. Inhibition of O2-reducing activity of horseradish peroxidase by diphenyleneiodonium. Phytochemistry. 1998;48:223–227. doi: 10.1016/s0031-9422(98)00004-1. [DOI] [PubMed] [Google Scholar]
- Gajhede M, Schuller DJ, Henriksen A, Smith T, Poulos TL. Crystal structure of horseradish peroxidase C at 2.15 A resolution. Nat Struct Biol. 1997;4:1032–1038. doi: 10.1038/nsb1297-1032. [DOI] [PubMed] [Google Scholar]
- Goldberg R, Catesson A, Czaninsky Y. Some properties of syringaldizine oxidase, a peroxidase specifically involved in the lignification process. Z Pfanzenphysiol. 1983;110:267–279. [Google Scholar]
- Hahn M, Darvill A, Albersheim P, Bergmann C, Cheong J-J, Koller A, Lò V-M. Preparation and characterization of oligosaccharide elicitors of phytoalexin accumulation. In: Gurr S, McPherson M, Bowles D, editors. Molecular Plant Pathology. II: A Practical Approach. Oxford: Oxford University Press; 1992. pp. 103–147. [Google Scholar]
- Hain R, Reif HJ, Krause E, Largebartels R, Kindle H, Vornam B, Weis W, Schmelxer E, Schreir P, Stöcker R. Disease resistance results from foreign phytoalescin expression in a novel plant. Nature. 1993;361:153–156. doi: 10.1038/361153a0. [DOI] [PubMed] [Google Scholar]
- Hirsinger C, Parmentier Y, Durr A, Fleck J, Jamet E. Characterization of a tobacco gene and regulation of its gene family in healthy plants and under various stress conditions. Plant Mol Biol. 1997;33:279–289. doi: 10.1023/a:1005738815383. [DOI] [PubMed] [Google Scholar]
- Jackson P, Paulo S, Brownleader MD, Freire P, Ricardo CPP. An extensin peroxidase is associated with white-light inhibition of lupin (Lupinus albus) hypocotyl growth. Aust J Plant Physiol. 1999;26:313–326. [Google Scholar]
- Jackson P, Ricardo CPP. Planta 311–317. 1994. An examination of the peroxidases from Lupinus albusL. hypocotyls. [Google Scholar]
- Kawalleck P, Schmelzer E, Hahlbrock K, IE S. Two pathogen-responsive genes in parsley encode a tyrosine-rich hydroxyproline-rich glycoprotein (hrgp) and an anionic peroxidase. Mol Gen Genet. 1995;247:444–452. doi: 10.1007/BF00293146. [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structured proteins during the assembly of heat proteins of bacteriophage T4. Nature. 1970;227:670. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Magliano TMA, Casal JJ. In vitrocross-linking of extensin precursors by mustard extracellular isoforms of peroxidase that respond either to phytochrome or to wounding. J Exp Bot. 1998;49:1491–1499. [Google Scholar]
- Memelink J, Swords KMM, de Kam RJ, Schilperoort RA, Hoge JHC, Staehelin LA. Structure and regulation of tobacco extensin. Plant J. 1993;4:1011–1022. doi: 10.1046/j.1365-313x.1993.04061011.x. [DOI] [PubMed] [Google Scholar]
- Mirza O, Henriksen A, Ostergaard L, Welinder KG, Gajhede M. Arabidopsis thalianaperoxidase N: structure of a novel neutral peroxidase. Acta Crystal Sect D. 1999;56:372–375. doi: 10.1107/s0907444999016340. [DOI] [PubMed] [Google Scholar]
- Otte O, Barz W. Characterization and oxidative in vitro cross-linking of an extensin-like protein and a proline-rich purified from chickpea cell walls. Phytochemistry. 2000;53:1–5. doi: 10.1016/s0031-9422(99)00463-x. [DOI] [PubMed] [Google Scholar]
- Papadakis A, Roubelakis-Angelakis K. The generation of active oxygen species differes in tobacco and grapevine mesophyll protoplasts. Plant Physiol. 1999;121:197–205. doi: 10.1104/pp.121.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier Y, Durr A, Marbach J, Hirsinger C, Criqui M, Fleck J, Jamet E. A novel wound-inducible extensin gene is expressed early in newly isolated protoplasts of Nicotiana sylvestris. Plant Mol Biol. 1995;29:279–292. doi: 10.1007/BF00043652. [DOI] [PubMed] [Google Scholar]
- Qi X, Behrens BX, West PR, J MA. Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures. Plant Physiol. 1995;108:1691–1701. doi: 10.1104/pp.108.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport DT. Isolation of pI 4.6 extensin peroxidasefrom tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- Schuller DJ, Nenad B, van Huystee RB, McPherson A, Poulos TL. The crystal structure of peanut peroxidase. Structure. 1996;4:311–321. doi: 10.1016/s0969-2126(96)00035-4. [DOI] [PubMed] [Google Scholar]
- Shirsat A, Wieczorek D, Kozbial P. A gene for Brassica napusextensin is differentially expressed on wounding. Plant Mol Biol. 1996;30:1291–1300. doi: 10.1007/BF00019559. [DOI] [PubMed] [Google Scholar]
- Showalter A, Zhou J, Rumeau D, Worst S, Varner J. Tomato extensin and extensin-like cDNAs: structure and expression in response to wounding. Plant Mol Biol. 1991;16:547–565. doi: 10.1007/BF00023421. [DOI] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell walls. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder KG. Amino acid sequence studies of horseradish peroxidase. Eur J Biochem. 1979;96:483–502. doi: 10.1111/j.1432-1033.1979.tb13061.x. [DOI] [PubMed] [Google Scholar]
- Wojtaszec P, Trethowan G, Bolwell G. Reconstitution in vitro of the components and conditions required for the oxidative cross-linking of extracellular proteins in French bean (Phaseolus vulgarisL.) FEBS Lett. 1997;405:95–98. doi: 10.1016/s0014-5793(97)00166-x. [DOI] [PubMed] [Google Scholar]