Abstract
Enveloped viruses enter cells by binding to their entry receptors and fusing with the membrane at the cell surface or after trafficking through acidic endosomal compartments. Species-specific virus tropism is usually determined by these entry receptors. Because mouse mammary tumor virus (MMTV) is unable to infect Chinese hamster cells, we used phenotypic screening of the T31 mouse/hamster radiation hybrid panel to map the MMTV cell entry receptor gene and subsequently found that it is transferrin receptor 1. MMTV-resistant human cells that expressed mouse transferrin receptor 1 became susceptible to MMTV infection, and treatment of mouse cells with a monoclonal antibody that down-regulated cell surface expression of the receptor blocked infection. MMTV, like vesicular stomatitis virus, depended on acid pH for infection. MMTV may use transferrin receptor 1, a membrane protein that is endocytosed via clathrin-coated pits and traffics through the acidic endosomes, to rapidly get to a compartment where acid pH triggers the conformational changes in envelope protein required for membrane fusion.
In recent years, a large number of viral receptors have been identified using a variety of approaches. These include the biochemical purification of virus-binding molecules, the generation of antibodies to cell surface proteins that block infection, and the introduction of genomic or cDNA libraries from susceptible into resistant cells, followed by infection with the virus of interest, usually engineered to encode an identifiable marker such as a drug resistance gene. Although such techniques are extremely powerful and have been successful in many cases, the identification of some virus receptors has been refractive to these methods.
Recently, Rai and colleagues (1) showed that phenotypic screening of radiation hybrid panels could be used for receptor identification. These investigators tested the Stanford G3 Chinese hamster/human radiation hybrid (RH) panel for infection with Jaagsietke sheep retrovirus (JSRV) pseudoviruses and mapped the receptor gene. Historically, somatic cell hybrid panels between Chinese hamster cells, which are refractive to infection by many viruses including mouse mammary tumor virus (MMTV), and cells from different species have been used to map receptor genes. These mapping studies resulted in the chromosomal placement of the virus receptor genes, but not their identification. RH panels, in which the cell line from the species of interest are X-irradiated to generate small chromosomal fragments before fusion with the hamster cells, have been one of the major tools used in generating high-resolution maps of different organisms and have resulted in maps that integrate genetic, physical, and expressed sequence tag (EST) data (2). The ability to precisely map the JSRV receptor gene on this RH panel in conjunction with the availability of cosmid/P1 contig maps and genomic sequence from the region led to cloning of the entry receptor, HYAL2 (3).
We undertook this approach to identify the receptor for MMTV, a betaretrovirus that causes breast carcinomas in mice. Previous work using vesicular stomatitis virus (VSV)-pseudotyped with the MMTV envelope (Env) protein had mapped a receptor to mouse Chr 16 (4). We also identified a potential MMTV receptor that mapped to Chr 19, termed MTVR2, using virus binding as an assay for expression cloning (5). To determine whether the bona fide MMTV receptor mapped to Chr 16, Chr 19, or elsewhere in the mouse genome, we screened the T31 mouse/Chinese hamster RH cell panel (6) with Moloney murine leukemia virus (MoMLV) pseudotypes bearing the MMTV Env protein on their surface. As described here, we found that the receptor gene mapped to Chr 16. This placement allowed us to test bacterial artificial chromosomes (BACs) from the candidate region for receptor activity and then to identify mouse transferrin receptor 1 (mTfR1) as the MMTV cell entry receptor.
TfR1 is a type II, single membrane-spanning glycoprotein with a short cytoplasmic domain and is the major means by which most cells take up iron (7). The receptor has a high affinity for iron-loaded transferrin at neutral pH, and upon binding ligand traffics to the early acidic endosome, where it releases iron, recycles back to the cell surface, and releases transferrin. The transferrin binding site has been determined by mutagenesis and binding studies, and maps to an RGD-containing sequence in the extracellular domain that is highly conserved between species (8, 9). In addition to transferrin, peptides that bind at other sites on TfR1 cause it to be endocytosed and traffic to the acidic endosome (10).
The identification of a protein that traffics to the acidic endosome upon ligand binding is interesting in light of a previous observation that treatment of MMTV-infected mammary tumor cells with acid pH induced cell–cell fusion (11). We show here that MMTV pseudovirus infection is also dependent on acidified endosomes. Taken together, these results suggest that MMTV binds mTfR1 at the cell surface and then traffics to an acidic endosomal compartment where virus/cell membrane fusion occurs.
Materials and Methods
Infection of the RH Panel.
MMTV-Env pseudotyped Mo-MuLV vectors were prepared as described (5). A23 hamster cells and A23-derived RH clones (gifts from P. N. Goodfellow, GlaxoSmithKline Pharmaceuticals, Stevenage, U.K., and L. C. McCarthy, Glaxo Wellcome Medicines Research, Stevenage, U.K.) were grown in hypoxanthine/aminopterin/thymidine (HAT)-DMEM, supplemented with 10% fetal bovine sera. The RH cells were plated at 5 × 104 cells per well in a 6-well plate and infected with the MMTV-Env pseudotypes the following day. After 2 days, the cells were stained for β-galactosidase activity. Wells containing greater than 25 β-galactosidase colonies were counted as positive.
Recombinant DNAs.
BAC clones were obtained from BACPAC Resources (Oakland, CA) or Research Genetics (Birmingham, AL) and were purified according to standard techniques. An insert containing the mouse TFR1 cDNA (a gift from N. Andrews, Howard Hughes Medical Institute, The Children's Hospital, Boston) was transferred from the pSPORT to the pcDNA3.1 mammalian expression vector (mTfR1). Human TFR1 in pCDM8 was a gift from M. Marks (University of Pennsylvania) and mouse TFR2 (IMAGE clone 4196597; pCMV-SPORT6 vector) was obtained from Research Genetics.
Transfection of 293T Cells and Pseudovirus Infection.
Two micrograms of BAC or plasmid DNA was transiently transfected into 293T cells plated in 6-well dishes, using calcium phosphate precipitation. Twenty-four hours after transfection, the cells were infected with MMTV pseudotypes containing the lacZ gene under the control of the MoMLV long terminal repeat (pHIT60) (12) and the cells were stained for LacZ activity 48 hr later as described (5). Normal murine mammary gland (NMuMG) cells were infected as positive controls. A20 cells were infected with 100 μl of pseudovirus stock in 96-well dishes at 1 × 105 cells per well for 2 hr; five wells for each treatment (1 × 106 cells in total) were pooled and diluted to 5 ml final volume with media. Two days after infection, cell extracts were made and β-galactosidase assays were performed using a commercial kit, according to the manufacturer's instructions (Promega). Data are presented as milliunits/μg protein. In some cases, duplicate wells were treated with 1 μl/ml goat anti-MMTV antisera (13) at the time of infection or 1 μg/ml rat anti-mouse CD71 monoclonal antibody C2 (MOPC-315; RDI, Flanders, NJ) for 10 min at room temperature before the addition of pseudovirus.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
NMuMG-, A20-, 293T-, and mTfR1-transfected cells were stained with rat-anti mouse CD71 conjugated with PE (PharMingen). A20 cells were also incubated with MMTV or VSV pseudovirus or anti-mTfrF1 antibody by using spinnoculation [2-hr centrifugation at 1,200 × g at room temperature (14)], washed extensively with cold PBS/1% fetal calf sera/0.2% sodium azide, and then stained with the PE-conjugated anti-CD71 antibodies. Data were acquired on a FACScan cytometer (Becton Dickinson) and analyzed using cellquest software (Becton Dickinson Immunocytometry Systems).
Western Blots.
After LacZ staining, the cells were washed extensively with PBS, and then lysed in SDS-sample buffer. The lysates were electrophoresed on 10% SDS-polyacrylamide gels, transferred to nitrocellulose, and then subjected to Western blot analysis, using polyclonal rabbit anti-transferrin receptor 1 antisera that recognizes both the mouse and human receptors (Santa Cruz Research, Santa Cruz, CA).
Ammonium Chloride and Bafilomycin-A Treatment of Cells.
NMuMG cells were treated for 1 hr with bafilomycin A (Sigma-Aldrich) or NH4Cl at the indicated concentrations in DMEM buffered to pH 7.4 with 20 mM Hepes and 10% FCS. Virus infections with MMTV, MuLV, or VSV pseudotypes were carried out in the same media and concentrations of NH4Cl or bafilomycin A. After 2 hr incubation with virus, the media was removed and the cells were washed and re-fed. The cells were stained for LacZ activity 48 hr later.
Results and Discussion
The T31 RH panel was created from a fusion between the Chinese hamster fibroblast A23 cell line and 129 embryonic stem (ES) cells (6). We first established that A23 cells were resistant to infection by MMTV pseudotypes (Table 1); we did not have the 129 ES cell line available, but we have found that all tested murine tissue culture lines were susceptible (13). We then screened 98/100 T31 RH panel cell lines for infection by these pseudotypes. The infection results for the 100 cell lines were: 000010110010110100011100001101001110010110000000100100020010011012100000110100100001001000100100111 [0 scored as uninfected (<10 LFU/ml), 1 scored as positive for infection (>25–2,000 LFU/ml), and 2 indicates that the cell line was unavailable for analysis]. The data were submitted to The Jackson Laboratory Mouse Radiation Hybrid Database server (www.jax.org/resources/documents/cmdata/rhmap/RHIn-tro.html; ref. 15).
Table 1.
BAC transfection to test for MMTV receptor activity
| Cell line | BAC/plasmid | LFU/ml | LFU/ml + anti-MMTV |
|---|---|---|---|
| 293T | 320J17 | 0 | 0 |
| 219I11 | 0 | 0 | |
| 342H15 | 0 | 0 | |
| 248H7 | 1 | 0 | |
| 27L21 | 0 | ND | |
| 121N24 | 0 | 0 | |
| 308K23 | 3 | 0 | |
| 73P12 | 77 | 1 | |
| 322N13 | 84 | 0 | |
| 324J8 | 1 | 0 | |
| 342H15 | 4 | 0 | |
| mTfR1 | 7,300 | 0 | |
| hTfR1 | 0 | ND | |
| — | 0 | 0 | |
| A23 | — | 0 | ND |
| NMuMG | — | 3,461 | 0 |
293T cells were transfected by the CaPO4 method in duplicate with 2 ug of each DNA in 6-well dishes. Twenty-four hours after transfection, the cells were infected with MMTV Env-pseudotyped Mo-MLV vectors in the absence or presence of goat polyclonal anti-MMTV antisera (+ anti-MMTV) (Quality Biotech, Camden, NJ). The normal murine mammary gland cell line, NMuMG, served as a positive control for MMTV infection (13). Shown are the lacZ-forming units (LFU) per milliliter of virus supernatant. ND, not determined.
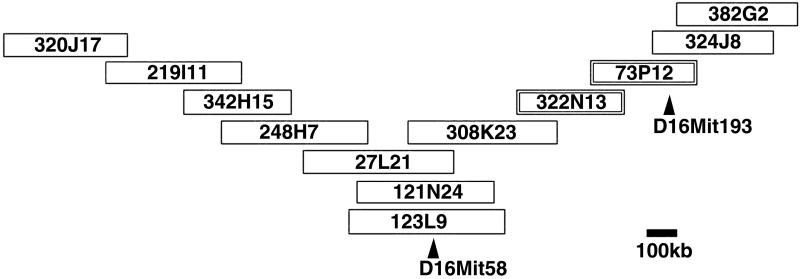
We found that the MMTV-susceptibility locus mapped on Chr 16 near to simple sequence length polymorphic (SSLP) markers D16Mit58 [logarithm of odds (lod) score 7.1] and D16Mit193 (lod score 7.2). Several other markers found near the receptor (lod scores in the range of 7 to 9) mapped to the BACs presented in Fig. 1 (S.R.R., unpublished work). These markers provided a link between the position of the susceptibility locus on the RH map and the BAC map of the mouse genome (http://mouse.ensembl.org). For example, D16Mit58 is found on BAC clone, RPCI-23 123L9, which mapped to contig 992 of the Ensembl Mouse Assembly. To refine the map position of the susceptibility locus, we transiently transfected eleven overlapping BAC clones surrounding RPCI-23 123L9 into 293T human embryonic kidney cells, which are resistant to MMTV infection (Fig. 1). Clones RPCI-23 73P12 and RPCI-23 322N13 conferred the ability to be infected with MMTV pseudotypes to 293T cells (Table 1). The infection depended on the MMTV Env protein, because pretreatment of the pseudovirus with goat anti-MMTV polyclonal antisera (Table 1) or infection with pseudovirus without Env protein did not result in β-gal positive clones (S.R.R., unpublished work).
Fig 1.
BAC contig map of MTVR1 region. The eleven BACs mapping to this region of Chr 16 that were used in transfection studies (Table 1) were identified using the Ensembl database. D16Mit58 maps to RPCI-23 123L9, whose sequence is in the public domain. D16Mit193 was placed on RPCI-23 73P12 by screening the eleven BACs for this marker. BACs RPCI-23 322N13 and RPCI-23 73P12 encode MTVR1 activity (Table 1).
Analysis of the Celera mouse genome sequence and Ensembl BAC assembly showed that the two BACs covered approximately 339 kb of sequence and that there was about 100 kb of overlap between the clones. The region of overlap encoded three genes: transferrin receptor 1 (mTfR1), an unknown gene with homology to RIKEN clone AK010304, and p21cdc42-associated tyrosine kinase ACK1. We excluded ACK1 from further analysis because it is a cytoplasmic protein and thus it was unlikely to encode receptor activity. The sequence of the unknown gene with homology to AK010304 revealed that it was a pseudogene, because it had lost all introns and part of the N terminus. We therefore tested whether mTfR1 encoded the MMTV receptor by transiently transfecting 293T cells with an expression vector containing the cDNA for this protein.
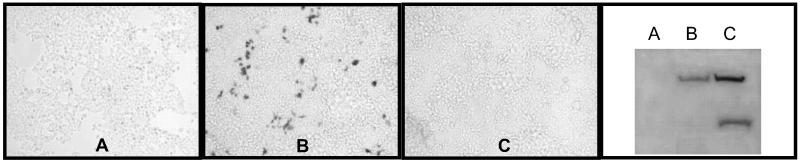
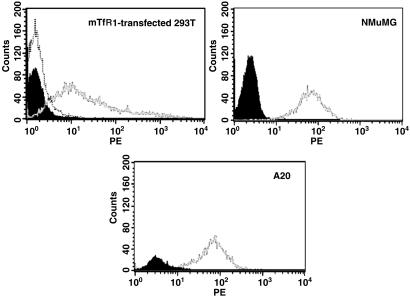
We found that 293T cells transfected with the mouse (mTfR1) cDNA-expression plasmid were highly susceptible to infection with the MMTV pseudotypes (Fig. 2, Table 1). We also tested cloned human TfR1, to exclude the possibility that it was the level of receptor expression; these transfected cells were not infected with MMTV (Fig. 2, Table 1). Both the mouse and human TfR1 proteins were expressed at equivalent levels in the transfected cells, as shown by Western blot analysis of cell lysates (Fig. 2). Cell surface staining of the transiently transfected 293T cells with anti-mTfR1 antibodies (Fig. 3) demonstrated they expressed levels of the receptor that were equivalent or lower than mouse A20 (B lymphoma) or NMuMG cells (Fig. 3). Transfection of mTfR2, a transferrin-binding molecule with 45% amino acid sequence identity to mTfR1 that maps to mouse Chr 5 (16), also did not make 293T cells susceptible to MMTV pseudovirus infection (Fig. 2, mTfR2).
Fig 2.
Mouse transferrin receptor 1 is the entry receptor for MMTV. Human 293T cells were transfected with the mouse TfR2 (A), mouse TfR1 (B), or human TfR1 (C) cDNAs under the control of the CMV promoter and then infected with MMTV pseudotypes containing the lacZ reporter gene. After staining for lacZ, extracts were prepared from the cells and subjected to Western blot analysis with anti-TfR1 antibodies. Endogenous hTfR1 levels are below detection in this analysis. The polyclonal anti-TFR1 antibodies do not recognize TfR2.
Fig 3.
Cell-surface expression of mTfR1. Fluorescence-activated cell sorter (FACS) analysis of mTfR1 transiently transfected 293T, NMuMG, and A20 cells. In each panel, the filled curve represents unstained cells. Untransfected 293T cells also showed no staining with the rat anti-mouse TfR1 antibodies (dotted curve).
To confirm that mTfR1 was the receptor for MMTV, we tested whether a rat-anti mTfR1 monoclonal antibody (C2) that is known to down-regulate cell surface receptor would block infection (ref. 17; see Fig. 4). Infection of mouse cells (NMuMG, A20) or 293T cells transfected with mTfR1 was blocked by 97–100% (Table 2). Thus, infection of mouse B and mammary gland cells was also dependent on the presence of mTfR1 on the surface.
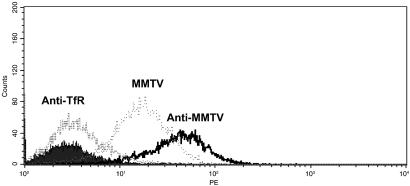
Fig 4.
MMTV binding down-regulates cell surface expression of mTfR1. A20 cells were incubated with MMTV pseudovirus (MMTV), MMTV pseudovirus plus neutralizing polyclonal anti-MMTV antisera (Anti-MMTV), or anti-mTfR1 monoclonal antibodies (AntiTfR) as described in Materials and Methods, washed extensively and analyzed for cell surface expression of mTfR1. The filled curve shows unstained cells.
Table 2.
Rat anti-mTfR1 monoclonal antibody blocks infection
| Cells | Antibody | LacZ activity | % inhibition |
|---|---|---|---|
| NMuMG | None | 2,160 | — |
| Rat anti-mTfR1 | 140 | 93.5 | |
| Goat anti-MMTV | 0 | 100 | |
| 293T/mTfR1 | None | 6,050 | — |
| Rat anti mTfR1 | 366 | 94 | |
| Goat anti-MMTV | 0 | 100 | |
| A20 | None | 24.6 | — |
| Rat anti-mTfR1 | 0 | 100 | |
| Goat anti-MMTV | 0.6 | 97.6 |
Shown are the data from one experiment for each cell line. The experiments were performed two to three times with similar results.
Colony assay; data are expressed as LFU per milliliter of pseudovirus stock.
Soluble assay; data is presented as milliunits of β-galactosidase per microgram of protein × 10−3.
We next demonstrated that binding of MMTV to A20 mouse cells induced loss of mTfR1 from the cell surface. A20 cells were incubated with pseudovirus for 2 hr at room temperature by using spinnoculation (14), then shifted to 4°C and washed with buffer containing sodium azide to prevent further internalization. As controls, the cells were incubated with pseudovirus in the presence of goat anti-MMTV antisera or the C2 anti-mTfR1 monoclonal antibodies. The cells were then examined by fluorescence-activated cell sorter (FACS) analysis for mTfR1 expression on the surface. There was a more than 2-fold decrease in TfR1 expression on A20 cells incubated with virus compared with cells incubated with virus plus anti-MMTV antisera (Fig. 4) or with no virus (not shown); incubation of A20 cells with VSV G-pseudotyped virions also had no effect on surface expression of TfrR1 (data not shown). The C2 antibody caused almost complete down-regulation of the receptor on the surface, as reported (17).
Taken together, these data indicate that mTfR1 is the bona fide MMTV receptor that was originally mapped on Chr 16 (4). We had previously attempted to clone this receptor by using an expression approach and identified another molecule, MTVR2, which appeared to function as a virus receptor. However, this molecule did not work in transient transfection assays and it conferred levels of infection to virus-resistant cells well below that seen in mouse cells even when expressed at high levels (5, 13). One possibility is that MTVR2 represents a low-affinity virus receptor that does not function when expressed at low levels, such as would occur on the RH panel. Alternatively, because MTVR2 was selected for its ability to confer binding and not infection, it may concentrate virus on the cell surface when expressed at high levels in stable transfectants, thereby occasionally allowing entry by a non-receptor-mediated pathway.
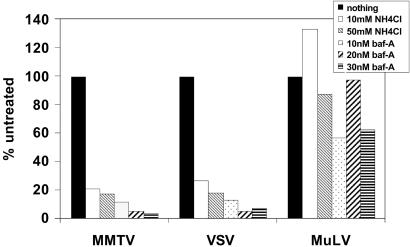
The identification of the TfR1 as the MMTV receptor has the potential to greatly increase our understanding of virus/receptor interactions, both at the molecular and cellular levels. Much is known about the movement of TfR1 between different cellular compartments. TfR1 rapidly traffics from the surface after binding transferrin to an early acidic endosomal compartment, where it dissociates from transferrin and then recycles back to the surface (7). Treatment of MMTV-infected cultured mammary tumor cells with acid had previously been reported to induce cell–cell fusion (11); we have also found that cells expressing both the Env and receptor can be induced to fuse with brief low pH treatment (J.J.S. and S.R.R., unpublished work). We therefore tested whether treating MMTV-susceptible NMuMG cells with bafilomycin A1, a specific inhibitor of the vacuolar H+-ATPase that prevents the acidification of endosomes and lysosomes (18), or NH4Cl, which also neutralizes the acidic endosomal compartment, inhibited infection with MMTV Env-pseudotyped Mo-MLV. As a control, we used pseudotypes packaged with the pH-independent Mo-MLV Env (19) or viruses packaged with the pH-dependent VSV-G protein (20). MMTV pseudotype infection was inhibited by bafilomycin A and NH4Cl to the same extent as VSV-G pseudotypes, whereas both of these treatments had very little effect on Moloney murine leukemia virus (MoMLV) infection (Fig. 5). Similar inhibition of MMTV infection with these reagents was seen with mTfR1-transfected 293T cells (S.R.R., unpublished work).
Fig 5.
MMTV infection is pH dependent. NMuMG cells or 293T cells transiently transfected with mTfR1 24 hr before infection were treated for 1 hr with the indicated concentrations of NH4Cl or bafilomycin A. Shown are the infection levels relative to untreated cells. This experiment was repeated three times with similar results in both NMuMG and mTfR1-transfected 293T cells.
For viruses that enter cells at the plasma membrane, binding to the cell entry receptor is thought to trigger a conformational change that exposes the fusion peptide (20). In contrast, low pH is the trigger for this conformational change during infection by pH-dependent viruses. TfR1 has been widely studied for its ability to traffic and reshuttle through the acidic endosomal compartment via clathrin-coated pits after transferrin binding (7). Moreover, it has been shown that TfR1 traffics to this compartment when bound with peptides that bind outside the transferrin-binding domain (17). The sequences that dictate internalization of the receptor and shuttling to the early acid endosomes are found in the short cytoplasmic tail and transmembrane domain (21, 22). Thus, it is possible that MMTV enters the acidic endosome bound to mTfR1. Even though much is known about the effects of low pH on influenza virus hemagglutinin conformation and membrane fusion activity, it is not known how binding to receptor, sialic acid, leads to trafficking to the acidic endosomes (20). Our identification of TfR1 as the cell entry receptor for MMTV could provide a mechanism for the efficient migration of pH-dependent, enveloped viruses to this compartment.
Pretreatment of some viruses that are pH-dependent causes premature exposure of the fusion peptide and renders them noninfectious. MMTV is stable at low pH, because as a milk-transmitted virus it encounters acid pH in the newborn stomach and yet is still infectious. This stability, coupled with the ability of acid to trigger cell–cell fusion when both SU and TfR1 are expressed on the surface and the pH dependence for infection, suggests that receptor binding and trafficking to the endosome may both be required for MMTV Env-induced membrane fusion. It has recently been shown that the alpharetrovirus avian leukosis virus (ALV) is also dependent on acid pH and receptor binding, although in this case it is also not known how it traffics to the acidic endosomal compartment (19).
The crystal structure of the human TfR1 ectodomain has been solved (23) and the transferrin-binding domain characterized (8). The mouse, human, rat, and hamster TfR1 proteins show about 90% identity at the amino acid level. Rat cells are also susceptible to infection with MMTV pseudotypes (S.R.R., unpublished work). We examined the primary sequence of the TfR1 proteins from these four species and found that there are fewer than 15 unconserved amino acids between the MMTV-susceptible and -resistant species; many of the these amino acids are unlikely to function in Env interaction because they are not on the surface of TfR1. Changing these variant amino acids in the context of the mouse and human receptors should identify which function in virus infection.
It is well known that TfR1 functions to deliver iron-bound transferrin to cells. Recently, the human and feline TfR1 have been shown to function as cell entry receptors for the nonenveloped canine and feline parvoviruses (24). That TfR1 proteins can function as virus receptors points to a role that would not have been predicted based on their biology. The identification of TfR1 as the MMTV receptor emphasizes the power of phenotypic screening of RH panels for both gene discovery and for finding new functions for already characterized proteins (2).
Acknowledgments
We thank Lorraine Albritton, Micky Marks, and Glen Gaulton for helpful discussions and reagents, Kay Swanson (Research Genetics) for help in accessing the cell lines, Lucy Rowe for analyzing the RH data, and Nancy Andrews for the mouse TFR1 cDNA. C.J.F. is a Medical Research Council Senior Research Fellow. This work was supported by National Institutes of Health Grant CA73746 (to S.R.R.).
Abbreviations
MMTV, mouse mammary tumor virus
NMuMG, normal murine mammary gland
Chr, chromosome
BAC, bacterial artificial chromosome
Env, envelope
RH, radiation hybrid
mTfR, mouse transferrin receptor
LFU, lacZ-forming units
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rai S. K., DeMartini, J. C. & Miller, A. D. (2000) J. Virol. 74, 4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ross S. R. (2001) Mamm. Genome 12, 879-881. [DOI] [PubMed] [Google Scholar]
- 3.Rai S. K., Duh, F. M., Vigdorovich, V., Danilkovitch Yphenmiagkovaqq, A., Lerman, M. I. & Miller, A. D. (2001) Proc. Natl. Acad. Sci. USA 98, 4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilkens J., van der Zeust, B., Buijs, F., Kroezen, V., Bluemink, N. & Hilgers, J. (1983) J. Virol. 45, 140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golovkina T. V., Dzuris, J. L., van den Hoogen, B., Jaffe, A. B., Wright, P. C., Cofer, S. M. & Ross, S. R. (1998) J. Virol. 72, 3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy L. C., Terrett, J., Davis, M. E., Knights, C. J., Smith, A. L., Critcher, R., Schmitt, K., Hudson, J., Spurr, N. K. & Goodfellow, P. N. (1997) Genome Res. 7, 1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponka P. & Lok, C. N. (1999) Int. J. Biochem. Cell Biol. 31, 1111-1137. [DOI] [PubMed] [Google Scholar]
- 8.West A. P., Giannetti, A. M., Herr, A. B., Bennett, M. J., Nangiana, J. S., Pierce, J. R., Weiner, L. P., Snow, P. M. & Bjorkman, P. J. (2001) J. Mol. Biol. 313, 385-397. [DOI] [PubMed] [Google Scholar]
- 9.Dubljevic V., Sali, A. & Goding, J. W. (1999) Biochem. J. 341, 11-14. [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J. H., Engler, J. A., Collawn, J. F. & Moore, B. A. (2001) Eur. J. Biochem. 268, 2004-2012. [DOI] [PubMed] [Google Scholar]
- 11.Redmond S., Peters, G. & Dickson, C. (2984) Virology 133, 393-402. [DOI] [PubMed] [Google Scholar]
- 12.Soneoka Y., Cannon, P. M., Ramsdale, E. E., Griffiths, J. C., Romano, G., Kingsman, S. M. & Kingsman, A. J. (1995) Nucleic Acids Res. 23, 628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzuris J. L., Zhu, W., Golovkina, T. V. & Ross, S. R. (1999) Virology 263, 418-426. [DOI] [PubMed] [Google Scholar]
- 14.O'Doherty U., Swiggard, W. J. & Malim, M. H. (2000) J. Virol. 74, 10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowe L. B., Barter, M. E. & Eppig, J. T. (2000) Genomics 69, 27-36. [DOI] [PubMed] [Google Scholar]
- 16.Fleming R. E., Migas, M. C., Holden, C. C., Waheed, A., Britton, R. S., Tomatsu, S., Bacon, B. R. & Sly, W. S. (2000) Proc. Natl. Acad. Sci. USA 97, 2214-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp J. D., Thorson, J. A., McAlmont, T. H., Horowitz, M., Cowdery, J. S. & Ballas, Z. K. (1987) J. Immunol. 138, 2422-2426. [PubMed] [Google Scholar]
- 18.Drose S. & Altendorf, K. (1997) J. Exp. Biol. 200, 1-8. [DOI] [PubMed] [Google Scholar]
- 19.Mothes W., Boerger, A. L., Narayan, S., Cunningham, J. M. & Young, J. A. (2000) Cell 103, 679-689. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez L. D., Hoffman, L. R., Wolfsberg, T. G. & White, J. M. (1996) Annu. Rev. Cell Dev. Biol. 12, 627-661. [DOI] [PubMed] [Google Scholar]
- 21.Collawn J. F., Lai, A., Domingo, D., Fitch, M., Hatton, S. & Trowbridge, I. S. (1993) J. Biol. Chem. 268, 21686-21692. [PubMed] [Google Scholar]
- 22.Zaliauskiene L., Kang, S., Brouillette, C. G., Lebowitz, J., Arani, R. B. & Collawn, J. F. (2000) Mol. Biol. Cell 11, 2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence C. M., Ray, S., Babyonyshev, M., Galluser, R., Borhani, D. W. & Harrison, S. C. (1999) Science 286, 779-782. [DOI] [PubMed] [Google Scholar]
- 24.Parker J. S., Murphy, W. J., Wang, D., O'Brien, S. J. & Parrish, C. R. (2001) J. Virol. 75, 3896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]