Abstract
Hemolytic anemia is a forme fruste of systemic lupus erythematosus (SLE), being observed months or even years before the onset of other clinical manifestations in some patients. We hypothesized that hemolytic anemia in those SLE-affected patients would identify a group of SLE pedigrees that share a high degree of genetic homogeneity. From 160 multiplex SLE pedigrees, we sought evidence for linkage in 35 (16 African-American, 17 European-American, and 2 Hispanic) who had at least one SLE-affected patient with hemolytic anemia. Significant linkage was present at 11q14 in the 16 African-American pedigrees, yielding a maximum two-point logarithm of odds (LOD) score of 4.5 at D11S2002. The segregation pattern of SLE in these African-American pedigrees suggested a dominant mode of inheritance and, when maximized across penetrance and disease allele frequencies, produced a multipoint LOD of 4.7. Multipoint analysis yielded a multipoint heterogeneity LOD score of 3.6 (α = 0.63), again with maximum LOD at D11S2002. Finally, markers typed 7 centimorgans to either side of D11S2002 achieved LOD scores of 3 or better by using the maximized model, supporting linkage to 11q14. Clearly, pedigree ascertainment based on select clinical manifestations is an important tool, capable of revealing otherwise cryptic genetic linkages in complex genetic diseases. Thus, we show strong evidence for an SLE susceptibility gene, SLEH1, near D11S2002 in African-American pedigrees multiplex for SLE that have at least one SLE-affected patient with hemolytic anemia.
Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterized by autoantibody production and tissue injury. SLE has a prevalence rate of ≈40 cases per 100,000 individuals with onset typically occurring in women of child-bearing age (female/male, ratio 9:1) (1). African Americans are three times more likely to be affected than European Americans, manifest SLE at an earlier age, and have a clinically more severe phenotype than other American racial groups (2–4).
Although the etiology of SLE is still undetermined, both genetic and environmental factors are involved. Evidence of a genetic component includes familial clustering (5), λs estimates between 10 and 20 (6, 7), and higher concordance rates between monozygotic twins (>20%) relative to dizygotic twins and other full siblings (2–5%) (8). Case-control association studies implicate numerous candidate gene loci including multiple alleles from the HLA region (reviewed in ref. 9), Fcγ receptors (10–13), and complement components (14–17). In addition, SLE studies using inbred mouse strains suggest multiple susceptibility loci (18–26).
SLE classification is based on an individual presenting any 4 of the 11 criteria set forth by the American College of Rheumatology (27, 28). Consequently, the SLE phenotype is extremely heterogeneous. Nevertheless, several genome scans have identified SLE linkages suggesting SLE susceptibility genes (29–35); significant evidence of linkage [logarithm of odds (LOD ≥ 3.3)] has been reported to seven different chromosomal regions (35, 36, ‖). However, for any particular study, there is a wide variation in the chromosomal locations identified. No doubt, genetic heterogeneity is at least partly responsible for the variations between studies and the evident genetic complexity of SLE.
Multiple genes are usually involved in complex diseases with alleles at any one locus making only small-to-moderate contributions to the total risk, thereby impeding their identification (37, 38). Various combinations of contributing alleles at multiple genes may be important for individual patients, resulting in apparent disease phenocopies when any one gene is considered in isolation. Likewise, complexity may arise from a suboptimal definition of the phenotype (at least from the perspective of the capacity to identify genetic effects). A simpler genetic model involving fewer genetic effects may operate in a subset of the phenotype. In the present study, we chose to examine pedigrees containing at least one member affected with both SLE and hemolytic anemia to select a set of pedigrees with a more homogeneous phenotype and thereby increase our ability to detect significant evidence of linkage to genes that contribute to SLE susceptibility.
Hematologic abnormalities (hemolytic anemia, leukopenia, lymphopenia, and thrombocytopenia) are common manifestations in patients with SLE. Most patients exhibit anemia at some point during their disease course (39). The causes of anemia in these patients may be of immune or nonimmune pathogenesis. Nonimmune hemolytic anemias include those anemias that are caused by chronic disease and present as normocytic or normochromic anemias with adequate stores of iron in the bone marrow (40) and are rarely life-threatening in SLE patients (41).
Autoimmune hemolytic anemias (AIHAs) are a cause of anemia in 7–15% of SLE patients (42–45). Several clinical syndromes constitute the AIHAs, each being mediated by different autoantibodies (IgG or IgM) against red blood cells (46). As a result of these autoantibodies, the red blood cells are destroyed prematurely in patients with AIHA, resulting in an inadequate number of circulating red blood cells. AIHA usually develops gradually in most patients, but on occasion may result in a rapidly progressive hemolytic crisis (47, 48).
Several studies suggest that AIHA is a forme fruste, the first isolated clinical presentation, of SLE in these patients (44, 47, 48). Dubois first suggested this in 1952 based on three lupus patients presenting with hemolytic anemia as the initial manifestation of what later became unmistakable SLE. Videbaek et al. (48) later observed that AIHA was present for several months and in some cases for years before other clinical manifestations were observed in 50% of their SLE patients. More recently, Nossent and Swaak (44) observed hemolytic anemia as the presenting symptom of SLE in 11 of 16 (68.8%) hemolytic anemia patients studied.
Studies of testicular cancer, multinodular goiter, and nonmedullary thyroid carcinoma already have revealed the power stratifying pedigrees by clinical manifestations has to detect previously unknown genetic linkages (49–52). Kokori et al. (53) suggested that AIHA may identify a particular subgroup of SLE patients because of an observed association with certain characteristic serologic and clinical manifestations. We used pedigree stratification to test the hypothesis that the presence of hemolytic anemia would identify a group of genetically homogeneous pedigrees in which significant genetic linkage to genes implicated in SLE would be identified. Two such linkages, at 1q24 and at 11q14, along with fine mapping support of the linkage to 11q14 are reported by using this approach.
Materials and Methods
Patients and Pedigrees.
SLE patients and their families were enrolled in the lupus genetics study as described (30). Medical records were obtained on each potential patient who also completed an extensive questionnaire and interview with a trained physician's assistant or registered nurse to determine clinical manifestations and establish diagnosis. A less comprehensive questionnaire was completed by unaffected family members to screen for the presence of SLE.
Multiplex SLE pedigrees that contained at least one member affected with both SLE and AIHA were selected from the collection of 160 pedigrees enrolled in the lupus genetic studies at the Oklahoma Medical Research Foundation. SLE-affected patients were considered to have AIHA if (i) there was evidence of red blood cell destruction noted in the medical records (a hemoglobin level below the normal range in the testing laboratory (usually <12 g/dl) and a reticulocyte count above the upper normal limit for the testing laboratory) or (ii) a reticulocyte count could not be found in the available medical records, but the investigators considered the patient to have definite AIHA through the presence of a decreased hemoglobin level, a positive Coombs test, and diagnosis of AIHA by a physician. The resulting sample consisted of 35 pedigrees containing 195 individuals, 92 of whom were classified as people with SLE. Forty of the 92 SLE-affected patients had a positive history of AIHA (Table 1).
Table 1.
Composition of 35 multiplex SLE pedigrees with at least one patient exhibiting hemolytic anemia
| Composition | Ethnicity, no. (%)
|
Total | ||
|---|---|---|---|---|
| AA* | EA* | Other | ||
| Pedigrees | 16 (45.7) | 17 (48.6) | 2 (5.7) | 35 |
| Individuals | 89 (45.7) | 96 (49.2) | 10 (5.1) | 195 |
| SLE-affected | 41 | 45 | 5 | |
| Unaffected | 48 | 51 | 5 | |
| Sib pairs (affected and unaffected) | 62 (51.7) | 49 (40.8) | 9 (7.5) | 120 |
| Affected relative pairs | 28 (41.8) | 35 (52.2) | 4 (6.0) | 67 |
| % full-sib pairs | 32.1 | 48.6 | 75.0 | |
| % half-sib pairs | 7.1 | 2.9 | 0.0 | |
| % other pairs† | 60.8 | 48.5 | 25.0 | |
AA, African American; EA, European American.
Avuncular (e.g., uncle/niece), grandparent/grandchild, cousins.
DNA Isolation.
After obtaining informed consent, blood samples and/or buccal swabs were collected from each of the participants. Genomic DNA was isolated from peripheral blood mononuclear cells, buccal cell swabs, or Epstein–Barr virus-transformed cell lines by using standard methods.
Genotyping.
A total of 307 microsatellite markers were typed from the Version 8 Weber screening set (http://research.marshfieldclinic.org/genetics/sets/Set8ScreenFrames.htm), which has an average marker spacing of 11 centimorgans (cM). Fine mapping was performed in the 16 African-American pedigrees with an additional 14 microsatellite markers with an average spacing of 4.2 cM across the region of interest at 11q14. Polymerase chain reactions were performed as described (33).
Linkage Analysis.
Before linkage analysis, sibling, half-sibling, and parent–offspring relationships were confirmed by using RELTEST (54, 55). We chose to analyze the data by using parametric LOD score analysis and a conditional logistic regression technique for affected relative pairs because of their distinctive properties for detecting genetic linkages.
Parametric Linkage Analysis.
Two-point LOD scores were calculated by using FASTLINK 4.1P and ANALYZE (56–58). Six different inheritance models were assumed for the screening analysis: a dominant and recessive inheritance model, each using penetrance values of 50% in all participants, a dominant model using 90% penetrance in all participants, a recessive model using 100% penetrance in all participants, and a dominant and recessive mixed inheritance model, each with sex-specific penetrance values of 92 and 49% in females and males, respectively. LOD scores were maximized further over the penetrance and disease allele frequency functions to generate multipoint LOD scores.
Nonparametric Linkage Analysis.
The multipoint conditional logistic analysis, as implemented in LODPAL (55, 59), was performed on the affected relative pairs. This analysis was executed on all 35 pedigrees as well as the separate African-American and European-American subsets of pedigrees. Additional nonparametric linkage analysis was performed on all chromosome 11 markers in the 16 African-American pedigrees by using GENEHUNTER-PLUS (60) as described (35) to obtain heterogeneity LOD (HLOD) scores.
Statistical Analysis.
A conservative reverse Bonferroni correction was used to correct for the multiple comparisons of using three ethnic groupings in the analysis. The original P value was multiplied by three to obtain the conservative, adjusted P value.
Results
Linkage Findings.
Parametric analysis revealed one significant genetic linkage (LOD = 4.5) and seven genetic linkages that surpassed the recommended threshold for suggestive linkage (LOD ≥ 1.9) (ref. 37; Table 2). Several of the genetic linkages were racially specific. Two genetic effects at 1q21-22 (at FcγRIIA) and 13p11 (at D13S787) were present in the 17 European-American pedigrees, whereas the 16 African-American pedigrees were linked to 5p15 (at D5S817) and 11q14 (at D11S2002). The effect at 1q24 (at D1S1589) was not racially specific.
Table 2.
Regions identified by the genome scan to have LOD ≥ 1.9
| Region | Marker | cM | Population | LOD* | LODPAL* |
|---|---|---|---|---|---|
| 1q21-22 | FcγRIIA | 173 | EA | 3.0 | — |
| 1q21-22 | FcγRIIA | 173 | All | 2.8 | 2.3 |
| 1q24 | D1S1589 | 200 | EA | — | 4.0† |
| 1q24 | D1S1589 | 200 | All | 2.0 | 2.6 |
| 2q34 | D2S1384 | 226 | All | — | 1.9 |
| 5p15 | D5S817 | 23 | AA | 2.1 | — |
| 6p12 | D6S2410 | 74 | EA | — | 2.3 |
| 11q14 | D11S2002 | 82 | AA | 4.5† | — |
| 12q13 | D12S398 | 57 | EA | — | 1.9 |
| 13p11 | D13S787 | 10 | EA | 2.3 | 2.8 |
| 13p11 | D13S787 | 10 | All | 2.6 | — |
| 17p13 | D17S1298 | 10 | All | — | 2.3 |
Nonparametric analysis using the 67 affected relative pairs yielded eight regions that surpassed the threshold for suggestive linkage. Again, racial differences were observed. Four linkages (1q24, 6p12, 12q13, and 13p11) were present predominantly in the European Americans, whereas linkages to 1q21-22, 2q34, and 17p13 were present in all pedigrees (Table 2). None of the identified linkages were specific to the African Americans when using the affected relative pair approach.
Identification of New Linkages.
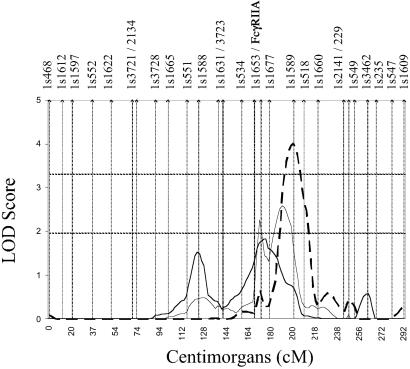
Most of the linkages identified in the present study have not been identified explicitly elsewhere. The linkage to 1q24 at D1S1589 (LOD = 4.0) was one of the two most powerful linkage effects in this study and was identified in the 35 European-American-affected relative pairs (Fig. 1). Linkage to 1q22-24 was identified previously at D1S1679 (33), but this marker is 20 cM centromeric to D1S1589 and was identified in African Americans. How many genes for SLE occur in this region cannot be discerned from the data. However, that these effects are reasonably far apart and were detected in different racial groups is consistent with the possibility that these two linkages may identify different genes. In any case, this region seems to have complicated associations with SLE. The linkages to 2q34, 12q13, and 13p11 are also new to the present study.
Figure 1.
Multipoint effect on chromosome 1 at D1S1589. Dashed horizontal lines, threshold for significant and suggestive linkage (LOD = 3.3 and 1.9); dashed line, European-American pedigrees; thick solid line, African-American pedigrees; thin solid line, all pedigrees. Approximate marker placements are represented as vertical bars along the x axis.
Evidence for Linkage to 11q14.
The linkage to 11q14 at D11S2002 (LOD = 4.5) in the 16 African-American pedigrees was the most significant result identified. This is one of the largest effects reported to date using SLE as the phenotype (Figs. 2 and 3). The two-point LOD scores were obtained by using our standard screening dominant model with sex-specific penetrances (92 and 49% in females and males, respectively). Maximizing the parameters of this dominant model produced a multipoint LOD of 4.7, with penetrance values of 99 and 35% in the females and males, respectively, at θ = 0 with 100% homogeneity, and a disease allele frequency of 2%. Because three different racial groupings were used in the analysis, we performed a reverse Bonferroni correction to correct for multiple comparisons. The original LOD of 4.7 (P = 1.64 × 10−6), once corrected (P = 4.93 × 10−6), still surpassed the threshold for significance in a genome-wide scan (36).
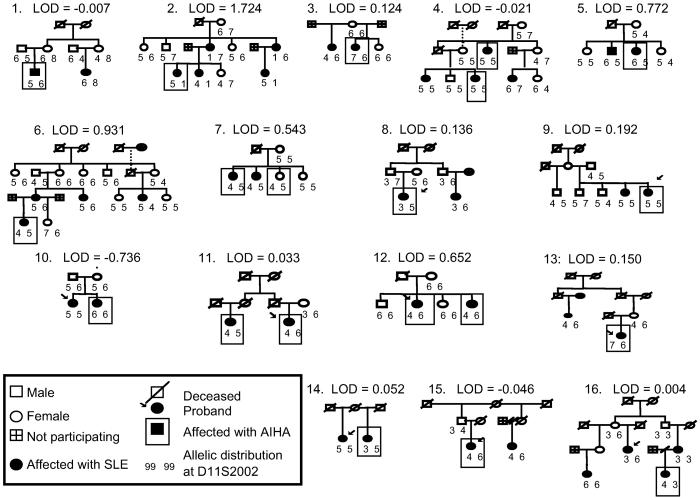
Figure 2.
Pedigree diagrams of the 16 African-American pedigree subset with their contribution to linkage in 11q14 at D11S2002.
Figure 3.
Chromosome 11 showing maximum two-point LOD at D11S2002. Dashed horizontal lines, threshold for significant and suggestive linkage (LOD = 3.3 and 1.9); dashed line, European-American pedigrees; thick solid line, African-American pedigrees; thin solid line, all pedigrees. Approximate marker placements are represented as vertical bars along the x axis.
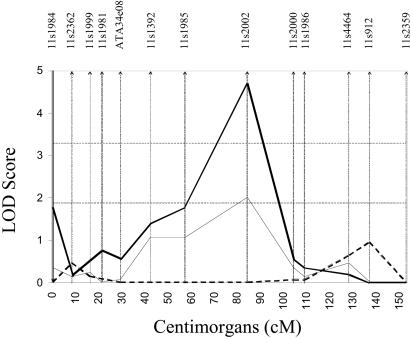
Coincidently, the markers placed in the 11q14 region were separated widely (≈20 cM between markers). We therefore typed an additional 14 markers with an average spacing of 4.2 cM in the 16 African-American pedigrees (Fig. 4). The maximum two-point LOD score remained at D11S2002. The two markers flanking D11S2002, D11S937 telomeric to and D11S901 centromeric to the main linkage effect, both achieved LOD scores of 3 using the maximized model (LOD = 3.1 and 3.0, respectively), further strengthening the evidence for an SLE susceptibility gene in this region.
Figure 4.
Fine mapping on chromosome 11. Dashed horizontal lines, threshold for significant and suggestive linkage (LOD = 3.3 and 1.9); thick sold line, the screening dominant model with penetrance values of 92 and 49% in females and males, respectively; thin double lines, the maximized dominant model with penetrance values of 99 and 35% in females and males, respectively. Only those markers used for fine mapping were maximized by using this model. Approximate marker placements are represented as vertical bars along the x axis.
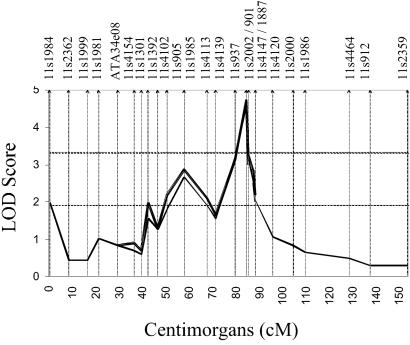
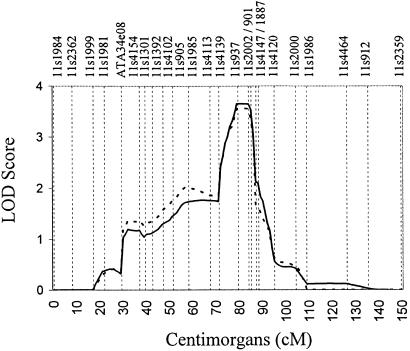
Multipoint HLOD score analysis provided further evidence of a susceptibility gene residing in this region. By using the maximized dominantly inherited model, the multipoint HLOD score was maximized at D11S2002 (HLOD = 3.6) with 63% of the families estimated to be linked to this region (Fig. 5). The one-unit-stepdown 95% support interval for the linked region reduced the region of interest from >20 cM in the initial screening of chromosome 11 to a 13-cM region at 11q14.
Figure 5.
Multipoint HLOD scores on chromosome 11 within the 16 African-American pedigrees (HLOD = 3.56). Thick solid line, the screening dominant model with penetrance values of 92 and 49% in females and males, respectively; thick dashed line, the maximized dominant model with penetrance values of 99 and 35% in females and males, respectively. Approximate marker placements are represented as vertical bars along the x axis.
We used a resampling study to determine whether the results obtained for the 16 African-American pedigrees selected for AIHA were coincidental. We performed two simulation experiments directed at assessing the stratification strategy. We first randomly selected 16 pedigrees from the 160 available pedigrees 10,000 times without regard to ethnicity and calculated the LOD score at D11S2002 for each resampled set of 16 to determine an empirical distribution of LOD score results. We repeated this except the 16 were selected randomly from the 56 available African-American pedigrees. For the resampling regardless of ethnicity, we observed a mean LOD = 0.6 (SD ± 0.60). For the resampling specifically of African-American pedigrees, we observed a mean LOD = 0.9 (SD ± 0.86). When comparing the mean LOD of 0.9 to the LOD of 4.5 in the African-American hemolytic anemia pedigrees, a statistically significant difference was obtained (χ2 = 19.0, 1df, P = 1.3 × 10−5). The results show that the screening LOD of 4.5 is unlikely to have occurred by chance and support the possible biological importance of stratification on race and hemolytic anemia.
Discussion
We selected a group of pedigrees based on the presence of hemolytic anemia with the purpose of decreasing genetic heterogeneity and identifying significant linkages involved in SLE susceptibility. Two significant linkages were identified: at 1q24 in the 17 European-American pedigrees and 11q14 in the 16 African-American pedigrees. The evidence of linkage on chromosome 11 was refined and strengthened by fine mapping. The data suggest the presence of an SLE susceptibility locus (SLEH1) on chromosome 11q14 in African-American pedigrees with at least one SLE-affected patient with hemolytic anemia.
AIHA is present in 7–15% of SLE patients (42–45). The present study identified 40 SLE patients with hemolytic anemia from a total of 374 (10.7%). Thus, the frequency of hemolytic anemia is not different from that reported in sporadic lupus.
Using the 35 pedigrees characterized by hemolytic anemia, we identified 15 linkage effects (using both maximum-likelihood model-based methods and the conditional logistic regression analysis) that surpassed the threshold of suggestive linkage (LOD ≥ 1.9), with two surpassing the threshold for established linkage (LOD ≥ 3.3). Only 2 of the 15 linkage effects were identified in all 35 pedigrees rather than dominating in one racial subset.
Three linkages identified in the present scan support linkages identified by other independent studies using SLE as the phenotype. The linkage effect at chromosome 1q21-22 with FcγRIIA was one of the most significant linkages identified using both the maximum-likelihood model-based method (LOD = 2.3) as well as the affected relative pair analysis (LOD = 3.0). Linkage to FcγRIIA was reported previously in 31 African-American pedigrees (30) and, in largely the same pedigrees, was observed later in a region of linkage identified by Gray-McGuire et al. (33). In the present study, the linkage to FcγRIIA was identified predominately in the 17 European-American pedigrees but also was highly supported by all pedigrees when analyzed together.
A modest linkage effect at 5p15 was reported first by Gray-McGuire et al. (33) in all pedigrees. However, the linkage with D5S817 (LOD = 2.1) in the present study is predominantly in the 16 African-American pedigrees. Gray-McGuire et al. (33) also reported epistasis between this marker and one at 4p16-15. Linkage at D5S1492 was identified independently in a collection of Swedish pedigrees (34), but this effect is of weaker magnitude and is 14 cM telomeric to the linkage effect observed in the present study on chromosome 5.
A possible linkage to 6p12 at D6S2410 (LOD = 2.3) was identified in the European-American-affected relative pairs. This effect is just centromeric to the HLA region, a region intensively studied by the lupus genetics group in Minnesota (29, 32). Although not a marker explicitly studied by the Minnesota group, D6S2410 is only 0.5 cM from their linkage effect (D6S257 with LOD = 3.1), thereby providing further evidence for linkage to this very interesting region of the genome.
Suggestive linkage to D11S2002 (LOD = 2.1) was observed first by Moser et al. (30) in 31 African-American multiplex pedigrees that were not selected for a particular clinical manifestation. However, when 25 additional pedigrees were added, the evidence for linkage in 11q14 decreased to LOD = 1.3 at D11S2002. Two comparisons support the important contribution made by pedigree stratification in the present study. First, the 40 African-American pedigrees not containing an SLE-affected patient with hemolytic anemia produced no evidence for linkage (LOD = −12.6, as the highest LOD score produced under the six screening models). This result is in stark contrast to the LOD of 4.5 produced by the 16 African-American pedigrees containing an SLE-affected patient with hemolytic anemia. Second, the LOD = 3.2 difference between the LOD of 1.3 in the total 56 African-American pedigrees and the LOD of 4.5 in the 16 African-American pedigrees stratified by hemolytic anemia is substantial. Indeed, both comparisons strongly suggest that the stratification strategy presented herein was successful in revealing a previously unidentified linkage at the 11q14 region.
Two genes mapped to the 11q14 region have mechanisms similar to genes already known to be involved in SLE pathophysiology: BCL1 and FADD. BCL1, a gene involved in chronic lymphatic leukemia (CLL), is an interesting candidate gene, because CLL has been associated with SLE as well as other autoimmune diseases (61). FADD, or FAS-associated via death domain, interacts with FAS, a cell surface cytokine receptor involved in apoptosis. Overexpression of FADD in mammalian cells induces apoptosis (Online Mendelian Inheritance in Man accession no. for FADD is MIM 602457; see http://www.ncbi.nlm.nih.gov/omim), and apoptosis has long been implicated in the pathogenesis of SLE (62).
The linkages identified herein are lupus genes, because they were identified in pedigrees multiplex for lupus, and the analysis is performed by using lupus as the phenotype. Nonetheless, hemolytic anemia must be related to these genes in some way, because the pedigrees were selected based on an SLE-affected patient having a history of hemolytic anemia. None of the regions demonstrating linkage in the present study contained genes known to be linked to hemolytic anemia (AK1 at 9q31.12, G6PD at Xq28, HUS at 1q32, PFKL at 21q22.3, and PGK1 at Xq13.3; see www.citi2.fr/cgi-bin/mug?tex1=hemolytic+anemia&category=0&phenotype=0&classification=0&cloned= 0&linked=0). This is not surprising, however, because lupus was used as the phenotype.
In conclusion, we present evidence of an SLE susceptibility locus at 11q14 in African-American SLE pedigrees stratified on hemolytic anemia. The strategy presented herein demonstrates that pedigree selection used to decrease clinical heterogeneity may increase the genetic homogeneity in a complex phenotype and substantially improve the likelihood of identifying significant genetic effects.
Acknowledgments
We thank all the patients and family members who participated in this study as well as the many referring physicians. The comments of two anonymous reviewers substantially improved the resulting paper. This work was supported by National Institutes of Health Grants AR12253, AI24717, AR42460, AR45231, AI31584, and RR15577, and a grant from the U.S. Department of Veterans Affairs. Some of the results of this paper were obtained by using the program package S.A.G.E., which is supported by U.S. Public Health Service Resource Grant 1 P41 RR03655 from the National Center for Research Resources. Ninety pedigrees (cohorts A, B, and C) were obtained from the Lupus Multiplex Registry and Repository (AR-1-2253; see http://omrf.ouhsc.edu/lupus).
Abbreviations
- SLE
systemic lupus erythematosus
- LOD
logarithm of odds
- AIHA
autoimmune hemolytic anemia
- cM
centimorgan(s)
- HLOD
heterogeneity LOD
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Kelly, J., Gray-McGuire, C., Salmon, J., Neas, B. R., Bruner, G. R., Behrens, T., Moser, K. L. & Harley, J. B. (2001) Lupus 10, Suppl. 1, 39 (abstr.).
References
- 1.Hochberg M C. In: Dubois' Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 49–65. [Google Scholar]
- 2.Fessel W J. Arch Intern Med (Moscow) 1974;134:1027–1035. [PubMed] [Google Scholar]
- 3.Kaslow R A, Masi A T. Arthritis Rheum. 1978;21:473–479. doi: 10.1002/art.1780210412. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence R C, Helmick C G, Arnett F C, Deyo R A, Felson D T, Giannini E H, Heyse S P, Hirsch R, Hochberg M C, Hunder G G, et al. Arthritis Rheum. 1998;41:778–799. doi: 10.1002/1529-0131(199805)41:5<778::AID-ART4>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Sestak A L, Shaver T S, Moser K L, Neas B R, Harley J B. J Rheumatol. 1999;26:1495–1499. [PubMed] [Google Scholar]
- 6.Hochberg M C. J Rheumatol. 1987;14:867–869. [PubMed] [Google Scholar]
- 7.Vyse T J, Todd J A. Cell. 1996;85:311–318. doi: 10.1016/s0092-8674(00)81110-1. [DOI] [PubMed] [Google Scholar]
- 8.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack T M. Arthritis Rheum. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 9.Arnett F C. In: Dubois' Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 77–117. [Google Scholar]
- 10.Duits A J, Bootsma H, Derksen R H, Spronk P E, Kater L, Kallenberg C G, Capel P J, Westerdaal N A, Spierenburg G T, Gmelig-Meyling F H, et al. Arthritis Rheum. 1995;39:1832–1836. doi: 10.1002/art.1780381217. [DOI] [PubMed] [Google Scholar]
- 11.Salmon J E, Millard S, Schachter L A, Arnett F C, Ginzler E M, Gourley M F, Ramsey-Goldman R, Peterson M G, Kimberly R P. J Clin Invest. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmon J E, Ng S, Yoo D H, Kim T H, Kim S Y, Song G G. Arthritis Rheum. 1999;42:818–819. doi: 10.1002/1529-0131(199904)42:4<818::aid-anr28>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Edberg J C, Redecha P B, Bansal V, Guyre P M, Coleman K, Salmon J E, Kimberly R P. J Clin Invest. 1997;100:1059–1070. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walport M J, Lachmann P J. Curr Opin Rheumatol. 1990;2:661–663. doi: 10.1097/00002281-199002040-00018. [DOI] [PubMed] [Google Scholar]
- 15.Bowness P, Davies K A, Norsworthy P J, Athanassiou P, Taylor-Wiedeman J, Borysiewicz L K, Meye P A, Walport M J. Q J Med. 1994;87:455–464. [PubMed] [Google Scholar]
- 16.Ratnoff W D. Rheum Dis Clin N Am. 1996;22:75–94. doi: 10.1016/s0889-857x(05)70263-5. [DOI] [PubMed] [Google Scholar]
- 17.Schur P H. In: Dubois' Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 245–261. [Google Scholar]
- 18.Drake C G, Babcock S K, Palmer E, Kotzin B L. Proc Natl Acad Sci USA. 1994;91:4062–4066. doi: 10.1073/pnas.91.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kono D H, Burlingame R W, Owens D G, Kuramochi A, Balderas R S, Balomenos D, Theofilopoulos A N. Proc Natl Acad Sci USA. 1994;91:10168–10172. doi: 10.1073/pnas.91.21.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morel L, Rudofsky U H, Longmate J A, Schiffenbauer J, Wakeland E K. Immunity. 1994;1:219–229. [PubMed] [Google Scholar]
- 21.Drake C G, Rozzo S J, Hirschfeld H F, Smarnworawong N P, Palmer E, Kotzin B L. J Immunol. 1995;154:2441–2447. [PubMed] [Google Scholar]
- 22.Vyse T J, Rozzo S J, Drake C G, Izui S, Kotzin B L. J Immunol. 1997;158:5566–5574. [PubMed] [Google Scholar]
- 23.Hogarth M B, Slingsby J H, Allen P J, Thompson E M, Chandler P, Davies K A, Simpson E, Morley B J, Walport M J. J Immunol. 1998;161:2753–2761. [PubMed] [Google Scholar]
- 24.Vidal S, Kono D H, Theofilopoulos A N. J Clin Invest. 1998;101:696–702. doi: 10.1172/JCI1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morel L, Tian X H, Croker B P, Wakeland E K. Immunity. 1999;11:131–139. doi: 10.1016/s1074-7613(00)80088-6. [DOI] [PubMed] [Google Scholar]
- 26.Haywood M E, Hogarth M B, Slingsby J H, Rose S J, Allen P J, Thompson E M, Maibaum M A, Chandler P, Davies K A, Simpson E, et al. Arthritis Rheum. 2000;43:349–355. doi: 10.1002/1529-0131(200002)43:2<349::AID-ANR14>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 27.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F, Schaller J G, Talal N, Winchester R J. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 28.Hochberg M C. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 29.Gaffney P M, Kearns G M, Shark K B, Ortmann W A, Selby S A, Malmgren M L, Rohlf K E, Ockenden T C, Messner R P, King R A, et al. Proc Natl Acad Sci USA. 1998;95:14875–14879. doi: 10.1073/pnas.95.25.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser K L, Neas B R, Salmon J E, Yu H, Gray-McGuire C, Asundi N, Bruner G R, Fox J, Kelly J, Henshall S, et al. Proc Natl Acad Sci USA. 1998;95:14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shai R, Quismorio F P, Jr, Li L, Kwon O J, Morrison J, Wallace D J, Neuwelt C M, Brautbar C, Gauderman W J, Jacob C O. Hum Mol Genet. 1999;8:639–644. doi: 10.1093/hmg/8.4.639. [DOI] [PubMed] [Google Scholar]
- 32.Gaffney P M, Ortmann W A, Selby S A, Shark K B, Ockenden T C, Rohlf K E, Walgrave N L, Boyum W P, Malmgren M L, Miller M E, et al. Am J Hum Genet. 2000;66:547–556. doi: 10.1086/302767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray-McGuire C, Moser K L, Gaffney P, Kelly J, Yu H, Olson J M, Jedrey C M, Jacobs K B, Kimberly R P, Neas B R, et al. Am J Hum Genet. 2000;67:1460–1469. doi: 10.1086/316891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindqvist A K, Steinsson K, Johanneson B, Kristjansdottir H, Arnasson A, Grondal G, Jonasson I, Magnusson V, Sturfelt G, Truedsson L, et al. J Autoimmun. 2000;14:169–178. doi: 10.1006/jaut.1999.0357. [DOI] [PubMed] [Google Scholar]
- 35.Nath S K, Kelly J A, Namjou B, Lam T, Bruner G R, Scofield R H, Aston C E, Harley J B. Am J Hum Genet. 2001;69:1401–1406. doi: 10.1086/324470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander E, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 37.Lander E S. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 38.Chakravarti A. Nat Genet. 1999;21, Suppl. 1:56–60. doi: 10.1038/4482. [DOI] [PubMed] [Google Scholar]
- 39.Quismorio F P., Jr . In: Dubois' Lupus Erythematosus. Wallace D J, Hahn B H, editors. Baltimore: Williams & Wilkins; 1997. pp. 793–816. [Google Scholar]
- 40.Shearn M A, Pirofsky B. Arch Intern Med (Moscow) 1952;90:790–807. doi: 10.1001/archinte.1952.00240120065007. [DOI] [PubMed] [Google Scholar]
- 41.Budman D R, Steinberg A D. Arch Intern Med (Moscow) 1977;86:220–229. doi: 10.7326/0003-4819-86-2-220. [DOI] [PubMed] [Google Scholar]
- 42.Alger M, Alarcon-Segovia D, Rivero S J. J Rheumatol. 1977;4:351–357. [PubMed] [Google Scholar]
- 43.Rothfield N F. Dis Mon. 1982;29:1–62. doi: 10.1016/0011-5029(82)90009-8. [DOI] [PubMed] [Google Scholar]
- 44.Nossent J C, Swaak A J G. Q J Med. 1991;80:605–612. [PubMed] [Google Scholar]
- 45.Reeves W H, Richards H B, Satoh M. In: Textbook of the Autoimmune Diseases. Lahita R G, Chiorazzi N, Reeves W H, editors. Williams & Wilkins, Philadelphia: Lippincott; 2000. pp. 81–100. [Google Scholar]
- 46.Engelfriet C P, Overbeeke M A, von dem Bourne A E. Semin Hematol. 1992;29:3–12. [PubMed] [Google Scholar]
- 47.Dubois E L. Am J Med. 1952;22:197–204. doi: 10.1016/0002-9343(52)90212-x. [DOI] [PubMed] [Google Scholar]
- 48.Videbaek A. Acta Med Scand. 1962;171:187–194. doi: 10.1111/j.0954-6820.1962.tb04180.x. [DOI] [PubMed] [Google Scholar]
- 49.Bignell G R, Canzian F, Shayeghi M, Stark M, Shugart Y Y, Biggs P, Mangion J, Hamoudi R, Rosenblatt J, Buu P, et al. Am J Hum Genet. 1997;61:1123–1130. doi: 10.1086/301610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neumann S, Willgerodt H, Ackermann F, Reske A, Jung M, Reis A, Paschke R. J Clin Endocrinol Metab. 1999;84:3750–3756. doi: 10.1210/jcem.84.10.6023. [DOI] [PubMed] [Google Scholar]
- 51.Rapley E A, Crockford G P, Teare D, Biggs P, Seal S, Barfoot R, Edwards S, Hamoudi R, Heimdal K, Fossa S D, et al. Nat Genet. 2000;24:197–200. doi: 10.1038/72877. [DOI] [PubMed] [Google Scholar]
- 52.McKay J D, Lesueur F, Jonard L, Pastore A, Williamson J, Hoffman L, Burgess J, Duffield A, Papotti M, Stark M, et al. Am J Hum Genet. 2001;69:440–446. doi: 10.1086/321979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokori S I G, Ioannidis J P A, Voulgarelis M, Tzioufas A G, Moutsopoulos H M. Am J Med. 2000;108:198–204. doi: 10.1016/s0002-9343(99)00413-1. [DOI] [PubMed] [Google Scholar]
- 54.Olson J M. Am J Hum Genet. 1999;64:1464–1472. doi: 10.1086/302360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Department of Epidemiology and Biostatistics. S.A.G.E., Statistical Analysis for Genetic Epidemiology. Cleveland: Case Western Reserve University; 1999. , Release 4.0, Beta 3. [Google Scholar]
- 56.Cottingham R W, Jr, Idury R M, Schaffer A A. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 57.Schaffer A A, Gupta S K, Shriram K, Cottingham R W., Jr Hum Hered. 1994;44:225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger J D. Am J Hum Genet. 1995;56:777–787. [PMC free article] [PubMed] [Google Scholar]
- 59.Olson J M. Am J Hum Genet. 1999;65:1760–1769. doi: 10.1086/302662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong A, Cox N J. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Conley C L, Misiti J, Laster A J. Medicine (Baltimore) 1980;59:323–334. doi: 10.1097/00005792-198009000-00001. [DOI] [PubMed] [Google Scholar]
- 62.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Nature (London) 1992;26:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]