Abstract
The hematopoietic transcription factor GATA-1 regulates erythropoiesis and β-globin expression. Although consensus GATA-1 binding sites exist throughout the murine β-globin locus, we found that GATA-1 discriminates among these sites in vivo. Conditional expression of GATA-1 in GATA-1-null cells recapitulated the occupancy pattern. GATA-1 induced RNA polymerase II (pol II) recruitment to subregions of the locus control region and to the β-globin promoters. The hematopoietic factor NF-E2 cooperated with GATA-1 to recruit pol II to the promoters. We propose that only when GATA-1 attracts pol II to the locus control region can pol II access the promoter in a NF-E2-dependent manner.
Eukaryotic DNA wraps ≈1.7 times around a core histone octamer to form nucleosomes, which in turn, fold into highly condensed chromatin. The role of chromatin structure in regulating gene expression is established through extensive analysis in diverse systems (1–8). An important consequence of chromatin folding is the regulation of cis-element accessibility, thereby preventing the constitutive loading of trans-acting factors and RNA polymerase II (pol II). Coactivators and corepressors are recruited to chromosomal sites via interactions with transcription factors and catalyze chromatin remodeling (9, 10). Although certain factors can recognize binding sites on nucleosomal DNA, others are occluded (11, 12). Factors capable of binding nucleosomal sites would not likely be able to access sites in condensed chromatin. Thus, site occupancy in vivo cannot be predicted by sequence analysis, but rather requires analysis of binding in living cells.
We use the murine β-globin locus to investigate how transcriptional control occurs within chromatin domains (13, 14). The β-globin locus consists of several genes arrayed in the order of their developmental expression (15). High-level transcription of these genes requires an upstream locus control region (LCR) (16–19), comprised of four DNaseI hypersensitive sites, HS1–HS4 (20, 21). Besides an erythroid-specific enhancer function, the LCR counteracts transgene silencing (16, 22).
A common feature of active chromatin is core histone acetylation (23). We hypothesized that LCRs function by recruiting histone acetyltransferases that establish broad acetylation patterns (24). Analysis of the human GH domain in transgenic mice has provided strong evidence for the hypothesis that LCRs can establish broad acetylation patterns (25). Analysis of acetylation at the murine β-globin locus in adult erythroid cells revealed enrichment of acetylated histones H3 and H4 at the LCR and the adult globin genes, βmajor and βminor (24, 26). Much less acetylation was evident over a ≈30-kb region spanning the silent embryonic β-globin genes Ey and βH1, and between the adult β-globin genes. In embryonic erythroid cells, acetylation was high at the LCR, the embryonic βH1 promoter and the inactive βminor promoter (24). Although the LCR confers high-level β-globin expression, deletion of HS1–HS4 from the murine locus did not abrogate hyperacetylation of the locus (27). In contrast, deletion of human HS1–HS4 reduced H3 acetylation at the adult promoters (26). Similarly, loss of the hematopoietic transcription factor and LCR component p45/NF-E2 (28, 29) in CB3 erythroleukemia cells (30), which have greatly reduced β-globin expression, decreased acetylation at the adult promoters 2- to 3-fold (31). As a 2-fold increase in acetylation prevents higher-order chromatin folding in vitro (32), changes of this magnitude are likely to be important.
To elucidate factors that establish the erythroid-specific chromatin structure of the β-globin domain, it is necessary to define proteins that occupy cis elements of the domain. Although many of these elements are predicted to be factor binding sites, and in vivo footprinting has provided evidence for occupancy of certain sites (33–35), only NF-E2 and GATA-1 have been shown to directly bind the endogenous domain. Chromatin immunoprecipitation (ChIP) analysis revealed NF-E2 crosslinking to the LCR (31, 36–38), strong at HS2 and weak at HS1, HS3, HS4 (31, 37), and the adult promoters (38). Transcriptional activation by NF-E2 requires the histone acetyltransferase CBP (CREB binding protein) (39–41) and results in pol II recruitment to the adult promoters in adult erythroid cells (31). Pol II also associates with the LCR in the absence of NF-E2, but this requires erythroid-specific factors (31).
Another hematopoietic factor that regulates β-globin transcription is GATA-1 (42, 43). Given its critical role in erythropoiesis (44–46) and its binding to the LCR (47, 48) and the Gγ-globin promoter in human K562 erythroleukemia cells (48), GATA-1 may be required for assembling the erythroid-specific β-globin domain. We asked whether GATA-1 discriminates between the abundant GATA-1 sites of the β-globin locus and describe the importance of GATA-1 vis-à-vis assembly of the β-globin domain.
Materials and Methods
Cell Culture and Transfection.
Mouse erythroleukemia (MEL) and CB3 cells were maintained in DMEM (Biofluids, Rockville, MD) containing 1% antibiotic/antimycotic (GIBCO/BRL) and 10% FCS. The p45/NF-E2 expression vector has been described (49). Stably transfected clones of CB3 cells expressing p45/NF-E2 were selected and maintained in 1 mg/ml G418 sulfate (Calbiochem). G1E cells (50) were maintained in Iscove's modified Dulbecco's medium (GIBCO/BRL) containing 2% penicillin-streptomycin (GIBCO/BRL), 2 units/ml erythropoietin, 120 nM monothioglycerol (Sigma), 15% FCS, and 0.6% conditioned medium from a kit ligand producing Chinese hamster ovary cell line. G1E-ER-GATA cells (51), which stably express GATA-1 as a fusion to the human estrogen receptor ligand binding domain, were maintained in 1 μg/ml puromycin.
ChIP Assay.
ChIP analysis of p45/NF-E2 and pol II was performed as described (14, 31). MEL and CB3 cells were incubated for 4 days with 1.5% DMSO (Sigma), and G1E and G1E-ER-GATA cells were incubated for 48 h with 1 μM tamoxifen (Sigma). ChIP analysis of GATA-1 binding required a rabbit anti-rat IgG antibody precoupled to the protein A-Sepharose (2 μg antibody/μl settled bed volume resin) to efficiently immunoprecipitate complexes with a rat anti-GATA-1 mAb. Antibodies used for ChIP are described in additional Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org. Immunoprecipitated DNA was analyzed by PCR with primers that generate 250- to 350-bp products. After 31–33 PCR cycles, products were resolved on 1.6% agarose gels containing ethidium bromide and quantitated by using National Institutes of Health IMAGE 1.61. Band intensities are expressed relative to signals obtained from 0.06% input. Signals were proportional to the amount of DNA input, and PCR primers (24, 52) amplified a single fragment of the expected size. Results were validated by real-time PCR (Applied Biosystems Prism 7000). Primers were designed by PRIMER EXPRESS 1.0 software (Perkin–Elmer Applied Biosystems) to amplify 50- to 150-bp subregions within the endpoints of the corresponding standard PCR primers. Primers (see the additional Materials and Methods) were based on Hbbd haplotype sequences (GenBank accession nos. Z13985, X14061, AF128269 and AF133300). Samples from three independent immunoprecipitations were analyzed in duplicate. Product was measured by SYBR green fluorescence in 25-μl reactions, and the amount of product was determined relative to a standard curve generated from a titration of input chromatin. Denaturation curves postamplification showed that primer pairs generated single products.
RNA and Protein Detection.
RNA and protein were prepared from the same cell cultures used for ChIP. Total RNA was purified with Trizol (GIBCO/BRL). Real-time reverse transcription–PCR was performed as described in the additional Materials and Methods. Protein isolation and analysis and antibodies are described in the additional Materials and Methods.
Results and Discussion
Selective Occupancy of GATA-1 Sites Throughout the Murine β-Globin Locus.
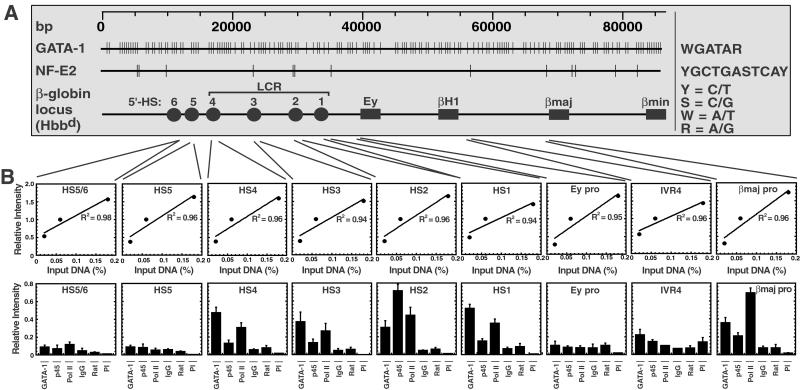
To understand the role of GATA-1 in long-range transactivation of the β-globin genes, it is necessary to identify cis elements occupied by GATA-1 in vivo. Because the consensus GATA-1 site is a simple hexanucleotide sequence (A/TGATAA/G) (53, 54), it occurs with high frequency throughout mammalian genomes. The murine Hbbd β-globin locus contains 271 consensus GATA-1 sites (see Fig. 2A). Given this formidable number of sites, one cannot predict which sites would be accessible in chromatin. Thus, we used ChIP analysis to define where GATA-1 binds within the endogenous β-globin locus of MEL cells (Fig. 1A).
Figure 2.
Occupancy of GATA-1 sites throughout the murine β-globin domain. (A) Distribution of GATA-1 and NF-E2 sites. Consensus sequences are defined on the right. Vertical lines indicate consensus GATA-1 sites and NF-E2 sites that conform to the consensus as well as those deviating from the consensus by 1 nt. Tandem sites within HS2 are the only consensus NF-E2 sites. (B) ChIP analysis of binding by GATA-1, p45/NF-E2, and pol II within the β-globin locus in MEL cells. (Upper) The linearity of input signals. (Lower) Signals obtained for immunoprecipitated samples expressed relative to the 0.06% input. Shown is the relative intensity of PCR products from 3–5 independent experiments (mean ± SEM). IgG, rabbit IgG; Rat, rat IgG; PI, rabbit preimmune sera.
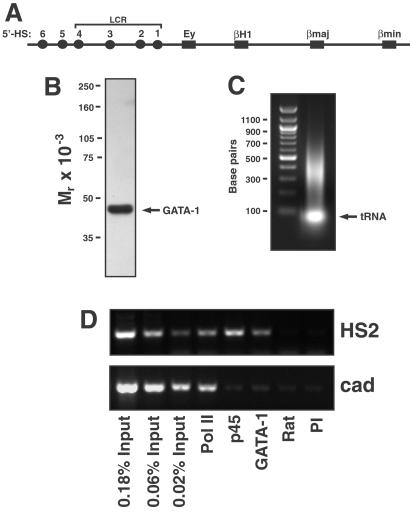
Figure 1.
GATA-1 binding to the endogenous β-globin locus of MEL cells. (A) Structure of the murine Hbbd β-globin locus. (B) Specificity of anti-GATA-1 antibody. Western blot analysis of total MEL cell lysate with anti-GATA-1 antibody. The sole reactive band, GATA-1, is indicated by the arrow. (C) Agarose gel analysis of sonicated chromatin. The representative ethidium bromide-stained agarose gel shows input DNA isolated from chromatin after reversal of crosslinks and deproteination. The average size of the chromatin is ≈400 bp. (D) ChIP analysis of pol II, p45/NF-E2 (p45), and GATA-1 binding to HS2 of the endogenous murine β-globin locus. The representative ethidium bromide-stained agarose gel shows PCR products obtained by ChIP from MEL cell lysates with HS2 and cad promoter primers. The numbers indicate the percent of input DNA used in the corresponding PCRs. Only HS2 DNA was specifically immunoprecipitated with anti-p45 and anti-GATA-1 antibodies, whereas HS2 and cad promoter DNA were specifically immunoprecipitated with anti-pol II antibody. Rat, rat IgG; PI, rabbit preimmune sera.
MEL cells express the GATA-1-regulated adult β-globin genes (βmajor and βminor) (55). The β-globin locus of MEL cells has a pattern of histone acetylation similar to 14.5 days postcoitum fetal liver (24), which is highly enriched in adult erythroid cells. Furthermore, pol II and p45/NF-E2 associate with the HSs of the LCR similarly in MEL cells and fetal liver (31). Thus, multiple parameters of the native structure of the β-globin locus are indistinguishable between MEL cells and primary, adult erythroid cells, thereby justifying the physiological relevance of this system.
An anti-GATA-1 mAb that reacted only with GATA-1 in Western blots of MEL cell lysates (Fig. 1B) was used to immunoprecipitate sonicated MEL cell chromatin with an average size of ≈400 bp (Fig. 1C). The amount of PCR product was proportional to the chromatin input (Fig. 1D). Immunoprecipitation with the anti-GATA-1 antibody resulted in recovery of HS2 DNA in an immune-specific manner. In contrast, the active cad promoter (56) lacking consensus GATA-1 sites was not recovered. Similarly, as described (31), an anti-p45/NF-E2 antibody immunoprecipitated HS2 but not cad DNA, whereas an anti-pol II antibody immunoprecipitated both HS2 and cad DNA.
Immunoprecipitated MEL cell chromatin was analyzed with primers spanning multiple regions of the β-globin locus to determine whether GATA-1 discriminates between the abundant GATA-1 sites (Fig. 2A). GATA-1 was crosslinked to chromatin at all four HSs of the LCR (HS1–HS4) and the adult βmajor promoter (Fig. 2B). GATA-1 was not crosslinked to the Ey promoter, and only very weak signals were detected at chromatin upstream of HS5 (HS5/6), HS5, and the intergenic site IVR4. These regions contained one, one, two, and one consensus GATA-1 sites, respectively, within the PCR product. By comparison, GATA-1 was strongly crosslinked to HS2, which contains one consensus GATA-1 site. Thus, clustered consensus sites are not required for strong crosslinking. This analysis shows that GATA-1 discriminates between the many sites, suggesting that a subset of the sites are occluded. As the central portion of the locus is hypoacetylated in adult erythroid cells, especially for histone H3 (24, 26), hypoacetylation might restrict site access. However, trichostatin A-induced hyperacetylation did not induce GATA-1 binding at the Ey promoter or IVR4 (unpublished data).
The GATA-1 occupancy pattern of MEL cells was different from human K562 cells (48), which express embryonic and fetal β-globin genes (57). Using a coupled ChIP-microarray chip method, GATA-1 was crosslinked only to HS2 and the Gγ promoter, one of the two fetal promoters. GATA-1 was not crosslinked to the other HSs, the embryonic ɛ promoter, nor the Aγ promoter. The unique pattern might reflect developmental stage or unidentified variables.
To define the nucleoprotein composition of the MEL cell β-globin domain in more detail, ChIP analysis was used to measure NF-E2 and pol II occupancy. Only two consensus NF-E2 sites (C/TGCTGAC/GTCAC/T) (28) exist within the β-globin locus: the tandem NF-E2 sites of HS2. However, 12 additional sites are present if a single nucleotide mismatch is allowed (Fig. 2A). p45/NF-E2 was shown via ChIP to occupy the tandem sites of HS2 in K562 and MEL cells (36, 37) and in 14.5 days postcoitum murine fetal liver (37). Some p45/NF-E2 crosslinking was observed at the βmajor promoter in MEL cells (58), which has a nonconserved, imperfect NF-E2 site. It was suggested that LCR-bound NF-E2 could be crosslinked to the promoter because of the close proximity of the two regions, or that p45/NF-E2 might interact directly with promoter-bound factors.
We tested whether p45/NF-E2 could be crosslinked to other functionally important regions of the locus. Weak crosslinking occurred at HS1, HS3, and HS4 (Fig. 2B). Although the recovery of these HSs was only slightly higher than background, the ratios of p45/NF-E2 to control signals were consistently higher than was detected at the hypoacetylated subdomain and the cad promoter. It was shown previously that p45/NF-E2 could be crosslinked to multiple HSs of the LCR in K562 cells (36). Although this was not done quantitatively, the highest crosslinking was at HS1, HS2, and HS4. p45/NF-E2 crosslinking was also seen at the βmajor promoter (Fig. 2B), analogous to the recent report (58). Thus, strong crosslinking of p45/NF-E2 occurred at HS2 containing consensus NF-E2 sites, whereas weaker crosslinking was apparent at other HSs and at the βmajor promoter.
Pol II associates with HS1, HS2, HS3, and the βmajor promoter, but not with the intergenic site IVR2 nor the Ey promoter in MEL cells (31). This analysis was extended to include HS4, HS5, and the additional intergenic sites, HS5/6 and IVR4. Pol II was crosslinked to HS1, HS2, HS3, and HS4, very weakly to HS5/6, but not to HS5, the Ey promoter, nor IVR4. Intriguingly, both GATA-1 and pol II localize to the four HSs of the LCR, suggesting a functional connection between these factors at the HSs.
GATA-1-Dependent Pol II Recruitment to the LCR and the Adult β-Globin Promoters.
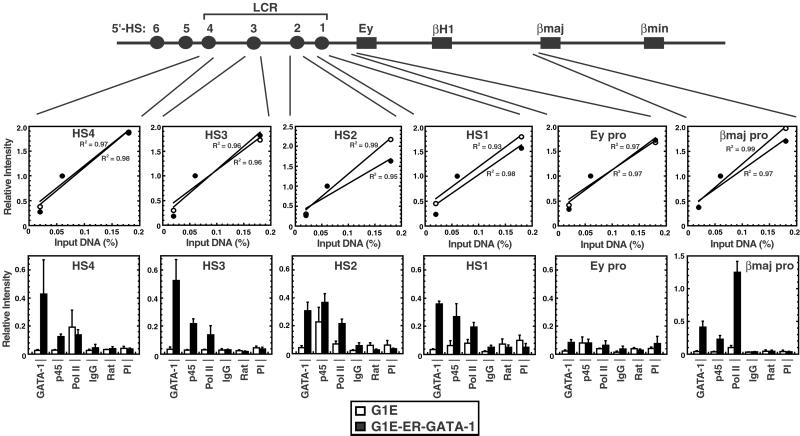
The GATA-1-null G1E erythroid cell line (50) was used to define the consequences of GATA-1 binding to the β-globin locus. G1E cells were derived from GATA-1−/− embryonic stem cells and express low levels of adult β-globin. Conditional expression of GATA-1 via tamoxifen activation of an estrogen receptor hormone binding domain-GATA-1 fusion (ER-GATA-1) induced erythroid differentiation (51, 59) and increased adult β-globin expression 1,090-fold as determined by real-time PCR (48 h postinduction). We asked whether activation of ER-GATA-1 induces a pattern of GATA-1 occupancy similar to MEL cells. As ER-GATA-1 was not entirely inactive in uninduced G1E cells (data not shown), we compared tamoxifen-treated G1E with tamoxifen-treated G1E-ER-GATA-1 cells. ER-GATA-1 activation resulted in GATA-1 crosslinking to HS1, HS2, HS3, HS4, and the βmajor promoter, but not to the Ey promoter (Fig. 3), recapitulating the MEL cell pattern (Fig. 2B). Thus, we asked whether GATA-1 regulates pol II and NF-E2 recruitment to the β-globin domain.
Figure 3.
GATA-1 regulates pol II recruitment to the adult β-globin promoters and the LCR. ChIP analysis of GATA-1, p45/NF-E2, and pol II binding to the β-globin locus in G1E and G1E-ER-GATA cells. (Upper) Graphs depict the linearity of input signals. (Lower) Signals obtained for immunoprecipitated samples are graphed and are expressed relative to the 0.06% input. The graphs show the relative intensity of PCR products from at least three independent experiments (mean ± SEM). IgG, rabbit IgG; Rat, rat IgG; PI, rabbit preimmune sera.
NF-E2 is not required to recruit pol II to the LCR, but is critical for pol II recruitment to the βmajor promoter (31). The lack of pol II binding to the LCR in nonerythroid cells suggested that other erythroid-specific factors attract pol II to the LCR, and therefore we tested whether GATA-1 is such a factor. Similar to MEL cells (Fig. 2B), pol II resided at all HSs of the LCR, at the βmajor promoter, but not at the Ey promoter, in G1E-ER-GATA-1 cells (Fig. 3). In G1E cells, no pol II crosslinking was detected at HS3, whereas considerably reduced crosslinking was apparent at HS1 and HS2. In contrast, GATA-1 deficiency had no impact on pol II crosslinking to HS4. Lastly, pol II crosslinking to the βmajor promoter was strongly GATA-1-dependent. Intriguingly, the analysis revealed distinctions between the association of pol II at different HSs. GATA-1 induced pol II recruitment to HS1, HS2, and HS3, whereas pol II recruitment to HS4 was GATA-1-independent. The HS4 result was surprising, as cis elements of HS4 bind GATA-1 in vitro and are required for maximal nuclease sensitivity of HS4 in stable transfection assays (60). Accordingly, GATA-1 deficiency might render HS4 less accessible to all factors. However, because GATA-1 was not required for pol II loading at HS4, this assumption seems unlikely.
GATA-1-Dependent NF-E2 Recruitment to Regions Lacking Consensus NF-E2 Sites.
Because GATA-1 induces crosslinking of pol II to subregions of the LCR (Fig. 3B), it was important to determine whether the recruitment of other factors was affected similarly. ChIP analysis of p45/NF-E2 binding to HS2 containing the consensus, tandem NF-E2 sites revealed only slightly less p45/NF-E2 crosslinking in G1E versus G1E-ER-GATA-1 cells (Fig. 3B). Thus, under conditions in which GATA-1 deficiency strongly reduced pol II crosslinking at HS2, the majority of p45/NF-E2 remained bound to the chromatin. In contrast, weaker crosslinking of p45/NF-E2 to regions lacking consensus NF-E2 sites (HS1, HS3, HS4, and βmajor promoter) was abolished upon loss of GATA-1. Of particular interest was HS4, in which GATA-1 deficiency abolished p45/NF-E2 but not pol II crosslinking. This result provides evidence that GATA-1 does not simply increase accessibility to all factors and reinforces our previous conclusion that NF-E2 is not essential for pol II recruitment to the LCR (31).
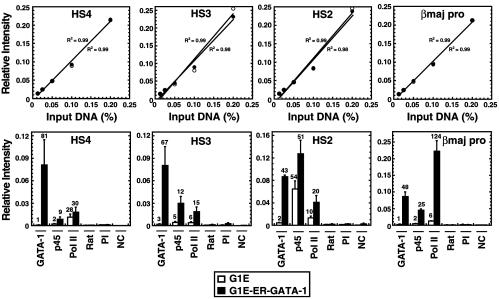
Real-time PCR, which considerably increased the signal-to-noise ratio, was used as an alternative mode of analysis. Analysis of GATA-1, p45/NF-E2, and pol II crosslinking to HS2, HS3, HS4, and the βmajor promoter revealed a pattern identical to the results of Figs. 2 and 3 (Fig. 4). The extent of GATA-1 crosslinking at HS2, HS3, HS4, and the βmajor promoter was comparable. Second, GATA-1 was critical for p45/NF-E2 crosslinking to regions lacking consensus NF-E2 sites (HS3, HS4, and βmajor promoter), whereas strong crosslinking of p45/NF-E2 to HS2 was largely GATA-1-independent. Lastly, GATA-1 increased pol II crosslinking to HS2, HS3, and the βmajor promoter, whereas pol II crosslinking to HS4 was GATA-1-independent.
Figure 4.
Quantitative analysis of GATA-1-dependent pol II and NF-E2 recruitment. Samples from three independent ChIP experiments that were included in the analysis of Fig. 3 were analyzed by real-time PCR. (Upper) The linearity of input signals. (Lower) The signals obtained for immunoprecipitated samples are graphed and are expressed relative to a standard curve generated from input samples (mean ± SEM). Values shown above the bars represent the ratio of signals obtained relative to the appropriate control antibody. Variations in background signals alter the ratios, although the variations did not exceed 2-fold. Rat, rat IgG; PI, rabbit preimmune sera; NC, no chromatin.
Although the mechanism of GATA-1-dependent p45/NF-E2 crosslinking to regions lacking consensus NF-E2 sites is unclear, it is instructive to consider four models. First, GATA-1 might induce a higher-order chromatin transition, bringing together physically segregated sites. Thus, HS2-bound p45/NF-E2 would gain access to HS1, HS3, HS4, and the βmajor promoter. Second, p45/NF-E2 might localize to the promoter via interaction with promoter-bound factors. These models were considered in a previous study describing p45/NF-E2 crosslinking to the promoter in MEL cells (58). Alternative possibilities are that GATA-1-containing complexes at the LCR increase accessibility of nonconsensus sites, leading to NF-E2 binding, or cooperative interactions enhance the affinity of NF-E2 for nonconsensus sites.
NF-E2-Dependent GATA-1 Recruitment to the βmajor Promoter.
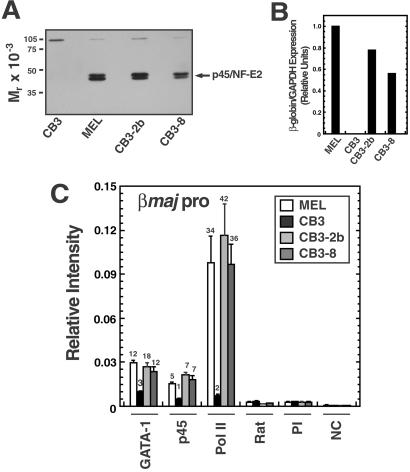
Because GATA-1 induced p45/NF-E2 crosslinking to certain sites within the locus, and both GATA-1 and NF-E2 were required for pol II recruitment to the promoter, we asked whether NF-E2 regulates GATA-1 occupancy at the adult promoters. Clonal cell lines were generated from p45/NF-E2-null CB3 cells that stably expressed physiological levels of p45/NF-E2 (Fig. 5A). p45/NF-E2 expression in CB3–2b and CB3–8 lines rescued β-globin transcription (Fig. 5B). p45/NF-E2 deficiency caused a 4-fold reduction in GATA-1 crosslinking to the βmajor promoter (Fig. 5C), but significant GATA-1 crosslinking (3-fold higher than controls) remained. Defective GATA-1 loading was rescued in the CB3–2b and CB3–8 lines, showing that NF-E2 induces GATA-1 binding to the promoter. CB3 cells also showed reduced GATA-1 binding to HS2 and HS3, but not to HS4 (Fig. 6, which is published as supporting information on the PNAS web site). As described (31), p45/NF-E2 deficiency nearly abrogated pol II recruitment to the promoter, and p45/NF-E2 expression rescued pol II recruitment (Fig. 5C). Because some GATA-1 occupancy was apparent without p45/NF-E2, and p45/NF-E2 deficiency had a much larger impact on pol II versus GATA-1 crosslinking (17- versus 4-fold, respectively), the association of GATA-1 with the promoter seems insufficient to induce pol II recruitment. An important implication of this result is that the GATA-1– NF-E2 cooperativity to recruit pol II to the promoter might not involve NF-E2-dependent loading of GATA-1 on the promoter, because GATA-1 occupancy of the promoter in the absence of NF-E2 does not suffice to strongly recruit pol II. However, because GATA-1 occupancy was not entirely NF-E2-independent, it remains possible that maximal GATA-1 occupancy is required for pol II recruitment.
Figure 5.
Quantitative analysis of NF-E2-dependent GATA-1 and pol II recruitment to the βmajor promoter. (A) Western blot analysis of p45/NF-E2 expression in total cell lysates from MEL and CB3 cells, and two independent CB3 cell lines stably expressing p45/NF-E2. (B) Real-time PCR analysis of β-globin RNA expression in the cell lines of A. β-globin RNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) Real-time PCR ChIP analysis of binding of GATA-1, p45/NF-E2, and pol II to the βmajor promoter. Signals obtained for immunoprecipitated samples from four independent experiments (mean ± SEM) are expressed relative to a standard curve generated from the input samples. Values shown above the bars represent the ratio of signals obtained relative to the appropriate control antibody. Rat, rat IgG; PI, rabbit preimmune sera; NC, no chromatin.
Cooperative Activities of Hematopoietic Regulators Recruit Pol II to the β-Globin Domain.
p45/NF-E2 deficiency resulted in a partial reduction in pol II binding to the LCR (HS1, HS2, and HS3) in CB3 cells, but a considerable amount of pol II remained associated (31). Although HS4 was not examined previously, the association of pol II with HS4 does not require NF-E2 (unpublished data). Work described herein showed that GATA-1 induces pol II recruitment to HS1, HS2, HS3, and the βmajor promoter. Thus, GATA-1–NF-E2 cooperativity is required for pol II loading at the βmajor promoter. A simple explanation for the cooperativity is that GATA-1 and NF-E2 might regulate the synthesis of each other. The murine p45/NF-E2 gene contains two promoters, one requiring a GATA-1 site for erythroid-specific activity in transient assays (61). We measured GATA-1 and p45/NF-E2 levels by Western blotting in CB3, MEL, G1E, and G1E-ER-GATA-1 cells (Fig. 7, which is published as supporting information on the PNAS web site). GATA-1 deficiency in G1E cells and activation of ER-GATA-1 did not alter p45/NF-E2 levels. Similarly, p45/NF-E2 deficiency in CB3 cells did not affect GATA-1 levels. Thus, cooperativity cannot be explained by changes in protein levels. Another model to explain cooperativity involves physical interactions between these factors. However, GATA-1 and p45/NF-E2 do not associate by coimmunoprecipitation analysis (Fig. 7). We propose that GATA-1 and NF-E2 regulate unique steps in a multistep pathway.
What might be the nature of such a multistep mechanism? Because pol II remains at the LCR but not at the promoter without NF-E2, and NF-E2 induces pol II loading on the promoter, we proposed the long-range pol II transfer model (31). The absence of pol II at the LCR in B cells suggests that erythroid-specific factors other than NF-E2 are required for pol II recruitment to the LCR. Long-range pol II transfer assumes that such factors would recruit pol II to the LCR, and NF-E2 would provide a signal to induce pol II relocalization to the promoter. Because the central region of the β-globin locus is hypoacetylated (24, 26) and is inaccessible to endonucleases (unpublished data), and because pol II cannot be detected within this region, we reasoned that pol II relocalization might require direct contact between the LCR and the promoter. As the promoter becomes enriched in acetylated H3 upon NF-E2 binding to HS2 and weaker binding to other sites in the locus, acetylated chromatin of the promoter might be the attracting signal. p45/NF-E2 crosslinking to the βmajor promoter in MEL cells has been described and discussed in the context of a model in which the LCR contacts the promoter (58).
Our current work provides strong evidence that GATA-1 recruits pol II to HS1, HS2, HS3, and the adult promoters in physiological chromatin. It is attractive to propose that GATA-1 recruits pol II to the LCR, and NF-E2 induces pol II relocalization to the promoter. If the GATA-1-dependent reaction is impaired, long-range pol II transfer would not occur, even in the presence of NF-E2. Such a mechanism would explain the GATA-1–NF-E2 cooperativity to recruit pol II to the promoters. Alternative models invoke novel activities of LCR-bound pol II, rather than relocalization to the promoters. These activities might involve transcription-dependent chromatin remodeling or the generation of regulatory transcripts (62, 63).
Intriguingly, pol II crosslinking to HS4 was GATA-1-independent. HS4 has typical high-affinity GATA-1 sites, and GATA-1 can be readily crosslinked to HS4. Studies in transgenic mice containing a human β-globin locus-yeast artificial chromosome (β-YAC) have implicated HS4 in conferring high-level β-globin expression during adult, but not embryonic, erythropoiesis (64). In another study, deletion of HS4 from a β-YAC strongly reduced β-globin expression at all developmental stages (65). Disruption of HS4 from the endogenous murine β-globin locus did not reveal a role for HS4 in stage-specific expression (66). Importantly, reductions in β-globin expression in mice containing single HS deletions (27% decrease upon deletion of HS4) suggest complex and redundant mechanisms to maintain high-level expression.
The GATA-1- and NF-E2-independent pol II crosslinking to HS4 supports an additional step in the transactivation mechanism, in which the loading or accumulation of pol II at HS4 is regulated. Hematopoietic factors implicated in the control of β-globin expression that might participate in this step include EKLF (67), TAL1 (68, 69), and GATA-2 (45), although these factors have not been shown to occupy the β-globin domain in living cells. One can speculate that HS4-bound pol II might gain access to other subregions of the LCR in a GATA-1-dependent manner, as an essential proximal regulatory step in long-range pol II transfer.
Supplementary Material
Acknowledgments
We thank Paul Ney for the p45/NF-E2 vector. We thank Moshe Sadofsky and Hogune Im for a critical review of the manuscript. We acknowledge support from the Milwaukee Foundation, the Leukemia and Lymphoma Society of America, and the National Institutes of Health (Grants DK50107 and DK55700 to E.H.B.). E.H.B. is a Leukemia and Lymphoma Society of America Scholar and a Shaw Scientist.
Abbreviations
- ChIP
chromatin immunoprecipitation
- HS
hypersensitive site
- LCR
locus control region
- MEL
mouse erythroleukemia
- pol II
polymerase II
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Felsenfeld G. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 2.Hager G L, Archer T K, Fragoso G, Bresnick E H, Tsukagoshi Y, John S, Smith C L. Cold Spring Harbor Symp Quant Biol. 1993;58:63–71. doi: 10.1101/sqb.1993.058.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg R D, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Grunstein M. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 5.Jones K A, Kadonaga J T. Genes Dev. 2000;14:1992–1996. [PubMed] [Google Scholar]
- 6.Brown C E, Lechner T, Howe L, Workman J L. Trends Biochem Sci. 2000;25:15–19. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- 7.Narlikar G J, Fan H-Y, Kingston R E. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Orphanides G, Reinberg D. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 9.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 10.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 11.Pina B, Bruggemeier U, Beato M. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 12.Archer T K, Cordingley M G, Wolford R G, Hager G L. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg E C, Bresnick E H. BioEssays. 2001;23:820–830. doi: 10.1002/bies.1117. [DOI] [PubMed] [Google Scholar]
- 14.Johnson K D, Bresnick E H. Methods. 2002;26:27–36. doi: 10.1016/S1046-2023(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 15.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 16.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 17.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 18.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 19.Navas P A, Li Q, Peterson K R, Swank R A, Rohde A, Roy J, Stamatoyannopoulos G. Hum Mol Genet. 2002;11:893–903. doi: 10.1093/hmg/11.8.893. [DOI] [PubMed] [Google Scholar]
- 20.Tuan D, London I M. Proc Natl Acad Sci USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrester W C, Thompson C, Elder J T, Groudine M. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 23.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 24.Forsberg E C, Downs K M, Christensen H M, Im H, Nuzzi P A, Bresnick E H. Proc Natl Acad Sci USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho Y, Elefant F, Cooke N, Liebhaber S. Mol Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- 26.Schubeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 27.Schubeler D, Groudine M, Bender M A. Proc Natl Acad Sci USA. 2001;98:11432–11437. doi: 10.1073/pnas.201394698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews N C, Erdjument-Bromage H, Davidson M B, Tempst P, Orkin S H. Nature (London) 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 29.Ney P A, Andrews N C, Jane S M, Safer B, Purucker M E, Weremowicz S, Morton C C, Goff S C, Orkin S H, Nienhuis A W. Mol Cell Biol. 1993;13:5604–5612. doi: 10.1128/mcb.13.9.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu S J, Rowan S, Bani M R, Ben-David Y. Proc Natl Acad Sci USA. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson K, Christensen H M, Zhao B, Bresnick E H. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 32.Tse C, Sera T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikuta T, Kan Y W. Proc Natl Acad Sci USA. 1991;88:10188–10192. doi: 10.1073/pnas.88.22.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy P M, Shen C K. Proc Natl Acad Sci USA. 1991;88:8676–8680. doi: 10.1073/pnas.88.19.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy P M, Stamatoyannopoulos G, Papayannopoulou T, Shen C K. J Biol Chem. 1994;269:8287–8295. [PubMed] [Google Scholar]
- 36.Daftari P, Gavva N R, Shen C K. Oncogene. 1999;18:5482–5486. doi: 10.1038/sj.onc.1202916. [DOI] [PubMed] [Google Scholar]
- 37.Forsberg E C, Downs K M, Bresnick E H. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- 38.Sawado T, Igarashi K, Groudine M. Proc Natl Acad Sci USA. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng X, Reginato M J, Andrew N C, Lazar M A. Mol Cell Biol. 1997;17:1407–1416. doi: 10.1128/mcb.17.3.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forsberg E C, Johnson K, Zaboikina T N, Mosser E A, Bresnick E H. J Biol Chem. 1999;274:26850–26859. doi: 10.1074/jbc.274.38.26850. [DOI] [PubMed] [Google Scholar]
- 41.Hung H L, Kim A Y, Hong W, Rakowski C, Blobel G A. J Biol Chem. 2001;276:10715–10721. doi: 10.1074/jbc.M007846200. [DOI] [PubMed] [Google Scholar]
- 42.Evans T, Felsenfeld G. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- 43.Tsai S F, Martin D I, Zon L I, D'Andrea A D, Wong G G, Orkin S H. Nature (London) 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 44.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S F, D'Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 45.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 46.Weiss M J, Orkin S H. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Z, Stamatoyannopoulos G, Li Q. Mol Cell Biol. 2001;21:3083–3095. doi: 10.1128/MCB.21.9.3083-3095.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horak C E, Mahajan M C, Luscombe N M, Gerstein M, Weissman S M, Snyder M. Proc Natl Acad Sci USA. 2002;99:2924–2929. doi: 10.1073/pnas.052706999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bean T L, Ney P A. Nucleic Acids Res. 1997;25:2509–2515. doi: 10.1093/nar/25.12.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiss M J, Yu C, Orkin S H. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregory T, Yu C, Ma A, Orkin S H, Blobel G A, Weiss M J. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- 52.Eberhardy S R, D'Cunha C A, Farnham P J. J Biol Chem. 2000;275:33798–33805. doi: 10.1074/jbc.M005154200. [DOI] [PubMed] [Google Scholar]
- 53.Ko L J, Engel J D. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merika M, Orkin S H. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nudel U, Salmon J E, Terada M, Bank A, Rifkind R A, Marks P A. Proc Natl Acad Sci USA. 1977;74:1100–1104. doi: 10.1073/pnas.74.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dean A, Ley T J, Humphries R K, Fordis M, Schechter A N. Proc Natl Acad Sci USA. 1983;80:5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francastel C, Magis W, Groudine M. Proc Natl Acad Sci USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shirihai O S, Gregory T, Yu C, Orkin S H, Weiss M J. EMBO J. 2000;19:2492–2502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatoyannopoulos J A, Goodwin A, Joyce T, Lowrey C H. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moroni E, Mastrangelo T, Razzini R, Moi P, Ottolenghi S, Giglioni B. J Biol Chem. 2000;275:10567–10576. doi: 10.1074/jbc.275.14.10567. [DOI] [PubMed] [Google Scholar]
- 62.Plant K E, Routledge S J, Proudfoot N J. Mol Cell Biol. 2001;21:6507–6514. doi: 10.1128/MCB.21.19.6507-6514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Navas P A, Peterson K R, Li Q, McArthur M, Stamatoyannopoulos G. J Mol Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]
- 65.Bungert J, Dave U, Lim K C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 66.Bender M A, Roach J N, Halow J, Close J, Alami R, Bouhassira E E, Groudine M, Fiering S N. Blood. 2001;98:2022–2027. doi: 10.1182/blood.v98.7.2022. [DOI] [PubMed] [Google Scholar]
- 67.Miller I J, Bieker J J. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown L, Cheng J-T, Chen Q, Siciliano M J, Crist W, Buchanan G, Baer R. EMBO J. 1990;9:3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Begley C G, Aplan P D, Denning S M, Haynes B F, Waldmann T A, Kirsch I R. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.