Abstract
The budding yeast Saccharomyces cerevisiae initiates polarized growth or budding once per cell cycle at a specific time of the cell cycle and at a specific location on the cell surface. Little is known about the molecular nature of the temporal and spatial regulatory mechanisms. It is also unclear what factors, if any, among the numerous proteins required to make a bud are involved in the determination of budding frequency. Here we describe a class of cdc42 mutants that produce multiple buds at random locations on the cell surface within one nuclear cycle. The critical mutation responsible for this phenotype affects amino acid residue 60, which is located in a domain required for GTP binding and hydrolysis. This mutation bypasses the requirement for the essential guanine-nucleotide-exchange factor Cdc24p, suggesting that the alteration at residue 60 makes Cdc42p hyperactive, which was confirmed biochemically. This result also suggests that the only essential function of Cdc24p is to activate Cdc42p. Together, these data suggest that the temporal and spatial regulation of polarized growth converges at the level of Cdc42p and that the activity of Cdc42p determines the budding frequency.
Cell polarization is crucial for the functions of many different cell types (1, 2). In the budding yeast Saccharomyces cerevisiae, the development of cell polarity (or budding) can be divided into four stages. First, cells of different mating types choose a particular pattern for budding (3, 4). Second, proteins that are central to the establishment of cell polarity are recruited to the cell cortex at the chosen site. These proteins include the essential small GTPase Cdc42p and those involved in Cdc42p-mediated pathways, including the p21-activated kinases, Ste20p and Cla4p (5–7); Gic1p and Gic2p (8–10); the formins, Bni1p and Bnr1p (11–13); and Msb3p and Msb4p (14, 15). Cdc42p is regulated positively by its guanine-nucleotide-exchange factor (GEF), Cdc24p, and negatively by its putative GTPase-activating proteins (GAPs), Bem3p, Rga1p, and Rga2p (refs. 16–18; Saccharomyces Genome Database, http://genome-www.stanford.edu/Saccharomyces). Next, polarization of F-actin at the presumptive bud site relays the polarity cues marked by Cdc42p in the cell cortex to the entire cell. Finally, post-Golgi vesicles are transported by a type V myosin through the actin cables to the bud tip (1, 2). Coordination of all four stages is critical for the establishment of cell polarity.
The fact that yeast cells bud only once per cell cycle—i.e., the singularity in budding—has been a mystery in yeast cell biology. Whether the budding frequency (the number of buds produced per mother per cell cycle) is mainly determined by a single cellular factor among many proteins involved in the budding process is not known. A temperature-sensitive mutation in CDC4 causes arrest as multibudded cells with a single bilobed nucleus (19). However, time-lapse analysis indicates that these buds are produced sequentially at the interval of a normal cell cycle time. Budding frequency is thus not altered in this mutant (19). A specific mutation in CDC42, cdc42D38E, causes consecutive initiation of budding in one cell cycle (20). But, at any given time, F-actin and polarized growth are restricted to a single bud, and only one bud can reach mature size per cell cycle (20). Thus, budding frequency is not altered in this mutant either.
Analyses of site-directed cdc42 mutants have led to several important conclusions. For example, Cdc42p may need to cycle between the GDP-bound state and the GTP-bound state to maintain cell viability (21). In addition, Cdc42p may play independent roles in actin and septin organization (22, 23). We reasoned that random mutagenesis of the entire CDC42 gene coupled with a functional selection of distinct morphological phenotypes might identify mutations that specifically affect one known process or mutations that reveal a previously unknown function of Cdc42p. Indeed, we have isolated a novel class of cdc42 mutations, detailed analysis of which has suggested that the temporal and spatial regulation of the budding process must converge at or upstream of Cdc42p and that Cdc42p activity is the crucial regulator of budding frequency.
Methods
Strains, Growth Conditions, and Plasmids.
Yeast strains are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Yeast cells were grown in standard synthetic or rich media (24, 25). To select for the loss of URA3-containing plasmids, 1 mg/ml 5-fluoroorotic acid (5-FOA) was added to the medium (26).
Plasmids pRS314-CDC42 and pRS314-CDC42–22 carry wild-type CDC42 and cdc42–22, respectively. pRS314-CDC42-G60X (X = A, D, or C) was constructed by a PCR-based, site-directed mutagenesis method using pRS314-CDC42 as the template. All mutations were confirmed by sequencing. pRS316-GAL1-CDC42 and pRS316-GAL1-CDC24 contain CDC42 and CDC24, respectively, under the control of a galactose-inducible promoter. pRS316-CDC3-GFP contains an in-frame GFP cassette inserted between amino acid 13 and 15 of Cdc3p. pSM217-GIC2-GFP contains GIC2-GFP. YEp352-CDC42 (for overexpressing Cdc42p in Fig. 4A) (15) and YEp13-RGA1 (27) were described previously. pB67 carries HA-CDC24 under an ADH promoter control (for overexpressing Cdc24p in Fig. 5B). pASF125 (integrative vector, URA3) carries GFP-TUB1.
Fig 4.
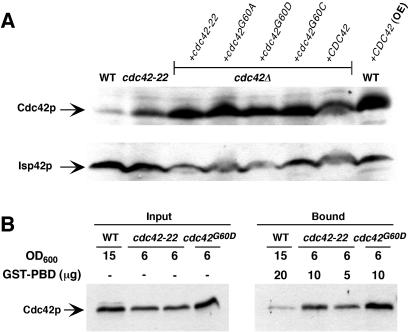
A single amino acid change in a GTP-binding and hydrolysis domain is responsible for the hyperactivity of cdc42-22 allele. (A) Cell lysates from YEF473A (WT), JPC250 [WT+CDC42 (OE)], YEF1963 (cdc42-22), and other cdc42Δ strains carrying different alleles of CDC42 (strains JPC152–156) were probed with antibodies against Cdc42p and, as a control, the mitochondrial outer membrane protein Isp42p. (B) Cell lysates from YEF473A (WT), YEF1963 (cdc42-22), and JPC241 (cdc42G60D) were assayed for the active portion of Cdc42p by GST-PBD pull downs.
Fig 5.
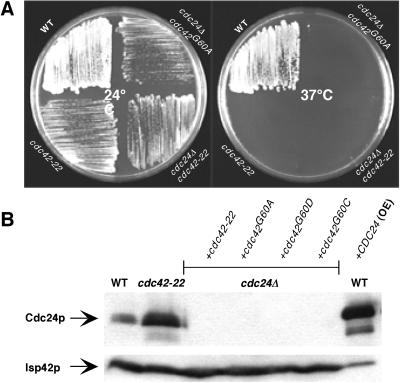
A single change of Cdc42p at glycine 60 allows the bypass of its GEF, Cdc24p. (A) Temperature-sensitive growth of cdc24Δ strains carrying a hyperactive cdc42 allele. Strains YEF473A (WT), YEF1963 (cdc42-22), JPC160 (cdc24Δ, pRS314-CDC42-22), and JPC162 (cdc24Δ, pRS314-CDC42-G60A) were streaked onto YPD plates and incubated for 2–4 days at 24°C and 37°C, respectively. (B) Western-blot analysis of cell lysates from YEF473A (WT), YEF1963 (cdc42-22), JCY403 [WT+CDC24 (OE)], and other cdc24Δ strains carrying different alleles of CDC42 (strains JPC160–164).
Strains YEF1963 (cdc42-22) and JPC241 (cdc42G60D) were constructed by replacing cdc42Δ:HIS3 in YEF1194 (cdc42Δ:HIS3, pRS316-GAL1-CDC42) with cdc42 mutant alleles. Strains JPC152–156 were generated by replacing the plasmid in YEF1194 with plasmids pRS314 carrying various alleles of CDC42. JPC83 (cdc24Δ:HIS3, pRS316-GAL1-CDC24) was constructed by replacing plasmid pMGF5 in YEF1201 (cdc24Δ:HIS3, pMGF5) (15) with pRS316-GAL1-CDC24. Strains JPC160–64 were generated by replacing plasmid pRS316-GAL1-CDC24 in JPC83 with plasmids pRS314 carrying various alleles of CDC42. Strains JPC40 and JPC46 were constructed by integrating GFP-TUB1 at the ura3 locus of YEF1963 (cdc42-22) and YEF473A (WT), respectively. All oligonucleotide primers were from Integrated DNA Technologies (Coralville, IA) and their sequences are listed in Table 2, which is published as supporting information on the PNAS web site. Detailed description of plasmid and strain constructions is also included in the supporting information.
Mutagenesis of CDC42.
We randomly mutagenized the entire CDC42 gene by a PCR-based method (28). The mutagenized PCR products were mixed with equal amount of linearized pRS314-CDC42, in which the entire CDC42 gene was removed by a restriction digestion with HpaI (67 bp upstream of the start codon) and NsiI (346 bp downstream of the stop codon). This mixture of DNA was used to transform YEF1194 (cdc42Δ:HIS3, pRS316-GAL1-CDC42) to allow gap repair between the PCR products and the linearized plasmid. Transformants were selected on SC-Trp plates at 24°C and then replicated onto SC-Trp plates containing 5-fluoroorotic acid to select for the loss of plasmid pRS316-GAL1-CDC42. The uracil-requiring colonies were replicated onto two sets of SC-Trp plates that were incubated at 24°C and 37°C, respectively, to allow the identification of temperature-sensitive mutants.
Immunoblotting and Cdc42p Activation Assay.
For immunoblotting, protein samples were prepared from exponentially growing cultures and separated by electrophoresis on an SDS/15% polyacrylamide gel. The blot was probed with rabbit anti-Cdc42 antibodies (Santa Cruz Biotechnology), rabbit anti-Isp42p antibodies, or affinity-purified rabbit anti-Cdc24p antibodies (29).
For Cdc42p activation assay, yeast cells were grown in 100 ml of YPD medium to mid-logarithmic phase and disrupted with glass beads in 500 μl of GPLB (G protein lysis buffer: 20 mM Tris⋅HCl, pH 7.4/150 mM NaCl/5 mM MgCl2/0.5% Nonidet P-40/5 mM glycerophosphate), containing a mixture of common protease inhibitors and 1 mM DTT plus various amounts (5–20 μg) of bacterially expressed GST-PBD (a GST fusion of the p21-binding domain from human Pak1). Cells were disrupted by Vortex mixing at 4°C 12 times for 30 sec with 30-sec intervals on ice, debris was spun down in a microcentrifuge at 4°C for 10 min, and supernatant was used for GST pull-down assays with 100 μl of glutathione-agarose beads (Molecular Probes). After gently rocking at 4°C for 1 h, beads were washed five times with GPLB. Proteins bound to the beads were eluted with 100 μl of SDS sample buffer. Aliquots of the input and bound fractions were separated by electrophoresis on a 12% polyacrylamide gel. The blot was probed with mouse anti-GST monoclonal antibody (Covance Research Products, Richmond, CA) or rabbit anti-Cdc42p polyclonal antibodies. Secondary antibodies including peroxidase-conjugated goat anti-mouse IgG and peroxidase-conjugated goat anti-rabbit IgG were purchased from Jackson ImmunoResearch.
Microscopy.
F-actin was stained with 20 units/ml rhodamine-conjugated phalloidin (Molecular Probes). DNA was stained with bisbenzamide (30). For monitoring polarized growth, yeast cells were grown in YM-P medium exponentially, collected by centrifugation, and resuspended in buffer P (10 mM sodium phosphate/150 mM NaCl, pH 7.2) at ≈5.0 × 107 cells per ml (30, 31). Then 0.9 ml of the cell suspension was mixed with 0.1 ml of FITC-conjugated Con A (stock: 1 mg/ml in buffer P) (Molecular Probes) and incubated at dark for 10 min. Cells were washed with buffer P and resuspended in fresh YM-P medium to allow new growth. Cells were fixed with formaldehyde at 24°C for 1 h and processed for observation of the growth pattern.
Time-lapse analysis of the budding process was performed with a computer-controlled microscope (E800, Nikon, Tokyo) with a digital camera (model 4742-95, Hamamatsu Photogenics, Bridgewater, NJ). Cells were grown to exponential phase in synthetic complete (SC) medium at 24°C and then spotted onto a thin layer of SC medium containing 25% gelatin (32). Five differential interference contrast (DIC) images along the z axis were acquired with Phase 3 Imaging Systems (Glen, PA) for strains YEF1963 (in 1.5-μm increments at a 10-min interval) and YEF473A (in 1.2-μm increments at a 5-min interval).
Results
Isolation of cdc42 Mutants That Form Multiple Buds During One Nuclear Cell Cycle.
From a random mutagenesis of CDC42, we isolated 82 temperature-sensitive mutants, 4 of which formed multiple buds per mother cell at the permissive temperature. In this report, we focus on the functional characterization of this class of mutants. Unless specified, all experiments were performed at 24°C.
We chose cdc42-22 for further studies, because it gave rise to a higher percentage of multibudded cells than the other alleles. Strains carrying cdc42-22 were temperature sensitive for growth at 37°C. This temperature sensitivity and the multibudded phenotype were complemented by a single copy of wild-type CDC42, suggesting that cdc42-22 is recessive. In an exponentially growing population, ≈45% of the cells (n = 400) were multibudded (defined as the same mother with two or more buds). Each bud neck was decorated by a ring of septins (Fig. 1A), suggesting that these are true buds, not simply surface protrusions from the mother. Individual multibudded cells were picked by micromanipulation and placed onto YPD plates to determine cell viability. Only 23% of the multibudded cells (n = 48) were viable, in comparison with 94% of budded cells (n = 48) from an isogenic WT strain. This poor viability may explain why the mutant strain grew so slowly even at the permissive temperature. In SC medium, the mass-doubling time for cdc42-22 cells was approximately 300 min, in comparison with 150 min for WT cells.
Fig 1.
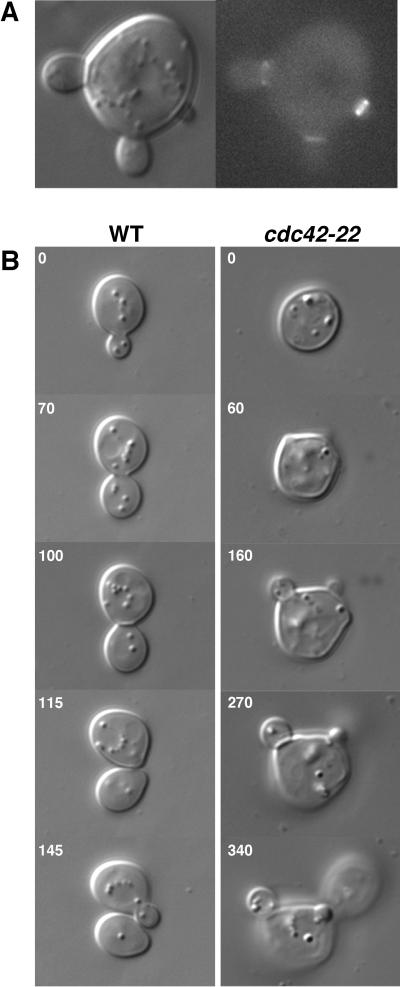
cdc42-22 cells produce multiple buds during one mass-doubling time. (A) Cells of YEF1963 (cdc42-22) carrying pRS316-CDC3-GFP were grown on a SC-Ura plate at 24°C for 16 h and then observed by DIC and fluorescence microscopy. (B) Time-lapse analysis of the budding process of YEF473A (WT) and YEF1963. All images in the same panel are at the same magnification.
To determine whether the multiple buds on the same mother were produced within one mass-doubling time, we performed time-lapse analysis on cdc42-22 cells. In a total of 14 time-lapse sequences with an average duration of 350 min, 10 of 45 cells were observed to form multiple buds. The interval between the appearance of two consecutive buds on the same mother varied greatly, from 0 to 270 min. In 6 of 14 documented cases, two buds emerged simultaneously or within a 10-min interval. In three cases, including the one shown in Fig. 1B (Right, 60 min), two buds emerged simultaneously and maintained active growth, albeit at different rates (compare the sizes of the two buds in Fig. 1B Right, 160 min). In some cases, a third bud emerged within the same mass-doubling time (Fig. 1B Right, 270 min. One of the old buds can be visualized as a bulged shadow just beneath the nascent one). Thus, cdc42-22 cells are able to produce multiple actively growing buds within one mass-doubling time. In contrast, WT cells invariably produced one bud per mass-doubling time (Fig. 1B Left).
cdc42-22 cells also appeared to have a delay in cell separation. Under our experimental conditions, the bud on a WT cell separated from its mother invariably 10–12 min after septum formation, which was manifested by a clear line across the bud neck on a DIC image (33) (Fig. 1B Left, 100 min). In contrast, the interval between septum formation and cell separation for the cdc42-22 cells was much longer, ranging from 35 to 90 min with an average of 54 min (n = 7). For example, at 270 min (Fig. 1B Right), a septum was clearly formed at one bud neck, but the bud remained on the mother even 70 min later. These data suggest that Cdc42p may play a role in septum maturation and/or cell separation.
In WT strain, septum formation occurs only after one nucleus is segregated into the daughter cell (33). To determine how septum formation was coordinated with the nuclear cycle in cdc42-22 cells, we examined 50 septated cdc42-22 cells and found that only 58% of the daughter cells had received a nucleus, suggesting that septum formation can occur without segregating a nucleus into the daughter cell.
To determine whether multiple buds on the same mother were formed within one nuclear cycle, we fixed cdc42-22 and WT cells carrying TUB1-GFP (a GFP-tagged α-tubulin) and stained the cells for F-actin and DNA. The status of the nuclear cycle in each cell was indicated by both the DNA staining and the spindle morphology. Sixty-four percent of the cdc42-22 cells with two or three small buds (n = 70; the size of the bud is 1/3 or smaller than that of the mother) had concentrated F-actin in more than one bud (Fig. 2A), suggesting that these buds on the same mother were growing simultaneously (concentrated F-actin is strictly correlated with active cellular growth in budding yeast). Among these cells, 87% had a single DNA mass and a single spindle (Fig. 2A). On the basis of the spindle morphologies, the nuclear cycle in many such cells had progressed to G1/S (Fig. 2A, bottom cell) or G2/M (Fig. 2A, top cell). The number and the size of the buds were not correlated with the progression of the nuclear cycle. As expected, 90% of the wild-type cells with a single small bud (n = 50) had concentrated F-actin in the bud, and all these cells had a single DNA mass and single spindle.
Fig 2.
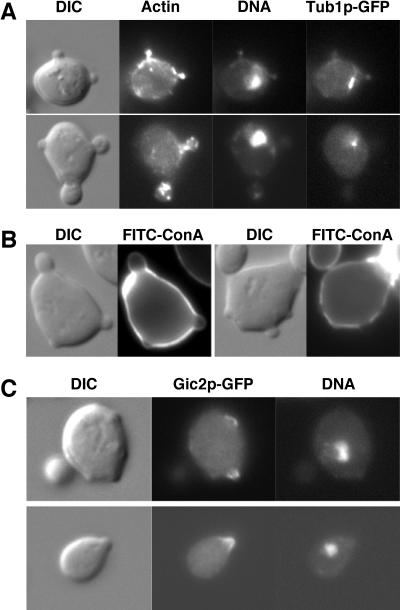
cdc42-22 cells produce multiple simultaneously growing buds within one nuclear cycle. (A) Cells of JPC40 (cdc42-22, TUB1:GFP) were fixed with ice-cold 70% ethanol and stained for F-actin and DNA. Microtubules were visualized with a GFP-tagged α-tubulin. (B) Cells of YEF1963 (cdc42-22) were pulse-labeled with FITC-Con A for 10 min and then released into fresh medium for 60 min to monitor the deposition of new cell wall materials (dark zones on the cell surface). (C) Cells of YEF1963 and YEF473A (WT) carrying pSM217-GIC2-GFP were fixed with ice-cold 70% ethanol and stained for DNA.
The simultaneous growth of multiple buds on a single mother in cdc42-22 cells was further supported by pulse-labeling experiments with FITC-Con A, which stains the glycoproteins in the cell wall. cdc42-22 cells were pulse-labeled with FITC-Con A for 10 min and then released to fresh medium to monitor the deposition of new cell wall materials. At 60 min after the release, multiple buds of the same mother had active polarized growth (indicated by the dark zone in the bud) (Fig. 2B). These data strongly suggest that cdc42-22 cells are able to produce multiple simultaneously growing buds within one nuclear cycle.
To determine whether Cdc42p is partitioned to multiple sites for budding, we localized the active form of Cdc42p by using one of its downstream effectors, Gic2p, as the reporter. Gic2p interacts with the active form of Cdc42p specifically, and this interaction is required for the localization of Gic2p to the presumptive bud site or the tip of a tiny bud (9). Many of the cdc42-22 cells carrying GIC2-GFP had multiple Gic2p caps in discrete regions of the cell cortex, which corresponded to the positions of the tiny buds (Fig. 2C, top cell). In contrast, WT cells always had one Gic2p cap at either the presumptive bud site or the tip of a small bud (Fig. 2C, bottom cell). Assuming that a given amount of active Cdc42p is required to initiate formation of one bud, the data above suggest that there is more active Cdc42p in cdc42-22 cells than that in WT cells (also see Fig. 4B).
The Hyperactive Cdc42p Overrides the Normal Spatial Control of Budding.
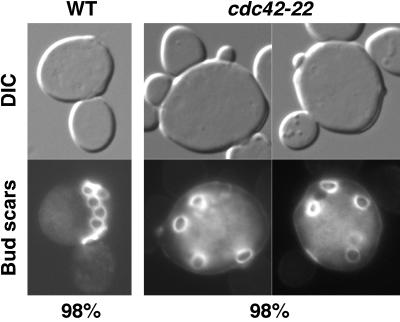
Time-lapse analysis also suggested that cdc42-22 cells could initiate budding from any point of the cell surface. To determine the budding patterns of the cdc42-22 mutant, the mutant strain and an isogenic WT strain were stained for bud scars. As expected, 98% of cdc42-22 cells (n = 100) budded randomly (cells with four or more bud scars counted) (Fig. 3 Right). In contrast, 98% of the WT cells (n = 100) budded axially (Fig. 3 Left). Thus, hyperactive mutation of CDC42 abolishes the spatial regulation of budding.
Fig 3.
cdc42-22 cells bud randomly. Cells of YEF473A (WT) and YEF1963 (cdc42-22) were stained for bud scars with Calcofluor.
A Single Amino Acid Change at Residue 60 Is Sufficient to Cause the Multibudded Phenotype and Makes Cdc42p Hyperactive.
To determine the mechanism underlying the multibudded phenotype of cdc42-22, we sequenced its entire coding region, along with the three other alleles of the same class, cdc42-109, cdc42-128, and cdc42-136. Remarkably, all four alleles had an amino acid change at residue 60 plus additional mutations (cdc42-22, G60A and I182T; cdc42-109, G60D and G107V; cdc42-128, V14A, I21T, and G60D; and cdc42-136, G60C, F98C, and V181D). To determine whether the mutation at residue 60 is sufficient to recapitulate the phenotype of the original alleles, we introduced different single amino acid changes at residue 60 identical to those in the original alleles. All of these single mutants were carried on a centromere-based plasmid and introduced into YEF1191 (cdc42Δ:HIS3, pRS316-pGAL1-CDC42). All single mutants were able to support the growth of the cdc42Δ strain as the sole source of CDC42 at 24°C, but not at 37°C. Thus, like the original alleles, these single mutants are temperature sensitive for growth. All single mutants produced the multibudded phenotype at a frequency similar to that of a cdc42-22 strain, suggesting that a single amino acid change at residue 60 is sufficient to cause the multibudded phenotype.
cdc42-22 and the single mutants that were carried on a centromere-based plasmid in a cdc42Δ background also appeared to express Cdc42p at levels moderately elevated (2- to 5-fold) relative to WT strains (Fig. 4A, lanes 3–7). The expression profile was similar when the mutant allele of CDC42 was integrated at its native locus (Fig. 4A, lanes 1 and 2). In either case, the increased expression alone cannot account for the multibudded phenotype, because a strain that expressed WT Cdc42p at a much higher level (Fig. 4A, lane 8) than any of the mutant strains did not produce any multibudded cells. Thus, it is the nature of the mutations that causes the multibudded phenotype.
Residue 60 is located in a GTP-binding and hydrolysis domain of Cdc42p (18). A mutation at residue 61 is known to render Cdc42p GTPase-deficient and constitutively active (21). GST-PBD pull-down assays were performed to examine the possibility that the alteration at residue 60 might have similar consequences. Because GST-PBD binds only to GTP-bound Cdc42p, this assay can be used to monitor the activity of Cdc42p-GTP in cell lysates (34, 35). Active Cdc42p was detected readily in extracts from cdc42-22 and cdc42G60D cells (Fig. 4B Right). In contrast, very little active Cdc42p was detected in the WT cell lysate, even though more input Cdc42p (Fig. 4B Left) and more GST-PBD (20 μg) (Fig. 4B Right) were included in the assay. This result provides direct biochemical evidence that cdc42-22 and cdc42G60D are hyperactive alleles.
The Hyperactive Cdc42p Is Able to Bypass the Requirement for the Essential GEF Cdc24p.
If cdc42-22 is indeed hyperactive, overexpression of the negative regulators of Cdc42p might reduce the proportion of the multibudded cells in a cdc42-22 strain. Indeed, overexpression of one of its GAPs, Rga1p, reduced the multibudded cells from 46% to 29% (n = 400). These data suggest that Cdc42-22p retains some degree of GTPase activity, and further, that it is still GAP sensitive.
The hyperactive cdc42-22 might also be able to bypass the requirement for its activator (GEF), CDC24, which is essential for cell viability (36). To examine this possibility, pRS314-derived plasmids carrying cdc42-22, the single-mutation alleles, and WT CDC42 were used to transform JPC83 (cdc24Δ:HIS3, pRS316-pGAL1-CDC24) and plated onto SC-Trp + glucose plates to determine whether transformants could be obtained in the absence of Cdc24p expression. All of the plasmids except for those carrying WT CDC42 yielded transformants; they grew slowly and displayed a multibudded phenotype similar to that of a cdc42-22 strain. We eliminated the plasmid pRS316-pGAL1-CDC24 from the host strain by replica-plating transformants onto plates containing 5-fluoroorotic acid. The resulting uracil-requiring strains were temperature sensitive for growth (Fig. 5A). The absence of Cdc24p in these strains was confirmed by Western-blot analysis of total protein from these strains (Fig. 5B). One interesting observation was that the level of Cdc24p in cdc42-22 cells was 2- to 3-fold higher than that of an isogenic WT strain (Fig. 5B). However, the increased expression of CDC24 in cdc42-22 cells is not responsible for the multibudded phenotype, because cdc24Δ strains carrying any of the hyperactive alleles of CDC42 displayed an identical multibudded phenotype. For example, a cdc24Δ strain carrying cdc42-22 produced 60% multibudded cells (n = 500), whereas a cdc42Δ strain carrying the same allele produced 58% multibudded cells (n = 500). These data suggest that the hyperactive alleles of CDC42 are able to completely bypass the need for Cdc24p and that the only essential function of Cdc24p is to activate Cdc42p.
The Hyperactive Cdc42p Requires Its Normal Effectors for Budding.
To determine whether the normal effector pathways are involved in the multibud formation, we examined whether deletion of some of the known effectors or proteins involved in the effector pathways would reduce the multibudded phenotype of cdc42-22 cells. Surprisingly, the cdc42-22 mutation was synthetically lethal with cla4Δ, bni1Δ, gic1Δ gic2Δ, and msb3Δ msb4Δ. The reasons for these synthetic lethalities are unclear, although it is possible that deletion of these genes reduces the pool of the downstream effectors of Cdc42p. In an otherwise WT background, the remaining effectors are sufficient to form a bud. In cdc42-22 cells, however, the limited pool of effectors is sequestered to multiple sites for bud emergence. As a result, at any given site, there is inadequate amount of effectors to initiate bud formation, resulting in cell lethality. These data suggest that these effector pathways are functional in cdc42-22 cells. In contrast, cdc42-22 was not synthetically lethal with ste20Δ. Deletion of STE20 reduced the multibudded cells of a cdc42-22 strain from ≈45% to ≈25% (n = 600), again suggesting that the hyperactive allele of CDC42 indeed uses the normal effector pathways for budding.
Discussion
Cdc42p Activity Regulates Budding Frequency.
This study analyzes the molecular basis for the singularity of the budding process in S. cerevisiae. Previously, a cdc4 mutant and a cdc42 mutant were shown to form multiple buds on the same mother, but detailed analyses indicated that these mutants produced only one normal-sized bud per cell cycle (19, 20, 37). In contrast, we have shown that a single amino acid change at residue 60 of Cdc42p leads to multiple budding events in the same cell cycle, in which buds on the same mother are growing simultaneously and reach mature size. These observations suggest that Cdc42p is the rate-limiting factor in bud formation and that the normal coordination between the budding cycle and the nuclear cycle must involve Cdc42p.
Multiple lines of evidence indicate that an amino acid change at residue 60 makes Cdc42p hyperactive. First, mutations of residue 60 can bypass the requirement for Cdc24p, the only known activator of Cdc42p. Second, GST-PBD pull-down assay indicates that Cdc42pG60X is indeed more active than WT Cdc42p. Third, residue 60 is located within a GTP-binding and hydrolysis domain of Cdc42p (18). Like the mutations at residue 61, a mutation at residue 60 might make Cdc42p hyperactive by decreasing its intrinsic GTPase activity (18). However, the GTPase activity of Cdc42pG60X must be higher than that of Cdc42pG12V or Cdc42pQ61L, which causes cell lethality (21). It has been hypothesized that Cdc42p may need to cycle between its active and inactive states to complete a budding cycle and/or maintain cell viability. When mutagenized at residue 12 or 61, Cdc42p may be locked in the active form, thus preventing cycling (21), whereas Cdc42p with a substitution at residue 60 may undergo enough cycling to support cell growth and division. Taken together, these results suggest that Cdc42p activity regulates budding frequency.
Cycling of Cdc42p Between Its GDP-Bound and GTP-Bound States Is Important for Its Function.
The fact that a single hyperactive mutation in CDC42 can bypass the requirement of Cdc24p for cell viability also suggests that the only essential function of Cdc24p is to activate Cdc42p. We know that the multibudded phenotype and the temperature-sensitive growth phenotype of cdc42-22 cells can be suppressed completely by a single copy of WT CDC42. However, a cdc24Δ strain carrying a WT CDC42 and additionally a single copy of cdc42-22 produces as many multibudded cells as a strain carrying cdc42-22 alone. This finding suggests that the WT Cdc42p protein produced in the cdc24Δ strain is totally inactive (presumably in the GDP-bound state) and that WT Cdc42p must cycle from GDP-bound state to GTP-bound state to suppress the phenotypes of cdc42-22. Thus, Cdc24p is absolutely required for the activation of WT Cdc42p in vivo, and is likely to be the only activator of Cdc42p in the cell. This finding also indicates that the GDP-bound WT Cdc42p produced in the cdc24Δ cells carrying both the WT and a hyperactive allele of CDC42 does not inhibit the multibud formation caused by the hyperactive allele.
The importance of Cdc42p cycling may help explain the surprising observation that the hyperactive allele, cdc42-22, is recessive and that overexpression of WT Cdc42p in the cell does not cause multibud formation (38). We can imagine that Cdc42p-GTP recruits its effectors and actin-binding proteins (ABPs) to the presumptive bud site. GTP hydrolysis results in the release of Cdc42p-GDP from its effectors and ABPs, which remain at the presumptive bud site. Repetitive cycling of Cdc42p causes rapid accumulation of its effectors and ABPs at the chosen site, which may be essential for the initiation and/or continuous growth of the bud. When WT Cdc42p is overexpressed, rapid GTP hydrolysis of Cdc42p (because of its relatively higher intrinsic GTPase activity and its multiple GAPs) may not allow the accumulation of enough active Cdc42p to cause multibud formation. When the WT and the hyperactive Cdc42p are present in the same cell, because the WT protein has a faster rate of GTP hydrolysis than the mutant protein, WT Cdc42p simply out-competes the mutant protein for recruiting the effectors and ABPs to the presumptive bud site, thus suppressing the mutant phenotypes.
Temporal and Spatial Regulation of Budding Must Converge at the Level of Cdc42p.
Our studies (Fig. 1B and Fig. 2) suggest that budding can occur during most of the cell cycle in cdc42-22 cells and that the size and number of the buds are not correlated with progression of the nuclear cycle. We also observed that cdc42-22 cells were able to initiate multiple buds when all three G1 cyclins were depleted (data not shown), suggesting that budding in cells carrying a hyperactive allele of CDC42 is no longer under the control of Cdc28p/G1 cyclins. This conclusion is supported by the observation that temporary expression of cdc42G12V, a dominant, constitutively active allele, can induce polarization of the actin cytoskeleton at several cortical sites in the cell in the absence of all G1 cyclins (39).
The hyperactive mutants of CDC42 exhibited a random budding pattern, suggesting that hyperactivation of Cdc42p overrides the spatial control of the budding process and affirms the notion that spatial regulation of budding proceeds through Cdc42p.
A Model on Cdc42p Clustering in the Absence of the Temporal and Spatial Signals.
How does a hyperactive Cdc42p mutant localize to multiple random sites in the cell cortex to initiate budding? One possibility is that the hyperactive Cdc42p mutant might be randomly distributed on the plasma membrane. Clustering of active Cdc42p (the site of budding) could arise through stochastic movement within the plasma membrane, which could be stabilized by interactions among Cdc42p effectors in the cell cortex. F-actin, which presumably interacts with some of the effectors, does not play a role in Cdc42p clustering (40). This hypothesis can explain many behaviors of different cdc42 mutants.
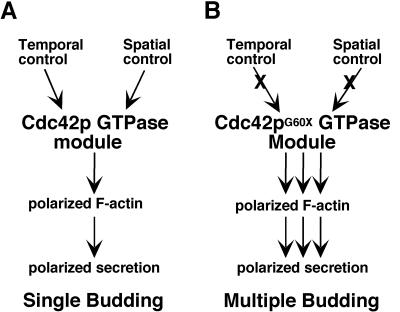
Detailed analysis of the cdc42-22 mutant has led us to several fundamental conclusions (Fig. 6). First, Cdc42p activity is the key determinant in regulating the frequency of budding. Second, the temporal and spatial regulation of budding must converge at the level of Cdc42p (Cdc42p itself or its upstream regulators). Because cdc42-22 defines a previously undescribed class of gain-of-function mutants, isolation and characterization of suppressors of this type of mutations may help us gain new insights into the regulation of Cdc42p function.
Fig 6.
A model for the role of Cdc42p in budding. (A) In WT cells, Cdc42p is subjected to temporal (CDK/cyclins) and spatial (bud-site selection) controls. The activity of Cdc42p is adequate for a single round of budding. (B) In cdc42G60X cells, hyperactive Cdc42p overrides the temporal and spatial controls, and the activity of Cdc42p is sufficient for multiple rounds of budding in a single cell cycle time. The Cdc42p GTPase module includes Cdc42p itself and its immediate regulators such as GEF and GAPs.
Supplementary Material
Acknowledgments
We thank Drs. C. Chan, J. Chant, D. Roof, M. Longtine, I.-c. Yu, and J. Pringle for providing plasmids; Drs. J. Chernoff and M. Chou for GST-PBD; and Dr. M. Douglas for anti-Isp42p antibodies. We also thank Drs. S. Zigmond, D. Lew, M. Chou, E. Vallen, and C. Burd for stimulating discussions. Particular thanks are given to Dr. J. R. Pringle for his encouragement throughout this work. This work was supported by National Institutes of Health Grant GM59216.
Abbreviations
GEF, guanine-nucleotide-exchange factor
GAP, GTPase-activating protein
PBD, p21-binding domain
DIC, differential interference contrast
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pruyne D. & Bretscher, A. (2000) J. Cell Sci. 113, 365-375. [DOI] [PubMed] [Google Scholar]
- 2.Pruyne D. & Bretscher, A. (2000) J. Cell Sci. 113, 571-585. [DOI] [PubMed] [Google Scholar]
- 3.Pringle J. R., Bi, E., Harkins, H. A., Zahner, J. E., De Virgilio, C., Chant, J., Corrado, K. & Fares, H. (1995) Cold Spring Harbor Symp. Quant. Biol. 60, 729-744. [DOI] [PubMed] [Google Scholar]
- 4.Chant J. (1999) Annu. Rev. Cell Dev. Biol. 15, 365-391. [DOI] [PubMed] [Google Scholar]
- 5.Cvrckova F., De Virgilio, C., Manser, E., Pringle, J. R. & Nasmyth, K. (1995) Genes Dev. 9, 1817-1830. [DOI] [PubMed] [Google Scholar]
- 6.Holly S. P. & Blumer, K. J. (1999) J. Cell Biol. 147, 845-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss E. L., Bishop, A. C., Shokat, K. M. & Drubin, D. G. (2000) Nat. Cell Biol. 2, 677-685. [DOI] [PubMed] [Google Scholar]
- 8.Chen G.-C., Kim, Y.-J. & Chan, C. S. M. (1997) Genes Dev. 11, 2958-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J. L., Jaquenoud, M., Gulli, M. P., Chant, J. & Peter, M. (1997) Genes Dev. 11, 2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaquenoud M., Gulli, M. P., Peter, K. & Peter, M. (1998) EMBO J. 17, 5360-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman S. (1998) Trends Cell Biol. 8, 111-115. [DOI] [PubMed] [Google Scholar]
- 12.Evangelista M., Pruyne, D., Amberg, D. C., Boone, C. & Bretscher, A. (2002) Nat. Cell Biol. 4, 32-41. [DOI] [PubMed] [Google Scholar]
- 13.Sagot I., Klee, S. K. & Pellman, D. (2002) Nat. Cell Biol. 4, 42-50. [DOI] [PubMed] [Google Scholar]
- 14.Albert S. & Gallwitz, D. (1999) J. Biol. Chem. 274, 33186-33189. [DOI] [PubMed] [Google Scholar]
- 15.Bi E., Chiavetta, J. B., Chen, H., Chen, G. C., Chan, C. S. & Pringle, J. R. (2000) Mol. Biol. Cell 11, 773-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y., Cerione, R. & Bender, A. (1994) J. Biol. Chem. 269, 2369-2372. [PubMed] [Google Scholar]
- 17.Stevenson B. J., Ferguson, B., De Virgilio, C., Bi, E., Pringle, J. R., Ammerer, G. & Sprague, G. F., Jr. (1995) Genes Dev. 9, 2949-2963. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D. I. (1999) Microbiol. Mol. Biol. Rev. 63, 54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwell L. H. (1971) J. Mol. Biol. 59, 183-194. [DOI] [PubMed] [Google Scholar]
- 20.Richman T. J. & Johnson, D. I. (2000) Mol. Cell. Biol. 20, 8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziman M., O'Brien, J. M., Ouellette, L. A., Church, W. R. & Johnson, D. I. (1991) Mol. Cell. Biol. 11, 3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozminski K. G., Chen, A. J., Rodal, A. A. & Drubin, D. G. (2000) Mol. Biol. Cell 11, 339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladfelter A. S., Moskow, J. J., Zyla, T. R. & Lew, D. J. (2001) Mol. Biol. Cell 12, 1239-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lillie S. H. & Pringle, J. R. (1980) J. Bacteriol. 143, 1384-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guthrie C. & Fink, G. R., (1991) Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology (Academic, San Diego). [PubMed]
- 26.Sikorski R. S. & Boeke, J. D. (1991) Methods Enzymol. 194, 302-318. [DOI] [PubMed] [Google Scholar]
- 27.Bi E. & Pringle, J. R. (1996) Mol. Cell. Biol. 16, 5264-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadwell R. C. & Joyce, G. F. (1992) PCR Methods Appl. 2, 28-33. [DOI] [PubMed] [Google Scholar]
- 29.Park H. O., Bi, E., Pringle, J. R. & Herskowitz, I. (1997) Proc. Natl. Acad. Sci. USA 94, 4463-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pringle J. R., Preston, R. A., Adams, A. E. M., Stearns, T., Drubin, D. G., Haarer, B. K. & Jones, E. W. (1989) Methods Cell Biol. 31, 357-435. [DOI] [PubMed] [Google Scholar]
- 31.Lew D. J. & Reed, S. I. (1993) J. Cell Biol. 120, 1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh E., Skibbens, R. V., Cheng, J. W., Salmon, E. D. & Bloom, K. (1995) J. Cell Biol. 130, 687-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bi E., Maddox, P., Lew, D. J., Salmon, E. D., McMillan, J. N., Yeh, E. & Pringle, J. R. (1998) J. Cell Biol. 142, 1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burbelo P. D., Drechsel, D. & Hall, A. (1995) J. Biol. Chem. 270, 29071-29074. [DOI] [PubMed] [Google Scholar]
- 35.Benard V., Bohl, B. P. & Bokoch, G. M. (1999) J. Biol. Chem. 274, 13198-131204. [DOI] [PubMed] [Google Scholar]
- 36.Broek D., Toda, T., Michaeli, T., Levin, L., Birchmeier, C., Zoller, M., Powers, S. & Wigler, M. (1987) Cell. 48, 789-799. [DOI] [PubMed] [Google Scholar]
- 37.Singer R. A., Bedard, D. P. & Johnston, G. C. (1984) J. Cell Biol. 98, 678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson D. I. & Pringle, J. R. (1990) J. Cell Biol. 111, 143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulli M.-P., Jaquenoud, M., Shimada, Y., Niederhauser, G., Wiget, P. & Peter, M. (2000) Mol. Cell 6, 1155-1167. [DOI] [PubMed] [Google Scholar]
- 40.Ayscough K. R., Stryker, J., Pokala, N., Sanders, M. & Crews, P. (1997) J. Cell Biol. 137, 399-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.