Abstract
A systematic proteomic analysis of rice (Oryza sativa) leaf, root, and seed tissue using two independent technologies, two-dimensional gel electrophoresis followed by tandem mass spectrometry and multidimensional protein identification technology, allowed the detection and identification of 2,528 unique proteins, which represents the most comprehensive proteome exploration to date. A comparative display of the expression patterns indicated that enzymes involved in central metabolic pathways are present in all tissues, whereas metabolic specialization is reflected in the occurrence of a tissue-specific enzyme complement. For example, tissue-specific and subcellular compartment-specific isoforms of ADP-glucose pyrophosphorylase were detected, thus providing proteomic confirmation of the presence of distinct regulatory mechanisms involved in the biosynthesis and breakdown of separate starch pools in different tissues. In addition, several previously characterized allergenic proteins were identified in the seed sample, indicating the potential of proteomic approaches to survey food samples with regard to the occurrence of allergens.
Although each cell in a given organism contains the same genome, the protein content of cells varies widely (1). Furthermore, numerous completed genome sequences contain ORFs that lack homology to any known proteins and whose protein products are not known to be expressed. Whereas oligonucleotide and cDNA arrays are excellent tools for the global analysis of mRNA expression, posttranscriptional regulatory mechanisms are factors that may result in a lack of correlation between mRNA and protein abundance (2–4). Thus, proteomics has evolved from the need to be able to directly and globally determine the protein expression patterns and their respective posttranslational modifications in cells, tissues, and whole organisms (5).
The analysis of highly complex protein mixtures, such as the proteome of whole organisms, requires the utilization of multidimensional separation methods (6). In the classical approach, two-dimensional PAGE (2D-PAGE), using a separation based on the isoelectric point in the first dimension and a separation by molecular weight in the second dimension, is used (7). Recently, an alternative technology, termed multidimensional protein identification technology (MudPIT), has been developed (8–10). MudPIT involves the generation of peptides from a complex protein mixture, followed by separation on a strong cation exchange phase in the first dimension and by reverse phase chromatography in the second dimension. Because 2D-PAGE and MudPIT use complementary and independent separation methods for the resolution of proteomic components, the integration of datasets obtained with both technologies should provide improved proteomic coverage.
Plant proteomics, when compared with proteomic analyses of prokaryotes, yeast, and humans, is in its infancy, which is partly due to the lack of availability of completed genomic or cDNA sequences from plants (11). The completion of the draft sequence of the rice (Oryza sativa) genome and subsequent prediction of the protein complement (12) provided us with the unique opportunity to study the metabolic pathways common to multiple tissues and those uniquely expressed in particular tissues. Here, we present a comprehensive comparative display analysis, using both 2D-PAGE and MudPIT, of protein expression in rice leaf, root, and seed.
Materials and Methods
Materials.
Urea, ammonium acetate, ammonium bicarbonate (AmBic), DTT, iodoacetamide (IAM), and EDTA were obtained from Sigma. Thiourea, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), and ampholytes were products of Bio-Rad. Poroszyme bulk immobilized trypsin was a product of Applied Biosystems. HPLC grade acetonitrile (ACN), HPLC grade methanol, and glacial acetic acid were purchased from Fisher Scientific. Endoproteinase Lys-C was purchased from Roche Diagnostics, heptafluorobutyric acid (HFBA) was obtained from Pierce, and SPEC Plus PT C18 solid phase extraction (SPE) pipette tips were purchased from Ansys Diagnostics (Lake Forest, CA).
Tissue Preparation.
Rice (O. sativa, Nipponbare, generally known as japonica) was grown in the greenhouse with a 12-h light (29°C)/12-h dark (21°C) regime. Humidity was maintained at 30%. Plants were grown in pots containing 50% sunshine mix and 50% nitrohumus. Leaf and root samples were collected 49 days after germination and were pooled. The seed samples were collected from the entire panicle at 14 days postanthesis (94 days after germination). Protein extracts from leaf, root, and seed were prepared from each tissue by acetone precipitation (13, 14).
2-D PAGE.
For 2D-PAGE, 10 mg of acetone powder of tissues were dissolved in 350 μl of sample buffer containing 7 M urea, 2 M thiourea, 4% CHAPS, and 0.5% ampholytes. Seventy-five micrograms of protein were used for one 2D gel. For the first dimension, a Bio-Rad Protean isoelectric focusing unit was used with 17 cm immobilized pH gradient (IPG) strips (Bio-Rad) according to the manufacturer's recommendation. Second-dimension electrophoresis was performed on 12% linear gels of 20 × 25 cm in a Bio-Rad Protean II XL gel cell, and gels were silver-stained (15). Protein spots were excised by using a Bio-Rad spot cutter according to the manufacturer's instructions, destained (16), and in-gel digested with trypsin (17) with a Massprep digestion robot (Micromass, Beverley, MA). Tryptic peptides were extracted from the gel pieces with 5% formic acid/5% ACN.
HPLC–Tandem Mass Spectrometry for Gel Spots.
Samples were introduced onto the analytical column by using a Surveyor autosampler (Surveyor, ThermoFinnigan, San Jose, CA), which first transferred the peptide extracts onto a C18 (300 μm × 5 mm) cartridge (LC Packings, San Francisco, CA). A switching valve was then used to transfer the eluted peptides onto the analytical column (5 cm × 100 μm i.d.), which was packed with 100 Å, 5 μm Zorbax C18 resin at 500 psi pressure into a fused silica capillary microcolumn (Agilent Technologies, Palo Alto, CA) prepared with a P-2000 laser puller (Sutter Instruments, Novato, CA) as described previously (18). The peptide separation protocol consisted of an initial wash step with buffer A (5% vol/vol ACN/0.1% formic acid) for 10 min, and subsequent elution of peptides with a linear gradient from 0–100% buffer B (90% vol/vol ACN/0.1% formic acid) over 20 min. The HPLC column eluent was directly transferred into the electrospray ionization source of a ThermoFinnigan LCQ-Deca ion trap mass spectrometer. Automated peak recognition, dynamic exclusion, and daughter ion scanning of the two most intense ions were performed by using the xcalibur software as described previously (19).
Digestion of Soluble Fraction for MudPIT Analysis.
Acetone powder from leaf, root, or seed protein extracts of rice (1 mg protein each) was placed into separate tubes, solubilization buffer (100 mM ammonium bicarbonate/8 M urea/1 mM DTT, pH 8.5) was added, and the mixture was incubated at 50°C for 20 min. The cysteines of each sample were then carboxyamidomethylated with iodoacetamide, each sample digested overnight with endoproteinase Lys-C, diluted to 2 M urea with 100 mM ammonium bicarbonate (pH 8.5), supplemented with CaCl2 to a final concentration of 1 mM, and digested overnight with Porosyzme-immobilized trypsin beads as described previously (9). The resulting complex peptide mixture from each sample was desalted, concentrated, loaded on SPEC-PLUS PTC18 cartridges according to the manufacturer's instructions as described previously (9), and stored at −80°C until further analysis.
Multidimensional Protein Identification Technology Analysis of Rice Proteome Extracts.
The MudPIT system was used as described previously (9, 10, 20). Briefly, a quaternary HP 1100 HPLC pump was interfaced with a Finnigan DECA LCQ ion trap mass spectrometer (Finnigan MAT, San Jose, CA). A 100 × 365 μm fused silica capillary microcolumn (Agilent Technologies) was prepared with a P-2000 laser puller (Sutter Instruments) as described previously (18). The fritless microcapillary column was first packed with 10 cm of 5 μm Zorbax Eclipse XDB-C18 and then with 4 cm of 5 μm Partisphere strong cation exchange (SCX) (Whatman). The column was connected to a PEEK microcross as described previously (18), and each sample was analyzed via a fully automated 13-cycle chromatographic run as described previously (20).
sequest Analysis of Tandem Mass Spectra.
The sequest algorithm was used to interpret MS/MS as described previously (21–24). The FASTA database used by sequest was generated by using the gene finding program fgenesh (25) to identify potential ORFs in the rice genome database (12). Matches to peptides identified by sequest were filtered according to their charge state, cross-correlation score (Xcorr), normalized difference in correlation score (ΔCn), and the tryptic nature of each peptide. The parameters used were conservative and chosen to filter the results to minimize the inclusion of false positive hits. In all datasets, all accepted results had a ΔCn ≥ 0.1. In both MudPIT and 2D-PAGE analyses, fully or partially tryptic singly charged peptides with Xcorrs ≥ 2.0 were accepted. For MudPIT, doubly charged peptides with Xcorrs of at least 2.7 and triply charged peptides with XCorrs above 3.8 were accepted regardless of their tryptic nature. For 2D-PAGE, doubly charged peptides with Xcorrs of at least 2.5 and triply charged peptides with XCorrs above 3.7 were accepted if they contained at least one tryptic end.
Results and Discussion
Protein Identification via 2D-PAGE and MudPIT.
Protein extracts from leaf, root, and seed were analyzed by two complementary approaches for proteomic resolution. First, proteins were separated and visualized by 2D-PAGE using immobilized pH gradient-SDS/PAGE, the proteins were excised and digested in-gel with trypsin, and the extracted peptide mixtures were separated and analyzed by using capillary liquid chromatography-tandem mass spectrometry (see Materials and Methods). Second, the soluble portions of the same protein extracts were analyzed by MudPIT where a complex peptide mixture was directly analyzed by using biphasic capillary liquid chromatography-tandem mass spectrometry (see Materials and Methods). The sequences of the peptides from both approaches were identified by using database searching of uninterpreted fragment ion mass spectra with the program sequest (21). The database used to search was constructed by using the gene finding program fgenesh (25) to identify potential ORFs in the rice genomic database (12). Matches to peptides identified by sequest were filtered according to their Xcorr factors, ΔCn, and the tryptic nature of the peptide (see Materials and Methods).
Analysis of protein extracts from rice leaf, root, and seed via 2D-PAGE and tandem mass spectrometry resulted in the detection and identification of 556 unique proteins (1,509 peptides), covering 348 different proteins from leaf, 199 different proteins from root, and 152 different proteins from seed (Table 1), constituting the most extensive rice proteomics effort thus far (13, 26–28). A repeated analysis of rice proteins by using MudPIT yielded 2,363 unique proteins (5,189 peptides), whereby 867 different leaf proteins, 1,292 different root proteins, and 822 different seed proteins were detected (Table 1). An integration of both datasets indicated that 2,528 unique proteins (6,296 peptides) were identified, with the detection of 1,022 different proteins from leaf, 1,350 different proteins from root, and 877 different proteins from seed (Table 1). This effort represents the most extensive proteomic coverage published (for a complete list of protein identifications, see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). A total of 165 proteins were detected only via 2D-PAGE (29.7% of all proteins identified via 2D-PAGE), whereas 1,972 proteins were uniquely detected via MudPIT, demonstrating the utility of increasing the proteomic coverage by integration of complementary multidimensional separation technologies. Of these 2,528 proteins, 2,251 proteins were identified with peptides that uniquely identify the proteins. The other 277 proteins (marked by an asterisk in Table 2) are not clear identifications, because the peptides that match to these proteins also match to other proteins. However, the proteins these peptides match to are isozymes of each other based on their protein similarity. Of the 6,296 unique peptides, however, 23 could not be matched to an individual protein or isozymes, each matching to completely different proteins, so therefore we dismissed these peptides altogether.
Table 1.
Numerical analysis of peptides and proteins detected and identified in the rice proteome
| 2D-PAGE
|
MudPIT
|
Combined*
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | Root | Seed | Total | Leaf | Root | Seed | Total | Leaf | Root | Seed | Total | |
| Total peptides† | 1,707 | 706 | 848 | 3,261 | 6,182 | 7,181 | 8,799 | 22,162 | 7,889 | 7,887 | 9,647 | 25,423 |
| Unique peptides‡ | 859 | 445 | 393 | 1,509 | 1,626 | 2,358 | 1,912 | 5,189 | 2,358 | 2,712 | 2,180 | 6,296 |
| Unique proteins§ | 348 | 199 | 152 | 556 | 867 | 1,292 | 822 | 2,363 | 1,022 | 1,350 | 877 | 2,528 |
The soluble protein extracts from each tissue of rice were analyzed via 2D-PAGE and MudPIT as described in Materials and Methods.
The section “Combined” details the integration of the datasets generated from both 2D-PAGE and MudPIT analysis of each tissue of the rice proteome.
The total number of sequest (21)-interpreted tandem mass spectra that passed Xcorr and ΔCn criteria (see Materials and Methods) are listed as peptides identified in each rice tissue from each method.
The total number of unique peptides identified from each tissue via each method is listed. In the subsections “Total” and in the section “Combined”, the total number of unique peptides is not additive because many peptides were identified in multiple tissues and/or with both methods.
The total number of unique proteins identified from each tissue via each method is listed. In the subsections “Total” and in the section “Combined”, the total number of unique proteins is not additive because many proteins were identified in multiple tissues and/or with both methods.
Functional Classification of Identified Proteins.
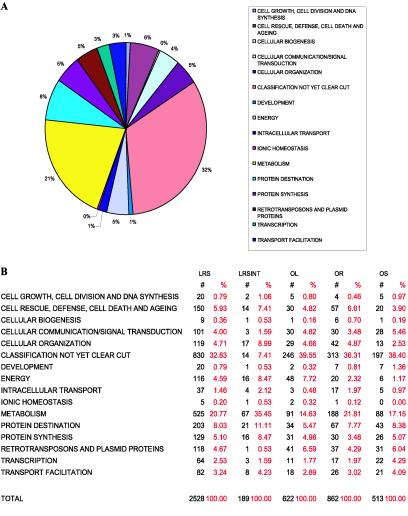
All protein sequences detected and identified as part of our rice proteomics effort were searched against the National Center for Biotechnology Information (NCBI) nonredundant protein database (29) by using the blast algorithm (30) and were sorted into functional categories (31) (Fig. 1).
Figure 1.
Functional class analysis of the identified proteins. Each of the combined total number of unique proteins (see Table 1) was functionally classified according to the categories described by Schoof et al. (31). After functional class assessment of each protein, the total number and percentage of proteins in each class were determined. (A) The percentage of proteins from each functional class from the total combined number of proteins from all tissues is shown. These values correspond to the Leaf Root Seed (LRS) column in B. (B) The table of numbers and percentages of proteins from each functional class from each grouping of tissue datasets is shown. LRS stands for the complete pool of proteins from Leaf, Root, and Seed. LRSINT are the shared proteins identified in Leaf, Root, and Seed. OL are the proteins found Only in Leaf. OR are the proteins found Only in Root. OS are the proteins found Only in Seed.
The most abundant category was occupied by proteins with as yet unidentified function and proteins with very low or no detectable homology to other predicted proteins in the database (792 proteins or 32.8% of the identified proteins; Fig. 1). Of these, 360 did not have any homologies to any proteins in the NCBI nonredundant protein database and are therefore considered to be rice-specific proteins. The second most abundant class of proteins was classified as being involved in metabolic processes (20.8% of the identified proteins), which is in accordance with the functional distribution of enzyme classes based on the rice genome sequence (12). As part of our proteomic survey, proteins involved in most cellular activities were detected, covering protein synthesis [ribosomal proteins (64 different proteins), translation elongation factors (14 unique proteins), and translation initiation factors (17 unique proteins)], protein degradation (28 unique proteins of the proteasome are present), and signal transduction (68 kinases or kinase-like proteins). Furthermore, essential components of the photosynthetic light-harvesting complexes have been detected, some of which are encoded in the nuclear genome (e.g., psaD, psaE, psaH, and psaK of photosystem I, psbP and a stability/assembly factor of photosystem II, the Rieske Fe-S protein of the cytochrome b6f complex, the photoreceptor phytochrome a, and the electron transport protein plastocyanin), whereas others are derived from genes transcribed from the plastidial genome (e.g., the D1 and D2 proteins of photosystem II, and cytochrome f of the cytochrome b6f complex). Additional identified proteins encoded in the chloroplast genome included the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, the Clp protease, and certain proteins localized to the thylakoid membrane (e.g., ATP synthase α and β chain).
Of the 2,528 detected proteins, 189 proteins (corresponding to 7.5%) were found to be expressed in all three tissues. The functional classification for these proteins indicated that they are involved in central metabolic pathways, in transcriptional control and mRNA biosynthesis, and protein biosynthesis (translational machinery, folding machinery including heat shock proteins and chaperonins, and degradation pathway including the proteins of the proteasome). However, the majority of proteins showed a tissue-specific expression pattern, as indicated by the detection of 622 leaf-specific proteins (60.9% of all proteins detected in leaves), 862 root-specific proteins (63.9% of all proteins detected in roots), and 512 seed-specific proteins (58.4% of all proteins detected in seeds). Leaf expressed the largest proportion of proteins classified as being involved in energy flow (leaf 8%, root 2%, seed 1%), which is largely due to the occurrence of photosynthetic processes in leaf plastids (Fig. 1). In roots, proteins belonging to the category “cell rescue, defense, cell death and aging” were quite abundant (root 7%, leaf 5%, seed 4%), with peroxidases being the most prominent representatives. However, a specific role for the 27 root-specific peroxidases (48% of all detected peroxidases) cannot be assigned based on the currently available datasets (32).
Tissue-Specific Expression of Metabolic Pathways in Rice.
The expression patterns of proteins identified and classified as being involved in metabolic pathways were visualized on a metabolic map to illustrate the contribution of these enzymes to tissue-specific metabolic pathways (Fig. 5, which is published as supporting information on the PNAS web site). It should be noted that this study focused mainly on a survey of soluble proteins, in which the metabolic enzymes embedded into or associated with membranes were not readily detected. Most enzymes involved in central metabolic pathways (glycolysis/gluconeogenesis, citric acid cycle, oxidative pentose phosphate pathway, and most pathways of amino acid biosynthesis) were identified in all tissues (Fig. 5). The ubiquitous presence of cytosolic and plastidial glycolytic isoenzymes (33) was confirmed for fructose 1,6-bisphosphatase, triosephosphate isomerase, glyceraldehyde 3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and phosphoglyceromutase. The expression of the majority of metabolic enzymes, however, was found to be tissue specific. For example, the enzymes involved in chlorophyll and carotenoid biosynthesis (components of light harvesting complexes) were expressed exclusively in leaves (34). The enzymes of phenylpropanoid metabolism, which produce, among other compounds, lignins as cell wall polymers and flavonoids as UV light protectants (35), were abundant in leaves and to a lesser extent in roots, but were, with the exception of phenylalanine ammonia lyase, not detectable in seeds.
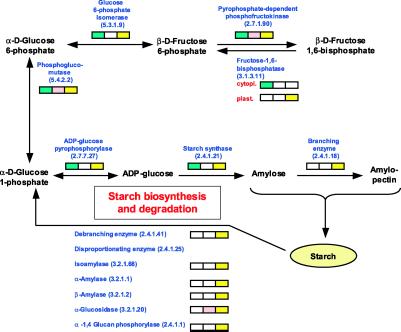
The proteomic dataset presented here also confirmed that biosynthesis and breakdown of starch, the most common storage polysaccharide in plants (36), are tissue-specific and subcellular compartment-specific processes. In leaf, starch is synthesized during the day directly from photosynthetically fixed carbon dioxide in the stroma of chloroplasts, where it serves as a short-term carbohydrate reserve termed transitory starch. During the night, this pool of starch is degraded to provide a carbon supply for sucrose synthesis and export, and for respiration (37). In seed, starch is synthesized in amyloplasts as a long-term storage form for carbohydrates. The starch biosynthetic pathway starts with the conversion of glucose 1-phosphate into ADP-glucose, a key step catalyzed by ADP-glucose pyrophosphorylase (AGPase). The ADP-glucose then serves as a glucosyl donor for α-glucan synthesis by the action of starch synthases and starch-branching enzymes (38, 39). Enzymes for the starch degradation pathway include debranching enzyme, disproportionating enzyme, isoamylase, α-amylase, β-amylase, α-glucosidase, and starch phosphorylase (40). With the exception of the disproportionating enzyme, all enzymes involved in starch metabolism were detected as part of our proteomic analysis of rice tissues. The starch biosynthetic enzymes were present in leaf and seed, whereas the starch degradation pathway enzymes were detected almost exclusively in seed (Fig. 2). The absence of the starch catabolic enzymes in the leaf sample is explained by the fact that the leaves were picked ≈4 h after dawn and were thus producing transitory starch.
Figure 2.
Tissue localization of proteins identified from the starch biosynthesis and degradation pathways. On functional classification of each uniquely identified protein, the tissue-specific protein expression was analyzed from a metabolic pathway perspective (Fig. 5). A focused portion of Fig. 5 is shown where the tissue-specific expression of the starch biosynthesis and degradation pathways is detailed. Enzymes involved in each pathway are in blue. The boxes show in which tissue the enzymes have been found, with green representing detection in leaf, red representing detection in root, and yellow representing detection in seed.
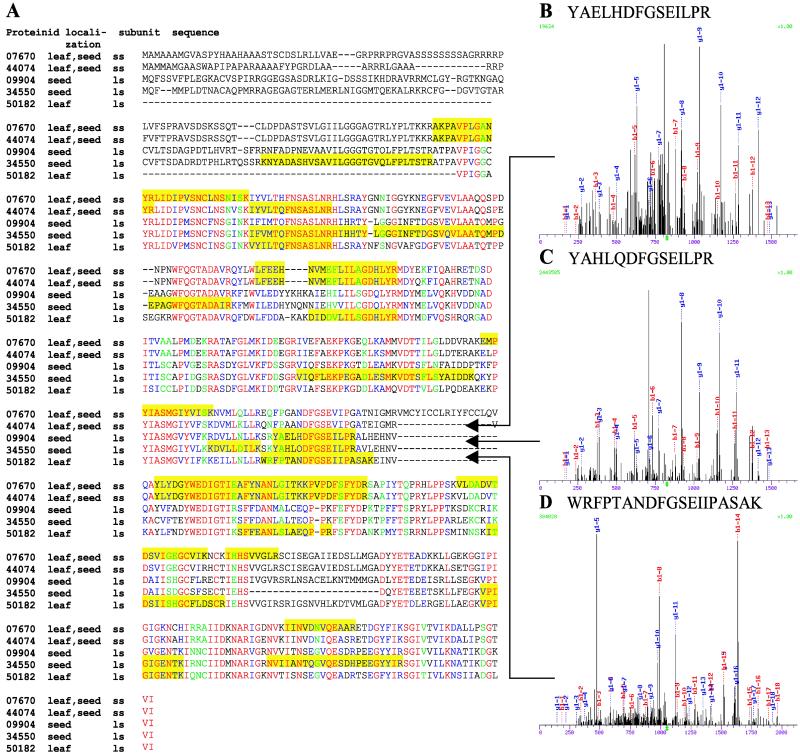
AGPase occurs as a heterotetramer, composed of two small and two large subunits (41). The small subunits are mainly responsible for the catalytic properties whereas the large subunits are of regulatory importance (42). The two isoforms of the small AGPase subunit, for which peptides identical to published rice sequences were identified (refs. 43–45; Fig. 3), were detected in both leaf and seed. Two isoforms of the large AGPase subunit were detected only in seed, whereas the third isoform was detected exclusively in leaf. Interestingly, based on an analysis of the subcellular compartmentation by ChloroP (46), the two seed-specific isoenzymes of the large subunit are devoid of a plastidial targeting sequence, which is in agreement with previously published reports indicating that AGPase activity is mainly localized to the cytosol of the graminaceous endosperm (38, 47). It has been speculated that the cytosolic AGPase may facilitate the partitioning of carbon from sucrose into starch when there is a sufficient supply of sucrose in the endosperm (38), which would require the import of cytosolic ADP-glucose into the plastids. This import has been proposed to be accomplished through the action of the brittle-1 protein, an adenylate translocator with a common ADP-glucose-binding domain (48), for which we detected a seed-specific expression, thus supporting the evidence of cytosolic AGPase pools in seed tissues.
Figure 3.
Tissue-specific expression of AGPase isozymes. (A) clustalw alignment (52) of the small subunits (ss under subunit) and large subunits (ls under subunit) of AGPase isozymes detected and identified in the current study. The tissues in which each enzyme were detected and identified are specified under the localization column. The yellow boxes designate the peptides that were detected and identified from each isozyme that passed the sequest (21) Xcorr and ΔCn criteria described in Materials and Methods. The proteinid listed for each AGPase isozyme corresponds to the proteinid listed in Table 2. A subset of the tandem mass spectra (MS/MS) that uniquely identified each of the AGPase isozymes is shown in B–D. (B) The sequest-interpreted MS/MS of the +2 peptide YAELHDFGSEILPR (m/z of 824.4; Xcorr = 2.97; ΔCn = 0.30) from the AGPase isozyme with the proteinid 09904. (C) The sequest-interpreted MS/MS of the +2 peptide YAHLQDFGSEILPR (m/z of 823.9; Xcorr = 3.28; ΔCn = 0.25) from the AGPase isozyme with the proteinid 34550. (D) The sequest-interpreted MS/MS of the +2 peptide WRFPTANDFGSEIIPASAK (m/z of 1,054.8; Xcorr = 3.95; ΔCn = 0.36) from the AGPase isozyme with the proteinid 50182.
Seed Storage Proteins.
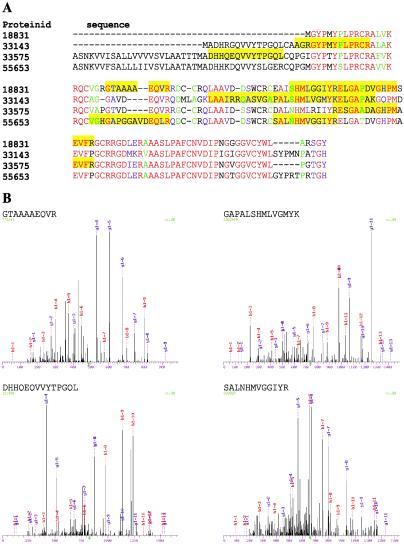
In addition to the chemical constituents present in all plant tissues, seed contain carbohydrates, fats, oils, and proteins as a source of food reserves to support early seedling growth (49). The storage proteins detected by our proteomic survey included 7 different globulins, 10 different prolamins, and 13 different glutelins. Whereas the globulins and prolamins were present only in seeds, 5 of the glutelins were also detected in leaf. Interestingly, the seed sample contained 4 known allergenic proteins (Fig. 4), which belong to the α-amylase/trypsin inhibitor gene family (50). The ability to detect known allergens illustrates the utility of proteomic approaches to proteotype food sources for the presence of allergenic proteins, which has become an important issue because public awareness of food-related allergies has increased tremendously (51).
Figure 4.
Proteomic identification of allergens in rice seed. (A) clustalw alignment (52) of the four allergens detected and identified in rice seed via proteomic methods in the current study. The proteinid listed for each allergen corresponds to the proteinid listed in Table 2. The yellow boxes designate the peptides that were detected and identified from each protein that passed the sequest (21) Xcorr and ΔCn criteria described in Materials and Methods. A tandem mass spectra (MS/MS) that uniquely identifies each allergenic protein is shown in B. (B) The sequest-interpreted MS/MS of the +2 peptide GTAAAAEQVR (m/z of 487.3; Xcorr = 3.17; ΔCn = 0.11) from the allergen with the proteinid 18831. The sequest-interpreted MS/MS of the +2 peptide GAPALSHMLVGMYK (m/z of 738.0; Xcorr = 3.47; ΔCn = 0.31) from the allergen with the proteinid 33143. The sequest-interpreted MS/MS of the +2 peptide DHHQEQVVYTPGQL (m/z of 826.2; Xcorr = 2.84; ΔCn = 0.23) from the allergen with the proteinid 33575. The sequest-interpreted MS/MS of the +2 peptide SALNHMVGGIYR (m/z of 616.3; Xcorr = 3.44; ΔCn = 0.34) from the allergen with the proteinid 55653.
Conclusions
In summary, this study indicates that large-scale proteomic analyses of plant tissues have become feasible provided that comprehensive genome databases are available. Using complementary multidimensional technologies, exciting insight is gained into the expression of tissue-specific metabolic pathways. In addition, we provide evidence that proteomic surveys are a useful tool to analyze food and feed sources for known allergens. The combination of multidimensional proteomic technologies with quantitative methods will allow the integration of mRNA and protein expression data, and will generate the foundation to understand complex metabolic networks.
Supplementary Material
Acknowledgments
We thank Derek Guist and Darrell Ricke for providing the protein database, Werner Bastian and Timothy Torchia for valuable discussions regarding the manuscript, and Steve Briggs for research support.
Abbreviations
- 2D-PAGE
two-dimensional PAGE
- MudPIT
multidimensional protein identification technology
- ACN
acetonitrile
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- Xcorr
cross-correlation score
- AGPase
ADP-glucose pyrophosphorylase
- ΔCn
normalized difference in correlation score
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 11564.
References
- 1.Collins F S. Genome Res. 2001;11:641–643. doi: 10.1101/gr.189801. [DOI] [PubMed] [Google Scholar]
- 2.Gygi S P, Rochon Y, Franza B R, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futcher B, Latter G I, Monardo P, McLaughlin C S, Garrels J I. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ideker T, Thorsson V, Ranish J A, Christmas R, Buhler J, Eng J K, Bumgarner R, Goodlett D R, Aebersold R, Hood L. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 5.Zivy M, de Vienne D. Plant Mol Biol. 2000;44:575–580. doi: 10.1023/a:1026525406953. [DOI] [PubMed] [Google Scholar]
- 6.Giddings J C. J High Resolut Chromatogr Chromatogr Commun. 1987;10:319–323. [Google Scholar]
- 7.Fey S J, Larsen P M. Curr Opin Chem Biol. 2001;5:26–33. doi: 10.1016/s1367-5931(00)00167-8. [DOI] [PubMed] [Google Scholar]
- 8.Link A J, Eng J, Schieltz D M, Carmack E, Mize G J, Morris D R, Garvik B M, Yates J R., 3rd Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 9.Washburn M P, Wolters D, Yates J R., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 10.Wolters D A, Washburn M P, Yates J R., 3rd Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 11.van Wijk K J. Plant Physiol. 2001;126:501–508. doi: 10.1104/pp.126.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goff S A, Ricke D, Lan T H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 13.Tsugita A, Kawakami T, Uchiyama Y, Kamo M, Miyatake N, Nozu Y. Electrophoresis. 1994;15:708–720. doi: 10.1002/elps.1150150198. [DOI] [PubMed] [Google Scholar]
- 14.Damerval C, de Vienne D, Zivy M, Thiellement H. Electrophoresis. 1986;7:52–54. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2013::AID-ELPS2013>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Blum H, Beier H, Gross H J. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 16.Gharahdaghi F, Weinberg C R, Meagher D A, Imai B S, Mische S M. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A, Chernushevich I, Wilm M, Mann M. Methods Mol Biol. 2000;146:1–16. doi: 10.1385/1-59259-045-4:1. [DOI] [PubMed] [Google Scholar]
- 18.Gatlin C L, Kleemann G R, Hays L G, Link A J, Yates J R., 3rd Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 19.Haynes P A, Fripp N, Aebersold R. Electrophoresis. 1998;19:939–945. doi: 10.1002/elps.1150190609. [DOI] [PubMed] [Google Scholar]
- 20.Washburn M P, Ulaszek R, Deciu C, Schieltz D M, Yates J R., 3rd Anal Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 21.Eng J, McCormack A L, Yates J R., 3rd J Am Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 22.Yates J R, 3rd, Eng J K, McCormack A L, Schieltz D. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 23.Link A J, Hays L G, Carmack E B, Yates J R., 3rd Electrophoresis. 1997;18:1314–1334. doi: 10.1002/elps.1150180808. [DOI] [PubMed] [Google Scholar]
- 24.Link A J, Robison K, Church G M. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- 25.Salamov A A, Solovyev V V. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsugita A, Kamo M, Kawakami T, Ohki Y. Electrophoresis. 1996;17:855–865. doi: 10.1002/elps.1150170507. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu S, Muhammad A, Rakwal R. Electrophoresis. 1999;20:630–636. doi: 10.1002/(SICI)1522-2683(19990301)20:3<630::AID-ELPS630>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 28.Rakwal R, Agrawal G K, Yonekura M. Electrophoresis. 1999;20:3472–3478. doi: 10.1002/(SICI)1522-2683(19991101)20:17<3472::AID-ELPS3472>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Hart K W, Searls D B, Overton G C. Comput Appl Biosci. 1994;10:369–378. doi: 10.1093/bioinformatics/10.4.369. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Schoof H, Zaccaria P, Gundlach H, Lemcke K, Rudd S, Kolesov G, Arnold R, Mewes H W, Mayer K F. Nucleic Acids Res. 2002;30:91–93. doi: 10.1093/nar/30.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiraga S, Yamamoto K, Ito H, Sasaki K, Matsui H, Honma M, Nagamura Y, Sasaki T, Ohashi Y. FEBS Lett. 2000;471:245–250. doi: 10.1016/s0014-5793(00)01409-5. [DOI] [PubMed] [Google Scholar]
- 33.Plaxton W C. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:185–214. doi: 10.1146/annurev.arplant.47.1.185. [DOI] [PubMed] [Google Scholar]
- 34.Formaggio E, Cinque G, Bassi R. J Mol Biol. 2001;314:1157–1166. doi: 10.1006/jmbi.2000.5179. [DOI] [PubMed] [Google Scholar]
- 35.Weisshaar B, Jenkins G I. Curr Opin Plant Biol. 1998;1:251–257. doi: 10.1016/s1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- 36.Smith A M, Denyer K, Martin C. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:67–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- 37.Zeeman S C, Rees T A. Plant Cell Environ. 1999;22:1445–1453. [Google Scholar]
- 38.Beckles D M, Smith A M, ap Rees T. Plant Physiol. 2001;125:818–827. doi: 10.1104/pp.125.2.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slattery C J, Kavakli I H, Okita T W. Trends Plant Sci. 2000;5:291–298. doi: 10.1016/s1360-1385(00)01657-5. [DOI] [PubMed] [Google Scholar]
- 40.Kossmann J, Lloyd J. Crit Rev Biochem Mol Biol. 2000;35:141–196. [PubMed] [Google Scholar]
- 41.Smith-White B J, Preiss J. J Mol Evol. 1992;34:449–464. doi: 10.1007/BF00162999. [DOI] [PubMed] [Google Scholar]
- 42.Fu Y, Ballicora M A, Preiss J. Plant Physiol. 1998;117:989–996. doi: 10.1104/pp.117.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson J M, Hnilo J, Larson R, Okita T W, Morell M, Preiss J. J Biol Chem. 1989;264:12238–12242. [PubMed] [Google Scholar]
- 44.Anderson J M, Larsen R, Laudencia D, Kim W T, Morrow D, Okita T W, Preiss J. Gene. 1991;97:199–205. doi: 10.1016/0378-1119(91)90052-d. [DOI] [PubMed] [Google Scholar]
- 45.Sikka V K, Choi S-B, Kavakli I H, Sakulsingharoj C, Gupta S, Ito H, Okita T W. Plant Sci. 2001;161:461–468. [Google Scholar]
- 46.Emanuelsson O, Nielsen H, von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denyer K, Dunlap F, Thorbjornsen T, Keeling P, Smith A M. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon J C, Pien F M, Cao H, Liu K C. Plant Physiol. 1998;117:1235–1252. doi: 10.1104/pp.117.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muntz K. Plant Mol Biol. 1998;38:77–99. [PubMed] [Google Scholar]
- 50.Alvarez A M, Adachi T, Nakase M, Aoki N, Nakamura R, Matsuda T. Biochim Biophys Acta. 1995;1251:201–204. doi: 10.1016/0167-4838(95)00075-6. [DOI] [PubMed] [Google Scholar]
- 51.Buchanan B B. Plant Physiol. 2001;126:5–7. doi: 10.1104/pp.126.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.