Abstract
Arabidopsis plants containing the ndr1-1 mutation are incapable of mounting a hypersensitive response to bacteria carrying avrRpt2, but show an exaggerated cell death response to bacteria carrying avrB (Century et al., 1995). We show here that ndr1-1 plants are severely impaired in induction of systemic acquired resistance and PR1-driven transcription of a reporter gene in response to Pseudomonas syringae strains carrying avrRpt2 but not in response to P. syringae carrying avrB. The ndr1-1 mutation also impaired salicylic acid (SA) accumulation in response to treatments that produced reactive oxygen species (ROS) and impaired induction of systemic acquired resistance in response to in situ production of ROS. Hydrogen peroxide accumulated in wild-type Arabidopsis leaves beginning 4 to 7 h postinoculation with P. syringae carrying either avrRpt2 or avrB. In ndr1-1 plants, P. syringae carrying avrRpt2 elicited no detectable hydrogen peroxide production. Hydrogen peroxide production in response to bacteria carrying avrB was similar to that of Columbia in kinetics but of lesser intensity at early time points. These data are interpreted to indicate that NDR1 links ROS generation to SA production and that the phenotypic consequences of the ndr1-1 mutation are caused by a reduced ability to accumulate SA upon pathogen infection.
Exquisite specificity is a hallmark of gene-for-gene disease resistance. Individual plant lines carry a specific complement of disease resistance (R) genes. Plants resist infection only if the pathogen carries a specific avirulence (avr) gene that is the matched cognate of one of these plant R genes. Mutant plants with nonfunctional alleles of a particular R gene fail to recognize a pathogen carrying the corresponding avr gene, and disease ensues (Parker et al., 2000; Staskawicz, 2001). With bacterial pathogens, this specificity of molecular recognition in at least some cases is associated with direct binding of the plant R gene product to the bacterial avr gene product (Scofield et al., 1996; Tang et al., 1996). Recognition takes place inside the plant cell (Gopalan et al., 1996; Leister et al., 1996) following export of the avr gene product from the bacteria via a type III secretion system (Pirhonen et al., 1996; Mudgett and Staskawicz, 1998).
In contrast to the specificity of upstream molecular recognition processes, downstream plant responses to pathogen infection often bear strong similarities despite being elicited by vastly different types of pathogen. Gene-for-gene disease resistance is usually accompanied by rapid cell death (the hypersensitive response [HR]; Klement, 1982) in plant cells that are in direct contact with pathogen (Turner and Novacky, 1974). Uninoculated regions of the plant are induced to display an immunity to further pathogen challenge termed systemic acquired resistance (SAR). SAR protects plants from a broad spectrum of pathogens including those very different from the original (Ryals et al., 1994). A set of genes termed “pathogenesis related” (PR) are induced both locally and systemically (Lotan and Fluhr, 1990). Some PR gene products have been shown to possess antimicrobial activity (van Loon, 1997).
Previous analysis of disease resistance signaling has identified two second messengers: reactive oxygen species (ROS) and salicylic acid (SA). An NADPH oxidase activity related to that of mammalian neutrophils is thought to produce superoxide in an oxidative burst early in the response to pathogen (Doke and Ohashi, 1988; Mehdy, 1994). Both enzymatic and nonenzymatic steps subsequently will convert the superoxide to other types of ROS (Sutherland, 1991). It is not known which type of ROS is critical for disease resistance. However, exposing plants to treatments such as UV light or ozone that generate multiple types of ROS induces most facets of the resistance response (Levine et al., 1994; Yalpani et al., 1994; Green and Fluhr, 1995; Sharma et al., 1996).
SA is found in plants mostly as a β-glucoside (Enyedi et al., 1992; Malamy et al., 1992). Coincident with the disease resistance response, levels of free SA, and subsequently the conjugated forms of SA, rise dramatically (Yalpani et al., 1993). Transgenic plants engineered with nahG, a gene that encodes salicylate hydroxylase that degrades SA, show enhanced pathogen growth and impaired induction of SAR and PR genes (Gaffney et al., 1993; Delaney et al., 1994). Transgenic nahG plants show impaired gene-for-gene resistance to disease caused by some pathogens (Gaffney et al., 1993), but not others (Brading et al., 2000). Arabidopsis nahG plants do not show an HR to bacteria carrying avrRpt2 but do show an HR to bacteria carrying avrRpm1 (Rate et al., 1999). Both bacterial strains are avirulent on the Columbia parental line.
It is thought that SA acts downstream of ROS production because the ability of exogenous ROS to induce resistance responses is dependent on SA accumulation (Bi et al., 1995; Neuenschwander et al., 1995). However, SA also appears to potentiate both ROS production and cell death in the response of cultured soybean cell suspensions to avirulent Pseudomonas syringae in what has been termed “agonist-dependent gain control” (Shirasu et al., 1997). Other experiments in Arabidopsis also support a role for SA in potentiating defense responses (Weymann et al., 1995; Mauch-Mani and Slusarenko, 1996; for discussion, see Shapiro, 2000). Nitric oxide has also been implicated in disease resistance signaling (Delledonne et al., 1998; Durner et al., 1998), and mammalian precedents suggest that it could function synergistically with ROS (Halliwell et al., 1999).
Mutations in NDR1 compromise resistance to numerous strains of P. syringae and Peronospora parasitica. Mutant ndr1 plants exhibit an exaggerated cell death response upon inoculation with bacteria carrying avrB, avrRpm1, or avrPphB despite being unable to restrict multiplication of these bacteria or resist disease caused by these bacteria. It is interesting that the HR of ndr1 mutant plants is strain specific in that bacteria carrying avrRpt2 do not elicit an HR (Century et al., 1995). NDR1 encodes a small, highly basic, putative integral membrane protein (Century et al., 1997). Transcription of NDR1 is induced by pathogen infection at very early time points. However, the sequence has not revealed a precise biochemical activity for NDR1.
Arabidopsis mutants are a valuable tool for understanding which of the myriad of responses correlated with gene-for-gene disease resistance make significant contributions to limiting pathogen growth and preventing disease (Shapiro, 2000). The strain specificity of the effects of the ndr1 mutations on the HR led us to examine the strain specificity of other responses correlated with gene-for-gene disease resistance. These data suggest that the ndr1-1 mutation blocks ROS-dependent SA accumulation. This hypothesis is further supported by results of experiments using in situ ROS generation. This reduced ability to accumulate SA appears to impair agonist-dependent gain control of the HR. These effects explain the phenotypic consequences of the ndr1-1 mutation.
RESULTS
PR1-Driven Transcription in ndr1-1 versus Columbia
The following approach was taken to determine the strain specificity of ndr1 effects on responses correlated with gene-for-gene disease resistance: P. syringae bacteria carrying avrB were chosen as a representative of the class of bacteria that gave an exaggerated cell death response (Century et al., 1995). These strains were compared with P. syringae carrying avrRpt2 for their effects on ndr1-1 and Columbia plants. The avr genes were carried on the same stable plasmid vector. When informative, multiple bacterial strain backgrounds were used in an experiment. The strain backgrounds P. syringae pv tomato DC3000 (DC3000) and P. syringae pv maculicola 4326 (P.s.m. 4326) are pathogenic strains that multiply in Arabidopsis leaves. P. syringae pv glycinea Race 5 (P.s.g. Race 5) is a soybean pathogen that does not multiply in Arabidopsis leaves or cause disease on Arabidopsis (Century et al., 1995). The ndr1-1 allele was chosen for these experiments because it was known to be a null allele (Century et al., 1997).
First, we investigated whether ndr1-1 mutant plants showed differences from Columbia in PR gene transcription in response to bacteria carrying avrRpt2 or avrB. The PR1 gene is the most tightly regulated of the Arabidopsis PR genes (Uknes et al., 1992). A transgenic line carrying the β-glucuronidase (GUS) reporter gene under the control of the PR1 gene promoter in a Columbia background was generated (see “Materials and Methods”). This line showed high uniform GUS expression in response to the same conditions that induce transcription of the native PR1 gene (Uknes et al., 1992). This transgene was crossed into the ndr1-1 background. These lines were designated Col-0:PR1/GUS and ndr1-1:PR1/GUS.
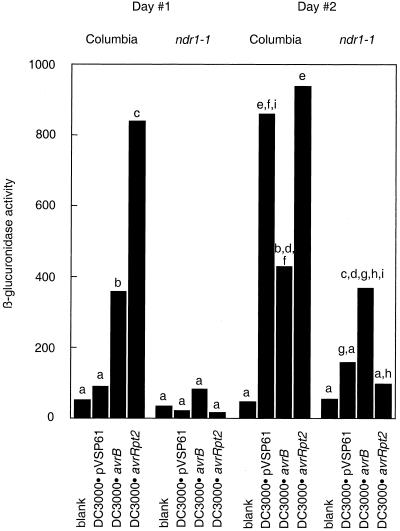
These lines were used to investigate PR1-driven transcription in response to bacterial inoculation. Leaves of either Col-0:PR1/GUS or ndr1-1:PR1/GUS plants were hand inoculated with 1 × 106 bacteria mL−1, carrying avrRpt2, avrB, or the empty pVSP61 vector. Blank inoculations were performed using 10 mm MgCl2. Bacteria-induced GUS activity was assessed using a fluorogenic substrate (see “Materials and Methods”). The data are presented in Figure 1.
Figure 1.
Bacteria-induced PR1-driven transcription in ndr1-1 versus Columbia. 1 × 106 bacteria mL−1 were hand inoculated into leaves of Arabidopsis carrying a transgene expressing GUS under control of the PR1 promoter in either the Columbia or the ndr1-1 background. Each bar represents a mean of data from 11 to 12 leaves. All data are expressed in arbitrary fluorescence units. Data are normalized in that an identically sized leaf area was sampled in each case. Differences between means were assessed for statistical significance using Student's t tests. Lowercase letters indicate which differences between means were judged significant at the P < 0.01 level. These comparisons were made separately for the two experimental time points. PR1-driven transcription induced by DC3000·avrB was significantly greater than that induced by DC3000·avrRpt2 on ndr1-1 plants at both time points if significance was judged at the P < 0.05 level (not pictured).
The most striking result from this experiment was that ndr1-1:PR1/GUS plants inoculated with bacteria carrying avrRpt2 showed negligible GUS activity. This impairment relative to Columbia plants was seen at both time points. Replicate experiments with P.s.g. Race 5 strains gave similar results (data not shown). All differences were judged significant at the P < 0.01 level using Student's t tests.
Experiments using bacteria carrying avrB had very different results. DC3000·avrB elicited comparable GUS activity in the two lines at the 2-d time point, although the ndr1-1:PR1/GUS response was significantly lower (Student's t test, P < 0.01) at the 1-d time point. Replicate experiments with P.s.g. Race 5 strains failed to show significant differences due to the ndr1-1 mutation at either time point (data not shown). Thus, the bacterial strains that did not elicit an HR on ndr1-1 plants caused negligible induction of PR1-driven transcription, whereas the bacterial strains that could give a vigorous cell death response on ndr1-1 induced PR1-driven transcription.
The behavior of the strains carrying the empty vector was also worthy of note. DC3000·pVSP61 induced only low-level GUS activity in Col-0:PR1/GUS plants at the 1-d time point, but high level activity by the 2-d time point. The data from both time points appear to show impairment in the response of the ndr1-1:PR1/GUS line. Differences between the Arabidopsis lines were significant at the P < 0.01 (Student's t test) level only with the data from the 2-d time point. P.s.g. Race 5·pVSP61 induced only low-level GUS activity in both lines at both time points (data not shown).
Biological Induction of SAR in ndr1-1 versus Columbia
Next, we investigated the relative ability of the two bacterial strains to induce SAR on ndr1-1 and Columbia plants. SAR can be assessed in Arabidopsis as the ability of a primary inoculation with bacteria to inhibit multiplication of bacteria introduced later as a secondary challenge (Cameron et al., 1994). P. syringae do not move systemically in Arabidopsis; therefore, excision of primary inoculated leaves prior to secondary challenge allows monitoring growth of only the challenge bacteria.
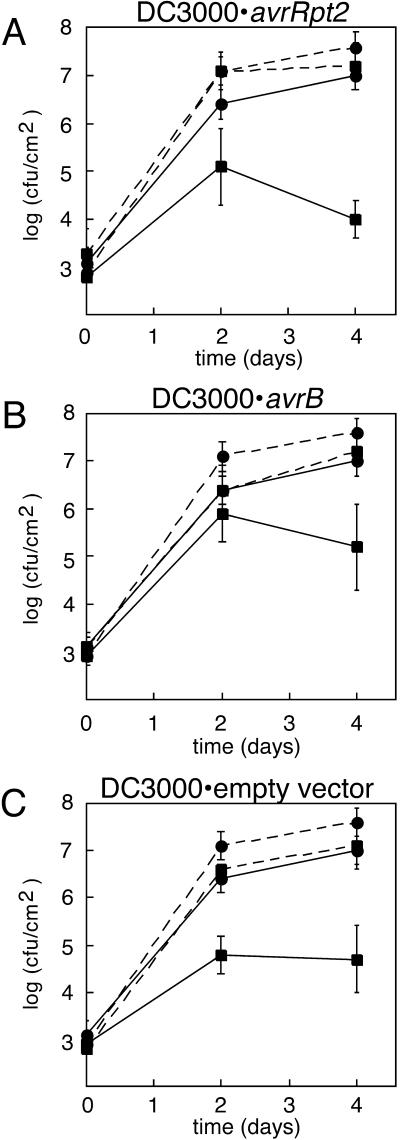
The data from one representative set of experiments is shown in Figure 2. In all cases, data from plants that had received a primary inoculation with bacteria (squares) is shown in comparison with data from plants where 10 mm MgCl2 was used as a blank primary inoculation (circles). SAR was judged to be occurring if comparisons of means showed, at minimum, significant differences (Student's t test, P < 0.1) at either the d-2 or day-4 time point in all replicates of the experiment. Growth of challenge bacteria in plants that received blank primary inoculations was statistically indistinguishable (Student's t test, P > 0.5) from growth of challenge bacteria in plants which did not receive any primary inoculation (data not shown).
Figure 2.
Biological induction of SAR in ndr1-1 versus Columbia. Columbia plants (solid lines) or ndr1-1 plants (dashed lines) were inoculated with bacteria (squares for data points) or a MgCl2 blank (circles for data points) 2 d prior to inoculation with P. syringae pv tomato DC3000 carrying only the empty pVSP61 vector. Primary inoculated leaves were excised prior to secondary inoculation. Data points represent means of triplicate determinations of in planta bacterial growth at specified time points. A, P.s. tomato DC3000 carrying avrRpt2 used for primary inoculation with bacteria. B, P.s. tomato DC3000 carrying avrB used for primary inoculation with bacteria. C, P.s. tomato DC3000 carrying empty vector used for primary inoculation with bacteria.
As evidenced by comparison of the solid lines in Figure 2A, DC3000·avrRpt2 elicited a strong SAR response on Columbia plants. Inoculation with this bacterial strain led to a 1.5 to 3 order of magnitude suppression of growth of the challenge bacteria. In contrast, inoculation of ndr1-1 mutant plants with this strain did not lead to SAR (compare dashed lines in Fig. 2A).
Inoculation of ndr1-1 mutant plants with DC3000·avrB elicited a SAR response comparable in magnitude to that shown by Columbia plants at the 2-d time point (Fig. 2B). The ndr1-1 response appears to be partially impaired relative to Columbia at the 4-d time point. In conclusion, the bacterial strain that does not elicit a HR on ndr1 mutant plants also does not elicit SAR on ndr1-1. However, the bacterial strain that elicited an exaggerated cell death response on ndr1 mutant plants elicited SAR on ndr1-1, albeit to a lesser degree than on Columbia plants. The same qualitative trends are seen if the experiment is performed using bacterial strains of a P.s.g. Race 5 background rather than DC3000-based strains in the primary inoculations (data not shown). All experiments shown in Figure 2 were repeated an additional one to three times with consistent results.
Figure 2C shows results of experiments in which the bacterial strain used in the primary inoculations carried only the empty pVSP61 vector without a cloned avr gene. Comparison of the solid lines Figure 2C indicates that the virulent bacterial strain (DC3000·pVSP61) elicited a strong SAR response on Columbia, comparable in magnitude with that elicited by DC3000·avrRpt2. A reduced SAR response was shown by ndr1-1 mutant plants (compare dashed lines). We conclude that SAR elicited by the virulent bacterial strain is partially NDR1-dependent.
In all experiments shown in Figure 2, DC3000·pVSP61 multiplies to a greater extent in ndr1-1 plants than in Columbia, irrespective of whether SAR has been induced. This behavior has been previously documented and is correlated with increased severity of disease symptoms (Century et al., 1995). Similar results have been obtained with npr1 and eds mutants (Cao et al., 1994; Rogers and Ausubel, 1997; Volko et al., 1998).
Bacteria-Induced Hydrogen Peroxide Production in ndr1-1 versus Columbia
Arabidopsis ndr1-1 plants did not show the HR, PR1-driven transcription, or SAR in response to bacteria carrying avrRpt2. These responses were elicited by bacteria carrying avrB. Therefore, signaling molecules implicated in eliciting these responses were investigated. Arabidopsis leaf tissue has been reported to release large amounts of ROS upon homogenization (Wolfe et al., 2000). We have confirmed using several published tissue homogenate methods for hydrogen peroxide quantitation that requisite signal-to-noise and reproducibility was not obtainable from these methods (data not shown). Instead, we have employed a recently published in vivo method for hydrogen peroxide quantitation (Wolfe et al., 2000). Leaves are infiltrated with 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), excised, exposed to 365 nm of UV light, and photographed. The use of UV light does not induce artifactual ROS production. The wavelength of UV light used to excite the dichlorofluorescein (DCF) is of much lower energy than that known to induce ROS production, this treatment lasts only 1 min, and photography is performed immediately afterward. This dye fluoresces only upon oxidation to DCF, and is known to be oxidized in Arabidopsis responding to P. syringae bacteria primarily by hydrogen peroxide (Bass et al., 1983; Wolfe et al., 2000). DCF fluorescence thus serves as a semiquantitative assay of hydrogen peroxide levels. Evidence has been presented that both DCF and DCFH will leak back into the apoplast following ester hydrolysis in the cytoplasm (Wolfe et al., 2000, and references therein). We have confirmed these observations via confocal microscopy (data not shown). As such, this assay does not distinguish between intracellular and extracellular pools of hydrogen peroxide.
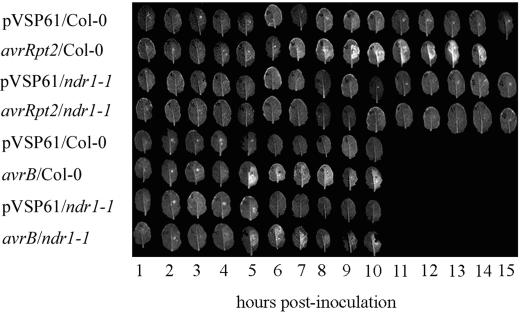
Time courses of increase in DCF fluorescence are presented in Figure 3. The low background fluorescence seen following inoculation with bacteria carrying the empty pVSP61 vector was exclusively red chlorophyll fluorescence. A similar background was seen on noninoculated half-leaves. As documented in Table I, green, DCF fluorescence was first seen in Columbia leaves at 4 to 7 h postinoculation, depending on bacterial strain. The first signs of the macroscopic HR were seen 2 to 4 h later with bacteria carrying avrB, but as much as 9 h later with bacteria carrying avrRpt2. The intensity of DCF signal at time points prior to onset of the macroscopically visible HR was greater in Columbia plants inoculated with bacteria carrying avrB than in those inoculated with bacteria carrying avrRpt2. This conclusion was confirmed in replicates of the experiment shown in Figure 3 where this comparison was made between leaves photographed at the identical time (data not shown). Differences were very large with the DC3000 or the P.s.m. 4326 backgrounds but were somewhat less pronounced with the P.s.g. Race 5 background.
Figure 3.
Biological induction of hydrogen peroxide accumulation in ndr1-1 versus Columbia. Columbia or ndr1-1 plants were inoculated with P. syringae pv tomato DC3000 carrying a plasmid-borne copy of avrRpt2, avrB, or only the empty pVSP61 vector. DCFH-DA was inoculated 15 to 30 min prior to excising of the leaves at the indicated time points for brief exposure to 365 nm of UV light followed immediately by photography.
Table I.
Kinetics of bacteria-induced hydrogen peroxide accumulation and the HR in ndr1-1 versus Columbia
| DC3000 avrB | DC3000 avrRpt2 | Psm 4326 avrB | Psm 4326 avrRpt2 | Psg Race 5 avrB | Psg Race 5 avrRpt2 | |
|---|---|---|---|---|---|---|
| Earliest DCF fluorescence in Columbia | 5 | 5 | 4 | 7 | 4–5 | 4 |
| Initial signs of leaf collapse in Columbia | 8 | 10 | 8 | 12 | 7–8 | 12–13 |
| Complete leaf collapse in Columbia | 10 | 14 | 10 | 15 | 10 | 15 |
| Earliest DCF fluorescence in ndr1-1 | 5 | None | 4 | None | 4–5 | None |
| Initial signs of leaf collapse in ndr1-1 | 7–8 | None | 8 | None | 7–8 | None |
| Complete leaf collapse in ndr1-1 | 9 | None | 9 | None | 10 | None |
Units are time in hours for all data.
By contrast, bacteria carrying avrRpt2 elicited no detectable signal above background in ndr1-1 mutant plants. DCF fluorescence of ndr1-1 mutant plants in response to bacteria carrying avrB was similar to that of Columbia in kinetics but of lesser intensity at early time points.
Bacteria-Induced SA Accumulation in ndr1-1 versus Columbia
Next, we investigated SA levels in pathogen-infected ndr1-1 versus Columbia plants. Preliminary attempts to use DC3000-based strains allowed measurements of SA levels up to 70 μg of total SA per gram fresh weight at 42 h postinoculation. All data precisely mirrored the HR phenotypes in that the highest levels of SA were seen only when programmed cell death was occurring (data not shown). However, there were obvious pathogen-induced changes in leaf water content during the experiment, even at the low levels of inoculum employed, with both virulent and avirulent strains of bacteria. Normalization based on fresh weight thus would not have allowed accurate comparisons between time points.
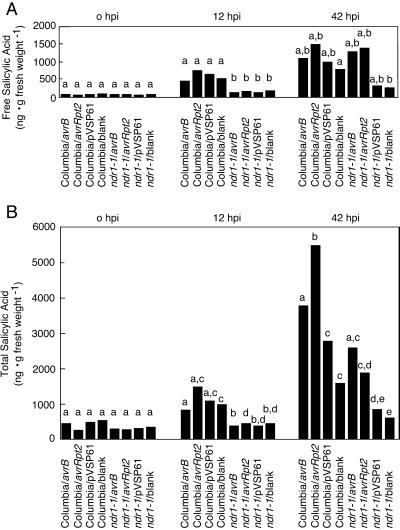
To obtain the most informative data possible, we switched to the Psg Race 5-based strains. Because these strains are not Arabidopsis pathogens, disturbances to leaf water content were minimized. An inoculum of 1 × 106 bacteria mL−1 was used throughout (this is sufficiently low to minimize HR-related loss of water content), and time points were taken at time zero, 12 h postinfiltration, and 42 h postinfiltration. Means of data from three replicate experiments are presented in Figure 4. Columbia plants accumulate both free and total SA in response to all treatments. An unexpectedly large response was seen to treatment with the MgCl2 blank and non-pathogen strain carrying only the empty vector (see below for explanation). Nonetheless, the accumulation of total SA was significantly greater in response to bacteria carrying either avr gene as compared with the other treatments. Two-way analyses of variance were used with data from the 42-h time point. Differences in means were found to be significant for comparisons of avrRpt2 versus empty vector (P < 0.001), avrB versus empty vector (P < 0.1), avrRpt2 versus blank (P < 0.01), and avrB versus blank (P < 0.1).
Figure 4.
Bacteria-induced SA accumulation in ndr1-1 versus Columbia. 1 × 106 bacteria mL−1 of P.s.g. Race 5 carrying the specified avirulence gene were inoculated into leaves of Columbia or ndr1-1 plants. Means of free (A) and total (B) SA determinations for identical time points from three separate experiments are presented. Lowercase letters indicate statistically significant differences between these means (ANOVA, P < 0.1 or in some cases greater significance). These comparisons were made separately for each time point.
It is striking that ndr1-1 mutant plants showed greatly reduced accumulation of both free and total SA at the 12-h time point relative to Columbia in response to all treatments. This impairment in SA accumulation was also seen at the 42-h time point in response to treatment with either the MgCl2 blank or the non-pathogen strain carrying only the empty vector. However, by the 42-h time point, accumulation of free SA in response to bacteria carrying either avr gene was comparable in ndr1-1 mutant plants with that in Columbia plants. Accumulation of total SA at 42 h in response to avirulent bacteria was less in ndr1-1 mutant plants than in Columbia. However, impairment in the response to avirulent bacteria was not seen to the same extent as impairment in the response to the MgCl2 blank or to the non-pathogen strain carrying only the empty vector. The differences seen at this time point in ndr1-1 mutant plants between the responses to bacteria carrying avrRpt2 versus avrB were not statistically significant (ANOVA, P > 0.2).
UV-C Induction of SA Accumulation in ndr1-1 versus Columbia
The production of SA in response to vacuum infiltration with the MgCl2 blank would be explained if the treatment, which involves exposure to anoxic conditions followed by rapid air reperfusion, generated oxygen radicals. The impairment of ndr1-1 plants in this response would then suggest that the ndr1 block lies downstream of ROS production and upstream of SA production. However, we could not detect hydrogen peroxide production induced by vacuum infiltration using the DCF-DA method (data not shown), most likely because it was low-level. The DCF-DA method can detect integral accumulation of hydrogen peroxide over time. It would be of greater sensitivity in experiments using a continuous inducer of signaling such as bacteria than in this experiment.
Therefore, to test this hypothesis and validate these conclusions, we used 254 nm of UV-C light as a noninvasive way to generate ROS in planta (Yalpani et al., 1994; Green and Fluhr, 1995). Plants were exposed to UV-C light for 10 min. Leaf samples were taken and processed for SA quantitation. To maximize precision with respect to time of sampling to facilitate comparisons, only single tissue samples were taken for each data point. The sampling process for a single time point was completed in less than 10 min.
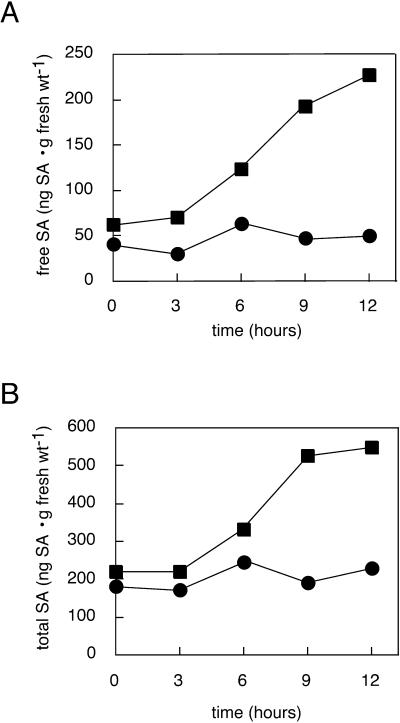
The results of the experiment are presented in Figure 5. Columbia plants showed increases in SA levels at the 6-h time point. Slight increases in total SA beyond that attributable to free SA were also seen at this time point. SA levels continued to increase with time. In contrast, SA levels in ndr1-1 mutant plants showed only a minor increase. Replicates of this experiment yielded similar results. These results support the placement of the ndr1-1 block between ROS generation and SA production. Obtaining the same qualitative results with both UV-C and anoxia/rapid air reperfusion supports the contention that it is ROS and not some other consequence of these treatments that is responsible for these effects.
Figure 5.
UV-C-elicited SA production in ndr1-1 versus Columbia. Tissue was harvested from ndr1-1 (circles for data points) or Columbia (squares for data points) plants at the indicated times following a 10 min of exposure to UV-C light. Free (A) and total (B) SA content of the samples were quantitated. To achieve maximum precision with respect to time of sampling, only one sample was taken for each time point. As such, no error bars are shown. Similar results were obtained in replicate experiments.
UV-C Induction of SAR in ndr1-1 versus Columbia
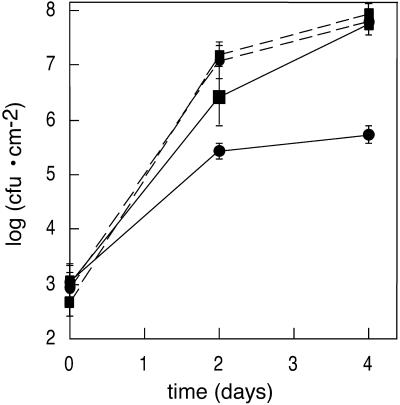
If NDR1 indeed acts to link ROS production to SA production, ndr1-1 plants should also be impaired in UV-C induction of SAR. We tested this prediction by treating ndr1-1 and Columbia plants with UV-C under identical conditions to those described above. Two days post-treatment, these plants and control ndr1-1 and Columbia plants that did not receive any treatment were inoculated with DC3000·pVSP61, and bacterial growth curves were performed. The data are presented in Figure 6. SAR was clearly induced by the UV-C treatment in Columbia plants (compare solid lines). In contrast, SAR was not induced by UV-C in ndr1-1 mutant plants (compare dashed lines). The slightly greater growth of bacteria in untreated ndr1-1 relative to untreated Columbia is consistent with previously documented results (Century et al., 1995) as discussed above.
Figure 6.
UV-C induction of SAR in ndr1-1 versus Columbia. Columbia plants (solid lines) or ndr1-1 plants (dashed lines) were exposed to UV-C light for 10 min (circles for data points) or left untreated (squares for data points) 2 d prior to inoculation with P. syringae pv tomato DC3000 carrying only the empty pVSP61 vector. Data points represent means of triplicate determinations of in planta bacterial growth at specified times. This experiment was repeated three times with similar results.
Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) Induction of SAR in ndr1-1 versus Columbia
If the ndr1-1 block is upstream of SA production, exogenous application of a SA analog should elicit SAR regardless of whether the plants are mutated in the NDR1 gene. BTH is metabolized in planta into a structural analog of SA and elicits effects similar to application of exogenous SA (Lawton et al., 1996). BTH treatment of plants does not induce SA production in either Columbia or ndr1-1 (data not shown).
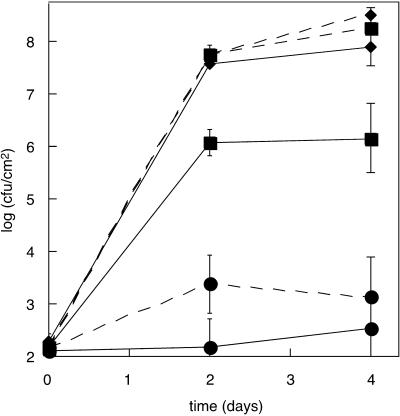
The response of ndr1-1 and Columbia plants to BTH was compared. Plants were vacuum infiltrated with either 0.12 mm BTH or water. Two days later, these plants and control plants that did not receive any treatment were inoculated with DC3000·pVSP61, and bacterial growth curves were performed. The data are presented in Figure 7. Columbia (solid lines) and ndr1-1 (dashed lines) plants both displayed BTH-induced SAR (compare curves where data points are diamonds or squares with curves where data points are circles). Three replicate experiments gave consistent results (data not shown). These results are consistent with the ndr1 block being upstream of the action of BTH.
Figure 7.
BTH induction of SAR in ndr1-1 versus Columbia. Columbia plants (solid lines) or ndr1-1 plants (dashed lines) were infiltrated with BTH (circles for data points) or a water blank (squares for data points) or were not treated (diamonds for data points). Two days later, plants were inoculated with P. syringae pv tomato DC3000 carrying only the empty pVSP61 vector. Data points represent means of triplicate determinations of in planta bacterial growth at specified times. This experiment was performed three times with similar results.
Columbia plants also displayed SAR in response to vacuum infiltration with a water blank (compare solid line with squares for data points with solid line with diamonds). The ndr1-1 plants were completely impaired in infiltration-induced SAR (compare dashed line with squares for data points with dashed line with diamonds). This result is consistent with the results presented in Figure 6 showing impairment of UV-C-induced SAR in ndr1-1 mutant plants. These results are also consistent with the impairment of vacuum infiltration-induced SA production in ndr1-1 mutant plants (Fig. 4).
DISCUSSION
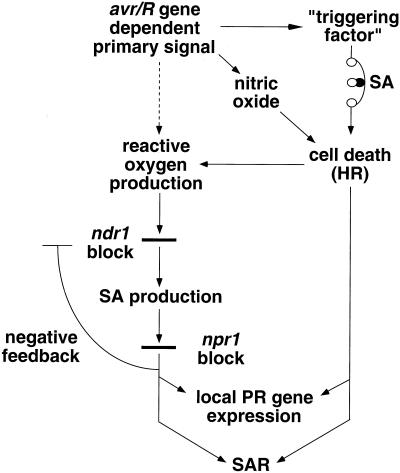
A model for disease resistance signal transduction is shown in Figure 8. Bacteria carrying avrRpt2 do not elicit PR gene transcription or SAR on ndr1-1 mutant plants, whereas bacteria carrying avrB can elicit both, albeit not as strongly as on wild-type plants. Yet, SA levels induced by these bacteria were shown to be reduced to similar levels in the ndr1-1 mutant. These results imply that at least two pathways contribute to PR gene transcription and SAR. One branch shown in Figure 8 is the well-characterized SA-dependent pathway that depends on the NPR1 gene (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). We have shown a correlation between which bacterial strains elicit cell death and which elicit PR gene transcription and SAR on ndr1-1 plants. This correlation suggests that the other branch is directly dependent either on cell death or on something closely correlated with the cell death response. The two branches can make independent contributions to PR gene transcription as shown by the induction of PR gene transcription in npr1 mutant plants by bacteria carrying avrRpt2 (Glazebrook et al., 1996; Shah et al., 1997). The two branches make a synergistic contribution to SAR as shown by the inability of npr1-1 plants to manifest SAR in response to avirulent, HR-inducing bacteria (Cao et al., 1994). However, SAR and PR gene expression can be induced by stimulation of only the NDR1/NPR1 branch (e.g. by exogenous SA or BTH).
Figure 8.
Model for disease resistance signal transduction. The dashed line leading from avr/R gene-dependent primary signal to reactive oxygen production is meant to indicate that this pathway is quantitatively much less significant than the cell death-induced reactive oxygen production. SA potentiation of cell death is represented with a symbol for a rheostat. Although pictured as being distinct, “triggering factor” could be nitric oxide, reactive oxygen, both, or something different. The SA-dependent negative feedback loop's precise target is not known. NDR1-independent pathways for SA production discussed in the text are not pictured.
Multiple lines of evidence indicate that NDR1 functions downstream of ROS production and upstream of SA production. Mutant ndr1-1 plants are impaired in SA production elicited by bacteria or by producing ROS in situ in response to either UV-C light or anoxia/rapid air reperfusion. Mutant ndr1-1 plants are also impaired in induction of SAR by bacteria or ROS but not in induction of SAR by BTH. However, ndr1-1 plants can still make SA as evidenced by comparable uninduced SA levels to wild-type plants and by the delayed rise in SA levels seen 42 h post-inoculation with P.s.g. Race 5 carrying avrB or avrRpt2. Because ndr1-1 is a null allele, NDR1-independent pathways for SA synthesis must exist.
Bacteria carrying avrRpt2 did not elicit hydrogen peroxide production on ndr1-1 mutant plants as assayed by DCF fluorescence. In contrast, bacteria carrying avrB did elicit DCF fluorescence in ndr1-1 mutant plants, albeit at levels lower than in Columbia plants at early time points. SA levels paradoxically were comparable on ndr1-1 mutant plants regardless of which avr gene was carried by the bacteria. Because ROS increases are known to lead to SA production, these results taken together argue strongly that the DCF fluorescence assay is not measuring an early oxidative burst.
We propose instead that most of the DCF fluorescence is due to hydrogen peroxide produced close to the time of cell death in primary responding cells or in cells surrounding the dying cells. We suspect that the first cell death events occur prior to the first macroscopically visible signs of the HR. The first cell death events probably occur either at the time of onset of detectable DCF fluorescence or just after this time. Wolfe et al. (2000) have documented low-level cell death assayed as decreases in chlorophyll fluorescence beginning at times comparable with those for the onset of DCF fluorescence in Columbia plants inoculated with DC3000·avrRpt2. Most plant cells, however, did not die until several hours later. The first detectable cell death events in the HR elicited by P. syringae pv. phaseolicola on an incompatible lettuce cultivar preceded macroscopic evidence of tissue collapse by 5 h (Bestwick, 1995).
The kinetics presented in Table I are consistent with this interpretation. The initial time of onset of DCF fluorescence is similar in most cases. However, bacteria eliciting slower increases in DCF fluorescence also elicit more delayed HRs. We suggest that the lack of DCF fluorescence in ndr1-1 mutant plants inoculated with bacteria carrying avrRpt2 is a consequence rather than a cause of the absence of programmed cell death.
If this interpretation of the data is correct, then the effects of the ndr1-1 mutation are explicable as a manifestation of agonist-dependent gain control. SA would function in gain control by lowering the threshold of a “triggering factor” required to see programmed cell death. Gain control might occur both in primary responding cells and in more distal cells because there is evidence that hydrogen peroxide, SA, and perhaps other signals can be transported intercellularly (Levine et al., 1994; Mölders et al., 1996).
We conclude based on data presented herein that this HR-inducing “triggering factor” accumulates to a higher level in response to bacteria carrying avrB than in response to those carrying avrRpt2. We have demonstrated a similar degree of impairment of SA production by ndr1-1 plants in response to bacteria carrying avrB or avrRpt2. This similar level of impairment could set the threshold value for the triggering factor at a level that the HR is seen in response to bacteria carrying avrB but not those carrying avrRpt2. This mechanism could also explain the ability of the nahG transgene to prevent the HR caused by bacteria carrying avrRpt2 but not that caused by bacteria carrying avrRpm1 (Rate et al., 1999). SA levels would be irrelevant to the plant's “decision” to mount a HR if the primary signal is sufficiently strong.
We cannot rule out the possibility that there are differences in very low-level, early production of hydrogen peroxide that would not be detectable with the DCF assay. It is formally possible that it is a signal produced as a consequence of very low-level ROS accumulation that is potentiated by SA to lead to programmed cell death. Nonetheless, we favor an alternative model that something other than hydrogen peroxide is the “triggering factor” that must reach a critical threshold value. One possibility for what this factor might be is nitric oxide. It is also possible that both ROS and nitric oxide (and perhaps other signals) contribute to the plant cell's decision to undergo programmed cell death (McDowell and Dangl, 2000).
An alternative explanation for the differences we have documented between signaling induced by bacteria carrying avrRpt2 as opposed to avrB is suggested by recent results concerning the virulence function of avrRpt2 (Chen et al., 2000). It has been suggested that avrRpt2 in its role as a virulence factor can suppress defense responses. Could the signaling impairments seen with ndr1-1 plants in response to bacteria carrying avrRpt2 be because of exaggeration of suppression of defense responses?
A strong prediction of this alternative explanation would be that defense responses of ndr1-1 plants elicited by bacteria carrying avrRpt2 would be impaired to a greater extent at higher levels of bacterial inoculum. In fact, the opposite is seen. When ndr1-1 plants are inoculated with 1 × 109 cfu mL−1 P.s.g. Race 5·avrRpt2, an HR is seen (Shapiro, 2000). Inoculation of ndr1-1 plants with this concentration P.s.g. Race 5 carrying only the empty vector elicited no response, validating the use of this assay (Shapiro, 2000). We feel these results are consistent with ndr1-1 phenotypes in response to bacteria carrying avrRpt2 being caused by limited production of a “triggering factor” and inconsistent with an explanation based on more efficacious suppression of defense responses.
HRs seen in response to bacteria carrying avrB, avrRpm1, avrPphB, or avrRps4 are more severe on ndr1-1 mutant plants than on Columbia plants (Century et al., 1995; Shapiro, 2000). Yet, we have shown that ndr1-1 is a loss-of-function mutation with respect to ROS-induced SA production. This more vigorous cell death response might be explained if the ndr1 mutation delayed or prevented induction of a feedback loop that negatively regulated the HR. This delay might be a consequence of the documented delay in SA accumulation (Fig. 4). Evidence has been presented for SA-dependent negative feedback (Cao et al., 1994). Whether the negative feedback loop identified in this study is identical to that which is responsible for regulating cell death is not certain.
The model in Figure 8 presents a framework for generating testable hypotheses concerning the action of these and other disease resistance signaling genes for which mutants are available. We expect that further work will clarify the uncertain aspects of this model and that the model will be a useful guide to using these signaling genes in attempts to engineer plant disease resistance.
MATERIALS AND METHODS
Bacteria and Plant Growth
Pseudomonas syringae, Agrobacterium tumefaciens, and Escherichia coli strains were cultured according to published methods (Whalen et al., 1991). Arabidopsis was grown in an MTR-30 growth chamber (Conviron, Winnipeg, MN) set for 8 h of light, 16 h of darkness, at 22°C, with 70% relative humidity for experiments involving bacterial growth curves or SA quantitation. For all other purposes, Arabidopsis growth was as described by Whalen et al. (1991). Columbia seed used in experiments was obtained from self-fertilized plants grown from seeds from the same lot used as the parental in the mutagenesis from which ndr1-1 was isolated.
Construction of Col-0:PR1/GUS
The PR1 cDNA (Uknes et al., 1992) was used as a probe to isolate the hybridizing yeast artificial chromosome yUP12F7. A sub-library was made in the A. tumefaciens cosmid vector pCLD04541 (Jones et al., 1992) using published methods (Bent et al., 1994). The PR1 cDNA was used again as a probe of colony lifts to identify a cosmid that included 3.2 kb of DNA 5′ to the PR1 coding region. This DNA was amplified with Pyrococcus furiosus polymerase using the T7 forward primer, which annealed to the vector immediately adjacent to the insert DNA, and a reverse primer designed from the known PR1 sequence (5′-CGAGAATAGCCAGTAGAATTCCTTTTTCTAAG-3′). The reverse primer introduced an EcoRI site and removed the PR1 start codon. The PCR product was cut with XbaI and EcoRI and ligated into the polylinker of pBluescript KS+ between the XbaI and EcoRI sites. The entire upstream region except the proximal 104 bp just upstream of the original start codon was then replaced with DNA from the cosmid (which had never been subject to PCR) to make pADS3.
The 3.2 kb of DNA 5′ to the PR1 coding region (hereafter, the PR1 promoter) was next subcloned upstream of the E. coli GUS gene and the nopaline synthase terminator (NOS). An EcoRI linker was blunt-end ligated into the ClaI site of pSLJ4K1 (Jones et al., 1992) to make pADS5. GUS and NOS were then excised together from pADS5 as an EcoRI/HindIII fragment and ligated between the EcoRI and HindIII sites of pCLD04541 (Jones et al., 1992) to make pADS6. The SacI/EcoRI fragment of pADS3 was then cloned between the SacI and EcoRI sites of pADS6 to make pADS7.
pADS7 was conjugated into A. tumefaciens strain GV3101 (Schell and Koncz, 1986) via triparental mating using the helper plasmid pRK2013 (Ditta et al., 1980). Plant transformation was performed using vacuum infiltration (Bechtold et al., 1993; Bent et al., 1994). Forty-three independent transgenic lines were isolated. A line homozygous for a single locus (two copies of the transgene integrated head-to-head as determined by Southern blotting) insertion, which showed strong GUS induction under all conditions predicted by northern analysis of PR1 expression (Uknes et al., 1992) and in all tissues, was chosen. This line was designated Col-0:PR1/GUS.
Construction of ndr1-1:PR1/GUS
Col-0:PR1/GUS was used as the male parent in a cross to ndr1-1. A line homozygous for both the ndr1-1 mutation and the transgene was identified in the F2. This line displayed lack of an HR in response to DC3000·avrRpt2 and an exaggerated HR in response to DC3000·avrRpm1 (Century et al., 1995). Southern-blot analysis showed the presence of only the ndr1-1 allele at the NDR1 locus [ndr1-1 contains a 1.2-kb deletion (Century et al., 1997)]. This line also showed nonsegregation of kanamycin resistance (the selectable marker associated with the transgene) in the F3.
Assay of GUS Activity
GUS activity was assessed by standard procedures (Jefferson, 1987). Single 0.125-cm2 punches of leaf tissue were taken using a number one cork borer and used for GUS assay with the fluorogenic substrate 4-methyl umbelliferyl β-d-glucuronide (Rose Scientific, Edmonton, AB). Data is shown from leaves harvested either 1 or 2 d postinoculation. Reactions were incubated precisely 2 h at 37°C and then quenched. The data is presented as the raw fluorescence units obtained via excitation at 365 nm and measuring emission at 450 nm using a fluorescence microplate reader (Dynex, Chantilly, Virginia). No attempt was made to derive rates of GUS production because preliminary experiments showed increases with time to be nonlinear. All values were within the linear range of the instrumentation and corresponded to the range within which fluorescence increases in direct proportion to concentration of pure methyl-umbelliferone.
Inoculation of Plants and in Planta Bacterial Growth Curves
Hand inoculation of plants utilized 1-mL tuberculin syringes to inject approximately 10 μL per cm2 into leaf tissue through wounds made at one site per half-leaf with a 22-G needle. Vacuum infiltration was according to published methods (Whalen et al., 1991). In planta bacterial growth curves were performed according to published methods (Whalen et al., 1991), except that the initial inoculum was 5 × 104 bacteria mL−1 and the plating of bacteria was done on nutrient yeast growth agar plates supplemented with rifampicin, kanamycin, and cycloheximide. Independent replicates of growth curves gave differences of equivalent statistical significance to those pictured.
Hydrogen Peroxide Quantitation
Hydrogen peroxide quantitation was according to a published procedure (Wolfe et al., 2000) except that hand infiltration rather than vacuum infiltration was used to inject the dye into plant leaves 15 to 30 min prior to excising the leaves. 2 × 107 bacteria mL−1 was used in inoculations of DC3000- or P.s.m. 4326-based strains. 1 × 108 bacteria mL−1 was used in inoculations of P.s.g. Race 5-based strains. These concentrations had previously been established as optimal for assay of the HR (Century et al., 1995, 1997). These levels of inoculum were also used in primary inoculations for SAR experiments.
Following bacterial inoculations, plants were transferred to a Percival Ar-75 growth chamber set for continuous light prior to assay of DCF fluorescence or observation of the HR. Leaves appeared totally dry 15 min postinoculation. Leaf excision did not result in a DCF signal; however, fluorescence was occasionally seen surrounding the inoculation site needle hole. Severely wilted or dead areas of leaves in the process of mounting a HR often could not be infiltrated with dye and appeared dark. Leaves displaying a confluent HR (from late time points in experiments using bacteria carrying avrB) could not be infiltrated with dye and thus are not pictured. Treatment with 0.12 mm SA or 0.12 mm BTH did not elicit DCF fluorescence (data not shown). All treatments were performed on triplicate leaves, and each entire experiment was repeated three to five times.
Confocal microscopy was used to establish that DCFH-DA entered the plant cytoplasm and that increases in fluorescence seen postinoculation were at least partially intracellular. Use of DCFH-DA diacetoxy methyl ester, a derivative of DCFH-DA that did not enter the cytoplasm and remained in the apoplast, led to kinetics of increase in fluorescence that were indistinguishable from those using DCFH-DA (data not shown). These results are consistent with hydrogen peroxide being generated initially in the apoplast with subsequent rapid movement into the cytoplasm.
SA Quantitation
Vacuum infiltration of bacteria was as described for growth curves except that the level of inoculum was 1 × 106 bacteria mL−1. Control experiments were performed to rule out the possibility that vacuum infiltration-induced SA production was caused by inadvertent inoculation with bacteria or fungi normally present on the leaf surface. In these experiments, benomyl was first dissolved in dimethyl sulfoxide (at 10 mg mL−1) and subsequently diluted 100-fold into 10 mm MgCl2. Rifampicin (100 μg mL−1) was dissolved directly in 10 mm MgCl2, and the solution was 0.2 μm filtered. Inclusion of either benomyl or rifampicin did not affect the level of vacuum infiltration-induced SA production (data not shown).
SA extraction was according to standard procedures (Gaffney et al., 1993). Ortho-anisic acid was used as an internal standard (Meuwly and Métraux, 1993). Chromatography was performed essentially according to published procedures (Gaffney et al., 1993). Two injections were made per sample, one to quantitate SA and one to quantitate ortho-anisic acid. SA detection was by excitation at 303 nm and monitoring emission at 437 nm. Detection of ortho-anisic acid was by excitation at 298 nm and monitoring emission at 350 nm. Detection used an LS30 fluorimeter (Perkin-Elmer, Norwalk, CT) connected in-line to the HPLC (Rainin, Woburn, MA).
UV-C and BTH/SA Treatment of Plants
UV-C (254 nm) treatments were for 10 min using a model UVGL-58 lamp (UV Products, Upland, CA) immobilized 24 cm above the plants in a closed, dark cabinet. BTH was introduced using vacuum infiltration for SAR induction experiments. BTH was introduced by hand infiltration in the control experiments documenting the lack of induction of hydrogen peroxide or SA production. SA was introduced by hand infiltration. BTH (0.12 mm; or SA, introduced as the sodium salt) in double-distilled water was used in all cases.
Statistics
Significance of differences between means in both SAR experiments and PR1-driven GUS activity determinations was determined using Student's t tests. Values of P were read from a t table. Differences between SA levels were assessed for significance using a two-way analysis of variance where either NDR1 allele (ndr1-1 or NDR1) or bacterial strain used was considered a fixed effect and experiment number was treated as a random factor.
ACKNOWLEDGMENTS
We thank Bernard Vernooij for his SA quantitation protocol; Kirk Czymmek for help with confocal microscopy and photography; John Ryals for the PR1 cDNA in Bluescript; Jeff Dangl for the Columbia parental seed; Sydney Kustu for use of her HPLC; Joseph Clarke and Kay Lawton for useful suggestions concerning the model for signal transduction; Peter P. Repetti for help with the BTH experiments; and Brian J. Staskawicz, Peter P. Repetti, and Cathy K. Worley for useful discussions and comments on drafts of this manuscript.
Footnotes
This project was initiated when A.D.S. was a National Institutes of Health postdoctoral fellow in the lab of Brian J. Staskawicz (University of California, Berkeley). This work was subsequently supported by the University of Delaware (start-up funds to A.D.S.) and by the College of Agriculture and Natural Resources, University of Delaware (predoctoral research assistantship to C.Z.). This is paper no. 1,694 in the Journal Series of the Delaware Agricultural Experiment Station.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010096.
LITERATURE CITED
- Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. Flow cytometric studies of oxidative product formation by neutrophils: a graded response to membrane stimulation. J Immunol. 1983;130:1910–1917. [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bestwick CS, Bennett MH, Mansfield JW. Hrp mutant of Pseudomonas syringae pv. phaseolicola induces cell wall alterations but not membrane damage leading to the hypersensitive reaction in lettuce. Plant Physiol. 1995;108:503–516. doi: 10.1104/pp.108.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y-M, Kenton P, Mur L, Darby R, Draper J. Hydrogen peroxide does not function downstream of salicylic acid in the induction of PR protein expression. Plant J. 1995;8:235–245. doi: 10.1046/j.1365-313x.1995.08020235.x. [DOI] [PubMed] [Google Scholar]
- Brading PA, Hammond-Kosack KE, Parr A, Jones JDG. Salicylic acid is not required for Cf-2- and Cf-9-dependent resistance of tomato to Cladosporium fulvum. Plant J. 2000;23:305–318. doi: 10.1046/j.1365-313x.2000.00778.x. [DOI] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ. Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J. 1994;5:715–725. [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ. NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci USA. 1995;92:6597–6601. doi: 10.1073/pnas.92.14.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1: a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278:1963–1965. doi: 10.1126/science.278.5345.1963. [DOI] [PubMed] [Google Scholar]
- Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN. The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant-Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci USA. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature. 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helinski DR. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke N, Ohashi Y. Involvement of an O2− generating system in the induction of necrotic lesions on tobacco leaves infected with tobacco mosaic virus. Physiol Mol Plant Pathol. 1988;32:163–175. [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA. 1998;95:10328–10333. doi: 10.1073/pnas.95.17.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyedi AJ, Yalpani N, Silverman P, Raskin I. Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc Natl Acad Sci USA. 1992;89:2480–2484. doi: 10.1073/pnas.89.6.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan S, Bauer DW, Alfano JR, Loniello AO, He SY, Collmer A. Expression of the Pseudomonas syringae avirulence protein avrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Fluhr R. UV-B-induced PR-1 accumulation is mediated by active oxygen species. Plant Cell. 1995;7:203–212. doi: 10.1105/tpc.7.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Zhao K, Whiteman M. Nitric oxide and peroxynitrite: the ugly, the uglier and the not so good. Free Radic Res. 1999;31:651–669. doi: 10.1080/10715769900301221. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- Jones JDG, Shlumukov L, Carland F, English J, Scofield SR, Bishop GJ, Harrison K. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- Klement Z. Hypersensitivity. In: Mount MS, Lacy GH, editors. Phytopathogenic Prokaryotes. New York: Academic Press; 1982. pp. 149–177. [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313x.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Leister RT, Ausubel FM, Katagiri F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Lotan T, Fluhr R. Function and regulated accumulation of plant pathogenesis-related proteins. Symbiosis. 1990;8:33–46. [Google Scholar]
- Malamy J, Hennig J, Klessig DF. Temperature-dependent induction of salicylic acid and Its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell. 1992;4:359–366. doi: 10.1105/tpc.4.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B, Slusarenko AJ. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of arabidopsis to Peronospora parasitica. Plant Cell. 1996;8:203–212. doi: 10.1105/tpc.8.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL. Signal transduction in the plant immune response. Trends Biochem Sci. 2000;25:79–82. doi: 10.1016/s0968-0004(99)01532-7. [DOI] [PubMed] [Google Scholar]
- Mehdy MC. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994;105:467–472. doi: 10.1104/pp.105.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwly P, Métraux J-P. ortho-Anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal Biochem. 1993;214:500–505. doi: 10.1006/abio.1993.1529. [DOI] [PubMed] [Google Scholar]
- Mölders W, Buchala A, Métraux J-P. Transport of salicylic acid in tobacco necrosis virus-infected cucumber plants. Plant Physiol. 1996;112:787–792. doi: 10.1104/pp.112.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB, Staskawicz BJ. Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr Opin Microbiol. 1998;1:109–115. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- Neuenschwander U, Vernooij B, Friedrich L, Uknes S, Kessmann H, Ryals J. Is hydrogen peroxide a second messenger of salicylic acid in systemic acquired resistance? Plant J. 1995;8:227–233. [Google Scholar]
- Parker JE, Feys BJ, Van Der Biezen EA, Noël L, Aarts N, Austin MJ, Botella MA, Frost LN, Daniels MJ, Jones JDG. Unravelling R gene-mediated disease resistance pathways in Arabidopsis. Mol Plant Pathol. 2000;1:17–24. doi: 10.1046/j.1364-3703.2000.00003.x. [DOI] [PubMed] [Google Scholar]
- Pirhonen MU, Lidell MC, Rowley DL, Lee SW, Jin SM, Liang YQ, Silverstone S, Keen NT, Hutcheson SW. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 1999;11:1695–1708. doi: 10.1105/tpc.11.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Uknes S, Ward E. Systemic acquired resistance. Plant Physiol. 1994;104:1109–1112. doi: 10.1104/pp.104.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell J, Koncz C. The promoter of T1-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Scofield SR, Tobias CM, Rathjen JP, Chang JH, Lavelle DT, Michelmore RW, Staskawicz BJ. Molecular basis of gene-for-gene specificity in Bacterial Speck Disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- Shah J, Tsui F, Klessig DF. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol Plant-Microbe Interact. 1997;10:69–78. doi: 10.1094/MPMI.1997.10.1.69. [DOI] [PubMed] [Google Scholar]
- Shapiro AD. Using Arabidopsis mutants to delineate disease resistance signaling pathways. Can J Plant Pathol. 2000;22:199–216. [Google Scholar]
- Sharma Y, Léon J, Raskin I, Davis KR. Ozone-induced responses in Arabidopsis thaliana: the role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc Natl Acad Sci USA. 1996;93:5099–5104. doi: 10.1073/pnas.93.10.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Nakajima H, Rajasekhar VK, Dixon RA, Lamb C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz BJ. Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 2001;125:73–76. doi: 10.1104/pp.125.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MW. The generation of oxygen radicals during host plant responses to infection. Physiol Mol Plant Pathol. 1991;39:79–93. [Google Scholar]
- Tang X, Frederick RD, Zhou J, Halterman DA, Jia Y, Martin GB. Initiation of plant disease resistance by physical interaction of avrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- Turner JG, Novacky A. The quantitative relation between plant and bacterial cells involved in the hypersensitive reaction. Phytopathology. 1974;64:885–890. [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon LC. Induced resistance in plants and the role of pathogenesis-related proteins. Eur J Plant Pathol. 1997;103:753–765. [Google Scholar]
- Volko SM, Boller T, Ausubel FM. Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics. 1998;149:537–548. doi: 10.1093/genetics/149.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner H-Y, Ryals J. Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell. 1995;7:2013–2022. doi: 10.1105/tpc.7.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen M, Innes R, Bent A, Staskawicz B. Identification of Pseudomonas syringae pathogens of Arabidopsis thaliana and a bacterial gene determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe J, Hutcheon CJ, Higgins VJ, Cameron RK. A functional gene-for-gene interaction is required for the production of an oxidative burst in response to infection with avirulent Pseudomonas syringae pv. tomato in Arabidopsis thaliana. Physiol Mol Plant Pathol. 2000;56:253–261. [Google Scholar]
- Yalpani N, Enyedi AJ, León J, Raskin I. Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta. 1994;193:372–376. [Google Scholar]
- Yalpani N, León J, Lawton MA, Raskin I. Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 1993;103:315–321. doi: 10.1104/pp.103.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]