Abstract
Tissues from rhesus monkeys were screened by PCR for the presence of sequences homologous to known adeno-associated virus (AAV) serotypes 1–6. DNA spanning entire rep-cap ORFs from two novel AAVs, called AAV7 and AAV8, were isolated. Sequence comparisons among these and previously described AAVs revealed the greatest divergence in capsid proteins. AAV7 and AAV8 were not neutralized by heterologous antisera raised to the other serotypes. Neutralizing antibodies to AAV7 and AAV8 were rare in human serum and, when present, were low in activity. Vectors formed with capsids from AAV7 and AAV8 were generated by using rep and inverted terminal repeats (ITRs) from AAV2 and were compared with similarly constructed vectors made from capsids of AAV1, AAV2, and AAV5. Murine models of skeletal muscle and liver-directed gene transfer were used to evaluate relative vector performance. AAV7 vectors demonstrated efficiencies of transgene expression in skeletal muscle equivalent to that observed with AAV1, the most efficient known serotype for this application. In liver, transgene expression was 10- to 100-fold higher with AAV8 than observed with other serotypes. This improved efficiency correlated with increased persistence of vector DNA and higher number of transduced hepatocytes. The efficiency of AAV8 vector for liver-directed gene transfer of factor IX was not impacted by preimmunization with the other AAV serotypes. Vectors based on these novel, nonhuman primate AAVs should be considered for human gene therapy because of low reactivity to antibodies directed to human AAVs and because gene transfer efficiency in muscle was similar to that obtained with the best known serotype, whereas, in liver, gene transfer was substantially higher than previously described.
Adeno-associated viruses (AAV) have been isolated from a number of species, including primates (1). They belong to the Parvoviridae family and require helper viruses such as adenovirus to replicate. Six primate AAVs have been isolated, and five have been determined to be distinct serotypes based on antibody crossreactivity studies (2–8). AAV6 appears to be a recombinant between AAV1 and AAV2 (9). All primate AAVs were isolated initially as contaminants in preparations of adenoviruses except for AAV5, which was recovered from a human condylomatous wart (2–8). Seroepidemiologic studies indicate that AAV serotypes 2, 3, and 5 are endemic to humans whereas AAV4 primarily infects nonhuman primates (2–8, 10). The reservoir for AAV1 (and the associated AAV6 species) is unclear because it has not been primarily isolated from tissues and reactive antibodies exist in both humans and nonhuman primates (10–13).
The isolation of a molecular clone for AAV2 in 1983 by Samulski et al. facilitated the development of recombinant vectors for somatic gene transfer (14). High titer stocks of AAV2-based vectors, devoid of all AAV ORFs, were created and evaluated in preclinical models of in vivo gene therapy. Several themes have emerged from these studies. In tissues such as liver, muscle, retina and the central nervous system, AAV2-mediated gene transfer confers extremely stable transgene expression (15). Recipient animals do not elicit T cell responses to most AAV-encoded transgene products, even if these proteins contain foreign epitopes. This phenomenon is believed to be due to an inability of AAV2 to infect antigen-presenting cells (16). The AAV genome appears to persist in a number of different integrated and nonintegrated forms after in vivo gene therapy (17–19).
Despite the impressive longevity of transgene expression obtained with AAV2, its application has been limited because of low levels of transgene expression. Blocks at the level of vector entry and post entry processing contribute to these inefficiencies (20, 21). Progress in overcoming these barriers has been made through the development of vectors based on other serotypes that enter the cell via receptors distinct from those that recognize AAV2. We showed that AAV1 vectors very efficiently transduce skeletal muscle (9) and retina (22) whereas others demonstrated high-level transduction of the central nervous system and lung with AAV5 vectors (23, 24).
In this report, we describe the isolation of two novel AAVs, AAV7 and AAV8, and their use as vectors for somatic gene transfer.
Methods
Isolation of AAV7 and AAV8 Sequences from Nonhuman Primate Tissues.
In an attempt to isolate novel AAV sequences from nonhuman primate tissues, published AAV sequences including primate AAV1–AAV6 and AAVs from duck and goose origins were aligned for comparison by using the clustal w program. A stretch of AAV sequence spanning 2,886 to 3,143 bp of AAV1 and corresponding sequences in AAV2–AAV6 and AAVs from duck and goose origins was selected as a PCR amplicon. This region is about 255 bp in length in which both 5′ and 3′ sequences are highly conserved, but the middle sequence is variable and unique to each known AAV serotype (called the “signature region”). A pair of universal primers that can anneal to the 5′ and 3′ ends of this signature region were designed for PCR to amplify from total DNAs isolated from various tissues of rhesus monkeys. Sequences for the primers are 5′-GGTAATGCCTCAGGAAATTGGCATT-3′ (19s) and 5′-GACTCATCAACAACAATTGGGGATTC-3′ (18as), respectively. The first novel AAV signature sequence isolated, named AAV serotype 7, was isolated by PCR amplification of heart DNA of a rhesus monkey (98E044) that was previously treated with an adenovirus vector for study of innate immune response (25). A second novel AAV sequence was obtained from heart DNA of another rhesus monkey (98E056) in the same adenovirus study, and was designated AAV serotype 8. Identical signature sequences were isolated from several tissues of rhesus monkeys from the facility at the University of Pennsylvania as well as animals derived from the Primate Center at Tulane University.
To extend cap gene sequences of AAV7 and AAV8, two other highly conserved regions were identified for use in PCR amplification. One region is ≈1.7 kb upstream of the signature region whereas the other is located 1.5 kb downstream. A primer within the upstream conserved region was selected (AV1Ns: 5′-gctgcgycaactggaccaatgagaac-3′) in combination with the 3′ primer (18as) in the signature region to extend AAV7 and AAV8 sequence amplification to 5′ of the cap gene and the junction of rep/cap genes, followed by Topo cloning of 1.7-kb fragments. Another primer located in a conserved region at the end of the cap gene (AV2Cas: 5′-cgcagagaccaaagttcaactgaaacga-3′) was used as 3′ primer to pair with the 5′ primer of the signature region (19s) to reamplify heart DNAs of the two monkeys, resulting in isolation of 1.5-kb fragments. Using the overlapping 255-bp signature sequence as an anchor, the 5′ fragment of 1.7-kb fragment was fused with the 3′ fragment of 1.5 kb at a DraIII site to form the fusion plasmids containing the complete cap genes of AAV7 and AAV8. The fusion plasmids were designated as pCRAAV7-Fusion and pCRAAV8-Fusion. A similar strategy was used to isolate the corresponding rep genes. The full sequences for rep and cap of AAV7 and AAV8 have been submitted to GenBank (accession numbers: AAV7, AF513851; and AAV8, AF513852).
Directly amplifying, cloning, and sequencing full-length cap sequences (i.e., 2.8-kb fragments) from the same monkey further confirmed cap structure for AAV7 and AAV8. The primers used for confirmation of AAV7 are 7VRF-1s, 5′-ccttcgagcaccagcagccgtt-3′ and 7VRF-2as, aggctcagagtaaacaccctggctgtca, whereas the primers for the AAV8 amplicon are 8VRF-2s, 5′-ccgcatctaccgcatcctcgct-3′ and 8VRF-2as, gttccaatttgaggagccgtgttttgct-3′.
Production of AAV Vectors.
A pseudotyping strategy was used to produce AAV vectors packaged with AAV7 and AAV8 capsid proteins (26). Recombinant AAV genomes equipped with AAV2 inverted terminal repeats (ITRs) were packaged by triple transfection of 293 cells with cis-plasmid, adenovirus helper plasmid and a chimeric packaging construct where the AAV2 rep gene is fused with cap genes of novel AAV serotypes. To create the chimeric packaging constructs, the XhoI site of p5E18 plasmid at 3,169 bp was ablated, and the modified plasmid was restricted with XbaI and XhoI in a complete digestion to remove the AAV2 cap gene and replace it with a 2,267-bp SpeI/XhoI fragment containing either AAV7 or AAV8 cap gene (9). A similar cloning strategy was used for creation of chimeric packaging plasmids of AAV2/1 and AAV2/5. All recombinant vectors were purified by the standard CsCl2 sedimentation method except for AAV2/2, which was purified by single step heparin chromatography.
Genome copy (GC) titers of AAV vectors were determined by TaqMan (Applied Biosystems) analysis by using probes and primers targeting SV40 poly(A) region as described previously (27).
Serologic Analysis.
C57BL/6 mice were injected with vectors of different serotypes intramuscularly (5 × 1011 GC), and serum samples were collected 34 days later and analyzed for the presence of neutralizing antibodies by assessing the ability of serum to inhibit transduction of 84-31 cells by reporter viruses (AAVCMVEGFP) of different serotypes (28). Serum samples were also kindly provided from J. Samulski (University of North Carolina at Chapel Hill) from animals that received two i.p. injections of vectors of different serotypes. Specifically, the reporter virus AAVCMVEGFP of each serotype (at multiplicity of infection equal to 105 genome copies/cell) was preincubated with heat-inactivated serum from animals that were immunized against different serotypes of AAV or from naive mice. After 1 h incubation at 37°C, viruses were added to 84-31 cells in 96-well plates for 48 or 72 h, depending on the virus serotype. The number of green fluorescent protein-expressing cells was assessed by fluorescent microscopy. Neutralizing antibody titers were reported as the highest serum dilution that inhibited transduction by 50% of that seen with serum from a naive animal.
In Vivo Evaluation of Different Serotypes of AAV Vectors.
In this study, seven recombinant AAV genomes, AAV2CBhA1AT, AAV2AlbhA1AT, AAV2CMVrhCG, AAV2TBGrhCG, AAV2TBGcFIX, AAV2CMVLacZ, and AAV2TBGLacZ were packaged with capsid proteins of different serotypes. In all seven constructs, minigene cassettes were flanked with AAV2 ITRs. cDNAs of human α-antitrypsin (A1AT; ref. 9), β-subunit of rhesus monkey choriogonadotropic hormone (CG; ref. 29), canine factor IX (30), and bacterial β-galactosidase (i.e., LacZ) genes were used as reporter genes. For liver-directed gene transfer, either mouse albumin gene promoter (Alb; ref. 9) or human thyroid hormone binding globulin gene promoter (TBG; ref. 30) was used to drive liver specific expression of reporter genes. In muscle-directed gene transfer experiments, either cytomegalovirus early promoter (CMV) or chicken β-actin promoter with CMV enhancer (CB) was used to direct expression of reporters.
For muscle-directed gene transfer, vectors were injected into the right tibialis anterior of 4- to 6-wk-old NCR nude or C57BL/6 mice (Taconic Farms). In liver-directed gene transfer studies, vectors were infused intraportally into 7- to 9-wk-old NCR nude or C57BL/6 mice (Taconic Farms). Serum samples were collected retroorbitally at different time points after vector administration. Muscle and liver tissues were harvested at different time points for cryosectioning and 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) histochemical staining from animals that received the LacZ vectors. For the readministration experiment, C56BL/6 mice initially received AAV2/1, 2/2, 2/5, 2/7, and 2/8 vectors expressing hA1AT driven by the CB promoter intramuscularly and followed for A1AT gene expression for 7 wk. Animals were then treated with AAV2/8TBGcFIX intraportally and studied for cFIX gene expression.
ELISA-based assays were performed to quantify serum levels of hA1AT, rhesus monkey CG (rhCG), and cFIX proteins as described previously (28–30). The experiments were completed when animals were killed for harvest of muscle and liver tissues for DNA extraction and quantitative analysis of genome copies of vectors present in target tissues by TaqMan using the same set of primers and probe as in titration of vector preparations (31).
Results
Isolation of AAV7 and AAV8 from Nonhuman Primate Tissues.
Our strategy was to use molecular techniques to isolate novel AAV genomes from tissues of latently infected nonhuman primates. A DNA sequence spanning 255 bp of the cap gene was selected as a PCR amplicon for the recovery of novel AAV genomes. This region has highly conserved 5′ and 3′ sequences and a middle variable sequence that is unique to each known serotype. The middle region was designated the signature region.
Rhesus monkey tissues were harvested at necropsy, and DNA was prepared and subjected to PCR amplification by using oligonucleotides spanning the conserved sequences of the signature region. Fragments amplified by using this strategy were cloned and sequenced. DNA representing two novel AAV genomes, called AAV7 and AAV8, were found in multiple tissues of several animals obtained from two different sources.
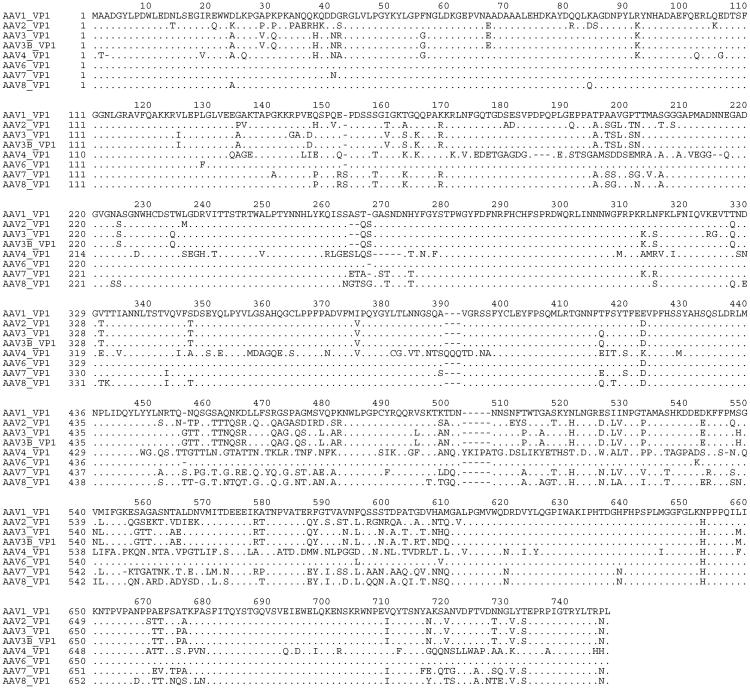
AAV7 and AAV8 genomes containing contiguous sequence across rep and cap ORFs were obtained by amplification of overlapping fragments using oligonucleotides to conserved regions within rep and cap. This sequence has been deposited into GenBank [accession nos. AF513851 (AAV7) and AF513852 (AAV8)]. The predicted amino acid sequences for capsid proteins of AAV7 and AAV8 are compared with the other known serotypes in Fig. 1. Comparisons of nucleotide and amino acid sequences between AAVs 1–8 revealed divergence primarily in capsid (Table 1). Comparisons of the capsid for AAV7 to the other serotypes (except serotype 5, which is highly divergent) revealed 63% to 85% identity for amino acid sequence and 68% to 84% identity in nucleotide sequence. Similar results were obtained for AAV8.
Figure 1.
Predicted amino acid sequences for capsid protein VP1 of AAV1, -2, -3A, -3B, -4, -6, -7, and -8. Amino acid sequences of capsid protein VP1 from AAV1 to -4 and AAV6 to -8 were aligned for comparison by using the clustal w program. AAV5 sequence was excluded from the alignment because of its strong divergence from other serotypes. The dots in the alignment represent the amino acids that are identical to those of AAV1 VP1. The dashes indicate the amino acids that are missing at the positions in the alignment.
Table 1.
Percentage of sequence similarity between VP1 coding regions of AAV serotypes 1, 2, 3, 4, 6, 7, and 8
| AAV1_VP1 | AAV2_VP1 | AAV3_VP1 | AAV3B_VP1 | AAV4_VP1 | AAV6_VP1 | AAV7_VP1 | AAV8_VP1 | |
|---|---|---|---|---|---|---|---|---|
| AAV1_VP1 | 100 | 83 | 86 | 87 | 63 | 99 | 85 | 84 |
| AAV2_VP1 | 80 | 100 | 88 | 88 | 60 | 83 | 82 | 83 |
| AAV3_VP1 | 80 | 82 | 100 | 99 | 62 | 86 | 84 | 85 |
| AAV3B_VP1 | 80 | 82 | 100 | 100 | 63 | 87 | 84 | 86 |
| AAV4_VP1 | 68 | 66 | 68 | 68 | 100 | 63 | 63 | 63 |
| AAV6_VP1 | 97 | 80 | 80 | 80 | 68 | 100 | 85 | 84 |
| AAV7_VP1 | 84 | 78 | 80 | 80 | 68 | 84 | 100 | 88 |
| AAV8_VP1 | 83 | 79 | 80 | 80 | 68 | 83 | 86 | 100 |
Multiple sequence alignments of nucleic acid and amino acid of VP1 of different serotype AAVs were performed using the VECTOR NTI 6.0 (Informax, Bethesda, MD) program. AAV5 sequence was excluded from the alignment because of its strong divergence from other serotypes. Bold numbers represent similarity in amino acid sequences whereas light face numbers indicate similarity in nucleic acid sequences. The underlined numbers represent similarity in both.
Creation of Vectors Based on AAV7 and AAV8 and Serologic Analyses.
Vectors based on AAV7 and AAV8 were developed as described for other alternative serotypes as follows. The AAV2 capsid was exchanged with capsid encoding AAV7 or AAV8 in an AAV2 rep-cap-packaging plasmid. This hybrid construct was cotransfected with a vector plasmid containing AAV2 ITRs flanking the transgene, together with a plasmid containing the necessary adenovirus helper genes. This procedure results in the production of chimeric virions, called AAV2/7 and AAV2/8 vectors, in which AAV2 vector genomes are packaged into heterologous capsids. Similar constructs were created for AAV1 (2/1), AAV5 (2/5), and AAV2 (2/2).
Recombinant virions were recovered by cesium chloride sedimentation in all cases except AAV2/2, which was purified by heparin chromatography. Vectors were constructed for each serotype for a number of in vitro and in vivo studies. Eight different transgene cassettes were incorporated into the vectors, and recombinant virions were produced for each serotype. The recovery of virus, based on genome copies, is summarized in Table 2. The yields of vector were high for each serotype, with no consistent differences between serotypes. Western blot analysis indicated that the ratio of capsid protein to genome copies was constant for all vector serotypes (data not shown). The relative transduction efficiency of the different serotypes on 293-based cells expressing E4 of Ad5 measured at GC/LacZ forming units varied as follows: AAV2 (6 × 102), AAV2/1 (1 × 104), AAV2/5 (3 × 104), AAV2/7 (2 × 104), and AAV2/8 (3 × 104).
Table 2.
Production of recombinant vectors
| AAV2/1 | AAV2/2 | AAV2/5 | AAV2/7 | AAV2/8 | |
|---|---|---|---|---|---|
| CMVLacZ | 7.30 ± 4.33 (n = 9) | 4.49 ± 2.89 (n = 6) | 5.19 ± 5.19 (n = 8) | 3.42 (n = 1) | 0.87 (n = 1) |
| CMVEGFP | 6.43 ± 2.42 (n = 2) | 3.39 ± 2.42 (n = 2) | 5.55 ± 6.49 (n = 4) | 2.98 ± 2.66 (n = 2) | 3.74 ± 3.88 (n = 2) |
| TBGLacZ | 4.18 (n = 1) | 0.23 (n = 1) | 0.704 ± 0.43 (n = 2) | 2.16 (n = 1) | 0.532 (n = 1) |
| Alb A1AT | 4.67 ± 0.75 (n = 2) | 4.77 (n = 1) | 4.09 (n = 1) | 5.04 (n = 1) | 2.02 (n = 1) |
| CB A1AT | 0.567 (n = 1) | 0.438 (n = 1) | 2.82 (n = 1) | 2.78 (n = 1) | 0.816 ± 0.679 (n = 2) |
| CMVrhCG | 8.78 ± 2.37 (n = 7) | 1.43 ± 1.18 (n = 2) | 1.63 ± 1.15 (n = 3) | 2.04 ± 0.67 (n = 3) | 1.32 ± 0.87 (n = 3) |
| TBGrhCG | 8.51 ± 6.65 (n = 6) | 3.47 ± 2.09 (n = 5) | 5.26 ± 3.85 (n = 4) | 6.52 ± 3.08 (n = 4) | 1.83 ± 0.98 (n = 5) |
| TBGcFIX | 1.24 ± 1.29 (n = 3) | 0.63 ± 0.394 (n = 6) | 3.74 ± 2.48 (n = 7) | 4.05 (n = 1) | 15.8 ± 15.0 (n = 5) |
Data presented in the table are average genome copy yields with standard deviation × 1013 of multiple production lots of 50 plate (150 mm) transfections. All recombinant vectors were purified by the standard CsCl2 sedimentation method except for AAV2/2, which was purified by single-step heparin chromatography. EGFP, enhanced green fluorescent protein; Alb, albumin gene promoter; CB, chicken β-actin promoter.
The availability of green fluorescent protein-expressing vectors simplified the development of an assay for neutralizing antibodies that was based on inhibition of transduction in a permissive cell line (i.e., 293 cells stably expressing E4 from Ad5). Antisera to all AAV serotypes (i.e., AAV1 to -8) were generated by intramuscular and/or i.p. injection of the recombinant viruses. The only antiserum that neutralized AAV7 was generated to AAV7, and AAV8 antiserum was the only one to neutralize AAV8 (Table 3). This result supports the proposal to designate AAV7 and AAV8 as new serotypes.
Table 3.
Neutralization of AAV2/7 and AAV2/8 with antisera directed to all serotypes of AAV
| Antisera | Vector
|
|
|---|---|---|
| AAV2/7 | AAV2/8 | |
| AAV1 | <1:20 | <1:20 |
| AAV2 | <1:20 | <1:20 |
| AAV3 | <1:20 | <1:20 |
| AAV4 | <1:20 | <1:20 |
| AAV5 | <1:20 | <1:20 |
| AAV6 | <1:20 | <1:20 |
| AAV7 | 1:160 | <1:20 |
| AAV8 | <1:20 | 1:320 |
Antisera was generated to all known serotypes in the mouse as described in Methods. The ability of these antisera to neutralize AAV2/7 and AAV2/8 vectors is presented in the table: antisera were generated at the University of Pennsylvania for serotypes 1, 2, 5, 7, and 8 and at the University of North Carolina for serotypes 1, 2, 3, 4, 5, and 6.
Human sera from 52 normal subjects were screened for neutralization against selected serotypes. A low level of neutralizing activity (i.e., ≈1:20) was detected in 3 of 52 subjects to AAV7 and 3 of 52 to AAV8, whereas substantially higher titer were observed in up to 20% of human subjects for AAV2. High titer neutralizing antibody to these serotypes was detected in rhesus monkey colonies that in the aggregate was 30% for AAV7 and 45% for AAV8.
In Vivo Performance of AAV Vectors.
The performance of vectors based on the new serotypes were evaluated in murine models of muscle and liver-directed gene transfer and compared with vectors based on the known serotypes AAV1, AAV2, and AAV5. Vectors expressing secreted proteins (A1AT, Fig. 2; and CG, Table 4) were used to quantitate relative transduction efficiencies between different serotypes through ELISA analysis of sera. The cellular distribution of transduction within the target organ was evaluated by using LacZ-expressing vectors and X-Gal histochemistry (Fig. 3).
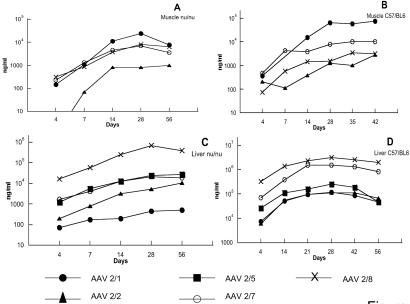
Figure 2.
Expression of a secreted protein A1AT, from AAV vectors injected into murine skeletal muscle and liver. Immune deficient (NCR nudes, A and C) and immune competent mice (C57BL/6, B and D) were treated with different serotypes of AAVCBA1AT (A and B) or AAVAlbA1AT (C and D) through intramuscular or intraportal injections, respectively, at a dose of 1 × 1011 GC per animal. Serum samples were collected retroorbitally at different time points post vector administration and assayed for A1AT concentration. The y axis represents A1AT concentration in the unit of nanogram per milliliter of serum. The x axis represents time points of different bleeds. The data were plotted as average A1AT concentrations of four animals per vector group. The A1AT level in the control animals that received PBS was under the detection limit of the assay.
Table 4.
Expression of β-unit of rhCG in mouse muscle and liver
| Vector | Muscle | Liver |
|---|---|---|
| AAV2/1 | 4.5 ± 2.1 | 1.6 ± 1.0 |
| AAV2 | 0.5 ± 0.1 | 0.7 ± 0.3 |
| AAV2/5 | ND* | 4.8 ± 0.8 |
| AAV2/7 | 14.2 ± 2.4 | 8.2 ± 4.3 |
| AAV2/8 | 4.0 ± 0.7 | 76.0 ± 22.8 |
CMVCG and TBGCG minigene cassettes were used for muscle and liver-directed gene transfer, respectively. Levels of rhCG were defined as relative units (rU × 103). The data were from assaying serum samples collected at day 28, post vector administration (four animals per group).
ND, Not determined in this experiment.
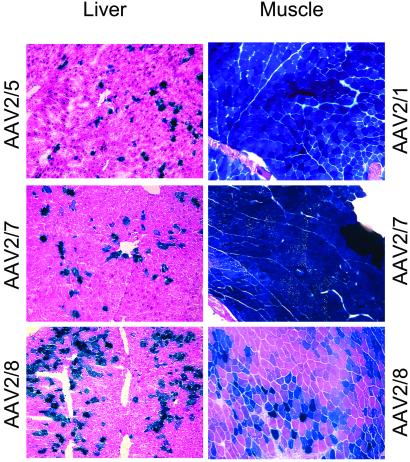
Figure 3.
X-Gal histochemistry of muscle and liver after injection of LacZ-expressing vectors. (Left, Liver) Representative tissue sections from NCR nude mice that received AAV vectors expressing LacZ driven by human thyroid hormone binding globulin gene promoter (TBG), a liver-specific promoter, at a dose of 1 × 1011 GC per animal. The liver tissues were harvested at day 28 post gene transfer for cryosections and X-Gal staining. (Right) Panels show LacZ gene transfer to skeletal muscle of NCR nude mice treated with 1 × 1011 GC of AAVCMVLacZ vectors of different serotypes. The animals were killed at day 10 after vector administration, and injected muscles were harvested for cryosections and X-Gal staining.
The performance of AAV vectors in skeletal muscle was analyzed after direct injection into the tibialis anterior muscles. Vectors contained the same AAV2-based genome, with the immediate early gene of CMV or a CMV-enhanced β-actin promoter driving expression of the transgene. A time course of expression of A1AT in nu/nu NCR and C57BL/6 mice is shown in Fig. 2. Previous studies indicated that immune competent C57BL/6 mice elicit limited humoral responses to the human A1AT protein when expressed from AAV vectors (9). In each strain, AAV2/1 vector produced the highest levels of A1AT and AAV2/2 vector the lowest, with AAV2/7 and AAV2/8 vectors showing intermediate levels of expression. Peak levels of CG at 28 days after injection of nu/nu NCR mice showed the highest levels from AAV2/7 and the lowest from AAV2/2 with AAV2/8 and AAV2/1 in between. Injection of AAV2/1 and AAV2/7 LacZ vectors yielded gene expression at the injection sites in all muscle fibers with substantially fewer LacZ positive fibers observed with AAV2/2 and AAV 2/8 vectors (Fig. 3). These data indicate that the efficiency of transduction with AAV2/7 vectors in skeletal muscle is similar to that obtained with AAV2/1, which is the most efficient in skeletal muscle of the previously described serotypes (9, 32, 33).
Similar murine models were used to evaluate liver-directed gene transfer. Identical doses of vector based on genome copies were infused into the portal veins of mice that were analyzed subsequently for expression of the transgene. Each vector contained an AAV2-based genome using previously described liver-specific promoters (i.e., albumin or thyroid hormone binding globulin) to drive expression of the transgene. The impact of capsid proteins on the efficiency of transduction of A1AT vectors in nu/nu and C57BL/6 mice and CG vectors in C57BL/6 mice was consistent (Fig. 3 and Table 4). In all cases, AAV2/8 vectors yielded the highest levels of transgene expression that ranged from 16 to 110 times greater than what was obtained with AAV2/2 vectors; expression from AAV2/5 and AAV2/7 vectors was intermediate, with AAV2/7 higher than AAV2/5. Analysis of X-Gal-stained liver sections of animals that received the corresponding LacZ vectors showed a correlation between the number of transduced cells and overall levels of transgene expression. DNAs extracted from livers of C57BL/6 mice who received the A1AT vectors were analyzed for abundance of vector DNA by using real time PCR technology. The amount of vector DNA found in liver 56 days after injection correlated with the levels of transgene expression (Table 5). These studies indicate that AAV8 is the most efficient vector for liver-directed gene transfer because of increased numbers of transduced hepatocytes.
Table 5.
Real-time PCR analysis for abundance of AAV vectors in nu/nu mouse liver after injection of 1 × 1011 genome copies of vector
| AAV vectors/dose | Genome copies per cell |
|---|---|
| AAV2/1AlbA1AT | 0.6 ± 0.36 |
| AAV2AlbA1AT | 0.003 ± 0.001 |
| AAV2/5AlbA1AT | 0.83 ± 0.64 |
| AAV2/7AlbA1AT | 2.2 ± 1.7 |
| AAV2/8AlbA1AT | 18 ± 11 |
A set of probe and primers targeting the SV40 poly(A) region of the vector genome was used for TaqMan PCR. Values shown are means of three individual animals with standard deviations. The animals were sacrificed at day 56 to harvest liver tissues for DNA extraction.
The serologic data described above suggest that AAV2/8 vector should not be neutralized in vivo after immunization with the other serotypes. C57BL/6 mice received intraportal injections of AAV2/8 vector expressing canine factor IX (1011 genome copies) 56 days after they received intramuscular injections of A1AT vectors of different serotypes. High levels of factor IX expression were obtained 14 days after infusion of AAV2/8 into naive animals (17 ± 2 μg/ml, n = 4), which were not significantly different from what was observed in animals immunized with AAV2/1 (31 ± 23 μg/ml, n = 4), AAV2/2 (16 μg/ml, n = 2), and AAV2/7 (12 μg/ml, n = 2). This result contrasts with what was observed in AAV2/8-immunized animals that were infused with the AAV2/8 factor IX vector in which no detectable factor IX was observed (<0.1 μg/ml, n = 4).
Discussion
The goal of this study was to isolate new AAV serotypes from a species other than humans and develop them as vectors for human gene therapy. The hope was that they would have improved efficiencies of gene transfer and would not be recognized by antibodies generated to AAV infections in humans. We recently pursued a similar strategy for the development of adenovirus-based vaccines where we were able to create vectors based on adenoviruses from chimpanzees that were shown to activate T and B cells in model systems but were not neutralized by antibodies generated to naturally acquired adenovirus infections in humans (34–36).
Our strategy for isolating unique AAVs differed from the traditional approach for isolating new parvoviruses in which cells that potentially contain AAV genomes are infected with helper viruses, such as adenovirus, to rescue latent AAV virus. We attempted to retrieve novel AAV sequences by PCR amplification of latent genomes by using primers to conserved regions of AAV that flank an area of divergence. Tissues from nonhuman primates were targeted for AAV isolation for a number of reasons. We believed that nonhuman primate AAVs should be sufficiently similar to human AAVs such that they retain tropism for human target cells and could be propagated with human adenoviruses and expanded as vectors by using standard techniques. However, it was hoped that there would be sufficient divergence from human AAVs that antibodies generated as a result of naturally acquired AAV infections in humans would not neutralize infection with the nonhuman primate AAVs.
Oligonucleotides to conserved regions of the cap gene did amplify sequences from rhesus monkeys that represented unique AAVs. Identical cap signature sequences were found in multiple tissues from rhesus monkeys derived from at least two different colonies. Full-length rep and cap ORFs were isolated and sequenced from single sources. Only the cap ORFs of the novel AAVs were necessary to evaluate their potential as vectors because we were able to generate vectors with the AAV7 or AAV8 capsids by using the ITRs and rep from AAV2. This strategy also simplified the comparison of different vectors because the actual vector genome is identical between different vector serotypes. In fact, the yields of recombinant vectors generated by using this approach did not differ between serotypes.
The objectives of our study were indeed realized. Vectors based on AAV7 and AAV8 do appear to be immunologically distinct (i.e., they are not neutralized by antibodies generated against other serotypes). Furthermore, sera from humans demonstrate very little neutralizing activity to AAV7 and AAV8 vectors, which is an advantage over the human-derived AAVs currently under development, for which a larger proportion of the human population has preexisting immunity that is high level and neutralizing (37).
The tropism of each new vector is indeed favorable for in vivo applications. AAV2/7 vectors appear to transduce skeletal muscle as efficiently as AAV2/1, which is the serotype that confers the highest level of transduction in skeletal muscle of the primate AAVs tested to date (9, 32, 33). Importantly, AAV2/8 provides a substantial advantage over the other serotypes in terms of efficiency of gene transfer to liver that until now has been relatively disappointing in terms of the numbers of hepatocytes stably transduced. AAV2/8 consistently achieved a 10- to 100-fold improvement in gene transfer efficiency as compared with the other vectors. The basis for the improved efficiency of AAV2/8 is unclear, although it presumably is due to uptake via a different receptor that is more active on the basolateral surface of hepatocytes. This improved efficiency will be quite useful in the development of liver-directed gene transfer where the number of transduced cells is critical, such as in urea cycle disorders and familial hypercholesterolemia. We described a novel approach for isolating new AAVs based on PCR retrieval of genomic sequences. The amplified sequences were easily incorporated into vectors and tested in animals. The relative lack of preexisting immunity to AAV7 and AAV8 and the favorable tropism of the vectors for muscle and liver, respectively, suggest that they should be considered for further development as vectors in human gene therapy.
Acknowledgments
The support of the Vector, Quality Control, and Cell Morphology Cores of the University of Pennsylvania was appreciated. TaqMan analysis provided by Julio Sanmiguel. Production of novel AAV vectors performed by Phoi Tran. We thank Dr. Jude Samulski for providing antisera to all of the known AAV serotypes. J.M.W. holds equity in Targeted Genetics, Inc. Support for this work was provided by the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases Grant P30 DK 47757-09 and National Heart, Lung, and Blood Institute Grant P01 HL 59407-03), the Juvenile Diabetes Research Foundation, the Cystic Fibrosis Foundation, and GlaxoSmithKline Pharmaceuticals.
Abbreviations
- AAV
adeno-associated virus
- ITR
inverted terminal repeat
- A1AT
α-1 antitrypsin
- CG
choriogonadotropic hormone
- rhCG
rhesus monkey CG
- GC
genome copy
- CMV
cytomegalovirus early promoter
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- TBG
thyroid hormone binding globulin gene promoter
- FIX
factor IX
Footnotes
References
- 1.Muzyczka N, Berns K I. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2327–2359. [Google Scholar]
- 2.Atchison R W, Casto B C, Hammon W M. Science. 1965;194:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 3.Melnick J L, Mayor H D, Smith K O, Rapp F. J Bacteriol. 1965;90:271–274. doi: 10.1128/jb.90.1.271-274.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoggan M D, Blacklow N R, Rowe W P. Proc Natl Acad Sci USA. 1966;55:1467–1474. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bantel-Schaal U, zur Hausen H. Virology. 1984;134:52–63. doi: 10.1016/0042-6822(84)90271-x. [DOI] [PubMed] [Google Scholar]
- 6.Georg-Fries B, Biederlack S, Wolf J, zur Hausen H. Virology. 1984;134:64–71. doi: 10.1016/0042-6822(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 7.Rutledge E A, Halbert C L, Russell D W. J Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bantel-Schaal U, Delius H, Schmidt R, zur Hausen H. J Virol. 1999;73:939–947. doi: 10.1128/jvi.73.2.939-947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao W, Chirmule N, Berta S C, McCullough B, Gao G, Wilson J M. J Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parks W P, Boucher D W, Melnick J L, Taber L H, Yow M D. Infect Immun. 1970;2:716–722. doi: 10.1128/iai.2.6.716-722.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blacklow N R, Hoggan M D, Rowe W P. J Natl Cancer Inst. 1968;40:319–327. [PubMed] [Google Scholar]
- 12.Rapoza N P, Atchinson R W. Nature (London) 1967;215:1186–1187. doi: 10.1038/2151186a0. [DOI] [PubMed] [Google Scholar]
- 13.Boucher D W, Parks W P, Melnick J L. J Immunol. 1970;104:555–559. [PubMed] [Google Scholar]
- 14.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowitz J E, Samulski J. Curr Opin Biotechnol. 1998;9:470–475. doi: 10.1016/s0958-1669(98)80031-1. [DOI] [PubMed] [Google Scholar]
- 16.Jooss K, Yang Y, Fisher K J, Wilson J M. J Virol. 1998;72:4212–4223. doi: 10.1128/jvi.72.5.4212-4223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakai H, Iwaki Y, Kay M A, Couto L B. J Virol. 1999;73:5438–5447. doi: 10.1128/jvi.73.7.5438-5447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z Y, Yant S R, He C Y, Meuse L, Shen S, Kay M A. Mol Ther. 2001;3:403–410. doi: 10.1006/mthe.2001.0278. [DOI] [PubMed] [Google Scholar]
- 20.Bals R, Xiao W, Sang N, Weiner D J, Meegalla R L, Wilson J M. J Virol. 1999;73:6085–6088. doi: 10.1128/jvi.73.7.6085-6088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Z, Zak R, Luxton G W, Ritchie T C, Bantel-Schaal U, Engelhardt J F. J Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auricchio A, Kobinger G, Anand V, Hildinger M, O'Connor E, Maguire A M, Wilson J M, Bennett J. Hum Mol Genet. 2001;10:3075–3081. doi: 10.1093/hmg/10.26.3075. [DOI] [PubMed] [Google Scholar]
- 23.Davidson B L, Stein C S, Heth J A, Martins I, Kotin R M, Derksen T A, Zabner J, Ghodsi A, Chiorini J A. Proc Natl Acad Sci USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabner J, Seiler M, Walters R, Kotin R M, Fulgeras W, Davidson B L, Chiorini J A. J Virol. 2000;74:3852–3858. doi: 10.1128/jvi.74.8.3852-3858.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnell M A, Zhang Y, Tazelaar J, Gao G P, Yu Q C, Qian R, Chen S J, Varnavski A N, LeClair C, Raper S E, Wilson J M. Mol Ther. 2001;3:708–722. doi: 10.1006/mthe.2001.0330. [DOI] [PubMed] [Google Scholar]
- 26.Hildinger M, Auricchio A, Gao G, Wang L, Chirmule N, Wilson J M. J Virol. 2001;75:6199–6203. doi: 10.1128/JVI.75.13.6199-6203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao G, Qu G, Burnham M S, Huang J, Chirmule N, Joshi B, Yu Q C, Marsh J A, Conceicao C M, Wilson J M. Hum Gene Ther. 2000;11:2079–2091. doi: 10.1089/104303400750001390. [DOI] [PubMed] [Google Scholar]
- 28.Gao G P, Yang Y, Wilson J M. J Virol. 1996;70:8934–8943. doi: 10.1128/jvi.70.12.8934-8943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoltick P W, Wilson J M. Mol Ther. 2000;2:657–659. doi: 10.1006/mthe.2000.0204. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Zoppe M, Hackeng T M, Griffin J H, Lee K F, Verma I M. Proc Natl Acad Sci USA. 1997;94:11563–11566. doi: 10.1073/pnas.94.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Chirmule N, Gao G P, Qian R, Croyle M, Joshi B, Tazelaar J, Wilson J M. Mol Ther. 2001;3:697–707. doi: 10.1006/mthe.2001.0329. [DOI] [PubMed] [Google Scholar]
- 32.Chao H, Monahan P E, Liu Y, Samulski R J, Walsh C E. Mol Ther. 2001;4:217–222. doi: 10.1006/mthe.2001.0449. [DOI] [PubMed] [Google Scholar]
- 33.Chao H, Liu Y, Rabinowitz J, Li C, Samulski R J, Walsh C E. Mol Ther. 2000;2:619–623. doi: 10.1006/mthe.2000.0219. [DOI] [PubMed] [Google Scholar]
- 34.Farina S F, Gao G P, Xiang Z Q, Rux J J, Burnett R M, Alvira M R, Marsh J, Ertl H C, Wilson J M. J Virol. 2001;75:11603–11613. doi: 10.1128/JVI.75.23.11603-11613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen C J, Xiang Z Q, Gao G P, Ertl H C, Wilson J M, Bergelson J M. J Gen Virol. 2002;83:151–155. doi: 10.1099/0022-1317-83-1-151. [DOI] [PubMed] [Google Scholar]
- 36.Xiang Z, Gao G, Reyes-Sandoval A, Cohen C J, Li Y, Bergelson J M, Wilson J M, Ertl H C. J Virol. 2002;76:2667–2675. doi: 10.1128/JVI.76.6.2667-2675.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chirmule N, Propert K, Magosin S, Qian Y, Qian R, Wilson J. Gene Ther. 1999;6:1574–1583. doi: 10.1038/sj.gt.3300994. [DOI] [PubMed] [Google Scholar]