While the genome contains the coded information that allows an organism to live and reproduce, the essential functions of living cells are accomplished by gene products. Those structures—mainly proteins, although ribonucleic acids are also essential—provide the scaffold, regulatory, and catalytic functions that drive metabolism. Proteomics seeks to measure the expression of all proteins within an organism and monitor changes in response to developmental and environmental cues in health and disease. Because the 30,000 or so human genes sustain life through a considerably larger variety of mature proteins, the technological challenge dramatically exceeds that of genomics. Ultimately, we would like a computer model that mimics life in silico, allowing accurate projections for metabolic engineering experiments in medicine and the life sciences. This grand experiment is just starting and the race is on to develop high-throughput technology to provide proteome-scale insights, as well as computational systems that allow realistic modeling of simple cells. Yates and coworkers (1) present the results of recent attempts to map the rice proteome and compare metabolism in three different functional states of the organism—that is, leaf, root, and seed. Although central metabolic pathways were present in all tissues, metabolic specialization was detected, confirming the existence of divergent regulatory mechanisms in starch biosynthesis and degradation in different tissues, as well as the presence of allergenic proteins in seed.
We would like a computer model that mimics life in silico, allowing accurate projections for metabolic engineering experiments.
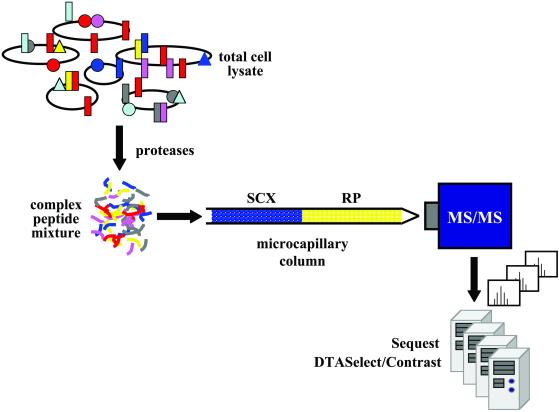
A central facet of proteomics is the matching of protein data to a corresponding gene providing a direct readout of expression for functional genomics, as opposed to inferences drawn from measurements of messenger RNA that can be misleading, as well as allowing for the measurement of posttranslational modifications. Rapid advances in mass spectrometry technology have driven proteomics to what is clearly the most dramatically expanding arena in the life sciences today. The recent 50th annual meeting of the American Society for Mass Spectrometry (www.asms.org) featured a number of new proteomics sessions to accommodate this interest. Accurate measurement of peptide masses and tandem mass spectrometry (MS–MS) experiments that produce peptide sequence data allow correlation with genomic data using software that translates genes and calculates peptide mass and/or fragment mass data. Measurement of intact protein masses is insufficient to allow assignment of all proteins (2), and thus enzymatic (trypsin) or chemical (CNBr) cleavage is used to break, in a sequence-dependent fashion, whole gene products into manageable pieces, some of which completely match a portion of a translated gene. Early proteomics studies relied on separation of proteins by two-dimensional (2D) gel electrophoresis, for example, followed by identification of individual protein spots after excision from the gel, cleavage reactions, extraction of peptides, and mass spectrometry with database searches (3–7). More recently, Yates and others (8, 9) have pioneered a “shotgun” approach whereby whole-cell protein extracts are immediately cleaved and the peptide mixture subjected to separation before mass spectrometry to generate peptide sequence data. Multidimensional chromatography is used to enhance fractionation of the complex peptide mixture from a whole-cell digest, giving rise to the MudPIT acronym (Multidimensional Protein Identification Technology; Fig. 1). The paper by Koller et al. (1) compares the 2D-gel approach to MudPIT, demonstrating the superior detection efficiency of the latter technique, while confirming the complementary nature of the methods.
Figure 1.
Multidimensional Protein Identification Technology (MudPIT). The complex mixture of proteins present in a whole cell lysate is fragmented first with lysine-specific endoproteinase lysC in the presence of 8 M urea and then with immobilized trypsin, after dilution to 2 M urea, generating a highly complex mixture. The peptides are collected on a strong cation exchange (SCX) column that is positioned immediately upstream of a reverse-phase (RP) column. Successive peptide fractions are released, depending on their isoelectric point, with salt steps of increasing concentration at low organic solvent concentrations and captured by the second-dimension reverse-phase column. The reverse-phase column is eluted with a gentle gradient of increasing organic solvent concentration between each salt step to displace the peptides, depending on their hydrophobicity, into the mass spectrometer. The ion-trap mass spectrometer (LCQ-DECA, ThermoFinnigan, San Jose, CA) employs data-dependent acquisition software to limit the time spent sequencing any particular peptide, so that as many different peptides as possible are sequenced, regardless of their abundance. sequest software correlates experimental sequence with genomic data (courtesy of Christine Wu, The Scripps Institute, La Jolla, CA).
The rice genome includes an as yet unknown number of genes currently reported to be in the range 32–56,000 (http://genome.sinica.edu.tw/irgsp.htm; http://rgp.dna.affrc.go.jp/index.html; refs. 10 and 11), although not all are expressed at the same time. The combined 2D-gel/MudPIT approach detected 2,528 unique proteins, many of which were tissue specific. Only 189 proteins were detected in all three tissues, highlighting the fact that the central metabolic pathways require relatively minor genetic commitment. Emphasizing this point, Tomita (12) has modeled a virtual self-surviving cell, based on a nonreplicating prokaryote, that has just 105 protein-coding genes and 22 RNA-coding genes. The increased complexity of eukaryotic central metabolism including glycolysis/gluconeogenesis, citric acid cycle, oxidative pentose phosphate pathway, amino acid biosynthesis, transcription, translation, and protein degradation still only accounts for a tiny fraction of the genome. A much greater number of genes are expressed in a tissue-specific fashion, and the proteomic analysis revealed 622 leaf-specific, 862 root-specific, and 512 seed-specific proteins. In some cases the origin of this specialization is obvious, such as the expression of photosynthesis genes in leaves, whereas other distributions, such as that observed for the large subunit of ADP-glucose pyrophosphorylase, are not at all intuitive. The assignment of function reported by Koller et al. (1) is based on blast (www.ncbi.nlm.nih.gov/BLAST/) homology to proteins from other species of “known function” according to current annotation. Of the proteins detected, 360 had no homology to any other protein in the NCBI nonredundant database and were thus assigned as rice-specific proteins. Because the only other higher plant whose complete genome sequence is available in the public domain is that of Arabidopsis thaliana, a dicotyledonous mustard that split from monocotyledonous rice around 200 million years ago, it seems likely that some of these proteins may be found in other cereals. However, the availability of the rice genome and now proteomic data establishes a base for comparative genomics and proteomics within the cereals and beyond (13). Soon, the forthcoming genome of the eukaryotic unicellular green alga Chlamydomonas reinhardtii (www.biology.duke.edu/chlamy_genome/) will help link green plant phylogeny with other eukaryotes and prokaryotes such as the cyanobacteria (www.kazusa.or.jp/cyanobase/) that have harbored evolving genes and in some cases shared them.
The importance of plant metabolic engineering stems from the diversity of secondary metabolism that results in biosynthesis of a myriad of compounds of known and unknown potential uses. A fine example is the recent engineering of rice plants that accumulate high levels of β-carotene in their seed. By engineering the overexpression in seed of phytoene synthase, lycopene β-cyclase, and phytoene desaturase, Ye et al. (14) engineered a crop plant producing “golden rice” that has the potential to address world vitamin A deficiency. However, other attempts to elevate accumulation of a desirable product have been thwarted by parallel acceleration of product degradation despite flux increase. Experimental hypotheses would be beneficially preinvestigated using virtual cell/organism modeling software (www.nrcam.uchc.edu; ref. 15) to more thoroughly test the metabolic consequences of proposed engineering and thereby avoid costly and time-consuming practical mutagenesis experiments with unforeseen results. Japan has initiated its own program to model rice (the Rice Simulator Project; ref. 16) and a number of U.S. programs embrace the concept, including the Department of Energy's Genomes to Life program (www.ornl.gov/hgmis/), which includes the Microbial Cell Project, the National Science Foundation's 2010 Arabidopsis initiative (www.arabidopsis.org/workshop1.html), and the National Institute of General Medical Sciences' Alliance for Cellular Signaling (www.cellularsignaling.org/). With substantial funds being directed toward such modeling efforts, it is certainly not too soon to be considering the technologies that will drive the testing of such models.
Technology that allows us to monitor global protein accumulation and turnover under physiological conditions for direct testing of virtual-cell technologies is needed and several criteria must be addressed if proteomics is to realize the expectations that have piqued our interest. Absolute and relative quantitation of cellular protein is of central importance, and a number of attractive approaches are under serious consideration (17). Coverage of “difficult” classes of proteins focuses on issues such as alternative splicing that can generate hundreds of protein sequence variants per gene (18), complex glycoproteins that can be decorated with hypervariable posttranslational modifications (19), and the integral membrane proteins that constitute one-third of the proteome but often elude us with their propensity to aggregate, precipitate, and generally fall from view when removed from their native environment (20). Measurement of protein turnover rate will allow us to incorporate protein flux into our cellular models and allow consideration of metabolic cost/benefit analysis in systems biology. Dynamic regulation of physiological adaptation and signal transduction pathways will test our ability to accurately monitor what can be chemically labile posttranslational modifications. A variety of proteomics technologies are being developed to supplement the shortfalls of 2D-gel approaches; aside from shotgun global peptide approaches, including MudPIT and the accurate mass tag approach described by Smith and coworkers (9), these include chromatographic 2D separation techniques for intact proteins and, in some cases, mass spectrometry of the intact protein as well. While the mass spectrum of an intact protein defines the native covalent form of a gene's product, as well as associated heterogeneity (20, 21), the “top-down” approach to proteomics described by McLafferty (22) faces a through-put challenge, just as occurred in genomic sequencing technologies.
MudPIT is shaping up to be a major contributor in the proteomics arena and the work of Koller et al. (1) demonstrates the ability to generate large amounts of useful data addressing at least some of the factors mentioned above. Especially worthy of mention is the coverage of integral membrane proteins; a hunt through the extensive list of detected proteins reveals many that have one or more predicted transmembrane helix domains, as well as some that are known to be among the most challenging, such as the chloroplast psbA gene product, for example. The proportion of total proteins detected has allowed genomic annotation of a detailed metabolic map of the rice cell that will be central to modeling efforts and ongoing studies to more thoroughly understand the diverse aspects of a plant's functional genomics.
Footnotes
See companion article on page 11969.
References
- 1.Koller A, Washburn M P, Lange B M, Andon N L, Deciu C, Haynes P A, Hays L, Schieltz D, Ulaszek R, Wei J, et al. Proc Natl Acad Sci USA. 2002;99:11969–11974. doi: 10.1073/pnas.172183199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez S M, Nishio J N, Faull K F, White legge J P. Mol Cell Proteomics. 2002;1:45–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Henzel W J, Billeci T M, Stults J T, Wong S C, Grimley C, Watanabe C. Proc Natl Acad Sci USA. 1993;190:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James P, Quadroni M, Carafoli E, Gonnet G. Biochem Biophys Res Commun. 1993;195:58–64. doi: 10.1006/bbrc.1993.2009. [DOI] [PubMed] [Google Scholar]
- 5.Mann M, Hojrup P, Roepstorff P. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 6.Pappin D J C, Hojrup P, Bleasby A J. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 7.Yates J R, Speicher S, Griffin P R, Hunkapiller T. Anal Biochem. 1993;214:397–408. doi: 10.1006/abio.1993.1514. [DOI] [PubMed] [Google Scholar]
- 8.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 9.Smith R D, Anderson G A, Lipton M S, Pasa-Tolic L, Shen Y, Conrads T P, Veenstra T D, Udseth H R. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Goff S A, Ricke D, Lan T H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Hu S, Wang J, Wong G K, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 12.Tomita M. Trends Biotechnol. 2001;19:205–210. doi: 10.1016/s0167-7799(01)01636-5. [DOI] [PubMed] [Google Scholar]
- 13.Bennetzen J. Science. 2002;296:60–63. doi: 10.1126/science.1071402. [DOI] [PubMed] [Google Scholar]
- 14.Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 15.Loew L M, Schaff J C. Trends Biotechnol. 2001;19:401–406. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 16.Harris S B. EMBO Rep. 2002;3:511–513. doi: 10.1093/embo-reports/kvf125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gygi S P, Rist B, Aebersold R. Curr Opin Biotechnol. 2000;11:396–401. doi: 10.1016/s0958-1669(00)00116-6. [DOI] [PubMed] [Google Scholar]
- 18.Black D L. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 19.Feizi T. Glycoconjugate J. 2000;17:553–565. doi: 10.1023/a:1011022509500. [DOI] [PubMed] [Google Scholar]
- 20.Whitelegge J P, Gundersen C, Faull K F. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitelegge J P, le Coutre J, Lee J C, Engel C K, Privé G G, Faull K F, Kaback H R. Proc Natl Acad Sci USA. 1999;96:10695–10698. doi: 10.1073/pnas.96.19.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelleher N L, Lin H Y, Valaskovic G A, Aaserud D J, Fridriksson E K, McLafferty F W. J Am Chem Soc. 1999;121:806–812. [Google Scholar]