Abstract
The 3′ Ig heavy chain locus (Igh) regulatory region is the most downstream known element of the murine Igh gene cluster. We report here that the nearest non-Igh genes—Crip, Crp2, and Mta1—are located ≈70 kb further downstream and are beyond the end of the domain of Igh transcriptional regulation. We have localized an origin of replication in MEL cells to a 3-kb segment located between the 3′ Igh regulatory region and Crip. Sequences downstream of this origin are replicated by forks that move in both directions. Sequences upstream of this origin (Igh-C, -D, and -J) are replicated in a single direction through a 500-kb segment in which no active bidirectional origins can be detected. We propose that this origin may lie at or near the end of the Igh regulation domain.
Enhancers of the 3′ regulatory region (reviewed in ref. 1) are the most downstream known elements of the Ig heavy chain locus (Igh). These have been shown to have a role in class switching (2) and are implicated in the regulation of levels of Igh expression (3–6) and somatic hypermutation (7). Recent studies have suggested that the most distant enhancer, hs4, is near the terminus of a class switch regulatory region (8). However, studies of mice in which hs3B and hs4 have been deleted have suggested the presence of additional regulators downstream of hs4 (2). We have wanted to determine what sequences lie beyond hs4 to assess the possibility of additional regulators and to identify where B cell-specific regulation of the Igh locus ceases.
Our studies on Igh gene replication in B cell lines have raised the possibility that regulators of replication of the Igh gene locus are located downstream of the 3′ regulatory region (9–12). Two distinct patterns of replication within the Igh gene cluster are apparent (see Fig. 3a, which is published as supporting information on the PNAS web site, www.pnas.org). In non-B cell lines, such as the mouse erythroleukemic cell line, MEL, replication of different portions of the Igh locus occurs throughout S phase (10). Replication initiates early in S from a region located downstream of the Igh cluster, and proceeds opposite to the transcriptional orientation through 400 kb of Igh-C (constant), -J (joining),and -D (diversity) sequences, concluding in late S phase with the most proximal Vh genes. More distal Vh genes are all replicated late in S. In B and plasma cell lines, the replication pattern is the same as that of the MEL cells, despite the reduction in the length of the locus resulting from VDJ recombination and class switching (12). The temporal transition region between early- and late-replicating domains extends through rearranged Igh-C sequences and the VDJ gene into formerly distant 5′ unrearranged Vh genes. Replication forks move in the same single 3′ to 5′ direction through this temporal transition region with no apparent activation of any additional bidirectional origins (9, 10).
A contrasting pattern of replication is observed in pro-B and pre-B cell lines (Fig. 3a). Here, all Igh sequences replicate early in S phase (11, 12), and replication forks move in both directions throughout the Igh-C region (12). Latent origins of replication within the early to late temporal transition region in MEL apparently become active when there is transcription of unrearranged Vh sequences associated with VDJ joining. Importantly, these studies suggest that during B cell development, replication of Igh is coordinated with various steps in Igh gene expression and rearrangement. In contrast, replication of the region immediately 3′ to the transition region is regulated independently; sequences in this region are replicated early in non-B cells and in B cells at all stages of B cell differentiation (10, 12). In the non-B cell line, MEL, the unidirectional fork movement of the transition region replicon was previously detected as far as ≈45 kb downstream of Cα, but forks moved in both directions at a point ≈100 kb downstream (9, 10). This change in replication fork direction implies that a replication origin lies in this region downstream of hs4, which has not been completely sequenced in either mouse or human. Transcriptional activity of downstream non-Igh genes may account for origin activation.
We report here the sequence of the 125 kb long BAC CT7–199M11 (199M11) (Fig. 1a), which extends 3′ from the IgA constant region membrane exon through the 3′ regulatory region toward the centromere on mouse chromosome 12. At the centromeric end of this sequence are the genes Crip, Crp2, and Mta1; these are the nearest recognized genes downstream of the Igh locus and are widely expressed in nonlymphoid cell types. We used unique sequences derived from 199M11 as probes on neutral/alkaline two-dimensional (N/A 2D) gels to map the direction of replication fork progression, and have localized a transition region between unidirectional and bidirectional fork progression to a 3-kb segment that is located ≈50 kb downstream of the 3′ Igh regulatory region and ≈20 kb upstream of Crip. This 3-kb segment contains an initiation site, which is the nearest to the 3′ boundary of the developmentally regulated domain of Igh replication. The replication boundary is, therefore, in the same vicinity as the terminus of B cell-specific transcriptional regulation of the Igh locus.
Figure 1.
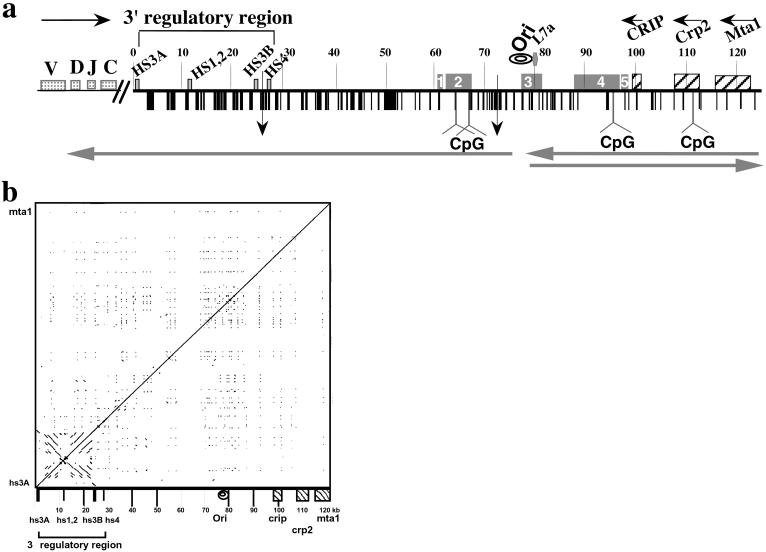
Structural features of the 125 kb DNA insert of 199M11. (a) Schematic of sequence features of BAC199M11 (GenBank accession no. AF450245). Position 1 in 199M11 is the telomeric end of the chromosome segment cloned in this BAC and corresponds to position 1242 of GenBank accession no. K00691 (Cα membrane exon). The Igh locus is in inverted transcriptional orientation on chromosome 12 so that the 5′ end of the locus is distal (telomeric). “Downstream” with respect to Igh is proximal on the chromosome, and we will use upstream, or 5′, and downstream, or 3′, to describe a position relative to Igh, not the chromosome. V, D, J, and C sequences are located upstream of BAC199M11 sequences, Although there are two small gaps in the 199M11 sequence (indicated by vertical arrows at 26,423, between hs3b and hs4 enhancers, and at position 71,578), each estimated to be <500 bp, we present the data as a continuous sequence 1 to 124,151 (GenBank Accession no. AF450245). Repetitive DNA sequences identified by Repeat Masker and by individual searches of DNA segments are shown with vertical black bars and specifically identified in GenBank accession no. AF450245. A higher density of repetitive sequences is apparent closer to the Igh gene cluster and a relatively lower density toward the downstream end of the BAC. Igh regulatory elements, L7a pseudogene, Crip, Crp2, and Mta1 genes and CpG islands (at 64180–65703; 67270–67539; 96721–97360; and 112328–113229) are indicated. Transcription direction is indicated by horizontal arrows above the sequence, and direction of replication fork movement is indicated by horizontal arrows below the sequence. A region within which the direction of replication fork movement changes is indicated by “ori”. An A/T-rich segment is located at 77,700–77,746. Locations in the 199M11 sequence are given as distance from the Igh end; e.g., 199M11:99 is 99 kb from the IgA membrane exon (5′) end of 199M11. ESTs are indicated by rectangular boxes containing nos. 1–5. EST region 1 (61,000–61,484) matches a single EST from heart-AA646361 (5′ end sequence of IMAGE clone 1021178; the 3′ sequence of this clone has not been determined). Much of EST region 2 (62,082–67,432) is contained in AK010454, which has been termed a full insert mRNA sequence that encodes a gene. Multiple ESTs matching this region have been identified as part of UniGene Cluster Mm. 133306 Mus musculus. EST region 3 (77,453–81,342) matches two IMAGE clones that appear to be identical, 641181 (5′ sequence, AI605834, and 3′ sequence, AI450404) and 3418382 (5′ sequence, BE848059, and 3′ sequence, BE853477). This region, identified in UniGene Cluster Mm. 32319 Mus musculus, contains the replicative boundary in MEL [199M11:76–79 and a previously unidentified L7a pseudogene (199M11:79)]. The gene encoding the L7a protein (Surf-3) (32) appears to be present in a single copy within the mouse surfeit locus on chromosome 2 (52); and there are 15–20 L7a-related family members, three of which have been analyzed and found to have the hallmarks of processed pseudogenes (53), i.e., lack of introns, presence of multiple defects that would prevent translation, and flanked by direct repeats of 13–15 bp. Immediately 3′ of the L7a homology region in 199M11, there is a B2/SINE element followed by a short poly(A) tract. EST region 4 extends ≈9 kb from 87,921–96,874, within which several ESTs have been arranged in two UniGene Clusters: Mm 87616 Mus musculus (87,921–88,550) and Mm. 90118 Mus musculus (88,807–95,766). Within UniGene Cluster Mm. 90118 is BC022617, which has been identified as an mRNA/gene sequence, although a predicted protein sequence is not clearly specified. Comparison of BC022617 to 199M11 suggests that the gene encoding BC022617 contains at least five exons. EST region 5 (97,382–98,337) shows three matches, including to the 5′ (BE624876) and 3′ (BE629214) ends of IMAGE clone 3370299. (b) DotPlot analysis (54) (http://bio.cse.psu.edu/pipmaker) of 199M11 to identify internal repetitive sequences shows unique features associated with the 3′ Igh regulatory region, namely an extensive palindrome and multiple repetitive sequences. Parallel lines indicate direct repeats and perpendicular lines indicate inverted repeats.
Materials and Methods
Sequence of 199M11.
BAC CT7–199M11, referred to here as 199M11, was purchased from Research Genetics, now Invitrogen; this bacterial artificial chromosome (BAC) is from the CitbCJ7 library made from the 129S1/SvImJ mouse strain. BAC DNA was purified according to a protocol from Qiagen with minor modifications (P1, 10 ml; P2, 11 ml; P3, 12 ml). The complete sequence of the 125 kb region included in 199M11 (GenBank accession no. AF450245) was determined by using direct end-sequencing, extension of end-sequences by cloning overlapping segments, extension of the end sequences of BACs CT7–145B12, CT7–397G20, and CT7–225H9, each of which terminates within 199M11, and extension of previously identified sequences in the 3′ Igh regulatory region. This created an extensive framework for the ultimate shotgun sequencing of 199M11 in pUC18.
Two internal contigs were extended from sequences we previously reported (AF005484, AF017981, and AF017982) from our analysis of a phage clone that extended downstream of the 3′ regulatory region (9). An additional contig was assembled by extending end sequences of BACs that overlapped 199M11, and the sequences of Crip cDNA and promoter. To complete the sequence, 199M11 was partially digested with Sau3A1, and 3- to 4-kb fragments were shotgun cloned into pUC18. Approximately 400 clones were obtained. Plasmid inserts were amplified by PCR with M13 forward and reverse primers. The same primers were used for sequencing. Contigs were generated by using sequencher software (Gene Codes, Ann Arbor, MI). Gaps between contigs were filled by primer walking using the plasmid DNA of the shotgun clones that bridge different contigs as a template. Remaining gaps, except for two ≈500-bp segments which contained intractable repetitive sequences, were covered with walking primers and PCR using BAC DNA as a template. Although utilization of partial digests as a template for sequence analysis provides some bias and not every segment was covered by sequencing of both strands, an average coverage of 3–4 genomic equivalents was achieved and the accuracy of the nucleotide sequence based on the total sequence analysis was estimated to be 99.9%.
Repeat elements in the sequence of 199M11 were identified and masked with Repeat Masker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). Gene and EST sequences were identified with blast (13, 14). CpG islands were identified by using a European Molecular Biology Laboratory program at www.ebi.ac.uk/emboss/cpgplot/index.html.
Isolation of Replicative Intermediates for N/A 2D Gel Electrophoresis (10).
MEL cells were grown in DME, supplemented with 10% FCS, and 1% penicillin-streptomycin. Cells from early or early middle S phase fractions, prepared by centrifugal elutriation (15), were used to isolate nuclear matrix-associated DNA enriched in replicative intermediates (16). DNA was digested by specific restriction enzymes, cleared of a high molecular weight fraction by sucrose gradient, chromatographed on BND-cellulose columns to further enrich replicative intermediates (17), and separated by N/A 2D gel electrophoresis and then transferred to HyBond N Plus membranes (18). We assess the direction of fork movement after hybridizing the blot to unique probes (Table 1, which is published as supporting information on the PNAS web site).
Results
The Sequence of 199M11.
To determine the DNA sequence of the region downstream of the mouse Igh 3′ regulatory region, we identified and sequenced a BAC extending from the 3′ end of IgA to the nearest known non-Igh genes. A schematic of the BAC sequence (GenBank accession no. AF450245) is shown in Fig. 1a. The locations of sequences previously identified in the region covered by 199M11 are indicated. For example, the 3′ regulatory region contains four enhancer sequences, hs3A, hs1,2, hs3B, and hs4, originally identified as DNase I hypersensitivity sites. Additionally, the 3′ regulatory region has a general palindromic nature that extends through hs3A, hs1,2 and hs3B and includes families of locally repeated sequences between enhancer segments (19–21). DotPlot analysis of 199M11 (Fig. 1b) shows the full extent of the palindromic region of the 3′ regulatory region from hs3A to hs3B. No palindromic sequences are evident elsewhere that would suggest additional downstream regulatory sites.
Crip, Crp2, and Mta1 Are the Nearest Non-Igh Genes Downstream of IgA.
To identify known genes in the 199M11 sequence, we performed blast comparisons of 199M11 to GenBank databases and identified several genes. The nearest gene to Igh is Crip (cysteine-rich intestinal protein) (22), located at 199M11:99 (Fig. 1a). Crip was reported to be closely linked to Igh (22, 23). Downstream of Crip, we detected significant matches with Crp2 (cysteine-rich protein 2) cDNA sequences from rat (GI:487283, D17512) (24) and human (GI:4503048) (25, 26) (Fig. 1a). Like Crip, CRP2 protein is a member of the LIM domain (cysteine-rich double zinc-finger) protein family. Based on the cDNA sequence for rat Crp2 and commonly used splice junctions, we were able to predict the exon/intron structure of the murine Crp2 gene and the expected mRNA product (GenBank accession no. AF450245). We found a murine cDNA for Crp2 in the EST database (GenBank accession no. BC002093) that matched our predicted sequence, suggesting that we have identified the mouse Crp2 gene.
The centromeric terminal portion of 199M11 matches the 3′ end of human and rat MTA1 (metastasis-associated protein) cDNA (27) (Fig. 1a). The MTA1 protein was originally identified because of its increased expression in highly metastatic rat and human mammary tumor cell lines (27). Subsequently, it was reported to be a component of the nucleosome remodeling and histone deacetylation (NURD) complex (28). Because neither the murine gene nor cDNA for Mta1 had previously been identified, we generated the murine cDNA for Mta1 by RT-PCR. Nested primers were developed based on sequences conserved at the 5′ and 3′ ends of rat and human Mta1. The sequence of Mta1 cDNA from the A20 cell line was determined (GenBank accession no. AF463504); this matched 199M11 sequences and encoded an ORF with extensive sequence identity to rat and human MTA1. In vitro transcription and translation revealed a single protein product commensurate with the 80-kD size previously reported for MTA1 from other species (see Fig. 3b). These data confirm the presence of the Mta1 gene in this location. Recent data have reported a cDNA for murine Mta1 (29) (GenBank accession no. AAK83044). Our predicted protein sequence matches AAK83044 except for two amino acid differences and a 17-aa insertion. Exon/intron boundaries were assigned in accordance with the sequence of splice sites (AT-exon-GT) for each exon of Crip and Crp2 (GenBank accession no. AF450245), and for the 3′ region of Mta1 contained within 199M11 (GenBank accession no. AF463504)
All three genes-Crip, Crp2, and Mta1-have the same transcriptional orientation, opposite to Igh sequences. Northern blots showed that both Crip and Mta1 were expressed in MEL (see Fig. 3c) in which Igh is silent. Previous studies have shown that all three genes are broadly expressed [Crp2 (24, 25) and Mta1 (30)], except that Crip was not detected in adult brain, kidney, or liver (22, 31). Thus, these neighboring genes are outside the domain of B cell-specific transcriptional regulation of the Igh locus.
Analysis of ESTs.
To identify any as yet unrecognized genes in the region downstream of Igh, we performed blast comparison of 199M11 with the GenBank EST database. This identified five intervals between the 3′ regulatory region and the Crip gene that match ESTs, the first of which is at ≈199M11:62, ≈30 kb downstream of the 3′ regulatory region (Fig. 1a). Only a few ESTs have been identified from intervals 1, 3, and 5, whereas intervals 2 and 4 are represented by many ESTs isolated from several different tissues. Both intervals 2 and 4 have recently been predicted to encode hypothetical proteins using a transcriptional orientation opposite to that of Igh and similar to Crip, Crp2, and Mta1; and CpG islands have been detected near the 5′ end of these predicted coding regions (Fig. 1a). ESTs from intervals 3 and 4, and possibly 2 as well, appear to have been derived by splicing from genomic DNA, as represented by BAC 199M11. The predicted intronic region of EST 3 contains a pseudogene for the L7a ribosomal protein (Surf-3) (32), which differs from other L7a pseudogenes in its sequence and its lack of flanking direct repeats. Interestingly, the two genes encoding the SCM-1 chemokine contain an intron in which there is an L7apseudogene (33). Therefore, the region between 199M11:62 and Crip contains putative genes that are oriented opposite to Igh and are expressed in multiple tissues. This suggests that the boundary of the Igh transcriptional domain is between hs4 and 199M11:62.
Localization of an Origin of the Igh Replicon for the Temporal Transition Region.
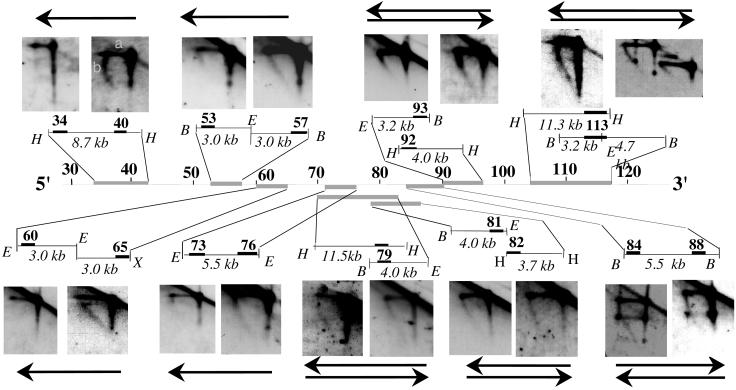
We wanted to locate the origin of this Igh replicon and identify any unusual sequence characteristics. We used N/A 2D gel electrophoresis to determine the direction of replication fork progression through Igh downstream sequences in MEL cells. Because this technique requires ≈5 × 109 cells for study, we are able to analyze cell lines like MEL, but not the smaller numbers of primary untransformed cells that can be purified at specific developmental stages. We have used 16 probes (Table 1) to examine 15 DNA segments to assess the direction of replication fork movement through an ≈80-kb region downstream of the 3′ Igh regulatory region (Fig. 2; see interpretation of N/A 2D gels in Fig. 4a, which is published as supporting information on the PNAS web site). This region extends from 199M11:34 (previously termed Cα-45) to 199M11:117 (previously termed Cα-100) and is where replication forks change direction in MEL (10). In the four sets of segments upstream of 199M11:76, i.e., at 199M11:34–40, 53–57, 60–65, and 73–76, 3′ end probes detected nascent strands of multiple sizes (ranging in size from very small to as large as the parental strand itself), whereas 5′ end probes detected only long nascent strands. These data indicate that replication forks move through these sequences in one direction, toward the telomere, and that initiation occurs downstream of 199M11:76.
Figure 2.
Analysis of replication fork direction in the region downstream of the 3′ regulatory region by N/A 2D gel electrophoresis. The first dimension of the agarose gel is run in neutral conditions in which restriction enzyme-digested DNA molecules enriched in replication intermediates are separated according to their mass. The second dimension is run in alkaline conditions to separate parental and nascent strands. The direction of replication fork movement (indicated by arrow) is determined by hybridization to probes (solid rectangles) located at the 5′ or 3′ end of a specific restriction fragment (probes 199M11:34 and 40, 73 and 76, and 84 and 88); or from 5′ and 3′ ends of two adjacent restriction fragments (probes 199M11:53 and 57, 60 and 65, 81 and 82, and 92 and 93); or by using a single probe located at the ends of two overlapping fragments (probes 199M11:79 and 113). The size of the restriction fragment is indicated. Note that 199M11:34 and 199M11:117 were previously termed Cα-45 and Cα-100, respectively. Each segment (indicated by gray lines) was analyzed by two or more 2D gels with similar findings. The topmost restriction fragment and the topmost arrow (if there are two) always refer to the left of the two panels. The horizontal line (a) represents the parental strands of the replication intermediates, which run at a single position in the second dimension because they have a single molecular weight. The diagonal line (b) emanating from the parental strands represents nascent strands of different sizes, ranging from very small to as large as the parental strands. If replication forks move from 3′ to 5′, a probe located at the 3′ end of a fragment will detect nascent strands of sizes ranging from small to parental-sized, whereas probes located at the 5′ end will detect nascent strands only of a relatively large size. If forks move in both directions through a segment, then nascent strands of multiple sizes will be detected with probes for both 5′ and 3′ ends of this segment. Because of limitations in the resolution of N/A 2D gel electrophoresis, we cannot rule out the possibility that <10% of the forks progress from the opposite direction in the region upstream of 199M11:76. The spots present on the nascent strand arcs and the linear molecules on the autoradiogram probed with 199M11:84or :88 are caused by the accumulation of a single-stranded break in this segment, produced by EcoRI digestion, which is released during alkaline treatment in the 2nd dimension. No spots were detected on neutral/neutral (N/N) 2D gels. Therefore, this single-strand break does not appear to form any replication barrier. H, HindIII; E, EcoRI; B, BamHI; X, XbaI.
Beginning at 199M11:79, we saw a change in the direction of replication fork progression (Fig. 2). Probes for both 5′ and 3′ ends of five sets of segments detected nascent strands of multiple sizes, indicating that replication forks proceed in both directions through these segments downstream of 199M11:79. That replication forks progress in opposite directions between 199M11:76 (3′ to 5′) and 199M11:79 (5′ to 3′) indicates that there must be an active origin located in between. This origin-containing segment, which begins immediately upstream of EST region 3, contains an A/T-rich segment of 47 bp flanked by B2 short interspersed repeated (SINE) sequences (Fig. 1). An A/T-rich segment is a feature of several origins of replication identified in yeast (discussed in refs. 34 and 35). Other sequences in this segment are unique and bear no distinctive homology to mammalian initiation segments that have previously been detected. Indeed, they are unlike any sequences in the recently assembled Whole Mouse Genome Sequence (www.ncbi.nlm.nih.gov/genome/seq/MmBlast.html).
Discussion
We have sequenced the BAC CT7-199M11 that links the mouse Igh locus to neighboring downstream genes and identified Crip, Crp2, and Mta1 as the nearest recognized genes downstream. Crip, the nearest of these genes, is located >70 kb downstream of hs4. Each of these genes is positioned in a transcriptional orientation opposite to that of the Igh gene cluster as are two putative genes identified by multiple ESTs, which are located between 30 and 60 kb downstream of hs4. The 3′ Igh regulatory region, therefore, lies 3′ of both Igh on one side and Crip, Crp2, and Mta1, and additional putative genes on the other. However, the latter genes and ESTs are not expressed in a B cell-specific manner. Mta1 is widely expressed, including in all cell lines that we have examined, and Crip is more narrowly expressed (Gene Expression Database, Mouse Genome Informatics Web Site, The Jackson Laboratory; www.informatics.jax.org/), but we find Crip mRNA expression in MEL. The expression of Crip and Mta1 genes is, therefore, not governed by Igh regulatory elements; and B cell-specific regulation of transcription of Igh-linked sequences stops somewhere in the interval between hs4 at199M11:28 and the widely expressed EST site at 199M11:62.
We used N/A 2D gel analysis in MEL, and have identified a 3-kb segment (199M11:76–79) that contains the active origin of DNA replication that is closest to the Igh-C gene cluster. This origin lies ≈50 kb downstream of the 3′ Igh regulatory region and ≈20 kb upstream of the neighboring non-Igh genes, Crip, Crp2, and Mta1. In all cells examined, sequences extending >200 kb downstream of this origin-containing segment replicate early in S (10, 12). We detect replication forks progressing in both directions through seven segments in this region, five in a 40-kb region downstream of 199M11:79, as reported here, and two segments located ≈140 and 220 kb further 3′ (10). Our data are most compatible with a zone of multiple initiation sites. Initiation zones have been described for the Chinese hamster dihydrofolate reductase locus (36), the DNA locus encoding ribosomal RNA (18), the chicken lysozyme gene (37) and the mouse β-globin locus (38). The ubiquitous expression of Mta1 may account for early replication of the Igh downstream region (for the relationship between transcription and replication, see review in ref. 39).
Although the Igh regulatory control of either transcription or replication does not influence the genes downstream of the origin located at 199M11:76–79, in the region extending upstream the replication of Igh sequences is developmentally regulated. In MEL, B and plasma cells, unidirectional fork movement produces a replicative temporal transition region that extends to the late replicating Vh genes. In B and plasma cell lines, the replicative temporal transition region appears to begin closer to Igh sequences than in MEL (12). This could reflect the activation of enhancers of the 3′ regulatory region in B and plasma cell lines, and also suggests that sequences between the 3′ regulatory region and 199M11:76–79 may acquire origin functionality at stages during B cell differentiation. It has been speculated (40) that origin usage is likely to be influenced by chromatin structure rather than specific sequence features of the origin itself. In fact, except for a 47 bp AT-rich region at 199M11:77, this active origin-containing region from MEL has no special structural characteristics, and these origins bear little homology to each other. Interestingly, beginning at 199M11:62, there are regions corresponding to EST transcripts and putative genes in a variety of tissue sources, including two B-lineage cell lines (N.A. and B.K.B., unpublished data). Potentially, replicative temporal transition regions might begin at somewhat different positions in different cells dependent on the expression of these DNA segments. We suggest that the domains of transcription and replication are coordinately regulated and may have a common boundary whose position in this region varies with the activation state of Igh and the expression of neighboring genes.
The Igh locus in pro- and pre-B cell lines was replicated early in S phase by replication forks that move in both directions (12). The region downstream of Igh also replicates early in S and forks progress in both directions through the segments analyzed by 199M11:34 (12) and 199M11:73 and 76 in the 70Z/3 pre-B cell line (J.Z. and C.L.S., unpublished data).
Associated with changes in Igh replication (12) and transcription, there are differences in the nuclear location of the Igh gene cluster (12, 41). In cells with a replicative temporal transition region, the Igh locus was positioned near the nuclear periphery. However, in pro-B and pre-B cells, the Igh locus was positioned away from the nuclear periphery (12, 41). Others have also noted differences in the nuclear location of the two Igh alleles relative to heterochromatin in LPS-activated splenic B cells (42).
Regulation of replication, transcription and nuclear location may be associated with changes in chromatin structure of the Igh locus during B cell development. Recent studies have reported an increase in DNase I sensitivity at both V and J segments of H and L loci before initiation of VDJ recombination, with a reduction in V region sensitivity after rearrangements are completed (43). Furthermore, there is an increase in the level of histone acetylation in Igh genes in pro-B and pre-B cells (44) and cell lines (43). However, no sequences downstream of Cγ2b have been analyzed. We hypothesize that the beginning of the replicative temporal transition region is located at the 3′ end of an Igh domain of related regulation of transcription, rearrangement, and replication. Based on identification of potential coding sequences beginning at 199M11:62, we predict that any additional regulators for the Igh locus are located between hs4 and this segment. Analysis of histone acetylation and DNase I sensitivity is underway to identify any demarcation in chromatin structure between 3′ Igh sequences and Crip, and to relate this to the replication boundary.
Recent studies have suggested various mechanistic links between replication of Igh genes and other processes that affect Igh genes during B cell differentiation. Among these are our observations that different Igh replication patterns are observed at different stages of B cell differentiation (10, 12). In addition, more subtle differences in the time of replication of the two Igh alleles have been identified in both non-B and B cells. The earlier replicating allele appears to be preferentially used for rearrangement and expression in B cells, thereby providing a mechanism for allelic exclusion (45). It is not known how mechanisms governing replication can be coordinated with DNA rearrangement and expression. However, the formation of a double-strand DNA break required for mating type switching in Schizosaccharomyces pombe depends on a specific direction of replication fork progression (46). Such (asymmetric) DNA breaks have also been observed to be associated with somatic hypermutation (47–50).
We propose that the Igh locus resides in a chromosomal region that is under B cell-specific regulation of transcription, rearrangement, and replication. The studies we present here identify 199M11:76–79 as a centromeric end of the replicative domain in MEL cells. This segment contains a replication origin from which a single replication fork progresses through the Igh-C locus throughout S phase. This origin is unlikely to be the sole initiation site because we do not detect termination in this region by N/N 2D gel electrophoresis (data not shown). Instead this origin is likely to be the most upstream of a series of origins for the Igh replication forks, and based on the intensity of the nascent strand arcs detected by using the same probe (199M11:79) on two overlapping fragments, is used in ≈15–50% of the cells. We have observed that genes located downstream of this origin and ESTs within close proximity are regulated independently of the Igh locus. Hence, the replication domain appears to correspond to a transcriptional regulatory domain. Temporal replication domains have been detected in the murine κ (51) and human Igh loci (P. Galgano and C.L.S., unpublished data). It will be important to determine whether these and other loci that are specifically expressed in lymphoid cells are regulated similarly to the mouse Igh locus.
Supplementary Material
Acknowledgments
We thank Alejandro Sepulveda, Paolo Norio, Moshe Sadofsky, and Alberto Martin for critical reading of the manuscript, and Alejandro Sepulveda for generating the PIP DotPlot figure. This work was supported by National Institutes of Health Grants AI13509 (to B.K.B.), GM45751 (to C.L.S.), and AI23548 (to R.R.). C.C. was supported by a fellowship from Institut National de la Santé et de la Recherche Médicale.
Abbreviations
- BAC
bacterial artificial chromosome
- N/A 2D
neutral/alkaline two-dimensional
Footnotes
References
- 1.Khamlichi A A, Pinaud E, Decourt C, Chauveau C, Cogne M. Adv Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- 2.Pinaud E, Khamlichi A A, Le Morvan C, Drouet M, Nalesso V, Le Bert M, Cogne M. Immunity. 2001;15:187–199. doi: 10.1016/s1074-7613(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 3.Gregor P D, Morrison S L. Mol Cell Biol. 1986;6:1903–1916. doi: 10.1128/mcb.6.6.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaelson J S, Giannini S L, Birshtein B K. Nucleic Acids Res. 1995;23:975–981. doi: 10.1093/nar/23.6.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X, Eckhardt L A. Int Immunol. 2001;13:1003–1012. doi: 10.1093/intimm/13.8.1003. [DOI] [PubMed] [Google Scholar]
- 6.Lieberson R, Ong J, Shi X, Eckhardt L A. EMBO J. 1995;14:6229–6238. doi: 10.1002/j.1460-2075.1995.tb00313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terauchi A, Hayashi K, Kitamura D, Kozono Y, Motoyama N, Azuma T. J Immunol. 2001;167:811–820. doi: 10.4049/jimmunol.167.2.811. [DOI] [PubMed] [Google Scholar]
- 8. Manis, J. P., Michaelson, J. S., Birshtein, B. K. & Alt, F. W. (2002) Mol. Immunol., in press. [DOI] [PubMed]
- 9.Michaelson J S, Ermakova O, Birshtein B K, Ashouian N, Chevillard C, Riblet R, Schildkraut C L. Mol Cell Biol. 1997;17:6167–6174. doi: 10.1128/mcb.17.10.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ermakova O V, Nguyen L H, Little R D, Chevillard C, Riblet R, Ashouian N, Birshtein B K, Schildkraut C L. Mol Cell. 1999;3:321–330. doi: 10.1016/s1097-2765(00)80459-1. [DOI] [PubMed] [Google Scholar]
- 11.Hatton K S, Dhar V, Brown E H, Iqbal M A, Stuart S, Didamo V T, Schildkraut C L. Mol Cell Biol. 1988;8:2149–2158. doi: 10.1128/mcb.8.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Ermakova O V, Riblet R, Birshtein B K, Schildkraut C L. Mol Cell Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown E H, Iqbal M A, Stuart S, Hatton K S, Valinsky J, Schildkraut C L. Mol Cell Biol. 1987;7:450–457. doi: 10.1128/mcb.7.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughn J P, Dijkwel P A, Mullenders L H, Hamlin J L. Nucleic Acids Res. 1990;18:1965–1969. doi: 10.1093/nar/18.8.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkwel P A, Vaughn J P, Hamlin J L. Mol Cell Biol. 1991;11:3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little R D, Platt T H, Schildkraut C L. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannini S L, Singh M, Calvo C-F, Ding G, Birshtein B K. J Immunol. 1993;150:1772–1780. [PubMed] [Google Scholar]
- 20.Chauveau C, Cogne M. Nat Genet. 1996;14:15–16. doi: 10.1038/ng0996-15. [DOI] [PubMed] [Google Scholar]
- 21.Saleque S, Singh M, Little R D, Giannini S L, Michaelson J S, Birshtein B K. J Immunol. 1997;158:4730–4787. [PubMed] [Google Scholar]
- 22.Birkenmeier E H, Gordon J I. Proc Natl Acad Sci USA. 1986;83:2516–2520. doi: 10.1073/pnas.83.8.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho M, Villani V, D'Eustachio P. Mamm Genome. 1991;1:30–36. doi: 10.1007/BF00350843. [DOI] [PubMed] [Google Scholar]
- 24.Okano I, Yamamoto T, Kaji A, Kimura T, Mizuno K, Nakamura T. FEBS Lett. 1993;333:51–55. doi: 10.1016/0014-5793(93)80373-3. [DOI] [PubMed] [Google Scholar]
- 25.Karim M A, Ohta K, Egashira M, Jinno Y, Niikawa N, Matsuda I, Indo Y. Genomics. 1996;31:167–176. doi: 10.1006/geno.1996.0028. [DOI] [PubMed] [Google Scholar]
- 26.Tsui S K, Chan P P, Cheuk C W, Liew C C, Waye M M, Fung K P, Lee C Y. Biochem Mol Biol Int. 1996;39:747–754. doi: 10.1080/15216549600201831. [DOI] [PubMed] [Google Scholar]
- 27.Toh Y, Pencil S D, Nicolson G L. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 28.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 29.Simpson A, Uitto J, Rodeck U, Mahoney M G. Gene. 2001;273:29–39. doi: 10.1016/s0378-1119(01)00563-7. [DOI] [PubMed] [Google Scholar]
- 30.Mazumdar A, Wang R A, Mishra S K, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi R K, Kumar R. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 31.Cousins R J, Lanningham-Foster L. J Infect Dis. 2000;182,(Suppl. 1):S81–S84. doi: 10.1086/315917. [DOI] [PubMed] [Google Scholar]
- 32.Giallongo A, Yon J, Fried M. Mol Cell Biol. 1989;9:224–231. doi: 10.1128/mcb.9.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida T, Imai T, Takagi S, Nishimura M, Ishikawa I, Yaoi T, Yoshie O. FEBS Lett. 1996;395:82–88. doi: 10.1016/0014-5793(96)01004-6. [DOI] [PubMed] [Google Scholar]
- 34.De Pamphilis M L. DNA Replication in Eukaryotic Cells. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 45–86. [Google Scholar]
- 35.Chuang R Y, Kelly T J. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dijkwel P A, Wang S, Hamlin J L. Mol Cell Biol. 2002;22:3053–3065. doi: 10.1128/MCB.22.9.3053-3065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phi-van L, Sellke C, von Bodenhausen A, Stratling W H. J Biol Chem. 1998;273:18300–18307. doi: 10.1074/jbc.273.29.18300. [DOI] [PubMed] [Google Scholar]
- 38.Aladjem M I, Rodewald L W, Lin C M, Bowman S, Cimbora D M, Brody L L, Epner E M, Groudine M, Wahl G M. Mol Cell Biol. 2002;22:442–452. doi: 10.1128/MCB.22.2.442-452.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert D M. Curr Opin Cell Biol. 2002;14:377–383. doi: 10.1016/s0955-0674(02)00326-5. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert D M. Science. 2001;294:96–100. doi: 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosak S T, Skok J A, Medina K L, Riblet R, Le Beau M M, Fisher A G, Singh H. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 42.Skok J A, Brown K E, Azuara V, Caparros M L, Baxter J, Takacs K, Dillon N, Gray D, Perry R P, Merkenschlager M, Fisher A G. Nat Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 43.Maes J, O'Neill L P, Cavelier P, Turner B M, Rougeon F, Goodhardt M. J Immunol. 2001;167:866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- 44.Chowdhury D, Sen R. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff B E, Chess A, Cedar H, Bergman Y. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 46.Dalgaard J Z, Klar A J. Nature. 1999;400:181–184. doi: 10.1038/22139. [DOI] [PubMed] [Google Scholar]
- 47.Sale J E, Calandrini D M, Takata M, Takeda S, Neuberger M S. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 48.Papavasiliou F N, Schatz D G. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 49.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 50.Haber J E. Nat Immunol. 2001;2:902–903. doi: 10.1038/ni1001-902. [DOI] [PubMed] [Google Scholar]
- 51.Hatton K S, Schildkraut C L. Mol Cell Biol. 1990;10:4314–4323. doi: 10.1128/mcb.10.8.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stubbs L, Huxley C, Hogan B, Evans T, Fried M, Duboule D, Lehrach H. Genomics. 1990;6:645–650. doi: 10.1016/0888-7543(90)90499-k. [DOI] [PubMed] [Google Scholar]
- 53.Huxley C, Williams T, Fried M. Mol Cell Biol. 1988;8:3898–3905. doi: 10.1128/mcb.8.9.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.