Abstract
Spt-Ada-Gcn5 acetyltransferase (SAGA) is a previously described histone acetyltransferase/transcriptional coactivator complex in yeast. At promoters of certain genes (HIS3 and TRP3), SAGA has an inhibitory function involving a nonproductive TATA-binding protein interaction mediated by the Spt3 and Spt8 subunits. Related to this, Spt8-less SAGA is a major form of the complex under activating conditions for these genes. In the present study, we purify this activation-specific complex, called SALSA (SAGA altered, Spt8 absent). Besides lacking Spt8, SALSA contains Spt7 subunit that is truncated. Examining the role of this subunit, we find that C-terminally truncated SPT7 resulted in derepressed HIS3 transcription. Furthermore, when grown in rich media (repressing conditions), wild-type cells yielded predominantly SAGA, but Spt7 C-terminal truncations resulted primarily in a form of complex similar to SALSA. Thus, SALSA-like structure and activating function can be partially recapitulated in yeast by truncating the C terminus of Spt7. Overall, these results lead to a model that for a subset of promoters SAGA is inhibitory through Spt3, Spt8, and an Spt8-interacting subdomain of Spt7, whereas SALSA is a form of complex for positive transcriptional regulation. These data clarify a mechanism by which a transcriptional regulatory complex can switch between positive and negative modulation.
Transcription by RNA polymerase II in eukaryotes is highly regulated and involves various proteins in addition to the general transcriptional machinery. Coactivator complexes, which bridge the interaction between activator proteins and the transcription complex, have been shown to harbor various regulatory functions (1). Among these are activities, such as histone acetyltransferases (HATs) (2), that counteract repressive chromatin structure (3). In Saccharomyces cerevisiae, the HAT complexes NuA4 and SAGA (Spt-Ada-Gcn5 acetyltransferase) are coactivators that have been specifically implicated in acetylation and transcriptional regulation in vivo and in vitro (4–8).
The SAGA complex, which is important for normal growth of yeast and transcription of various genes, contains several groups of previously identified transcription-related proteins (4, 9). Known subunits include the HAT Gcn5 (10), Ada adaptor proteins (Ada1 through 5; Ada4 is Gcn5) (11), the TATA-binding protein (TBP)-related subgroup of Spt proteins (Spt3, Spt7, Spt8, and Spt20, also known as Ada5) (12), a subset of TBP-associated factors (TafII17, 25, 60, 68, and 90) (13), and the essential protein Tra1 (14, 15), the yeast homolog of human coactivator TRRAP. Consistent with its diverse composition, SAGA is known to have a modular structure and contain several discrete regulatory functions (9, 16): defined subgroups within SAGA include Gcn5/Ada2/Ada3 (HAT/adaptor), Spt3/Spt8 (TBP-related function), and Ada1/Spt7/Spt20 (apparently required for SAGA structural integrity).
The multiple functions within SAGA and their roles in transcription have been examined experimentally in various studies. The HAT activity of Gcn5 has been extensively studied in vitro and in vivo, and in the context of SAGA it is known to have a positive effect on transcription of certain genes (reviewed in ref. 2). The activator-binding ability of SAGA [such as through Tra1 (17)] is another positive function shown to be critical for activation in transcription assays (5, 18), and recently SAGA was also demonstrated to have the positive effect of facilitating transcription complex assembly at GAL1 (7, 8). On the other hand, components of SAGA have also been shown to mediate an apparent negative effect on transcription at certain Gcn4-regulated genes such as HIS3 and TRP3 (19). Specifically, unscheduled derepression of these genes is promoted in vivo by mutation of SPT3, SPT8, or the TBP-encoding gene (spt15–21, a point mutant suppressable by point mutant spt3–401), and SAGA complexes lacking either Spt3 or Spt8 inhibit TBP binding to TATA box DNA in vitro (19), suggesting that a TBP-related repression mechanism is at work in SAGA at these promoters.
The coexistence of positive and negative regulatory features or the interconversion between these functions has also been observed in other complexes involved in transcriptional regulation. For example, TFIID generally supports transcription, and overall, its Taf subunits act as positive cofactors (20). However, TAF250 homologs (such as Drosophila TAF230 and yeast TafII145) have been shown to have a negative, autoinhibitory effect on TFIID by preventing TBP-TATA interaction (21, 22), but this may be counteracted by activators or TFIIA, possibly acting through TAF135 (23). Another example is that human coactivator PC4 can be converted from a repressive to an active form in a stepwise process that may involve phosphorylation by TFIIH and/or TAF250 (24). In addition, yeast or Drosophila transcription factor NC2 (Dr1-Drap1) can also either activate or repress transcription, in this case depending on the nature of the promoter it occupies (25, 26).
The structure and composition of certain regulatory complexes can also vary, influencing regulatory states. For example, human chromatin-remodeling complexes containing Brg1 can be purified in at least two different forms, largely similar but containing or lacking the regulatory protein mSin3A and several other subunits (27). In addition, the STAGA complex, a human homolog of SAGA, seems to exist in multiple forms with different functional moieties (28).
Yeast SAGA is also a modular complex with variable subunit composition, and these properties may be directly related to its positive and negative transcriptional effects, mentioned above. The negative regulation of HIS3 and TRP3 apparently occurs through nonproductive interaction with TBP, mediated by the Spt3 and Spt8 subunits. Moreover, there are at least two similar, but structurally and chromatographically distinct, forms of the SAGA complex in yeast cells: classically defined SAGA and an altered form of SAGA lacking Spt8 (19). The prevalence of these forms shows an interesting correlation with SAGA's inhibitory function in that SAGA is predominant in rich media (repressive for HIS3 and TRP3), whereas the altered form of SAGA is predominant under activating conditions for these genes. Because a major characteristic of this latter complex is lack of the Spt8 subunit, we therefore name it SALSA, for SAGA altered, Spt8 absent. In the present study, we purify the SALSA complex to near homogeneity and further characterize its structural and apparent functional differences with SAGA. Specifically, the Spt7 C terminus has a role in Spt8 interaction with SAGA and may be an important distinguishing feature between SAGA and activation-specific SALSA.
Materials and Methods
Yeast Strains and Media.
For large-scale purification of SAGA and SALSA, Ada2–2FLAG epitope-tagged strain SB345 (29) was used. Wild-type strain FY631 and spt7 mutants FY571 (spt7–217) and FY569 (spt7–223) were provided by the Winston lab (30) and used for S1 analysis and conventional purification (4) of complexes. SPT7-deleted strain FY963 was used for pY-SPT7 integrations and subsequent S1 analysis. For SPT7 integrations followed by anti-FLAG immunoprecipitations, haploid strains SB345 and SB303 (31) were crossed to create a diploid, which underwent spt7Δ∷LEU2 knockout (30) and was sporulated and tetrad-dissected, followed by identification of an spt7Δ Ada2–2FLAG-containing haploid. For anti-Myc immunoprecipitations, 13 tandem Myc tags were added to the C-terminal ends of the SPT3, SPT20, or SPT8 genes in the strain FY1531 (from F. Winston, Harvard Medical School, Cambridge, MA) by the method of Longtine et al. (32); URA3-plasmid-borne SPT7 was removed by growth on 5-fluoro-orotic acid media (33), and SPT7 wild-type or Δ213C were integrated as described below. Rich (yeast extract/peptone/dextrose) and synthetic complete media (34) were used for nonselective and selective growth, respectively, at 30°C.
Plasmids, Sequencing, and S1 Analysis of HIS3 Transcription.
To prepare SPT7 constructs for plasmid integration of spt7Δ strains, 3.1-kb and 5.2-kb ClaI/MluI fragments of pYES2 (Invitrogen) and pFW128 (30) were ligated to produce pY-SPT7; a modification (35) of the QuikChange mutagenesis method (Stratagene) was used to prepare C-terminal truncation constructs of this plasmid. These were digested with ApaI and transformed into FY963 for integration at the URA3 locus; for empty-vector integration (spt7Δ), NsiI-digested pRS306 (36) was used.
Sequencing of the spt7–217 and spt7–223 mutations was accomplished by preparing genomic DNA from the FY571 and FY569 strains and PCR cloning (TOPO-TA cloning kit, Invitrogen) SPT7 C-terminal regions into pCR2.1. Analysis of in vivo HIS3 transcription was carried out as described (19), by hybridization of 40 μg of total RNA with specific probes.
SAGA and SALSA Purification from Ada2–2FLAG Yeast Strain.
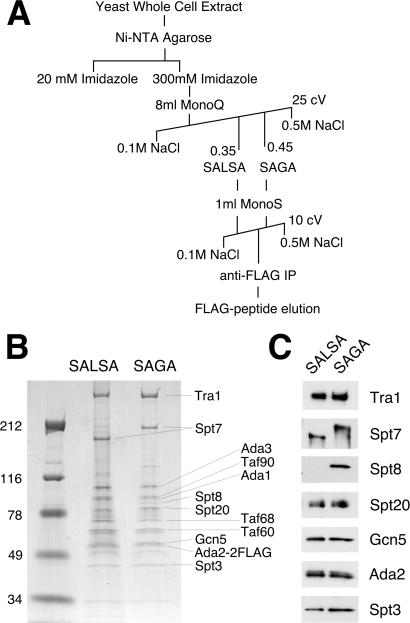
The complexes were purified as illustrated in Fig. 1A, a scheme adapted from previous methods (4, 19). Yeast strain SB345 was grown in 60 liters of synthetic complete medium at 30°C to an optical density (600 nm) of 1.5. Sixty milliliters of Ni2+-NTA agarose (Qiagen, Chatsworth, CA) was used for initial binding of whole-cell extract, and eluate was run on an 8-ml Mono Q HR 10/10 column (Amersham Pharmacia). Peak SALSA and SAGA fractions (monitored by Western blots with antibodies to known shared subunits) were pooled separately, dialyzed against the buffer containing 0.1 M NaCl and 40 mM Hepes, pH 7.5, and loaded onto a Mono S HR 5/5 column, followed by a 10 column volume, 0.1–0.5 M NaCl linear gradient. Peak fractions were adjusted to 0.25 M NaCl and incubated with anti-FLAG M2-Agarose (Sigma) overnight. The resin was extensively washed on a column, and bound proteins were eluted with FLAG peptide (Sigma) at 0.2 mg/ml. Samples of SALSA and SAGA were then run on an SDS/PAGE gradient gel (4–20%; Invitrogen) and stained with colloidal blue (Coomassie G-250; Invitrogen).
Figure 1.
Purification and subunit composition of the SALSA and SAGA complexes. (A) Purification scheme for the isolation of SALSA and SAGA from Ada2–2FLAG cells. IP, immunoprecipitation; cV, column volume. (B) Coomassie-stained gradient gel (SDS/PAGE) run with similar amounts of purified SALSA and SAGA. Molecular mass markers (kDa) are shown at left. Indicated at right are subunits identified by size and/or Western blotting. (C) Western blots of purified SALSA and SAGA with antibodies against various subunits in the complexes.
Immunoprecipitation of Complexes and Western Blotting.
Extracts of yeast extract/peptone/dextrose-grown pY-SPT7 integrants of the spt7Δ Ada2–2FLAG-tagged strain were immunoprecipitated with anti-FLAG antibodies and eluted with FLAG peptide as described (29). Immunoprecipitations of Myc-tagged strains were performed similarly, but with mouse anti-c-Myc mAb (9E10; Santa Cruz Biotechnology). A total of 5–20 μl of the elutions was used for Western blots with SAGA subunit antibodies as described (19). Additional antibodies used in this study were anti-Ada1 (1:1,000 dilution) from L. Guarente (Massachusetts Institute of Technology, Cambridge, MA), anti-Spt7 (1:250) from F. Winston, and anti-Tra1 (1:1,000) from J. Workman (Pennsylvania State University, State College, PA). It should be noted that Saleh et al. (15) previously performed an spt7Δ-extract immunoprecipitation experiment analogous to ours and observed Tra1 in the immunoprecipitate; however, the strain, epitope tags [hemagglutinin (HA)-Ada2 and Myc-Tra1], and immunoprecipitation conditions (anti-HA antibody, and 150 mM salt instead of 350 mM) differed significantly from ours.
Results
Purification and Subunit Characterization of SALSA.
Previously, we identified an altered form of the SAGA complex predominant under activating conditions for certain genes [amino acid biosynthesis derepression by 3-aminotriazole (3-AT)], and its primary observed difference with SAGA was lack of the Spt8 subunit and shifted elution from a Mono Q ion exchange column (19). In the present study, we seek to characterize this altered complex (SALSA) further. To do this, we prepared an extract from epitope-tagged Ada2–2FLAG cells grown in synthetic complete media [shown previously to result in approximately similar levels of SAGA and SALSA (19)] and purified the two complexes to near homogeneity by the purification scheme shown in Fig. 1A. These purifications resulted in bands mostly identifiable by size (Fig. 1B) and/or Western blot analysis (Fig. 1C) as known SAGA subunits. A subsequent Superose 6 gel filtration column of these preparations demonstrated that SALSA eluted in similar fractions to SAGA, and therefore the complexes are not discernibly different in size (1.8 MDa) within the resolution of the column (data not shown).
Besides the presence/absence of Spt8, the most apparent difference between SAGA and SALSA is the gel mobility of the Spt7 subunit: it has a significantly smaller apparent size in SALSA compared with SAGA. Multiple species of Spt7 had been observed previously (15), although it was not determined whether the smaller forms of Spt7 were associated with specific Gcn5-containing complexes. It should be noted that aside from Spt7, SALSA and SAGA also show differences in a few faint bands in Fig. 1B (particularly near the mobility of the 116-kDa marker), but at this point it is unclear whether these are nonspecific, substoichiometric contaminants, altered species of other subunits, or unknown subunits unique to the particular forms of complex.
Transcriptional Effects of SPT7 C-Terminal Truncation Mutants.
Because of initial evidence of the existence of C-terminally processed Spt7 in extracts (P.-Y. J. Wu and F. Winston, personal communication), we investigated the possibility that the faster gel mobility of Spt7 in SALSA represented a truncation of the protein. To explore this phenomenon further and determine whether it was related to our previous observations of Spt3/Spt8-mediated transcriptional repression, we characterized two existing C-terminal truncation mutants of SPT7—spt7–217 and spt7–223. Previous results had shown that both mutants gave Spt− phenotypes and that the mutant proteins were stable in extracts (30). Furthermore, spt7–223 was found to have a larger truncation and more severe phenotype than spt7–217.
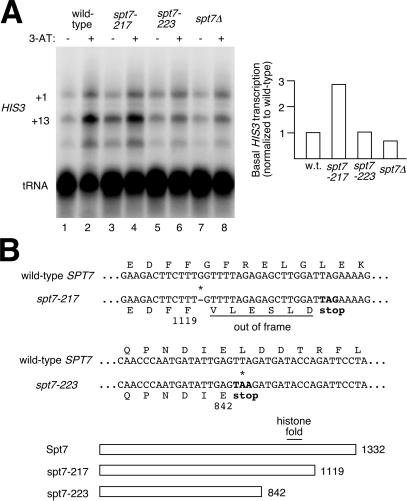
To investigate possible transcriptional effects of these truncations and potential relationship with SALSA, gene expression was tested at the SAGA-dependent HIS3 gene by isolating RNA from cells under noninducing and inducing conditions (− and + 3-AT) and performing S1 nuclease assays (Fig. 2A). As observed in ref. 19, wild-type cells show a significant amount of HIS3 transcription (focusing on the +13 start-site band) under inducing conditions (Fig. 2A, lane 2), but display a low basal level of transcription under noninducing conditions (Fig. 2A, lane 1). In the case of spt7Δ, transcription is low under both conditions (19) (Fig. 2A, lanes 7 and 8). The more dramatic truncation of Spt7, spt7–223, displays a transcription profile (Fig. 2A, lanes 5 and 6) very similar to that of spt7Δ, suggesting that this mutant has lost Spt7 function. However, a different profile is observed in the spt7–217 samples: basal transcription is elevated ≈3-fold (Fig. 2A, lane 3) compared with wild type (Fig. 2A, lane 1). This high-basal effect is a hallmark of spt3Δ, spt8Δ, and spt15–21 mutants, which disrupt the TBP-related negative regulation of SAGA, and the relative values are comparable to those of an spt8Δ strain (about 4-fold over wild-type basal) (19). The spt7–217 results therefore indicate that the C terminus of Spt7 may also be involved in the same process.
Figure 2.
HIS3 transcriptional analysis and sequencing of spt7 mutants. (A) (Left) S1 nuclease assays of RNA isolated from wild-type and spt7 mutant strains under repressing and derepressing conditions (− and + 3-AT). tRNA was probed as a control for normalization. (Right) The quantitation of basal transcription (minus 3-AT) from HIS3's +13 start site, relative to wild type. (B) (Upper) Sequencing of the spt7–217 and spt7–223 mutant genes. Partial DNA sequences and predicted protein sequences (amino acids as one-letter codes) indicate the positions of the mutations (*) and introduction of premature stop codons. (Lower) A schematic diagram of the truncated proteins and the number of residues they contain is shown.
Because the spt7–217 and spt7–223 alleles were not previously sequenced, we determined where these mutations occurred in SPT7, revealing the extent of the truncations (Fig. 2B). spt7–217 had a single-base deletion at codon 1120, leading to six out-of-frame residues followed by a stop codon. The net result is that the last 213 residues of Spt7 are no longer coded for in the altered ORF. In spt7–223, a single-base substitution occurred, changing codon 843 to a stop codon and removing the last 490 residues of Spt7. Notably, this removed the potential histone fold motif of Spt7, defined as between residues 975 and 1051 (37).
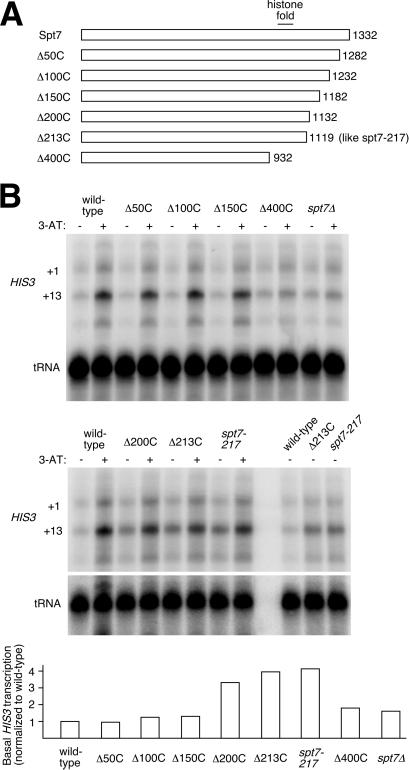
To map the critical regions of the Spt7 C terminus more precisely, we prepared a series of truncations of Spt7, deleting the C terminus at 50-residue intervals (Fig. 3A), and used them for S1 analysis. Truncations of 50, 100, or 150 residues led to no discernible effect on HIS3 transcription (Fig. 3B Upper). A 400-residue deletion mutant displayed a profile similar to that of spt7Δ, like spt7–223; although this truncation was slightly less extensive than spt7–223, it did also remove Spt7's histone-fold motif. Unlike the smaller deletions, truncations of 200 or 213 residues did give high-basal effects (Fig. 3B Lower). Δ213C was designed to recapitulate the spt7–217 mutation (it removes the same residues, albeit by deletion instead of frameshift), and this mutant does in fact display the same high-basal profile as spt7–217, indicating that this effect is specific to Spt7 truncation and not to anything else in the spt7–217 strain background (for example, the spt7–217 strain, FY571, contains four additional auxotrophies compared with the FY963 integrants). In the case of Δ200C, the basal transcription level was nearly as high as that of Δ213C, suggesting that the region responsible for the high-basal effect has its C-terminal border between 150 and 200, and the region may continue beyond 200.
Figure 3.
HIS3 transcriptional analysis of Spt7 C-terminal truncation mutant series. (A) Diagram of truncation mutations prepared in pY-SPT7 plasmid and integrated into spt7Δ strain FY963. (B) (Upper) S1 nuclease assays of RNA isolated from wild-type and spt7 mutant strains under repressing and derepressing conditions (− and + 3-AT). Lanes at right in the lower panel show direct comparison of +13 basal transcription (minus 3-AT) for repeated samples of wild type, Δ213C, and spt7–217. (Lower) Quantitation of basal transcription relative to wild type. tRNA was probed as a control for normalization.
Analysis of SAGA/SALSA Complexes from spt7 Mutants.
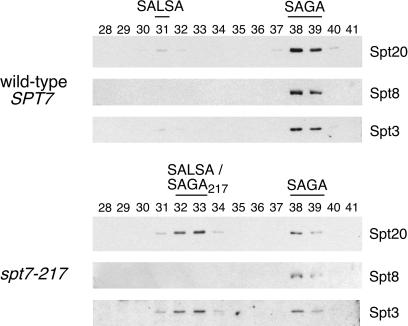
The above results suggest a link between Spt7 C-terminal truncation and the SALSA form of SAGA observed under activated conditions. To address this directly, we performed chromatographic preparations of HAT complexes (4) from yeast extract/peptone/dextrose-grown (noninducing conditions) wild-type and spt7–217 strains and analyzed the resulting Mono Q fractions for SAGA and SALSA (Fig. 4). The wild-type fractions displayed a typical profile for this strain: abundant SAGA in fractions 38–39 and a very small amount of SALSA about seven fractions earlier. spt7–217, however, led to a dramatically shifted profile with the predominant form of Spt-Ada complex now eluting at or near the SALSA position. SAGA was still present (Fig. 4, lanes 38 and 39), albeit at a significantly reduced level.
Figure 4.
Subunit analysis of conventionally purified complexes from wild-type and spt7–217 cells grown in rich media. Western blots of three Spt subunits show the relative amounts of the Spt/Ada complexes and the Mono Q fractions in which they elute. In the case of the earlier-eluting species from spt7–217, it is unclear whether it is SALSA itself, or a mutant-specific, Spt8-less derivative of SAGA, or a mixture of both.
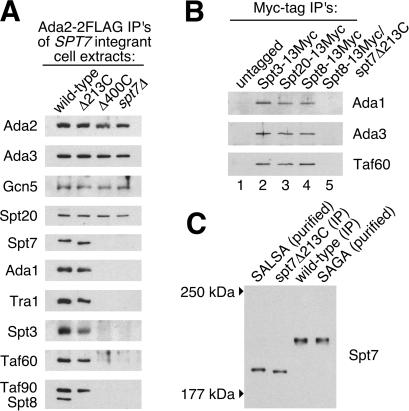
The original chromatographic study of SAGA (4) suggested that SPT7 deletion causes disruption of the complex, but it did not explore the issue of possible partial complexes in spt7 deletion or truncation mutant extracts. To investigate this, we immunoprecipitated Ada2–2FLAG-containing complexes (which should include at least SAGA, SALSA, and the non-Spt-containing complex ADA) (4, 38) from extracts of cells with wild-type, Δ213C, or Δ400C Spt7 or with SPT7 deleted, and characterized them by Western analysis (Fig. 5A). In the case of Spt7Δ213C, all tested SAGA subunits were present with the exception of Spt8, again supporting the structural and functional relationship between the Spt7 C terminus and the participation of Spt8 in the complexes. This is further supported by an alternative set of immunoprecipitations (Fig. 5B), where SAGA/SALSA could be pulled down by Spt8 in the presence of wild type (Fig. 5B, lane 4) but not Δ213C Spt7 (Fig. 5B, lane 5), demonstrating that Spt8 is disconnected from the complex in the truncation mutant, even in rich media. However, because the conventional purification from spt7–217 did show some Spt8-containing SAGA (Fig. 4), Spt8 may have a weak interaction with SAGA in that mutant.
Figure 5.
Subunit analysis of immunoprecipitated complexes from rich-media extracts of spt7 mutants. (A) Western blots of immunoprecipitated Ada2–2FLAG complexes (should include ADA, SAGA, and SALSA) showing various Ada, Spt, and Taf subunits and Tra1 recovered from the indicated strains. (B) Western blots of immunoprecipitated complexes from extracts of strains with Myc-tagged SAGA components as indicated. As shown, one strain had C-terminally truncated Spt7Δ213C in combination with Spt8–13Myc, whereas the others had wild-type Spt7. (C) Size comparison of truncated Spt7Δ213C with naturally processed Spt7 in SALSA. Purified SALSA and SAGA (side lanes) from Fig. 1 were run alongside Ada2–2FLAG-immunoprecipitated Δ213C and wild-type samples (center lanes) on a 6% SDS/PAGE gel, Western blotted, and visualized with anti-Spt7 antibodies. The migrations of molecular size markers are indicated at left.
To verify that the spt7Δ213C mutant does in fact approximate the state of Spt7 in SALSA, we performed a direct size comparison of mutant-derived Spt7 with Spt7 from purified wild-type SALSA. As shown in Fig. 5C, the truncation mutant does lead to an Spt7 species very similar in apparent size to naturally processed Spt7 in SALSA, but the migration of the mutant protein is slightly faster, indicating that the normal processing site is slightly downstream of the mutant's truncation point. These results are therefore consistent with the mapping of the transcriptional effect, where a 150-residue truncation had no effect, whereas a 200-residue truncation gave a result similar to that of Δ213C (Fig. 3B).
In the case of loss of Spt7 or its histone fold (Δ400C), SAGA/SALSA was disrupted, resulting in an adaptor complex containing just Ada2, Ada3, Gcn5, and Spt20/Ada5 (Fig. 5A). Thus, as suggested previously, Spt7 is critical for integrity of the complexes. It may be significant that loss of the Spt7 histone fold correlates with complex disruption, because several other histone fold proteins are contained in SAGA–Ada1, Spt3, and Tafs 17, 25, 60, and 68. One possibility is that loss of a potential histone octamer-like structure (37) could contribute to disruption of the complex. The data in Fig. 5A also point to a potentially integral relationship between Spt20/Ada5 and the adaptor/HAT module, as demonstrated by its apparent stable association with these subunits even in the absence of the rest of the complex. Spt7, on the other hand, seems to have a closer functional relationship through its C terminus to Spt3/Spt8, and a structural relationship with the TafIIs, Ada1, and Tra1 through Spt7's histone fold.
Discussion
In yeast, many highly inducible promoters are primed to be rapidly activated. Although the level of DNA-bound activators generally rises after induction, there is activator present at many promoters even in the repressed state. In addition, many coactivators are present constitutively in the nucleus. The paradox of needing rapid induction but at the same time preventing association with coactivators at repressed promoters has been solved by negative regulation inherent at many promoters. One mechanism is direct blockage of an activation domain, such as Gal80 for the activator Gal4 (39). A related mechanism is illustrated by Pho80 binding to the Pho4 activator, at a region distinct from the activation domain and inhibitory to activation (40). A different mechanism is to build negative regulation into coactivator complexes to prevent their activity, such as the TFIID complex's reversible inhibition of TBP-TATA interaction, described above.
We have identified a second example of this latter strategy, within the yeast SAGA complex. Our data indicate that Spt7 and Spt8 within SAGA participate in negative regulation at the HIS3 promoter. Under derepressing conditions, Spt7 within SAGA is processed (perhaps by proteolysis), and Spt8 is not associated, resulting in SALSA, an apparent activated form of the complex. That SALSA's Spt7 is C-terminally truncated is supported by a very recent study that examined Spt7 with epitope tags at both ends and demonstrated that the form that copurifies with SALSA lacks the C-terminal tag, but not the N-terminal tag (41). So far, it is unclear whether processing of the Spt7 C terminus is a cause or effect of Spt8 loss from the SAGA complex. However, it is clear that a SALSA-like complex and a partial activated state of transcription in cells can be recapitulated in repressing conditions by truncating the Spt7 C terminus. At the molecular level, one obvious possibility is that the C-terminal end of Spt7 may act as part of a docking site for Spt8 to maintain negative regulation of SAGA during gene repression, whereas the rest of the protein, including the histone fold motif, has a more integral function in holding modules of SAGA/SALSA together.
A recent study from Wu and Winston (41) characterized Spt7 C-terminal interaction with Spt8 and mapped Spt7's processing site (via an unprocessable, internal-deletion mutant), and these results agree well with ours. Their unprocessable Spt7 construct resulted in reduced amounts of SALSA and little effect on HIS3/TRP3 activation, interpreted as evidence of a subtle transcriptional role for SALSA. However, their spt7Δ strain also had only minor HIS3/TRP3 transcriptional effects compared with our observations. Thus, further investigation will be required to reconcile the different results and conclusions of these studies.
Recently, a human homolog of Spt7 has been described (28). The gene product is a component of STAGA, a human counterpart of yeast SAGA/SALSA, but encompasses just the C-terminal third of yeast Spt7, including the histone fold and putative Spt8-interacting region, suggesting that the C-terminal functional regions described above may be conserved throughout the eukaryotes. Although no human homolog of Spt8 (or Spt20) has yet been identified, STAGA's subunit composition and function remain to be fully characterized.
Overall, four yeast proteins have been implicated in SAGA-mediated negative regulation of HIS3—Spt3, Spt7, Spt8, and TBP (Spt15)—and mutation of their genes can lead to high-basal effects on transcription (ref. 19 and present study). However, it should be noted that transcription is only partially derepressed by engineered truncation of Spt7 or loss of Spt8, whereas disruption of Spt3, also centrally involved in the inhibitory mechanism, derepresses more fully (19). Therefore, we hypothesize that Spt3 is likely also to be altered in SALSA (perhaps by covalent modification) to contribute to activating function, although this is not yet apparent in the subunit analysis.
It is important to note that the relationship between SALSA and activated transcription has been studied here primarily just in the context of HIS3 and repression/derepression of amino acid biosynthetic genes. So far, only HIS3, and by extension TRP3, have been identified as having Spt7/Spt8-mediated repression, but potentially, any genes up-regulated by loss of Spt8 or C-terminal truncation of Spt7 may be candidates for this scheme of negative regulation. For example, these may include Swi5-dependent genes such as HO and PCL2, shown by S1 analysis to have heightened expression in spt3Δ or spt8Δ mutants (ref. 42 and D. Stillman, personal communication).
In actuality, the expression of many inducible yeast genes depends on Spt/Ada complexes, but different subsets of promoters seem to be regulated in different ways by them, so the question remains as to how these modes are reconciled in the cell. In cases such as GAL1–10 or PHO5, the complex may act simply as a coactivator, linking an activator with the transcriptional machinery. At GAL1–10, the HAT activity of Gcn5 is not required, and the Spt3 moiety (inhibitory in the case of HIS3/TRP3) interacts with TBP to positively, instead of negatively, affect transcription (43). Thus, SAGA may be the functional form of the complex at some promoters (7, 8), but at others (such as HIS3) the altered form SALSA is used for activation. The basis for this selectivity is unknown. One possible determinant (like for NC2) is the nature of the promoter: the sequence, position, or environment of the TATA box might dictate a productive or nonproductive interaction between TBP and SAGA. Another possibility is that there is yet another mode or level of regulation that has yet to be described, such as an unknown Spt3 modification, as mentioned above.
Alternatively, SALSA may be the active form at many Spt/Ada-complex-regulated promoters, but perhaps certain promoters are not subject to SAGA-mediated repression because they are negatively regulated by other, epistatic means (e.g., the inhibitor Gal80 at GAL1–10). It may be that many or most SAGA/SALSA-dependent genes (based on known examples such as HIS3, TRP3, GAL1, and PHO5) are inducible, and their standard state, absent of extreme conditions in the cell, is repression (hence the predominance of SAGA over SALSA in rich media). One way this model may be tested is to examine levels of SALSA versus SAGA from cells under inducing conditions other than 3-AT for various known genes (for example, low phosphate media for PHO5 or galactose media for GAL1–10), to see whether the SAGA/SALSA equilibrium changes in response to which form is needed at these promoters. Although much remains to be learned about transcriptional regulation by coactivator complexes such as SALSA and SAGA, it is clear that the flexibility offered by the multiple levels of their control is an important aspect of gene regulation.
Acknowledgments
We thank F. Winston and P.-Y. J. Wu for making this study possible by providing Spt7-related strains, plasmids, and antibody, and for sharing unpublished data and reviewing the manuscript. We also thank D. Stillman for sharing unpublished data, P. Melloy for technical help, J. Workman and lab for contributions to purification, J. Workman and L. Guarente for antibodies, and G. Moore and K. Henry for reviewing the manuscript. S.L.B. was supported by grants from the National Science Foundation and the National Institutes of Health, and D.E.S. was supported by a postdoctoral fellowship from the American Cancer Society.
Abbreviations
- SAGA
Spt-Ada-Gcn5 acetyltransferase
- SALSA
SAGA altered, Spt8 absent
- HAT
histone acetyltransferase
- TBP
TATA-binding protein
- 3-AT
3-aminotriazole
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hampsey M. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sterner D E, Berger S L. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolffe A P, Hayes J J. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, et al. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 5.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 6.Galarneau L, Nourani A, Boudreault A A, Zhang Y, Héliot L, Allard S, Savard J, Lane W S, Stillman D J, Côté J. Mol Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik S R, Green M R. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larschan E, Winston F. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts S M, Winston F. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 11.Berger S L, Piña B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 12.Winston F. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 1271–1293. [Google Scholar]
- 13.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, III, Workman J L. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 14.Grant P A, Schieltz D, Pray-Grant M G, Yates J R, III, Workman J L. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 15.Saleh A, Schieltz D, Ting N, McMahon S B, Litchfield D W, Yates J R, III, Lees-Miller S P, Cole M D, Brandl C J. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- 16.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown C E, Howe L, Sousa K, Alley S C, Carrozza M J, Tan S, Workman J L. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda K, Steger D J, Eberharter A, Workman J L. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belotserkovskaya R, Sterner D E, Deng M, Sayre M H, Lieberman P M, Berger S L. Mol Cell Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 21.Kokubo T, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Proc Natl Acad Sci USA. 1994;91:3520–3524. doi: 10.1073/pnas.91.9.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guermah M, Tao Y, Roeder R G. Mol Cell Biol. 2001;21:6882–6894. doi: 10.1128/MCB.21.20.6882-6894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik S, Guermah M, Roeder R G. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willy P J, Kobayashi R, Kadonaga J T. Science. 2000;290:982–984. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 26.Geisberg J V, Holstege F C, Young R A, Struhl K. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sif S, Saurin A J, Imbalzano A N, Kingston R E. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez E, Palhan V B, Tjernberg A, Lymar E S, Gamper A M, Kundu T K, Chait B T, Roeder R G. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sterner D E, Wang X, Bloom M H, Berger S L. J Biol Chem. 2002;277:8178–8186. doi: 10.1074/jbc.M108601200. [DOI] [PubMed] [Google Scholar]
- 30.Gansheroff L J, Dollard C, Tan P, Winston F. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trievel R C, Rojas J R, Sterner D E, Venkataramani R, Wang L, Zhou J, Allis C D, Berger S L, Marmorstein R. Proc Natl Acad Sci USA. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine M S, McKenzie A, III, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 34.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 35.Wang W, Malcolm B A. BioTechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- 36.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gangloff Y G, Sanders S L, Romier C, Kirschner D, Weil P A, Tora L, Davidson I. Mol Cell Biol. 2001;21:1841–1853. doi: 10.1128/MCB.21.5.1841-1853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saleh A, Lang V, Cook R, Brandl C J. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 39.Carrozza M J, John S, Sil A K, Hopper J E, Workman J L. J Biol Chem. 2002;277:24648–24652. doi: 10.1074/jbc.M201965200. [DOI] [PubMed] [Google Scholar]
- 40.Jayaraman P S, Hirst K, Goding C R. EMBO J. 1994;13:2192–2199. doi: 10.1002/j.1460-2075.1994.tb06496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu P-Y J, Winston F. Mol Cell Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belotserkovskaya R. Doctoral thesis. Philadelphia: University of Pennsylvania; 2000. [Google Scholar]
- 43.Dudley A M, Rougeulle C, Winston F. Genes Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]