Abstract
A map of 191 single-nucleotide polymorphism (SNPs) was built across a 5-Mb segment from chromosome 13q34 that has been genetically linked to schizophrenia. DNA from 213 schizophrenic patients and 241 normal individuals from Canada were genotyped with this marker set. Two 1,400- and 65-kb regions contained markers associated with the disease. Two markers from the 65-kb region were also found to be associated to schizophrenia in a Russian sample. Two overlapping genes G72 and G30 transcribed in brain were experimentally annotated in this 65-kb region. Transfection experiments point to the existence of a 153-aa protein coded by the G72 gene. This protein is rapidly evolving in primates, is localized to endoplasmic reticulum/Golgi in transfected cells, is able to form multimers and specifically binds to carbohydrates. Yeast two-hybrid experiments with the G72 protein identified the enzyme d-amino acid oxidase (DAAO) as an interacting partner. DAAO is expressed in human brain where it oxidizes d-serine, a potent activator of N-methyl-D-aspartate type glutamate receptor. The interaction between G72 and DAAO was confirmed in vitro and resulted in activation of DAAO. Four SNP markers from DAAO were found to be associated with schizophrenia in the Canadian samples. Logistic regression revealed genetic interaction between associated SNPs in vicinity of two genes. The association of both DAAO and a new gene G72 from 13q34 with schizophrenia together with activation of DAAO activity by a G72 protein product points to the involvement of this N-methyl-d-aspartate receptor regulation pathway in schizophrenia.
Schizophrenia affects almost 1% of the world's population, with a similar prevalence throughout diverse ethnic groups (1). Clinically, this disease, one of the largest causes of disability worldwide, comprises thought disorder, delusions, hallucinations, and blunted affect, exhibiting extensive individual variation. Dopaminergic and more recently glutamatergic neurotransmission systems abnormalities have both been considered in schizophrenia pathogenesis (2–6). Molecular mechanisms triggering schizophrenia remain elusive. Although several environmental risk factors have been considered (7, 8), risk studies with twins and other relatives suggest a highly heritable disease (9). Identification of schizophrenia genes is particularly challenging, because the disease may encompass several entities that have not yet been defined, limiting the accuracy of schizophrenia diagnosis as a phenotypic definition. In addition, the lack of conclusive linkage from genome scans could be due to the existence of numerous susceptibility genes having weak individual effects that are difficult to detect and replicate (10).
In an attempt to track such susceptibility genes, we focused on the human chromosome 13q22-q34 region that generated linkage scores in some but not all sets of affected families, ranging from suggestive to significant in several studies (11–15). The compiled linkage data pointed to a large 50-cM region that potentially harbors one or several predisposition genes (Fig. 1a). The distal 5-Mb part of this region around marker D13S174, showing after analysis of 54 affected families a nonparametric linkage score of 4.18 (13), was investigated by using linkage disequilibrium (LD)-based positional cloning (16).
Figure 1.
Strategy used to identify potential schizophrenia genes in chromosome 13q. (a) Schizophrenia linked region (50 cM) derived from several studies (11–15). The position of the 5-Mb region selected for association studies is marked. (b) Allele frequencies association tests performed by Armitage trend test are shown as –Log (P value) for 191 markers. Thresholds for P values of 5E-02 (straight line) and 1E-02 (dotted line) are indicated. Statistically significant markers (P value lower than 5E-02) obtained in this study are shown: 99-13064/328 (M-1), 99-62753/178 (M-2), 99-62654/305 (M-3), 99-26234/336 (M-4), 99-26189/164 (M-5) in Bin B and 99-16105/152 (M-12), 99-5919/215 (M-22) in Bin A. These last two markers present P values lower than 1E-02. (c) Location of genes found in the 5-Mb analyzed region. Predicted genes found in the proximal 2-Mb portion of the region are shaded red (1, CanIon; 2, IT6BL1; 3, FHF4; 4, TPP2; 5, PHSP8; 6, XPG; 7, SLC10A2); unpredicted genes found in the distal 3 Mb described in this study are shaded blue (8, G90; 9, G72; 10, G30).
Our strategy consisted of systematic single-point association analysis, replication in another population, haplotype analysis, gene discovery, and functional studies to provide the convergent lines of evidence necessary for the dissection of a common multifactorial disease such as schizophrenia.
Methods
Collections.
The French Canadian collection sample consisted of 213 cases and 241 controls recruited throughout the Province of Quebec. Research protocols, including ethical procedures to be followed during the study, were submitted to the Institutional Review Board of each institution. All patients and family members as well as controls signed consent forms. Patients had to be diagnosed as schizophrenic by Diagnostic and Statistical Manual of Mental Disorders (DSM)-IV criteria (17). A case was included if it was not too closely related to other cases (over seven meiotic steps), and if more than two-thirds of his ancestors could be identified in any generation up to and including the sixth generation to ensure French Canadian origin. Controls were recruited in the same geographical regions as the cases. Genetic homogeneity between cases and controls was assessed by using a set of 27 “random” markers from five different chromosomes by using the test derived by Pritchard and Rosenberg (18). The analysis of allelic frequencies of these markers shows total χ2 = 23.28, df = 27, P value = 0.71.
The Russian collection was composed of 183 cases and 183 controls. Cases were recruited from a psychiatric hospital in Moscow [Research Center of Mental Health (RCMH) of the Russian Academy of Medical Sciences]. The study was approved by the ethical committee of RCMH. Informed consent was obtained for all patients and controls. The structured interview (19) was administrated to each patient, and diagnosis was made in accordance with International Classification of Disorders 10 and DSM-III-R criteria. The population-based controls were recruited by questionnaire. Genetic homogeneity between cases and controls was also checked with a set of 20 “random” markers by using the same statistical test of Pritchard and Rosenberg applied to the French Canadian collection. The analysis shows total χ2 = 25.6, df = 20, P value = 0.18.
Subcloning and Sequencing the Candidate Region.
Fifty-four bacterial artificial chromosome clones were sequenced by using the random shotgun method. After contigation (3.7 Mb; GenBank accession nos. AE014293 and AE014294), two gaps remained. To fill the gaps, sequences were subjected to blast (20) analysis against publicly available databases and the Celera sequence database. Contigation of all these sequences resulted in a single sequence of 4,995,781 bp. The entire compiled sequence and its annotation are available on the Genset web site, www.genset.fr/schizophrenia-gene-paper. Annotation prediction was performed on this 5-Mb region by using blastn to compare DNA sequences (GenBank, dbEST) and blastx to compare protein sequences (SwissProt, Sptrembl), genscan (21), and grail2 gene prediction software with standard parameters. We also used online predictions with the help of fgenes, genemark, and otto software.
The synthenic mouse genomic bacterial artificial chromosome sequences have GenBank accession no. AY143165.
Single-Nucleotide Polymorphism (SNP) Identification and Genotyping.
SNPs were identified from pools of 100 unrelated French individuals through resequencing 500-bp amplicons covering the 5-Mb region as previously described. SNP genotyping was performed by allele-specific ddNTP termination of sequencing reactions. One hundred ninety-one SNP markers from chromosome 13, 8 markers from DAAO gene region on 12q24, and 27 whole-genome SNPs used for stratification homogeneity test were registered in dbSNP with aliases SS5105638–SS5105863.
Association Analysis.
Single-locus tests of association between either SNP allele frequencies or SNP genotype frequencies and case-control status were carried out via standard contingency χ2 tests, and P values were determined via χ2 tables (22). The null distributions of significant test statistics were validated empirically via permutation tests. We performed 20,000 permutations for most significant tests to determine empirical significance. The standardized measure of linkage disequilibrium (LD), termed D′, was computed in each group for alleles at pairs of SNP loci (23). Tests of departures from LD were performed by using the likelihood ratio test of linkage disequilibrium as implemented in arlequin (24). Haplotype frequencies were estimated via the method of maximum likelihood from the genotype data through the use of the Expectation–Maximization algorithm under the assumption of the Hardy–Weinberg equilibrium (25–28). Logistic regression analyses to examine genetic interactions were performed by using the commercial sas package, Ver. 8.1 (29).
Experimental Annotation.
Testis and brain “Marathon Ready” cDNAs from CLONTECH were used. Rapid amplification of cDNA ends experiments were carried out as indicated by the manufacturer. PCR products were sequenced by using the big dye terminator DNA sequencing reaction (ABI Perkin–Elmer) following the manufacturer's instructions. Sequences were assembled by using the staden package and aligned to the genomic sequences to detect exon/intron boundaries. Reverse transcription was carried out by using the Advantage reverse transcription for PCR kit (CLONTECH) according to the manufacturer's instructions. Full-length cDNAs were amplified by PCR by using primers in terminal 5′ and 3′ exons. PCR products were cloned in pGEM11Zf(+) (Promega) and sequenced as previously described. cDNA sequences have GenBank accession nos. AY138546–AY138548.
In Vitro Translation Assay.
Coupled in vitro transcriptions/translations of the pGEM-LG72 (G72 candidate gene) and pGEM-LG30 (G30 candidate gene) plasmids were performed as described by the manufacturer (TNT quick coupled Transcription/Translation systems, Promega).
Transient Transfection Experiments and Western Blotting.
Ten micrograms of pCMV-LG72 (+) (sense orientation), pCMV-LG72 (−) (antisense orientation) or pHM6-LG72 (Roche, Molecular Biochemicals) eukaryotic expression constructs were transfected into 2.105 COS-7 cells (American Type Culture Collection no. CRL-1651) according to the calcium phosphate procedure (Invitrogen, calcium phosphate transfection kit). One hundred twenty-five micrograms of each extract were fractionated on a 15% polyacrylamide gel and transferred to nitrocellulose membrane (Protran BA85, Schleicher & Schuell) at room temperature. Antibody staining was performed with the anti-pLG72 purified antiserum (SE3014, Eurogentech, Brussels) diluted 1:250. Immunocomplexes were detected with the enhanced chemiluminescence Western blotting analysis system (Amersham Pharmacia–Pharmacia Biotech).
Protein Purification and Enzymatic Activity.
The untagged recombinant pLG72 protein was purified from bacteria [E. coli BL21(DE3) Codon Plus RIL, Stratagene] transformed with the pET11a vector with the insertion of pLG72 coding sequence. Bacterial cells were extracted with BugBuster reagent (Novagen) in the presence of protease inhibitors mixture (setIII, Calbiochem) and 10 mM EGTA at pH 8. After ammonium sulfate fractionation, proteins were subjected sequentially to Ultragel AcA44, DEAE-Macroprep (Bio-Rad), and Superdex 75 (Pharmacia) chromatographies in PBS, pH 8, followed by ultrafiltration (BioMax membrane, 10-K cutoff). Pig kidney DAAO was purified to homogeneity from Sigma crude preparation. DAAO was assayed by the colorimetric method (30).
Yeast Two-Hybrid Screening.
To identify the physical partners of the G72 candidate gene product, a portion of it coding for the carboxyl terminal 89 amino acids was used as a bait in protein interaction experiments by using the Matchmaker Two-Hybrid System from CLONTECH (catalogue no. K1612-1). The pretransformed 5⋅106 independent clones of Human Brain Matchmaker cDNA library (CLONTECH, catalogue no. HY4004AH) was used as prey following experimental procedures recommended by CLONTECH. The physical interaction between the G72 protein fragment and the DAAO protein was confirmed by transfer into the opposite mating type as well as cotransfections of two-hybrid constructs containing complete G72 fusion and DAAO fragment.
Results and Discussion
The LD association strategy requires a high-density marker map. The vast majority of human polymorphic sites are single-nucleotide position variations (SNPs) that are mostly biallelic (31), with several millions of such potential markers spread over the genome (32). Because functionally relevant polymorphisms are expected to be associated with the disease, anonymous flanking SNPs in strong LD with such functional polymorphisms are also expected to be disease associated. Our marker map was constructed dynamically until, using an entire set of 191 markers in the region, the average LD between consecutive markers had reached at least 0.85 (>0.5 in 70% of the cases), as estimated by normalized D′ value. This 191-SNP marker map was built also in parallel with bacterial artificial chromosome physical mapping and sequencing followed by integration of available genomic data from various sources (33, 34). Disease association was tested by genotyping each marker in 213 unrelated cases and 241 unrelated controls originating from Quebec, which was shown experimentally to be genetically homogeneous.
Fig. 1b shows the plot of the statistical significance [−Log(P value)] of the allelic association with schizophrenia obtained for each marker in 13q34. At the nominal significance level of <0.05, two smaller regions containing associated markers were evident. We refer them as Bin A, 65.9 kb in length between M-12 and M-22 (Fig. 1b; Table 1), and Bin B, 1,380 kb in length between M-1 and M-5 (Fig. 1b). These results should be interpreted with caution because multiple testing implicated 191 markers. At the level of significance obtained here, a certain number of type one errors (false positives) are expected to occur. The number of these false positives can be diminished by performing correction of nominal values, taking into account the number of independent tests performed (Bonferroni correction) (35), or by performing a multiple permutation test. Although effectively controlling the frequency of type one errors (false positives), this type of correction can increase the probability of type II errors (false negatives) to an unacceptably high level. Moreover, the 191 tests were not independent. It is also important to note that, whereas associated markers caused by type one errors should be distributed randomly throughout the region, substantial clustering of independent associated markers is observed in our case. This is especially evident in the case of Bin A, where clustering is probably meaningful because more than two independent markers (not in a strong LD; Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org) exceed the nominal 1% level, and they are relatively close. Using haplotypes instead of individual markers for association could reduce the chance of false-positive findings by providing stronger statistical support for the region. We calculated the estimated frequencies of all 9,000 possible combinations of three-marker haplotypes for 20 SNP markers in and around Bin A (Table 1) separately for cases and controls. The vast majority of these haplotypes are not independent, because a strong LD exists in this bin (Fig. 6, which is published as supporting information on the PNAS web site). To reduce the number of haplotypes for further examination, we retained only haplotypes that are “common,” i.e., with allelic frequencies in controls of at least 4%, and provide stronger statistics than single markers (odds ratio of more than 1.5 with P values of <0.001). Thirty-seven resulting three-marker haplotypes meet these criteria (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org). The strongest nominal value we observed was 3.55⋅10−10. The null distributions of significant test statistics were approximated by performing 20,000 permutation tests. Some of these common haplotypes still show P values close to zero despite such correction. For two of them (Table 2, first and third lines), we performed further correction through 50,000,000 permutations and produced strong P values of 3⋅10−6 and 5⋅10−5.
Table 1.
Statistical analysis for SNPs in and around Bin A in French Canadian and Russian samples
Twenty markers have been selected in and around Bin A: 99-25965/399 (M-6), 99-25966/241 (M-7), 99-25989/398 (M-8), 99-16047/115 (M-9), 99-16052/214 (M-10), 99-15875/165 (M-11), 99-16105/152 (M-12), 99-16032/292 (M-13), 99-15880/162 (M-14), 99-16038/118 (M-15), 8-155/258 (M-16), 99-15870/400 (M-17), 99-5897/143 (M-18), 8-130/143 (M-19), 99-24644/194 (M-20), 99-24658/410 (M-21), 99-5919/215 (M-22), 99-5862/167 (M-23), 99-24634/108 (M-24), and 99-31941/320 (M-25). The distance between M-6 and M-25 is 265.7 kb. This table shows data for markers and results from univariate analysis. Position/Gene line indicates the relative position of G72 (red) and G30 (blue) in reference to SNPs. The type of polymorphism is indicated, and the allele with increased frequency in French Canadian cases compared to French Canadian controls is underlined. Significant P values (lower than 5E-02) are lighted in red. [−, P values higher than 1E-01; HW Da, Hardy–Weinberg disequilibrium; ns, not significant HW Da (exact P value higher than 5E-02); nd, not done.]
For replication, we genotyped, with a subset of 13 markers linked to Bin A, 183 schizophrenic cases and 183 controls from Moscow. The results are shown on Table 1. Marker M-23 gave a genotypic association with a P value of 0.0004 and a weaker allelic association with a P value of 0.017, comparable to what was observed in the larger French Canadian sample. Marker M-24, which gave significant univariate P values in the Canadian sample, is in LD (D′ = 0.99) with marker M-23. It also gives a P value of 0.006 for genotypic association in the Russian collection.
Gene Discovery in the 13q Subregion.
Computational gene annotation, performed across the entire 5-Mb region, showed the presence of several potential genes. Annotation of some of these genes was then completed experimentally by reverse transcription–PCR and rapid amplification of cDNA ends. As a result, seven genes were assigned to the proximal 2-Mb portion of the region (Fig. 1c). The distal 3 Mb could be referred as a “gene desert” (34), with only sparsely located expressed sequence tags. Both Bins A and B mapped within this “gene desert.” Nevertheless, we decided to perform an experimental annotation within and around Bins A and B. By this process, several potential genes were identified within the putative gene desert (Fig. 1c). Within Bin B, a large gene containing at least 20 exons spanning 591 kb was found. The experimental annotation of this gene, named G90, is still being undertaken. A 250-kb segment centered on 65-kb Bin A was extensively screened by experimental annotation. This segment harbors two genes named G72 and G30, both entirely located within associated Bin A. They are transcribed from the two opposite DNA strands and span, respectively, 29 and 47 kb. G72 is entirely included in G30 (Fig. 6). Interestingly, exons of these three genes cannot be predicted by any tested computational method. On the basis of reverse transcription–PCR full length transcript cloning and sequencing, both G72 and G30 generate numerous splice variants in various parts of human brain, spinal cord, and testis. The longest possible G72 ORF, named LG72, is coded by transcripts detected in amygdala, caudate nucleus, spinal cord, and testis. It encodes a 153-aa protein referred to as pLG72 (Fig. 2). Other G72 splice variants share a short common ORF encoding a 16-aa peptide and were detected in testis, different parts of brain, and spinal cord. The longest potential G30 ORF, named LG30, encodes a 71-aa-long putative protein referred to as pLG30. LG30-containing transcripts were detected in several brain regions. No statistically significant homology (E value <0.05) could be found between these G72 and G30 potential proteins and any confirmed or hypothetical protein. We isolated and sequenced a syntenic mouse bacterial artificial chromosome from mouse chromosome band 8A. It contains eight highly conserved blocks collinear in mouse and human genomes. None of these blocks (Fig. 6) overlap with potentially coding parts of G72 or G30. They do not code for human or mouse larger ORFs. No new human or mouse transcripts were detected when these blocks were used as entry points for experimental annotation. By using extended PCR primers, we were able to amplify and sequence coding exons and flanking genomic DNA of LG72 in rhesus monkey and ape genomes. The alignment of corresponding hypothetical pLG72 protein is shown in Fig. 7. In rhesus monkey, most of the exons contain stop codons, and the vast majority of splice sites are mutated. Potential pLG72 orthologues can be deduced for chimpanzee, gorilla, and gibbon genomes. The chimpanzee ORF length is almost half the length of the human LG72 ORF. This is surprising, because only 1% of nucleotide changes occur between chimpanzee and human (36), with most proteins nearly identical in both species. Therefore, G72 could represent a rare case (37) of a primate-specific gene with a rapidly changing protein structure presumably connected with a rapid evolution of underlying brain function.
Figure 2.
Structure of LG72 protein and evolution of its putative orthologues in primates. DNA from four different primates (chimpanzee, gorilla, gibbon, and rhesus monkey) were amplified by using specific primers from the human G72 gene. Amplicons were sequenced, and exons were defined by homology to the corresponding human exons. Alignment of theoretical polypeptide sequences corresponding to pLG72 in human and these different primate species is shown. pLG72 translation in chimpanzee and gorilla is altered by a 4-nt insertion causing a frameshift and a premature stop. In rhesus monkey, the insertion of one T at the end of Exon M causes a frameshift and a premature stop.
Because G72 and G30 reside entirely within the 65-kb-long Bin A and are covered by statistically significant haplotypes, they represent potential disease candidates.
Functional Analysis of Putative Protein Product.
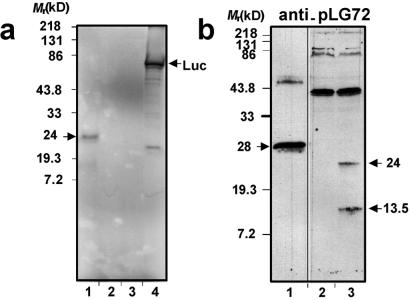
As a first step for functional analysis of G72 and G30 candidate genes, we performed coupled in vitro transcription/translation for each of them (Fig. 3a). Only LG72 gave a specific 24-kDa translation product. No product was detected for LG30, whose translation might require tissue-specific factors. Alternatively, the function of transcripts of this gene could reside in regulation of G72 expression by an antisense mechanism.
Figure 3.
In vitro and ex vivo translation of the G72 candidate gene. (a) Coupled in vitro transcription/translation assays in the presence of [35S]-Met for both LG72 (lane 1) and LG30 (lane 2) cDNAs, unprimed lysates (lane 3), and a luciferase cDNA control (lane 4). (b) Expression vectors harboring the nontagged LG72 cDNA either in the antisense orientation (−) (lane 2) or in the sense orientation (+) (lane 3) were transfected into COS-7 cells and the expressed proteins analyzed with a purified anti-pLG72 serum. A His-Xpress-pLG72 recombinant protein (5 ng) purified from bacteria was run as a control (time exposure for this lane is shorter than for the two others). The His-Xpress tag is 33 amino acids long.
Because of the lack of experimental evidence for a G30 protein, we proceeded further solely with LG72. Rabbit antibodies were raised against bacterially produced 6HIS-tagged pLG72 and were purified on immobilized recombinant pLG72 tagged by glutathione S-transferase. The purified antiserum was used for Western blot analysis of proteins produced by COS7 cells transiently transfected by a nontagged LG72-containing construct (Fig. 3b). Twenty-four- and 13.5-kDa specific products were clearly detected. However, the theoretical molecular mass of the 153-aa-long pLG72 is 18, not 24, kDa. We consistently observed this 6-kDa difference in apparent molecular mass for both tagged and nontagged pLG72 produced by in vitro translation, in bacteria, yeast, or eukaryotic cells. This might be due to a particular secondary structure persisting even after denaturation in reducing conditions. The 13.5-kDa product may result from a proteolytic cleavage from the full length protein.
Characterization of the properties of a protein coded by a given ORF may help to elucidate its function and to distinguish genuine proteins from possible artifacts. We found that in transiently transfected cells, this protein is localized specifically to Golgi (Fig. 7, which is published as supporting information on the PNAS web site). Bacterially produced pLG72 protein, in addition to the previously mentioned abnormal migration in SDS-containing gels, shows the ability to form dimers and oligomers and binds specifically to β-d-galactopyranoside residues (Fig. 8, which is published as supporting information on the PNAS web site), placing it functionally among galectin-like proteins.
Preliminary Mutation Search in 13q Gene G72.
Among 20 markers in and around associated Bin A, two common SNPs could have potential consequences for gene function. Marker M-15 (99–16038/118) is the only common SNP in the region that could influence directly the structure of LG72 protein (lysine/arginine polymorphism at position 29 of ORF). Despite the conservative nature of an arginine-to-lysine substitution, numerous examples of drastic functional changes are known for different classes of proteins (38, 39). This marker gives an allelic P value of association of 0.03 and is in high LD (P value of <10−5) with the mostly associated marker M-12. Six of 37 haplotypes presented in Table 2 contain marker M-15. Another potentially functional common SNP is marker M-19, situated 15 bp upstream of the 5′ end of protein-coding exon Qbis, the part of consensus for an acceptor splicing site. Although M-19 does not show association by itself, it is present in two haplotypes of Table 2. This polymorphism is changing one pyrimidine (C) for another (T) in a polypyrimidine-containing part of this consensus. We do not know for the moment what could be the consequences of such a change to the splicing of the G72 transcript. Although the nucleic acid polymorphisms of M-15 and M-19 could have some functional effect, it is clear that they cannot explain entirely the complex pattern of association in Bin A. Several haplotypes are giving lower P values than single markers, which could mean that as-yet-undiscovered disease-specific sequence variations are contained in strongly associated haplotypes. An alternative explanation may be that these common haplotypes are themselves functionally relevant, predisposing to the disease because of the synergistic effect of their constituent SNPs, as was recently described (40).
Analysis of G72 Functional Interactions.
To investigate whether pLG72 could influence a pathway implicated in schizophrenia, we attempted to identify potential interacting proteins from human brain. With this aim, we screened 500,000 independent clones from a human brain cDNA library in a two-hybrid yeast system (41). Screening the library, we found a nearly full length clone (300 amino acids of 347) of d-amino acid oxidase (DAAO). DAAO is able to oxidize d-serine, an allosteric activator of the NMDA-type glutamate receptor (42). NMDA receptors are ligand-gated ion channels, which have a modulatory site for the amino acids glycine, and d-serine (43). The modulatory site must be occupied by these ligands for glutamate to stimulate cation flow.
A physical interaction between pLG72 and DAAO was confirmed in vitro by column binding and glutathione S-transferase pull down (Fig. 9, which is published as supporting information on the PNAS web site). To evaluate the impact of this physical interaction on DAAO activity, we measured d-serine oxidation by DAAO in the presence of increasing concentrations of recombinant pLG72. Data shown in Fig. 4 suggest that pLG72 behaves as an activator of DAAO. This activation approaches its maximum value at pLG72/DAAO molar ratios of 5:1 calculated for the monomer form of these proteins. These in vitro results could suggest the participation of pLG72 in the regulation of NMDA-type glutamate receptors in specific parts of the human brain. If some individuals were to overproduce pLG72, they could exhibit a lower NMDA-type glutamate receptor activity predisposing them to schizophrenia, which could result in glutamate signaling hypofunction, a mechanism recently proposed in schizophrenia (44–48).
Figure 4.
Oxidation of d-serine by DAAO in the presence of LG72 protein. Activity is expressed as amount H2O2 generated as a result of oxidation of d-serine. The concentration of DAAO was constant (45 ng/μl), the concentrations of pLG72 varied from 0 to 680 ng/μl, and BSA was added to maintain the equal total protein concentration. The curves represent the mean of the three measures for each incubation point. Confidence interval (“error bars”) = mean (three measures) plus or less 1.96. *, Standard deviation (three measures).
Association Studies of DAAO and Its Genetic Interaction with Chromosome 13q Markers.
We attempted to verify genetically the hypothesis that G72 and DAAO operate in the same pathological pathway. We developed eight common SNPs, separated on average by 17 kb, covering the DAAO gene on human chromosome 12q24. They were then tested for disease association in the same French Canadian case/control samples that led to the identification of G72. Four intronic SNPs, in partial linkage disequilibrium (MDAAO-4, MDAAO-5, MDAAO-6, and MDAAO-7) gave significant association P values by univariate analysis as shown in Table 3, which is published as supporting information on the PNAS web site.
Finally, we tried to investigate whether the functional molecular interaction of pLG72 and DAAO detected in vitro can be confirmed through statistical analysis for interaction between these two genes. In principle, the concept of interaction is straightforward: the combined effect of two risk factors in increasing the risk for the disorder is higher than the effect of each single factor per se. In our case, the definition of the phenotype is qualitative, being either present or absent, and the synergic effect of the two genes can be measured by estimating the odds ratios (OR) for the combinations of genotypes at different polymorphisms in G72 and DAAO (49). We therefore examined the joint effect of M-22 and MDAAO-6, the markers showing the greatest evidence for association with schizophrenia at G72 and DAAO, by considering the genotypes at risk for both polymorphisms (Table 4, which is published as supporting information on the PNAS web site). The synergistic effect is calculated as genotype/genotype interaction in a logistic regression model: there are four possible genotypes at risk for M-22 and MDAAO-6, e.g., M-22_AA, M-22_AG, MDAAO-6_GT, and MDAAO-6_TT, leading to four possible meaningful interaction factors among those genotypes. The genotype M-22_AA/MDAAO-6_TT shows a significant interaction effect with an estimated OR of 5.02, P value of 0.04, which is greater than the “additive” effect of M-22_AA (OR = 1.89) and MDAAO-6_TT (OR = 1.04), consistent with a multiplicative/epistatic effect of the two polymorphisms jointly considered (Table 4).
Together, these various results provide convergent evidence for the interpretation that G72 and DAAO may both contribute risk for schizophrenia via the same pathological pathway, at least in some individuals. Future genetic and functional work is necessary to further validate these data converging toward the contribution of G72 and DAAO in schizophrenia and to exclude other possible genes in LD with them. First, the association results should be confirmed by independent groups in other populations. Association with Bin A markers was recently replicated in a familial trio sample by TDT analysis (D.W., unpublished results). Large-scale human brain mRNA and protein differential expression as well as d-serine local concentration measurements will provide important elements for such validation and will elucidate the relationship of genotype to expression profile. Because the NMDA receptor is critical for the development and modifiability of neuronal contacts, the specific expression of G72 and DAAO during both prenatal and postnatal development should also be addressed. At present, little is known about genes predisposing to schizophrenia and their interaction with environmental factors. The application of an LD-based association strategy may be less efficient if several susceptibility alleles for each gene exist. Moreover, the susceptibility to schizophrenia could be highly epistatic, with genes regulating each other in complex interconnected networks of interactions. The definition of disease itself is not homogeneous, and the number of predisposing genes is unknown but likely to be considerable. All these factors will result in relatively weak statistical values for individual SNP association based on LD. Identification of a subset of patients with modified expression of G72 or DAAO or with lower d-serine concentration could produce new molecular markers and may be a valid step toward etiological stratification of schizophrenia. The discovery of other predisposition genes and environmental factors acting on the same pathway may result from studies on this subset of patients.
Supplementary Material
Acknowledgments
We thank E. R. Kandel, J. T. Coyle, Ann Pulver, and Nick Schork for helpful discussions. These data were generated in part through the use of the Celera Discovery System and Celera Genomics' associated databases.
Abbreviations
- DAAO
d-amino acid oxidase
- LD
linkage disequilibrium
- NMDA
N-methyl d-aspartate
- SNP
single-nucleotide polymorphism
Footnotes
References
- 1.Bromet E J, Fennig S. Biol Psychiatry. 1999;46:871–881. doi: 10.1016/s0006-3223(99)00153-5. [DOI] [PubMed] [Google Scholar]
- 2.Breier A, Su T P, Saunders R, Carson R E, Kolachana B S, de Bartolomeis A, Weinberger D R, Weisenfeld N, Malhotra A K, Eckelman W C, et al. Proc Natl Acad Sci USA. 1997;94:2569–2574. doi: 10.1073/pnas.94.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abi-Dargham A, Gil R, Krystal J, Baldwin R M, Seibyl J P, Bowers M, van Dyck C H, Charney D S, Innis R B, Laruelle M. Am J Psychiatry. 1998;155:761–767. doi: 10.1176/ajp.155.6.761. [DOI] [PubMed] [Google Scholar]
- 4.Olney J W, Farber N B. Arch Gen Psychiatry. 1995;52:998–1007. doi: 10.1001/archpsyc.1995.03950240016004. [DOI] [PubMed] [Google Scholar]
- 5.Jentsch J D, Roth R H. Neuropsychopharmacology. 1999;20:201–225. doi: 10.1016/S0893-133X(98)00060-8. [DOI] [PubMed] [Google Scholar]
- 6.Lewis D A, Lieberman J A. Neuron. 2000;28:325–334. doi: 10.1016/s0896-6273(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 7.Verdoux H, Geddes J R, Takei N, Lawrie S M, Bovet P, Eagles J M, Heun R, McCreadie R G, McNeil T F, O'Callaghan E, et al. Am J Psychiatry. 1997;154:1220–1227. doi: 10.1176/ajp.154.9.1220. [DOI] [PubMed] [Google Scholar]
- 8.Mortensen P B, Pedersen C B, Westergaard T, Wohlfahrt J, Ewald H, Mors O, Andersen P K, Melbye M. N Engl J Med. 1999;340:603–608. doi: 10.1056/NEJM199902253400803. [DOI] [PubMed] [Google Scholar]
- 9.Tsuang M. Biol Psychiatry. 2000;47:210–220. doi: 10.1016/s0006-3223(99)00289-9. [DOI] [PubMed] [Google Scholar]
- 10.Pulver A E. Biol Psychiatry. 2000;47:221–230. doi: 10.1016/s0006-3223(99)00281-4. [DOI] [PubMed] [Google Scholar]
- 11.Lin M W, Curtis D, Williams N, Arranz M, Nanko S, Collier D, McGuffin P, Murray R, Owen M, Gill M, et al. Psychiatr Genet. 1995;5:117–126. doi: 10.1097/00041444-199505030-00004. [DOI] [PubMed] [Google Scholar]
- 12.Levinson D F, Holmans P, Straub R E, Owen M J, Wildenauer D B, Gejman P V, Pulver A E, Laurent C, Kendler K S, Walsh D, et al. Am J Hum Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blouin J L, Dombroski B A, Nath S K, Lasseter V K, Wolyniec P S, Nestadt G, Thornquist M, Ullrich G, McGrath J, Kasch L, Lamacz M, et al. Nat Genet. 1998;20:70–73. doi: 10.1038/1734. [DOI] [PubMed] [Google Scholar]
- 14.Shaw S H, Kelly M, Smith A B, Shields G, Hopkins P J, Loftus J, Laval S H, Vita A, De Hert M, Cardon L R, et al. Am J Med Genet. 1998;81:364–376. doi: 10.1002/(sici)1096-8628(19980907)81:5<364::aid-ajmg4>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 15.Brzustowicz L M, Honer W G, Chow E W, Little D, Hogan J, Hodgkinson K, Bassett A S. Am J Hum Genet. 1999;65:1096–1103. doi: 10.1086/302579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riley J H, Allan C J, Lai E, Roses A. Pharmacogenomics. 2000;1:39–47. doi: 10.1517/14622416.1.1.39. [DOI] [PubMed] [Google Scholar]
- 17.Am. Psychiatr. Assoc. Diagnostic and Statistical Manual. 4th Ed. Washington, DC: Am. Psychiatr. Assoc.; 1994. [Google Scholar]
- 18.Pritchard J K, Rosenberg N A. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. Schedules for Clinical Assessment in Neuropsychiatry. Geneva: World Health Organization; 1994. [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 22.Schlesselman J J. Case-Control Studies: Design, Conduct, Analysis. Oxford, U.K.: Oxford Univ. Press; 1982. [Google Scholar]
- 23.Lewontin R C. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider S, Roessli D, Excoffier L. arlequin, Ver. 2000: A Software (Genetics and Biometry Laboratory, University of Geneva, Geneva) 2000. [Google Scholar]
- 25.Excoffier L, Slatkin M. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 26.Fallin D, Schork N J. Am J Hum Genet. 2000;67:947–959. doi: 10.1086/303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fallin D, Cohen A, Essioux L, Chumakov I, Blumenfeld M, Cohen D, Schork N J. Genome Res. 2001;11:143–151. doi: 10.1101/gr.148401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Good P. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. New York: Springer; 1994. [Google Scholar]
- 29. sas Package Software (1999–2001) (SAS Institute, Incorporated, Cary, NC), Ver. 8.1.
- 30.Gabler M, Hensel M, Fischer L. Enzyme Microb Technol. 2000;27:605–611. doi: 10.1016/s0141-0229(00)00262-3. [DOI] [PubMed] [Google Scholar]
- 31.Wang D G, Fan J B, Siao C J, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J, et al. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 32.Sachidanandam R, Weissman D, Schmidt S C, Kakol J M, Stein L D, Marth G, Sherry S, Mullikin J C, Mortimore B J, Willey D L, et al. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 33.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 35.Matthew D E, Farewell V T. Using and Understanding Medical Statistics. 3rd Ed. Basel: Karger; 1996. pp. 182–186. [Google Scholar]
- 36.Chen F C, Li W H. Am J Hum Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson M E, Viggiano L, Bailey J A, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler E E. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Roy R. Biochemistry. 2001;40:13617–13622. doi: 10.1021/bi011053b. [DOI] [PubMed] [Google Scholar]
- 39.Agarwal S, Hattori Y, Gupta U R, Agarwal S S. Hemoglobin. 1999;23:263–265. doi: 10.3109/03630269909005707. [DOI] [PubMed] [Google Scholar]
- 40.Drysdale C M, McGraw D W, Stack C B, Stephens J C, Judson R S, Nandabalan K, Arnold K, Ruano G, Liggett S B. Proc Natl Acad Sci USA. 2000;97:10483–10488. doi: 10.1073/pnas.97.19.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fields S, Song O. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 42.Mothet Y P, Parent A T, Wolosker H, Brady R O, Jr, Linden D J, Ferris C D, Rogawski M A, Snyder S H. Proc Natl Acad Sci USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson J W, Ascher P. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 44.Mohn A R, Gainetdinov R R, Caron M G, Koller B H. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- 45.Carlsson A, Waters N, Carlsson M L. Biol Psychiatry. 1999;46:1388–1395. doi: 10.1016/s0006-3223(99)00117-1. [DOI] [PubMed] [Google Scholar]
- 46.Heresco-Levy U. Int J Neuropsychopharmacol. 2000;3:243–258. doi: 10.1017/S1461145700001978. [DOI] [PubMed] [Google Scholar]
- 47.Heresco-Levy U, Javitt D C, Ermilov M, Mordel C, Horowitz A, Kelly D. Br J Psychiatry. 1996;169:610–617. doi: 10.1192/bjp.169.5.610. [DOI] [PubMed] [Google Scholar]
- 48.Javitt D C. Curr Psychiatry Rep. 2001;3:413–417. doi: 10.1007/s11920-996-0036-9. [DOI] [PubMed] [Google Scholar]
- 49.Kleinbaum D G, Kupper L L, Morgenstern H. Epidemiologic Research. New York: Van Nostrand Reinhold; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.