Abstract
Excessive estrogen stimulation unopposed by progesterone strongly predisposes to endometrial cancer. Because the antiproliferative effect of progesterone requires the progesterone receptor (PR), which exists in two isoforms, PR-A and -B, we reasoned that variants in the PR gene may predispose to endometrial cancer. We found six variable sites, including four polymorphisms in the hPR gene and five common haplotypes. One promoter region polymorphism, +331G/A, creates a unique transcription start site. Biochemical assays showed that the +331G/A polymorphism increases transcription of the PR gene, favoring production of hPR-B in an endometrial cancer cell line. Using a case-control study nested within the Nurses' Health Study cohort, we observed a statistically significant association between the +331G/A polymorphism and the risk of endometrial cancer, which was even greater in overweight women carriers. After including a second population of controls, these associations remained intact. Our findings suggest that the +331G/A hPR gene polymorphism may contribute to endometrial cancer risk by increasing expression of the hPR-B isoform.
Although stimulation by exogenous and endogenous estrogens is critical in the development of endometrial cancer, a strong inherited component also exists. Population-based studies have demonstrated up to a 3-fold increased risk of endometrial cancer among first-degree relatives (1–3). Endometrial cancer is a feature of hereditary nonpolyposis colorectal cancer syndrome; however, this syndrome is too infrequent to account for the majority of cases. Genetic mechanisms for endometrial cancer are likely distinct from cancers in other hormonally responsive tissues such as breast cancer. For instance, mutations of the BRCA1 and BRCA2 genes are powerful risk factors for breast and ovarian cancer, yet not for endometrial cancer. These findings suggest that unique genetic determinants of risk for endometrial cancer exist, perhaps working in concert with environmental risk factors.
Overwhelming evidence supports the increased risk of endometrial cancer with estrogen replacement therapy in postmenopausal women (4–6). Increased endogenous estrogen from prolonged ovarian cycling and obesity also contributes to risk (4, 7). In contrast, progesterone potently counteracts estrogen-dependent endometrial cancer development (8–10). In searching for endometrial cancer susceptibility genes, components of the progesterone-dependent pathway are likely candidates. As demonstrated in progesterone receptor-deficient mice, the physiological effects of progesterone depend completely on the presence of the human progesterone receptor (hPR), a member of the steroid-receptor superfamily of nuclear receptors (11). The single-copy human (hPR) gene uses separate promoters and translational start sites to produce two isoforms, hPR-A and -B (12–14), which are identical except for an additional 165 amino acids present only in the N terminus of hPR-B (15, 16). Although hPR-B shares many important structural domains as hPR-A, they are in fact two functionally distinct transcription factors (17), mediating their own response genes and physiological effects with little overlap (18, 19). Selective ablation of PR-A in a mouse model, resulting in exclusive production of PR-B, unexpectedly revealed that PR-B contributes to, rather than inhibits, epithelial cell proliferation both in response to estrogen alone and in the presence of progesterone and estrogen (20). These results suggest that in the uterus, the PR-A isoform is necessary to oppose estrogen-induced proliferation as well as PR-B-dependent proliferation. Considering the overwhelming epidemiological evidence of the role of estrogen and progesterone in endometrial cancer causation, and the biological data demonstrating the selective contributions of the individual PR isoforms to endometrial hyperplasia, we hypothesized that variations in the hPR gene may result in a loss of progesterone-mediated tumor suppression and may predispose to endometrial cancer. We sequenced the eight exons, flanking splice sites, and the promoter regions of the hPR gene to identify polymorphisms. We inferred haplotypes and identified a polymorphism that is associated with a risk of endometrial cancer among women in the Nurses' Health Study (NHS) cohort. Using an in vitro system, we showed that this polymorphism results in a unique transcriptional start site and increased transcriptional activity, likely resulting in increased production of the hPR-B isoform.
Materials and Methods
Study Population.
The NHS began in 1976, when 121,700 female U.S. registered nurses between ages 30 and 55 yr completed and returned the initial NHS questionnaire. Information regarding endometrial cancer risk factors was obtained from biennial questionnaires and a questionnaire completed at the time of blood collection. Women were defined as postmenopausal at the time of blood collection if they reported having a bilateral oophorectomy or no menstrual cycle within the last 12 mo before blood draw. Menopausal status and postmenopausal hormone use, including the dose and duration of current use of conjugated estrogen or estrogen plus progestin, were updated until the date of diagnosis for cases and matched controls. First-degree family history of endometrial and colorectal cancer was assessed retrospectively from the 1996 follow-up questionnaire. Between 1989 and 1990, blood samples were collected from 32,826 women. Follow-up has been >90% in all of the subsequent questionnaire cycles for this subcohort.
In this study, we included both incident and prevalent cases of invasive endometrial carcinoma from the blood subcohort of the NHS. Eligible incident cases consisted of women with pathologically confirmed invasive endometrial cancer diagnosed anytime after blood collection up to June 1, 1996, with no previously diagnosed cancer except for nonmelanoma skin cancer. Prevalent cases had pathologically confirmed invasive endometrial cancer diagnosed between 1976 and the date of blood collection, with no previously diagnosed cancer except for nonmelanoma skin cancer. Controls were randomly selected participants who gave a blood sample, had not had a hysterectomy, and were free of diagnosed cancer (except nonmelanoma skin cancer) up to and including the interval in which the case was diagnosed. Controls were matched to cases on year of birth, menopausal status at blood draw and diagnosis, use of hormone replacement therapy at blood draw (current vs. not current users), as well as time of day, month, and fasting status at blood draw (21). The case-control study consisted of 187 invasive endometrial cancer cases and 397 matched controls. In addition, 506 women who were controls in a nested case-control study of breast cancer and who had not had a hysterectomy (21) were genotyped for the +331G/A and the AluIns polymorphism. The protocol was approved by the Brigham and Women's Hospital Committee on Human Subjects.
Single-Nucleotide Polymorphism (SNP) Discovery.
Genomic DNA from each subject was prepared by using a QiAmp 96 spin blood procedure (Qiagen, Chatsworth, CA). “Working draft” hPR genomic sequence (GeneIndex: 8570374) was obtained through the National Center for Biotechnology Information and assembled manually for primer design (see supporting information on the PNAS web site, www.pnas.org). Using germ-line DNA from the 68 incident endometrial cancer cases, we screened the eight exons, flanking splice sites, and 5′ UTR. Briefly, we screened for variants using Big Dye Terminator chemistry (PE Applied Biosystems) on the ABI 377X automated sequencer (PE Applied Biosystems). Base calling of the sample files was performed by using ABI sequence analysis software, version 3.1. sequencher; version 3.0 alignment software was used to mark potential heterozygous positions and display them for evaluation. Heterozygotes were called at positions where the secondary peak height was greater than or equal to 45% of the primary peak height in both forward and reverse sequence reads. Where possible, restriction digests with appropriate enzymes were performed to confirm the sequences.
Genotyping Assays.
Genotyping analysis was performed by various techniques: Pyrosequencing (Pyrosequencing, Uppsala, Sweden) (22), Single Base Extension and Fluorescence Detection (LJL Biosystems, Sunnyvale, CA) (23), and restriction fragment length polymorphism. Primers used in these assays are published in Tables 1 and 2 as supporting information on the PNAS web site, www.pnas.org. All of the genotyping was performed by laboratory personnel unaware of case-control status, and blinded quality control samples were inserted to validate genotypes. Concordance for blinded samples was 100%.
Statistical Analysis.
Student's t test and the χ2 test were used to evaluate differences in endometrial cancer risk factors between cases and controls. Odds ratios (ORs) and 95% confidence intervals were calculated by using conditional and unconditional logistic regression. In addition to the matching variables, we adjusted for endometrial cancer risk factors: body mass index (BMI) (kg/m2), weight gain since age 18, age at menarche, parity/age at first birth, duration of postmenopausal use, pack-years of smoking, first-degree family history of endometrial cancer, and first-degree family history of colorectal cancer. Indicator variables for all genotypes were created by using the wild-type hypothesized low-risk genotype as the reference category in the regression models. Because of the low prevalence of homozygote variants, we combined heterozygotes and homozygotes in the logistic regression analysis. Unconditional logistic regression models enabled controls already included in the nested breast cancer case-control study in which we have available genotypes to be included in these analyses. Interactions between genotypes and endometrial cancer risk factors were evaluated by including interaction terms between genotype and risk factor variables in unconditional logistic regression models. The likelihood ratio test was used to assess the statistical significance of these interactions. We used the sas (SAS Institute, Cary, NC) statistical package for all analysis (sas, version 8.0). We tested Hardy–Weinberg agreement by using a χ2 test. We calculated/D′/from phase-unknown data by calculating maximum likelihood estimates of the gametic proportions; an exact solution for the maximum likelihood proportions was used. We estimated haplotype frequencies from our observed genotypes by using the multilocus program (24) and arlequin 2.0 written by L. Excoffier, S. Schneider, and D. Roessli (http://anthro.unige.ch/arlequin). We ascertained differences in haplotype distributions between cases and controls by using the χ2 test.
Expression Assays.
Transcription factor-binding sites were identified by using matinspector, Ver. 2.2 (http://transfac.gbf.de/cgi-bin/matSearch/matsearch.pl) (25). Fragments of the human progesterone receptor promoter containing +331G or +331A were amplified from genomic DNA by using a sense primer (hPR–711, GGATCCATTTTATAAGCTCAAAGA) and an antisense primer (hPR+763 cgaagcttGCCTTCAGCTCAGTCATGA), then cloned into the Hind3 site of pGL2-Basic (Promega). Human endometrial carcinoma cells [Ishikawa, courtesy of S. Safe (26)] were cultured in DMEM supplemented with 10% FBS/100 units/ml of penicillin/100 μg/ml of streptomycin in a humidified atmosphere at 37°C with 5% CO2. pGL2-hPR(−711 to +763)+331G and +331A plasmids were transfected into human endometrial carcinoma cells by using the Superfect (Qiagen) reagent according to manufacturer's protocol. One day after transfection, total RNA was isolated, then digested with DNaseI, RNase free. Rapid amplification of cDNA ends (5′ RACE) analysis was performed by using the FirstChoice RNA ligase-mediated (RLM)-RACE (Ambion, Austin, TX) kit according to the manufacturer's protocol. The product of first-strand synthesis was amplified by using the 5′-linker outer primer and the GL2 primer, and that product was then amplified by using the 5′-linker inner primer and the hPR+763 primer. PCR products were separated on a 2% agarose gel. For reverse transcriptase (RT)-PCR, RT reactions of DNase-digested total RNA were performed by using the Superscript System (Invitrogen). The hPR-B, combined hPR-B+A transcripts, and β-actin were amplified by using previously described primers (27). For quantitation of isoform expression, the PCR reaction products were blotted onto nitrocellulose then probed with a P32-labeled hPR-A specific probe shared by both products. The hPR−B and hPR-B+A signals were quantified by using a Storm PhosphorImager (Molecular Dynamics). After subtracting background, the ratio of the hPR−B:hPR−B+A signal for each sample was determined, then normalized to the ratio obtained by +331G within each experiment to measure fold increase. Results from four experiments with five samples in each group were compiled for a composite analysis by using the statview program. For analysis of protein isoform expression, the hPR full-length cDNA (courtesy of Bert O'Malley, Baylor College of Medicine) was cloned in frame with either the wild-type (+331G) or the variant (+331A) hPR promoter, producing hPR(−711 to +763)-hPR. Cultured T47D cells were transfected with either pcDNA3.1-hPR-B, hPR(−711 to +763)-hPR + 331G, or hPR(−711 to +763)+hPR-331A by the calcium phosphate technique. Cellular extracts were separated by SDS/PAGE, transferred to nitrocellulose, and probed with anti-PR antibody (Santa Cruz Biotechnology).
Transient Transfection.
Ishikawa cells cultured in six-well dishes (200,000 cells per well) were transfected with 2 μg of pGL2-hPR(−711 to +763)+331G or +331A plasmid with 0.5 μg of cytomegalovirus-β galactosidase, which served as an internal control of transfection efficiency. Three hours after applying the plasmid–Superfect complexes, the medium was changed to F12/DMEM lacking phenol red with 10% FBS and l-glutamine supplemented with 17 β-estradiol (2.5 nM final concentration) or vehicle (ethanol). After 24 h, cellular extracts were harvested in Reporter Lysis Buffer (Promega), and luciferase activity was measured with a luminometer (Autolumat 953, EG & G, Gaithersburg, MD) by using the Promega luciferase system; β-galactosidase activity was measured by conversion of o-nitrophenyl β-d-galactopyranoside. Results from four experiments (n = 11 for each sample) were normalized to the average corrected luciferase activity of the +331G construct in the absence of estrogen. The mean and SEM were determined for replicate samples. Differences were determined by factorial analysis of variance with the statview program. A P value of less than 0.01 was considered significant.
Results
SNP Discovery.
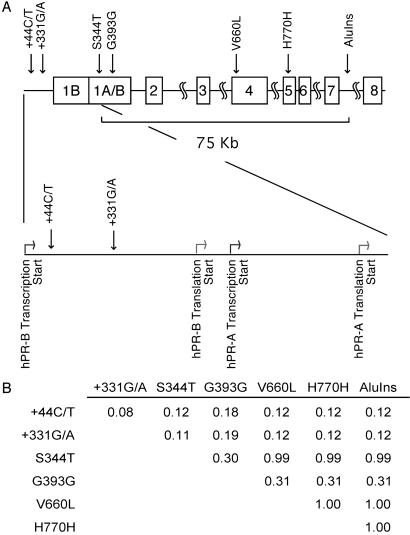
By sequencing hPR in germ-line DNA from 68 incident invasive endometrial cancer cases, we identified six variable sites. Variants were found in the promoter region, +44C/T and +331G/A, between the hPR-B (nucleotide + 1) and the hPR-A (nucleotide + 751) transcriptional start sites (Fig. 1A). There were also two variants in the coding region, S344T and G393G, and we confirmed the previously reported codonV660L, H770H polymorphisms, and the AluIns (Progin) allele in intron 7. In addition, we found one case with a silent mutation in exon 4 (G661G). To understand the degree to which these polymorphisms are linked, we performed standardized pairwise linkage disequilibrium (LD) (D′) tests (Fig. 1B). We observed that the V660L, H770H, and AluIns polymorphisms are in complete LD (D′ = 1.00), and that the S344T polymorphism was also linked with V660L, H770H, and AluIns (D′ = 0.99). LD in a U.S. population of Northern European descent extends 60 kb on average (28). In our population of mostly Caucasian women, the LD of the hPR gene extends approximately 70 kb (Fig. 1B). Given the high degree of LD, we grouped S344T, V660L, H770H, and the AluIns polymorphisms as a single genotype, which we call the framework haplotype for all statistical analysis.
Fig 1.
(A) Diagram of hPR gene. Darker and lighter arrows indicate transcriptional and translational start sites, respectively. Polymorphism sites are indicated. (B) Standardized pairwise LD (D′) for endometrial cancer cases and controls. D′ numbers were calculated for each combination of polymorphisms. A D′ of 1.00 indicates complete LD.
+331G/A SNP Produces a Transcriptional Start Site.
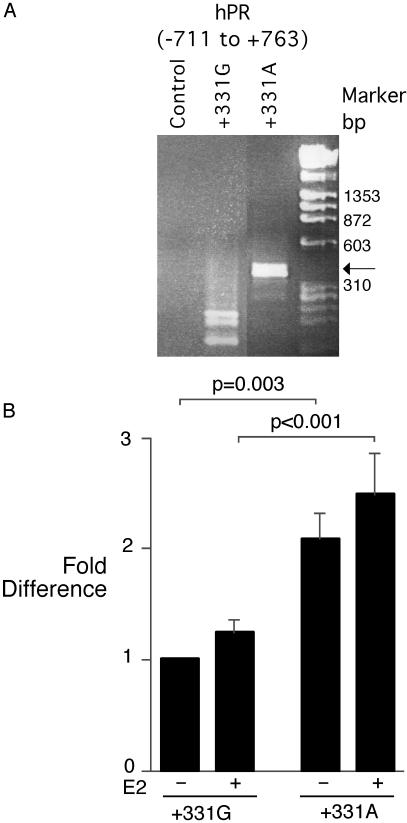
The S344T and G393G coding region polymorphisms produce conserved alterations, not likely to have a functional consequence. Of the promoter region polymorphisms, we first analyzed their potential effect on transcription factor binding by using the matinspector, Ver. 2.2, program (25). Computer modeling predicted that the +331G/A polymorphism would create a potential TATA-box. This prediction was of potential importance, because the human progesterone receptor promoter is a TATA-less promoter (12), and the introduction of a TATA-box, even lacking the complete consensus sequence, could create a new site for transcriptional initiation (29). To address this issue, we designed hPR promoter constructs [hPR (−711 to +763)] that included the hPR-B transcriptional start site (+1), the hPR-A transcriptional start site (+751), and either wild-type or variant nucleotides at position +331. After transfection of these reporter plasmids into Ishikawa endometrial cancer cells (26), RNA was harvested and subjected to 5′ RACE to identify sites of transcription initiation. After transfection of the hPR +331G promoter construct, three 5′ RACE products were found (Fig. 2A, lane 2), and sequence analysis confirmed their origin near the previously described hPR-A transcription initiation sites. Multiple closely spaced sites of transcription initiation, commonly seen in TATA-less promoters, have been found for hPR-A (12). After transfection with the hPR +331A promoter construct, a product was found (Fig. 2A, lane 3), and sequence analysis showed that the product began at nucleotide +398, which lies between the hPR-B and hPR-A transcriptional start sites, and which is 67 bp downstream of the +331G/A polymorphism. The finding of a new RNA species is consistent with the +331G/A polymorphism producing a unique transcriptional start site, producing an mRNA species with a 5′ untranslated region before the hPR-B initiator methionine, likely producing the hPR-B protein isoform. In this assay, we did not observe a 5′ RACE product derived from the hPR-B transcription start site, perhaps reflecting a low abundance of hPR-B message, inefficient copying by RT, or competition between hPR-A and hPR-B 5′ RACE products for amplification.
Fig 2.
(A) A 5′ RACE product is produced by the hPR promoter +331A polymorphism. RNA from endometrial cancer cells transfected with hPR(−711 to +763) regulated plasmids was subjected to 5′ RACE analysis. Control transfected cells produced no 5′ RACE product, whereas +331G produced fragments near the previously described hPR-A transcriptional start sites. The +331A construct produced a band (arrow) that began at nucleotide +398. (B) +331A polymorphism increases hPR promoter transcriptional activity. Endometrial cancer cells were transfected with hPR(−711 to +763)-luciferase reporter constructs, then stimulated with 2.5 nM 17-β-estradiol (+) or vehicle control (−). The mean +/−SEM corrected luciferase activity from four experiments (n = 11 for each group) is shown.
hPR+331A Has Increased Transcriptional Activity.
Because the introduction of a new transcriptional start site may alter promoter strength, we used the same wild-type and +331G/A SNP hPR-(−711 to +763) luciferase reporter constructs to analyze transcription activity. Because transcription from the hPR-B, hPR-A, or variant transcription start sites could produce the luciferase protein, this analysis is not isoform-specific. The corrected luciferase activity from cells transfected with the hPR(−711 to +763)+331A reporter plasmid was consistently stronger than the wild-type (+331G) plasmid (2.1 ± 0.2-fold, P < 0.003) (Fig. 2B). Stimulation of transfected cells with 17-β-estradiol in phenol red-free medium produced a mild (nonsignificant) increase in luciferase activity of both +331G and +331A (Fig. 2B). This finding is not surprising, because hPR promoters lack a complete estrogen-response element (30).
The hPR-B Encoding Transcript Is Increased by the hPR+331A SNP.
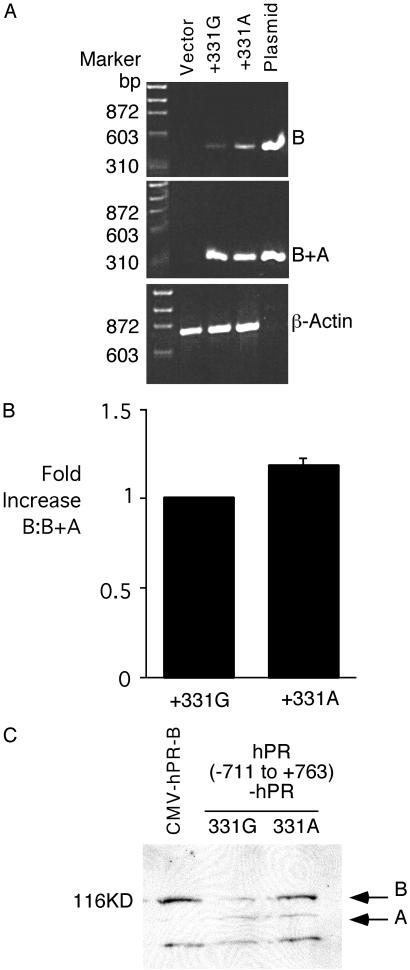
Because the hPR+331A SNP increased transcription and produced a new 5′ RACE product, we hypothesized that the balance of messenger RNA encoding hPR-B and -A would be altered. To analyze this issue, we cloned the hPR(−107 to +1450) promoter fragment into an expression plasmid. To analyze the effect of the +331 SNP on isoform-specific transcript production, RNA from transfected cells was isolated and copied by using a RT and then amplified with primer pairs specific for hPR-B and for sequence shared by hPR-B and -A (B+A). In three different cell lines, the +331A SNP was associated with increased production of hPR-B relative to hPR-B+A compared with the wild-type +331G construct (Fig. 3A). Relative amplification of hPR-B and hPR-B+A was measured by using Southern blotting with a hPR-A probe shared by both the hPR-B and hPR-B+A amplification products. The mean normalized hPR-B:hPR-B+A ratio from four experiments demonstrated significantly more hPR-B transcript produced by +331A compared with +331G (17.8 ± 3.8% increase, P = 0.002) (Fig. 3B). Because transcript levels may not predict protein isoform expression, we exchanged the luciferase cDNA with the hPR cDNA, thereby placing the hPR promoter, with or without the +331 variant, in front of the hPR cDNA. After transient transfection, relative hPR isoform expression was analyzed by Western blotting. Expression of hPR-B by the cytomegalovirus promoter produced just hPR-B (Fig. 3C Left). By contrast, the hPR(−711 to +763)-hPR constructs produced both hPR-A and -B. Three independent experiments have shown that hPR(−711 to +763)-hPR +331A produced a greater amount of hPR-B protein compared with the corresponding +331G construct. Taken together, these biochemical assays demonstrate that the +331A SNP alters transcription from the hPR promoter in favor of producing the hPR-B encoding transcript and protein.
Fig 3.
(A) +331A polymorphism increases production of hPR-B transcript. RT-PCR analysis of hPR isoform expression. RT products were amplified with primers specific for hPR-B (B) (Top), the shared region of hPR-B and -A (B+A) (Middle), and for β-actin (Bottom). Total RNA from control transfected endometrial cancer cells and plasmid DNA was used for control. Despite an equal PCR product with B+A amplification, the B product from +331A product was more intense than +331G, suggesting increased expression of hPR-B. Similar results were obtained in three different cell lines. (B) Bar graph demonstrating increased relative hPR-B:hPR-B+A ratio for the +331A variant compared with +331G. Relative isoform amplification was measured by using Southern blotting with a probe shared equally by hPR-B and -A. (C) Anti-hPR Western blot demonstrating increased expression of hPR-B compared with hPR-A after transfection of hPR(−711 to +763)-hPR + 331A compared with +331G. Protein size marker is shown (Left); Right, position of hPR-B (B) and -A (A).
SNPs and Endometrial Cancer Risk.
Having identified variants existing in promoter and coding regions of the hPR gene, a hPR gene framework haplotype, and the functional consequences of the +331G/A polymorphism, we used a nested case-control study to analyze the risk of endometrial cancer associated with these polymorphisms. In this study, we included both incident and prevalent cases of invasive endometrial carcinoma from the blood subcohort of the NHS. Eligible incident and prevalent cases consisted of women with pathologically confirmed invasive endometrial cancer with no previously diagnosed cancer except for nonmelanoma skin cancer. Controls were randomly selected participants who gave a blood sample, had not had a hysterectomy, and were free of diagnosed cancer, except nonmelanoma skin cancer. Controls were matched to cases on year of birth, menopausal and hormone replacement therapy status, as well as other matching factors (see Materials and Methods). The case-control study consisted of 187 invasive endometrial cancer cases and 397 matched controls. Compared with controls, cases had a significantly greater BMI at diagnosis and gained more weight since age 18 (Table 3, which is published as supporting information on the PNAS web site). Cases had children at a younger age (P = 0.01) and tended to give birth to fewer children (P = 0.04). Cases were more likely to have a family history of endometrial or colorectal cancer than controls, and cases smoked significantly less than controls. All 187 documented cases of invasive endometrial cancer from 1978 to 1996 and 397 matched controls were genotyped for all of the polymorphisms. The hPR genotype frequencies were similar between incident and prevalent cases, so they were combined for all statistical analyses. The prevalence of variant carriers was (cases vs. controls) 12% vs. 13% for +44C/T, 15% vs. 11% for +331G/A, 61% vs. 57% for G393G, and 27% vs. 30% for the framework haplotype (Table 4, which is published as supporting information on the PNAS web site). The distributions of the hPR genotypes were in accordance with the Hardy–Weinberg equilibrium. We observed a statistically significant association between the +331G/A polymorphism and endometrial cancer risk. Compared with the +331G/G wild-type genotype, the adjusted OR for women with the +331G/A and +331A/A was 1.90 (95% confidence interval, 1.10–3.29) (Table 4). We attempted to analyze the heterozygotes and homozygotes separately; unfortunately, there were too few homozygote variants for an informative analysis.
Considering our findings, we sought to further define this relationship by including a second population of control women from the same cohort. We genotyped 506 additional controls from a breast cancer study, which included women with no cancer other than nonmelanoma skin cancer. Comparison of population characteristics of the second control group with the first control group revealed no material differences in ethnicity, mean BMI, weight gain since age 18, first-degree family history of endometrial or colorectal cancer, age at first birth, menarche, and parity. The association of +331G/A with endometrial cancer was essentially the same with an adjusted OR 1.71 (95% confidence interval, 1.01–2.94). Considering the overall similarity of the two control groups, we do not believe our results are due to population admixture or other possible confounding factors. We did not observe an association between +44C/T, G393G, or the framework haplotype polymorphisms and endometrial cancer risk (Table 4).
Polymorphisms and Endometrial Cancer Risk Factors.
Given that BMI is a major endometrial cancer risk factor, we evaluated effect modification of the +331G/A genotype by BMI. As expected overall, there was an increasing risk of endometrial cancer with increasing BMI. We observed a statistically significant association between women who were overweight (BMI > 28 kg/m2) and carried at least one variant allele with endometrial cancer risk (Table 5, which is published as supporting information on the PNAS web site). Compared with wild-type (+331G/G) lean women (BMI < 25 kg/m2), the OR for overweight (BMI > 28 kg/m2) women variant carriers (+331A/G + 331A/A) was 4.71(1.87–11.87)-fold stronger than that for overweight women who did not carry the variants (OR = 1.20, 95% confidence interval, 0.72–2.00) (Table 5). This result included the second population of 506 control women. Hormone replacement therapy (HRT) is another well-established risk factor for endometrial cancer; however, we observed no significant association between any of the hPR polymorphisms, with never, past, or current HRT use (data not shown).
Haplotypes and Endometrial Cancer Risk.
The interactions of multiple SNPs within a haplotype can affect biological phenotypes (31). Therefore, in addition to analyzing the polymorphisms independently, we also analyzed them in the context of haplotypes. The haplotype frequencies were computed from phase-unknown genotypes using multiple locus haplotype analysis (mlocus) program (24) and arlequin 2.0. These programs estimate haplotype frequencies by using an expectation maximization algorithm. In our population of mostly Caucasian women, we estimated eight haplotypes of a theoretically possible 128 (27). Of these 128 possible combinations, five common haplotypes (frequency of >5%) accounted for 99% of the chromosomes at this locus. Only four SNPs are necessary to distinguish the ancestral haplotypes in our population: +44C/T for haplotype F (Table 6, which is published as supporting information on the PNAS web site), +331G/A for haplotype E, G393G for haplotype C, and any of S344T, V660L, H770H, or AluIns for haplotype B. By using the χ2 test, we found no significant differences in any haplotype combination between the cases and the controls.
Discussion
To our knowledge, this is the first population-based study that describes polymorphisms in the hPR gene and their potential contribution to endometrial cancer risk. We identified six variable sites within the hPR gene and constructed haplotypes. Given our SNP discovery strategy, we have missed polymorphisms in the introns, 3′ and 5′ untranslated regions; however, our haplotype analysis did not reveal an association with endometrial cancer, making a highly prevalent disease causing polymorphism in these regions unlikely. We anticipated that loss-of-function polymorphisms altering the hPR protein or its production may reduce the antiproliferative activity of progesterone and predispose to endometrial cancer. We identified two polymorphisms in the coding region, S344T and G393G, yet they were not associated with endometrial cancer risk. Considering that many functional polymorphisms affect promoter function, rather than protein function, we also screened for polymorphisms in the hPR gene promoter. The hPR gene has two promoters, regulating production of the hPR-B and -A protein isoforms, and we identified two polymorphisms, +44C/T and +331G/A, 3′ of the hPR-B transcriptional start site (+1). Importantly, the +331G/A SNP is upstream of the hPR-B translational start site (+744) and the hPR-A transcriptional start sites (12). The +44C/T polymorphism was not associated with an increased risk of endometrial cancer, whereas carriers of the variant +331G/A allele have almost a 2-fold increased risk for developing endometrial cancer compared with noncarriers.
To our surprise, the biochemical studies suggested that the +331G/A polymorphism was not a classical loss-of-function polymorphism reducing hPR gene transcription. Rather, the +331G/A polymorphism increased transcription (Fig. 2B). This finding posed a paradox, because increased hPR protein production would logically antagonize endometrial cancer formation by promoting progesterone-dependent endometrial cell differentiation and reducing proliferation. However, the antineoplastic effects of progesterone depend on tight regulation of the hPR-A and -B isoform balance (27, 32, 33). By altering the balance of hPR-A and -B, the +331G/A polymorphism may predispose to endometrial cancer.
The hPR-B and -A proteins are identical, except hPR-B has an additional 165 amino acids in its N terminus (34). hPR-B is a more potent transcriptional activator than hPR-A, because the hPR-B N terminus can selectively recruit transcriptional coactivators (17, 35, 36). By comparison, hPR-A is transcriptionally inactive and functions as a strong transdominant repressor of hPR-B (15, 37). Finally, through a nontranscriptional mechanism, hPR-B alone can promote cell growth by interacting with the estrogen receptor and stimulate the Src/p21ras/Erk pathway (38). Through these transcriptional and nontranscriptional mechanisms, the hPR isoforms are in fact functionally distinct. Microarray analysis of human breast cancer cells expressing either hPR-B or -A have confirmed that each hPR isoform has a unique set of target genes, with little overlap (19). Genes selectively up-regulated by hPR-B predisposing to endometrial cell survival and proliferation include IAP homolog C and cyclin D3, as well as the antiapoptotic protein BcL-XL (19, 39). Further evidence implicating the PR-B isoform in endometrial cell proliferation comes from the PR-A-deficient mouse model, which demonstrated marked endometrial cell proliferation in response to progesterone and estrogen stimulation (20). Therefore, although PR-A is necessary for progesterone-mediated repression of endometrial cell proliferation, PR-B antagonizes this function. Consequently, the increased production of hPR-B by the +331G/A polymorphism may predispose to cancer development through increased hPR-B-dependent stimulation of endometrial cell growth.
There is conflicting evidence regarding the role of specific hPR isoforms in endometrial cancer. Highly malignant forms of endometrial, cervical, and ovarian cancer have been correlated with overexpression of hPR-B, suggesting causation (40, 41). However, this mechanism stands in contrast to the observation that the hPR-A isoform is often found to be overexpressed in human endometrial cancer cells (27, 33). It should be noted that in our population the +331G/A polymorphism is present in a subset (15%) of endometrial cancer cases. Using an unbiased genomic approach to understanding the genetic basis of endometrial cancer risk, our work suggests that hPR-B, rather than hPR-A, contributes to cancer development. These observations can be reconciled if: (i) any imbalance of hPR isoforms can predispose to endometrial cancer; or (ii) the increased expression of hPR-A in well-established endometrial cancer is an epiphenomenon unrelated to the causative events in cancer formation. A detailed mechanistic understanding of the role of PR isoform balance in cancer development will require endometrial overexpression of specific PR isoforms in vivo.
Genetic risk factors of endometrial cancer likely work in concert with environmental stimulants, particularly exogenous and endogenous estrogens. Extraovarian estrogen derived from androgens aromatized in adipose tissue is a well-established risk factor for endometrial cancer. We observed evidence of an interaction between a BMI > 28 kg/m2 and the +331G/A polymorphism. Using lean (BMI < 25 kg/m2) wild type + 331G as reference, we found that overweight (BMI > 28 kg/m2) variant carriers had a 4.71-fold risk of endometrial cancer, whereas their overweight wild-type (+331G) counterparts had a 1.20-fold risk. Thus, the effect of the hPR genotype may be modified by BMI.
In this study, we identified six polymorphisms in the hPR gene, one of which is modestly associated with endometrial cancer. The +331G/A polymorphism may predispose to endometrial cancer by altering the balance of hPR-B and -A. There are several potential mechanisms that may explain the increased transcriptional activity and overexpression of hPR-B associated with the +331G/A polymorphism, and this is the subject of active work. Further mechanistic studies using transgenic mice are required to better understand the role of hPR isoform balance and endometrial cancer. Taken together, our work supports screening for polymorphisms in progesterone pathway genes as a strategy for identifying candidate markers of endometrial cancer risk.
Supplementary Material
Acknowledgments
We thank the participants of the NHS for continuing exceptional cooperation. We thank Lisa Li, Lori Arbietman, and Jay Boltax for technical assistance and Steven Orzack for assistance with LD estimations. We thank Frank Speizer, principal investigator of the NHS, for support. This work is supported by National Institutes of Health Grants CA82838 (I.D.), CA87969, CA49449, and a grant from the American Cancer Society (RPG-00-061-01 CCE to I.D.). G.S.H. is supported by Mentored Clinical Scientist Development Award K08 HL03667-01A1.
Abbreviations
hPR, human progesterone receptor
OR, odds ratio
BMI, body mass index
NHS, Nurses' Health Study
LD, linkage disequilibrium
SNP, single-nucleotide polymorphism
RACE, rapid amplification of cDNA ends
RT, reverse transcriptase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Parazzini F., La Vecchia, C., Moroni, S., Chatenoud, L. & Ricci, E. (1994) Int. J. Cancer 59, 460-462. [DOI] [PubMed] [Google Scholar]
- 2.Sandles L., Shulman, L., Elias, S., Photopulos, G., Smiley, L., Posten, W. & Simpson, J. (1992) Gynecol. Oncol. 47, 167-171. [DOI] [PubMed] [Google Scholar]
- 3.Gruber S. & Thompson, W. (1996) Cancer Epidemiol. Biomarkers Prev. 5, 411-417. [PubMed] [Google Scholar]
- 4.Kelsey J., LiVolsi, V., Holford, T., Fischer, D., Mostow, E., Schwartz, P., O'Connor, T. & White, C. (1982) Am. J. Epidemiol. 116, 333-342. [DOI] [PubMed] [Google Scholar]
- 5.Mack T., Pike, M., Henderson, B., Pfeffer, R., Gerkins, V., Arthur, M. & Brown, S. (1976) N. Engl. J. Med. 294, 1262-1267. [DOI] [PubMed] [Google Scholar]
- 6.Hulka B., Fowler, W., Kaufman, D., Grimson, R., Greenberg, B., Hogue, C., Berger, G. & Pulliam, C. (1980) Am. J. Obstet. Gynecol. 137, 92-101. [DOI] [PubMed] [Google Scholar]
- 7.Key T. & Pike, M. (1988) Br. J. Cancer 57, 205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ace C. & Okulicz, W. (1995) Mol. Cell Endocrinol. 115, 95-103. [DOI] [PubMed] [Google Scholar]
- 9.Ehrlich C., Young, P. & Cleary, R. (1981) Am. J. Obstet. Gynecol. 141, 539-546. [DOI] [PubMed] [Google Scholar]
- 10.Persson I., Adami, H., Bergkvist, L., Lindgren, A., Pettersson, B., Hoover, R. & Schairer, C. (1989) Br. Med. J. 298, 147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lydon J., DeMayo, F., Funk, C., Mani, S., Hughes, A., Montgomery, C., Jr., Shyamala, G., Conneely, O. & O'Malley, B. (1995) Genes Dev. 9, 2266-2278. [DOI] [PubMed] [Google Scholar]
- 12.Kastner P., Krust, A., Turcotte, B., Stropp, U., Tora, L., Gronemeyer, H. & Chambon, P. (1990) EMBO J. 9, 1603-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conneely O., Maxwell, B., Toft, D., Schrader, W. & O'Malley, B. (1987) Biochem. Biophys. Res. Commun. 149, 493-501. [DOI] [PubMed] [Google Scholar]
- 14.Conneely O., Kettelberger, D., Tsai, M.-J., Schrader, W. & O'Malley, B. (1989) J. Biol. Chem. 264, 14062-14064. [PubMed] [Google Scholar]
- 15.Wen D., You-Feng, X., Mais, D., Goldman, M. & McDonnell, D. (1994) Mol. Cell. Biol. 14, 8356-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sartorius C., Melville, M., Hovland, A., Tung, L., Takimoto, G. & Horwitz, K. (1994) Mol. Endocrinol. 8, 1347-1360. [DOI] [PubMed] [Google Scholar]
- 17.Giangrande P., Kimbrel, E., Edwards, D. & McDonnell, D. (2000) Mol. Cell. Biol. 20, 3102-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horwitz K. (1992) Endocr. Rev. 13, 146-163. [DOI] [PubMed] [Google Scholar]
- 19.Richer J., Jacobsen, B., Manning, N., Abel, M., Wolf, D. & Horwitz, K. (2002) J. Biol. Chem. 277, 5209-5218. [DOI] [PubMed] [Google Scholar]
- 20.Mulac-Jericevic B., Mullinax, R., DeMayo, F., Lydon, J. & Conneely, O. (2000) Science 289, 1751-1754. [DOI] [PubMed] [Google Scholar]
- 21.Haiman C. A., Hankinson, S. E., Colditz, G. A., Hunter, D. J. & De Vivo, I. (2001) Cancer Res. 61, 3955-3960. [PubMed] [Google Scholar]
- 22.Ronaghi M. (2001) Genome Res. 11, 3-11. [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Levine, L. & Kwok, P. (1999) Genome Res. 9, 492-498. [PMC free article] [PubMed] [Google Scholar]
- 24.Long J. C., Williams, R. C. & Urbanek, M. (1995) Am. J. Hum. Genet. 56, 799-810. [PMC free article] [PubMed] [Google Scholar]
- 25.Wingender E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I. & Schacherer, F. (2000) Nucleic Acids Res. 28, 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wormke M., Castro-Rivera, E., Chen, I. & Safe, S. (2000) J. Steroid Biochem. Mol. Biol. 72, 197-207. [DOI] [PubMed] [Google Scholar]
- 27.Kumar N., Richer, J., Owen, G., Litman, E., Horwitz, K. & Leslie, K. (1998) Cancer Res. 58, 1860-1865. [PubMed] [Google Scholar]
- 28.Reich D., Cargill, M., Bolk, S., Ireland, J., Sabeti, P., Richter, D., Lavery, T., Kouyoumjian, R., Farhadian, S., Ward, R. & Lander, E. (2001) Nature (London) 411, 199-204. [DOI] [PubMed] [Google Scholar]
- 29.Martinez E., Ge, H., Tao, Y., Yuan, C.-X., Palhan, V. & Roeder, R. (1998) Mol. Cell. Biol. 18, 6571-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petz L. & Nardulli, A. (2000) Mol. Endocrinol. 14, 972-985. [DOI] [PubMed] [Google Scholar]
- 31.Drysdale C., McGraw, D., Stack, C., Stephens, J., Judson, R., Nandabalan, K., Arnold, K., Ruano, G. & Liggett, S. (2000) Proc. Natl. Acad. Sci. USA 97, 10483-14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnett-Mansfield R. & deFazio, A. (2001) Cancer Res. 61, 4576-4582. [PubMed] [Google Scholar]
- 33.Sasaki M., Dharia, A., Oh, B., Tanaka, Y., Fujimoto, S.-I. & Dahiya, R. (2001) Cancer Res. 61, 97-102. [PubMed] [Google Scholar]
- 34.Bain D. & Franden, M. (2001) J. Biol. Chem. 276, 23825-23831. [DOI] [PubMed] [Google Scholar]
- 35.Tung L. & Shen, T. (2001) J. Biol. Chem. 276, 39843-39851. [DOI] [PubMed] [Google Scholar]
- 36.Liu Z. & Wong, J. (2001) Proc. Natl. Acad. Sci. USA 98, 12426-12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vegeto E., Shahbaz, M., Wen, D., Goldman, M., O'Malley, B. & McDonnell, D. (1993) Mol. Endocrinol. 7, 1244-1255. [DOI] [PubMed] [Google Scholar]
- 38.Migliaccio A., Piccolo, D., Castoria, G., Di Domenico, M., Bilancio, A., Lombardi, M., Gong, W., Beato, M. & Auricchio, F. (1998) EMBO J. 17, 2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore M. & Conover, J. (2000) Biochem. Biophys. Res. Commun. 277, 650-654. [DOI] [PubMed] [Google Scholar]
- 40.Farr C., Easty, D., Ragoussis, J., Collignon, J., Lovell-Badge, R. & Goodfellow, P. (1993) Mamm. Genome 4, 577-584. [DOI] [PubMed] [Google Scholar]
- 41.Fujimoto J., Ichigo, S., Hirose, R., Sakaguchi, H. & Tamaya, T. (1997) J. Steroid Biochem. Mol. Biol. 62, 449-454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.