Abstract
T cell receptor (TCR)-mediated activation of CD4+ T cells is known to require multivalent engagement of the TCR by, for example, oligomeric peptide–MHC complexes. In contrast, for CD8+ T cells, there is evidence for TCR-mediated activation by univalent engagement of the TCR. We have here compared oligomeric and monomeric Ld and Kb peptide–MHC complexes and free peptide as stimulators of CD8+ T cells expressing the 2C TCR. We found that the monomers are indeed effective in activating naïve and effector CD8+ T cells, but through an unexpected mechanism that involves transfer of peptide from soluble monomers to T cell endogenous MHC (Kb) molecules. The result is that T cells, acting as antigen-presenting cells, are able to activate other naïve T cells.
Keywords: CD8+ T lymphocytes‖T cell receptor‖major histocompatibility complex‖antigen presentation‖lymphocyte activation
Antigen-driven activation of cells of the immune system generally involves multivalent antigen engagement that leads to clustering or “crosslinking” of cell-surface receptors. This concept arose largely from observations of antigen-induced capping of surface immunoglobulins on B cells (1, 2) and multivalent antigen triggering of histamine release from mast cells (3). Whether multivalent engagement of T cell antigen receptors (TCR) is required to initiate T cell activation has been difficult to determine, primarily because the antigens are peptide–MHC complexes normally found as integral membrane proteins on the surfaces of other cells. However, experiments using soluble recombinant MHC molecules lacking transmembrane domains and carrying defined synthetic peptides have shown clearly that peptide–MHC oligomers, but not monomers, can activate CD4+ T cells (4–7). Thus, for CD4+ T cells, multivalent engagement of TCR is sufficient to initiate cellular activation, whereas monovalent engagement is apparently nonproductive.
For CD8+ T cells, however, the issue remains unclear. Several studies have found that multivalent engagement is required to activate these cells (4, 8–10), but some evidence suggests the sufficiency of monovalent TCR engagement to trigger a cytolytic response, such as the requirement for such a small number of peptide–MHC complexes per target cell in cytolytic assays, to imply that a single complex can trigger the cytolytic response in CD8 T cells (11). Activation of CD8+ T cells by soluble peptide–MHC monomers has been clearly observed in the presence of CD8 molecules (12, 13). It was suggested from that result that monovalent engagement of TCR can serve as the initiating event when it results in TCR association with CD8 (12). However, in another study, TCR-CD8 heterodimerization induced by peptide–MHC monomers was observed but was found not to activate the cells (9).
In this study, we investigated the requirements for initiating signaling processes in naïve and activated CD8+ T cells carrying the 2C TCR. The 2C TCR recognizes various peptide–MHC complexes, including (i) the potent allogenic activator QLSPFPFDL-Ld (QL9-Ld), which binds with the highest affinity so far measured for any TCR/peptide–MHC interaction [Kd ≈ 0.07–0.1 μM (14)]; and (ii) an agonist isolated from a random peptide library (15) SIYRYYGL-Kb (SIY-Kb), which binds almost as strongly to 2C cells if cell-surface CD8 is present (Kd ≈ 0.3 μM) but more weakly if cells do not express CD8 [Kd > 3 μM (16)]. Using soluble monomeric and oligomeric forms of these class I MHC–peptide complexes, we find that monomers can induce a variety of characteristic T cell activation processes in naïve and effector CD8+ T cells, but only if the T cells express a class I MHC molecule able to form peptide–MHC complexes with peptide released from the offered peptide–MHC monomers. In addition to clarifying how MHC monomers activate CD8+ T cells, these results show that CD8+ T cells can act as antigen-presenting cells (APCs) able to stimulate naïve T cells to undergo a variety of activation and development processes.
Materials and Methods
Soluble Peptide–Kb Complexes.
Soluble murine class I MHC proteins H-2 Kb and Ld were produced by expression in Escherichia coli and folded in vitro in the presence of peptide, as described for human class I MHC proteins (17), except that the heavy chains carried a C-terminal biotinylation signal peptide (bsp; refs. 18 and 19). E. coli expression vectors coding for Kb-bsp, Ld-bsp, and human β2M were a gift from J. Lippolis and J. Altman (National Institutes of Health Tetramer Facility). Peptides were synthesized by using 9-fluorenylmethoxycarbonyl chemistry and verified by using mass spectrometry: OVA (SIINFEKL), SIY (SIYRYYGL), and QL9 (QLSPFPFDL). For preparation of oligomers, folded peptide–MHC complexes were biotinylated at the bsp sequence by using birA enzyme (Avidity, Denver). Biotinylated complexes were isolated by gel filtration and oligomerized by stepwise addition of streptavidin to one-fourth of the molar ratio of the MHC (20). Oligomers were characterized by gel filtration and nondenaturing SDS/PAGE. Protein concentration was estimated by UV absorbance (ɛ280 nm = 90,000 M−1⋅cm−1 for Kb, 108,000 M−1⋅cm−1 for Ld). Immediately before use, peptide–MHC complexes were repurified by gel filtration chromatography (Superdex 200, Pharmacia) to remove aggregates and any free peptide released during storage.
Fluorescent Labeling of Class I MHC Complexes.
In some experiments, the association of MHC complexes with cells was monitored by flow cytometry, by using fluorescein labels incorporated into the Kb and Ld heavy chain sequences at the free cysteine present at position 122. Cysteine 122 is not very reactive and generally was found in the reduced state after conventional isolation, so that treatment with reducing agent was not required to generate a free thiol before labeling. For labeling, purified MHC–peptide complex (100 μM) in PBS, pH 7.4, was treated with a 20-fold molar excess of fluorescein maleimide (Molecular Probes, 20 mM in DMSO). The mixture was incubated for 1 h at room temperature, and peptide–MHC complexes were reisolated by gel filtration to remove unreacted fluorescein. Labeling efficiency, determined by UV absorbance, was ≈99%.
T Cell Clones.
Murine CD8+ T cell clones 4G3 (specific for OVA-Kb) (21) and L3.100 (expressing the 2C TCR, specific for SIY-Kb and QL9-Ld) (22) were cultured at 37°C, 5% CO2, in RPMI medium with 10% FCS; cells were used 6–7 days after weekly stimulation by using irradiated APCs (EG7OVA and P815, respectively).
Naïve T Cell Purification.
Rag−/− 2C TCR transgenic mice (H-2b) (23) were used as a source of naïve T cells. Lymph nodes were isolated, and naïve T cells were purified from APCs by depletion of CD44+ cells by using magnetic sorting. At least 99% of the resultant cell population expressed the 2C TCR, with 60–70% of the 2C TCR-positive population CD8+ and 30–40% CD8−. In some experiments, Rag−/− 2C TCR transgenic mice, crossed onto a constitutive GFP transgenic background, were used as the source of naïve T cells (24, 25). Naïve H-2Kb−/−Db−/− 2C T cells were obtained by adoptive transfer of 107 bone marrow cells from mice carrying the 2C transgene on an H-2Kb−/−Db−/− background (26) into sublethally irradiated RAG−/−, H-2b+ C57BL/6 recipients. This procedure allows the H-2Kb−/−Db−/− T cells to undergo positive selection in an H-2b+environment. Eight weeks after transfer, T cells isolated from the recipients were mostly 2C TCR+ H-2Kb−/−Db−/− and were further purified to ≈98% by depletion of (host) CD44+ cells using magnetic sorting.
T Cell Activation Assays.
Activation assays were carried out in 96-well plates with 50,000–100,000 cells per well in 50 μl. Plates were preblocked by overnight incubation with 1% BSA in PBS solution at 4°C. Cells were incubated with dilutions of peptide–MHC complexes or free peptide in culture medium for 2, 3, or 6 h at 37°C, then washed in cold FACS buffer (PBS with 1% BSA and 0.1% sodium azide) and stained with fluorescent antibodies (PharMingen) to cell surface markers such as TCR (α-pan-TCRβ), CD69, CD25, and CD8, for analysis using a Becton Dickinson FACS Calibur flow cytometer. Data from flow cytometry experiments are shown in units of mean fluorescence intensity (MFI). For experiments with soluble or plate-bound QL9-Ld complexes, an excess of purified QL9 peptide was included to prevent degradation or peptide loss from the complex. QL9 peptide alone does not activate 2C (H-2b) cells up to concentrations of at least 10 μM. For antigen representation assays, naïve 2C T cells (GFP−) were incubated with peptide–MHC complexes or free peptide for 3 h at 37°C, washed four times, and resuspended in media. Naïve GFP+ 2C T cells were added, and the GFP+ and GFP− cells were incubated together for an additional 3 h at 37°C before staining, as described above. Responses by GFP+ and GFP− cells were separated by gating on GFP fluorescence. For proliferation assays, naïve 2C T cells were incubated in the presence of soluble or plate-bound stimuli in 96-well plates for 3 days at 37°C, after which [3H]thymidine (1 μCi/well) was added, and the cells were incubated at 37°C for an additional 12 h before harvest and assay for 3H incorporation. For IFN-γ assay, naïve 2C T cells were treated with immobilized anti-CD3, soluble peptide–MHC complex, or peptide alone for 3 days at 37°C, then restimulated with SIY peptide in the presence of brefeldin-A. After 6 h, the cells were stained for CD8 and TCR, fixed with paraformaldehyde, and stained with anti-IFN-γ antibodies in the presence of 0.1% saponin, and then analyzed by flow cytometry as described above.
Results
Activation of CD8+ T Cell Clones by Class I MHC Monomers.
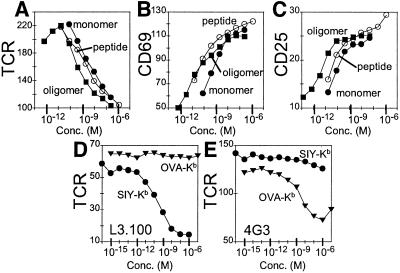
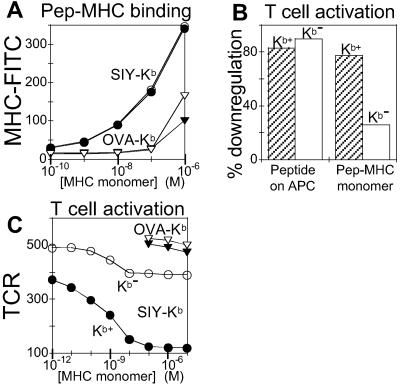
Soluble peptide–MHC complexes carrying a biotinylation signal sequence at the heavy chain C terminus were produced by a standard in vitro folding method (17). Streptavidin-mediated oligomerization (19) yielded primarily tetramers (≈70%), with small amounts of trimers (≈20%), dimers (≈3%), and monomers (7%) (as detected by high-resolution gel filtration; see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). We prepared monomers and oligomers of the 2C agonist SIY-Kb and the nonactivating control complex OVA-Kb and tested their ability to stimulate the CD8+ T cell clone L3.100, which carries the 2C TCR (22). Several conventional activation markers were observed after incubation of L3.100 cells with SIY-Kb complexes, including down-regulation of TCR (Fig. 1A) (27), up-regulation of the early activation marker CD69 (Fig. 1B) (28), and up-regulation of the high-affinity IL-2 receptor α subunit CD25 (Fig. 1C) (29). SIY-Kb oligomers (Fig. 1 A–C, squares) were more potent than monomers (filled circles) and consistently induced equivalent activation responses at lower concentrations than the corresponding monomer. Activation depended on the specific peptide–MHC combination, because OVA-Kb complexes, which carry a different peptide, did not activate L3.100 cells (Fig. 1D), although they did activate 4G3 T cells, which are specific for OVA-Kb (Fig. 1E) (21).
Figure 1.
Activation of CD8+ T cell clones by class I MHC monomers and oligomers. (A–C) Response of a T cell clone expressing the 2C TCR (L3.100) to MHC monomers, oligomers, and free peptide. (A) TCR down-regulation in response to 3-h incubation with SIY-Kb oligomer (filled squares), SIY-Kb monomer (filled circles), and SIY peptide alone (open circles). Mean fluorescence intensity (MFI) shown on y axis. (B) CD69 up-regulation (MFI) and (C) CD25 up-regulation (MFI) in response to the same treatment as A. (D and E) Specificity of the response. (D) TCR down-regulation (MFI) is observed for L3.100 incubated for 3 h with SIY-Kb monomers (circles) but not for nonspecific OVA-Kb (inverted triangles). (E) Response of a T cell clone specific for OVA-Kb complex (4G3) shows TCR down-regulation (MFI) after 3 h of incubation with OVA-Kb monomers (inverted triangles), but not with SIY-Kb (circles).
We previously have used these same activation markers in studies of the response of CD4+ T cells, where we observed activation by class II MHC oligomers but not by monomers (7). In contrast, the CD8+ T cells clearly were activated in the presence of class I MHC monomers. Given this result, we performed several control experiments to verify that no multivalent protein was present in the experiments. Because immobilized monomeric SIY-Kb is a potent T cell stimulus, we verified that the activation was not due to adventitious immobilization of the soluble molecules on the BSA-blocked assay wells. Removal of the medium containing SIY-Kb before addition of T cells completely abrogated activation (not shown). Activation by the monomeric preparations was not likely to be the result of contaminating MHC oligomers or aggregates, as none were observable by gel filtration (detection limit ≈0.5%) (Fig. 7) or by native gel electrophoresis (not shown), and a miniscule amount of oligomer contamination would not be expected to account for the observed activity of the monomer preparations, because oligomers were only ≈6-fold more potent activators than the monomers (1.5-fold per peptide–MHC complex; Fig. 1 A–C, filled symbols).
Because SIY peptide alone was able to induce activation processes in the T cells (Fig. 1 A–C, open circles), we wanted to evaluate any contribution of released peptide to the observed monomer activation. Peptide–MHC monomers and oligomers routinely were separated from free peptide by gel filtration immediately before the activation assay. We incubated the peptide–MHC preparations under assay conditions and separated free peptide by centrifugal ultrafiltration. Bioassay of the filtrates using peptide-induced cytotoxic T lymphocyte-mediated lysis of target cells indicated that very little peptide was released during the 3-h assay period [the approximate half-time for peptide release was 27 h, consistent with earlier studies (30)]. Moreover, in many experiments, the response to peptide–MHC monomers was significantly greater than that induced by an equivalent amount of peptide alone (see below). These details suggest that the observed activation induced by MHC monomers could not be accounted for by release of peptide into the solution.
Activation of Naïve T Cells by Peptide–MHC Monomers.
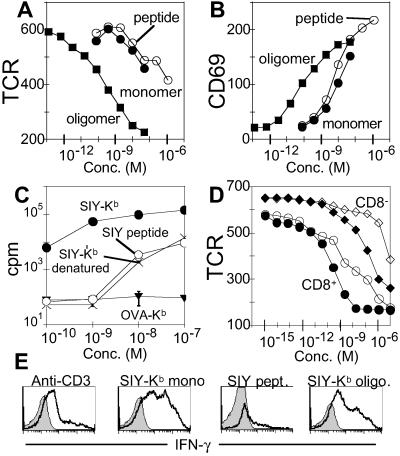
To investigate whether naïve T cells could be activated by incubation with monomeric peptide–MHC complexes, we used T cells freshly isolated from 2C TCR+/+ RAG−/− (H-2b) transgenic mice. We observed that the purified naïve 2C T cells exhibited TCR down-regulation (Fig. 2A), CD69 up-regulation (Fig. 2B), and CD25 up-regulation (not shown), in response to peptide-Kb monomers and oligomers. Activation levels and half-maximal activation concentrations were similar to those observed with the T cell clones, with tetramers activating substantially more potently than monomers. In addition to these activation markers, we also observed that naïve 2C cells that proliferated in response to soluble SIY-Kb (Fig. 2C). In this assay, the SIY-Kb monomers were substantially more active than free peptide (compare open and filled circles), further indicating that the monomer activation was not due to simple peptide release into the medium. The T cell proliferation due to denatured (boiled) SIY-Kb was similar to that induced by the corresponding concentration of purified peptide alone, suggesting that endotoxins or other bacterial products potentially present in the SIY-Kb preparations were not contributing significantly to the observed activation (Fig. 2C).
Figure 2.
Activation of naïve CD8+ T cells by class I MHC monomers and oligomers. (A and B) Response of naïve T cells expressing the 2C TCR to MHC monomers, oligomers, and free peptide. (A) TCR down-regulation in response to 2-h treatment with SIY-Kb oligomer (filled squares), SIY-Kb monomer (filled circles), and SIY peptide alone (open circles). Mean fluorescence intensity (MFI) shown on y axis. (B) CD69 up-regulation (MFI) in response to the same treatment. (C) Proliferation of naïve 2C T cells measured by 3H-thymidine incorporation 3 days after treatment in response to SIY-Kb monomer (filled circles), SIY peptide alone (open circles), denatured (boiled) SIY-Kb monomer (X), and nonspecific OVA-Kb monomer (inverted triangles). (D) TCR down-regulation (MFI) of CD8+ (circles) or CD8− (diamonds) populations present in purified naïve 2C T cells in response to SIY-Kb monomers (filled symbols) or SIY peptide alone (open symbols). (E) Maturation of naïve 2C T-cells in response to MHC monomers, oligomers, and free peptide. Naïve 2C T cells were stimulated with immobilized anti-CD3 antibody (10 μg/ml), SIY-Kb oligomers, SIY-Kb monomers, or free SIY peptide for 3 days. Intracellular IFN-γ production was measured after 6-h restimulation by SIY peptide (bold line). The shaded area corresponds to isotype control staining.
To evaluate the importance of CD8 in the 2C-Kb system, we analyzed the substantial CD8− subpopulation present in naïve T cells isolated from 2C TCR transgenic mice and compared it to the corresponding CD8+ population. Both CD8+ and CD8− subpopulations exhibited activation responses after treatment with specific SIY-Kb but not control OVA-Kb monomers, although the CD8− population required ≈103-fold greater concentration to induce comparable activation levels (Fig. 2D and Fig. 8, which is published as supporting information on the PNAS web site).
Finally, we evaluated the ability of Kb monomers and oligomers to drive conversion of the naïve cells to effector cells. We relied on the ability of effector cells, but not naïve cells, to produce IFN-γ after stimulation (39). After treatment with SIY-Kb monomers or oligomers, the 2C cells responded to antigenic stimulation with production of IFN-γ (Fig. 2E). Although less effective than SIY-Kb, free SIY peptide alone also was able to elicit a small degree of conversion to effector cells (Fig. 2E and data not shown), a response that conventionally is believed to require the participation of costimulatory molecules.
Activation Mechanism: Direct Engagement or Uptake/Representation?
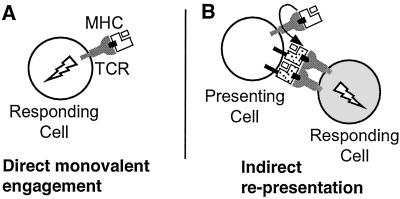
The results presented above indicate that addition of peptide–MHC monomers to CD8+ T cells is sufficient to induce T cell activation. Such activation could occur by direct monovalent engagement of TCR by soluble MHC (Fig. 3A), which conventionally is the interpretation given to such results (12). However, another process could also lead to the observed activation. Peptide–MHC complexes that bind to TCR on one cell could release peptides that are then transferred onto that T cell's endogenous MHC proteins for presentation to other T cells (Fig. 3B). Although T cells generally are not thought to be antigen-presenting cells, we observed potent activation of such cells after treatment with peptide alone (Figs. 1 and 2), and T cell fratricide has been described previously (31). Participation of T cell costimulatory and adhesion molecules in the activation process could help to explain the observed high sensitivity of the L3.100 and naïve 2C T cell response to soluble MHC monomers (ED50 ≈ 1 nM), which contrasts with the relatively weak binding affinity measured for the corresponding MHC–TCR interaction [Kd ≈ 300 nM (16)]. To distinguish between these two mechanisms, we performed additional activation studies, as described below.
Figure 3.
Two potential mechanisms for activation of T cells by MHC monomers. (A) Binding of monomeric peptide–MHC complexes to cell-surface T cell receptors directly leads to activation (direct engagement model). (B) Transfer of peptide from the soluble MHC molecules to endogenous MHC molecules expressed by the T cell indirectly leads to activation of another T cell through interaction with peptide–MHC complexes on the surface of the first cell (representation model). Binding of soluble peptide–MHC to cell-surface TCR (or CD8) could facilitate peptide transfer, although other mechanisms are possible.
Activation by Soluble Monomers Requires the Presence of Endogenous MHC Proteins.
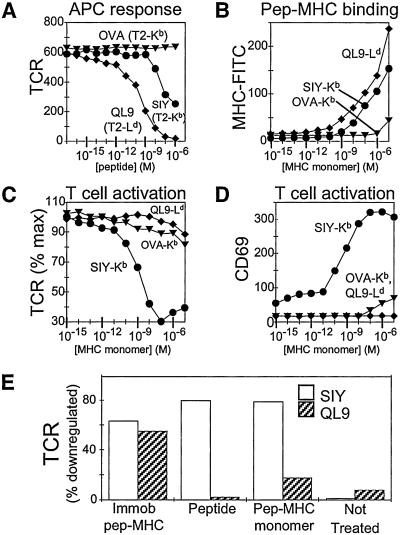
To evaluate the ability of a soluble monomer to activate 2C cells without contribution from the peptide representation pathway, we made use of the allogeneic agonist (QL9-Ld) that has been described for the 2C TCR. QL9 peptide pulsed onto cell-surface Ld of transfected APCs (T2-Ld) was a potent stimulator for 2C cells (Fig. 4A), confirming previous work using other activation markers; and soluble QL9-Ld monomer bound tightly to 2C (Fig. 4B), consistent with previous measurements (14, 16). The 2C T cells express Kb but not Ld molecules on their surface. By comparing the response to soluble SIY-Kb, which can access the representation pathway, with that of QL9-Ld, which cannot, we were able to evaluate the role of representation by endogenous T cell MHC molecules to the observed activation processes. In contrast to SIY-Kb monomers, QL9-Ld monomers were not able to stimulate 2C TCR down-regulation and CD69 up-regulation (Fig. 4 C and D). To demonstrate that the recombinant QL9-Ld complexes were functionally active, we used plate-bound QL9-Ld to stimulate 2C T cells: both TCR down-regulation (Fig. 4E) and CD69 up-regulation (not shown) were observed. Taken together, these results show that the allogeneic QL9-Ld monomer bound to the 2C TCR but did not induce activation processes, whereas the syngeneic SIY-Kb monomer induced substantial activation despite being an intrinsically weaker-binding ligand. Thus, no detectable activation was induced by direct binding of peptide–MHC monomers to TCR, and activation of 2C T cells induced in the presence of peptide–Kb monomers requires the participation of endogenous (Kb) MHC molecules.
Figure 4.
CD8+ T cell activation by MHC monomers requires endogenous MHC proteins. 2C T cells expressing Kb, but not Ld, are activated by soluble monomers of the syngeneic SIY-Kb but by not the potent alloantigen QL9-Ld. (A) Response of naïve 2C T cells to 3 h of incubation with peptide-pulsed APCs. T2 cells expressing Kb were pulsed with OVA (inverted triangles) and SIY (circles) peptides, and T2 cells expressing Ld were incubated with QL9 peptide (diamonds). TCR mean fluorescence intensity (MFI) shown on y axis. (B) Binding of fluorescein-labeled SIY-Kb (circles), OVA-Kb (inverted triangles), or QL9-Ld (diamonds) to naïve 2C T cells. Cell-associated fluorescence was measured after incubation with MHC monomers for 30 min at 4°C, washing, and fixation with paraformaldehyde. (C and D) Response of naïve 2C T cells to 3 h incubation with SIY-Kb (circles), OVA-Kb (inverted triangles), or QL9-Ld (diamonds) monomers. (C) TCR down-regulation (percentage of maximum TCR expression), and (D) CD69 up-regulation (MFI). (E) TCR down-regulation of naïve 2C T cells in response to 6-h incubation with plate-bound and soluble monomers or free peptide. Open bars represent SIY or SIY-Kb; shaded bars represent QL9 or QL9-Ld.
To further evaluate the role of endogenous MHC in activation by soluble peptide–MHC monomers, we used 2C T cells that lacked the classical class I MHC molecules Kb and Db (26) (see Materials and Methods). We compared the activation of 2C TCR+ Kb−/−Db−/− cells and normal (Kb+) 2C T cells after incubation with soluble peptide–MHC monomers. Binding curves for fluorescent SIY-Kb monomers were nearly identical for normal naïve and Kb−/−Db−/− 2C cells (Fig. 5A, open and filled circles), as were TCR expression levels. Activation by SIY-pulsed T2-Kb APCs was nearly indistinguishable between the two cell types, as measured by TCR down-regulation (Fig. 5B). However, activation by soluble SIY-Kb monomer was severely reduced for the Kb−/−Db−/− 2C cells (Fig. 5B). The response was not completely eliminated, and the low level of monomer-induced TCR down-regulation observed for the Kb−/−Db−/− cells (Fig. 5C) probably resulted from the small fraction of contaminating Kb+ cells in the T cell preparations. Nonetheless, these results show clearly that T cell activation by soluble SIY-Kb relies largely or completely on the participation of endogenous Kb.
Figure 5.
Lack of activation of H2-Kb−/−Db−/− T cells by soluble MHC monomers. T cells expressing the 2C TCR, but not H2-Kb/Db, are unable to be efficiently activated by soluble MHC monomers. (A) Binding of fluorescein-labeled MHC monomers to naïve 2C T cells (filled symbols) and 2C+, Kb−/−Db−/− T cells (open symbols). Binding is shown for SIY-Kb (circles) and OVA-Kb (inverted triangles). MFI shown on y axis. (B) Bar graph showing the percent of TCR down-regulation for treatment of naïve 2C T cells (shaded bars) and 2C+, Kb−/−Db−/− T cells (open bars) with peptide-pulsed T2-Kb cells and soluble SIY-Kb monomer. (C) Concentration dependence of TCR down-regulation (MFI) response in Kb+/+Db+/+ (filled symbols) and Kb−/−Db−/− (open symbols) naïve 2C T cells after 3-h incubation with soluble SIY-Kb (circles) and OVA-Kb (inverted triangles).
Triggering by Soluble MHC Monomers Does Not Require Direct Exposure.
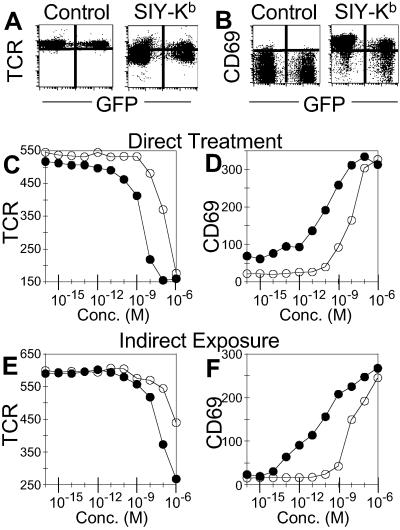
In the proposed representation mechanism (Fig. 3B), T cells that have not been directly exposed to antigen are activated by contact with other T cells that present peptide antigens on their surface. To determine whether our results stemmed from this mechanism, we used naïve 2C T cells expressing GFP (24, 25) to track antigen exposure in a mixed T cell population. The presence of GFP does not interfere with T cell activation, and GFP+ 2C T cells exhibited Kb monomer and peptide activation profiles that were essentially indistinguishable from their GFP− counterparts (see Fig. 9, which is published as supporting information on the PNAS web site). In the experiments shown in Fig. 6, GFP− 2C cells were incubated with specific Kb monomers for 3 h at 37°C, then washed and cultured with untreated GFP+ 2C cells for an additional 3 h before analysis of activation markers by flow cytometry. Untreated GFP+ cells indirectly exposed to antigen, as well as GFP− cells that had been directly exposed to antigen, exhibited characteristic TCR downmodulation (Fig. 6A) and CD69 up-regulation (Fig. 6B) responses. Thus, activation of naïve 2C T cells did not require direct exposure to monomeric SIY-Kb complexes. To evaluate the efficiency of the uptake/representation mechanism, we compared the titration profiles for GFP+ T cells that were directly exposed to monomer or peptide (Fig. 6 C and D), with the profiles for GFP+ T cells that had been cocultured with GFP− monomer- or peptide-pulsed counterparts for 3 h (Fig. 6 E and F). The profiles were similar, with only ≈10-fold decreased sensitivity for the indirectly exposed cells. The difference is likely due to more effective activation in the continuous presence of stimulators for the directly exposed GFP+ cells (Fig. 6 C and D), as compared with the indirectly exposed GFP+ cells (Fig. 6 E and F). These results indicate that uptake/representation can provide an efficient antigenic stimulus able to activate untreated T cells, and that such a process is sufficient to account for all or nearly all of the T cell activation induced by soluble class I peptide–MHC monomers (and perhaps also for soluble oligomers, to some extent).
Figure 6.
T cell activation by uptake and representation of antigen. Naïve GFP+ 2C T cells were activated by exposure to GFP− 2C T cells that previously had been treated with MHC monomers. (A and B) GFP− cells were treated with control media or SIY-Kb for 3 h, washed, and then GFP+ cells were added to the GFP− cells in fresh media. The cells were incubated together for an additional 3 h at 37°C. (A) TCR down-regulation and (B) CD69 up-regulation. The horizontal scale is GFP fluorescence level which distinguishes the directly treated (GFP−) and indirectly exposed (GFP+) T cells. (C and D) Response of directly treated GFP+ 2C T cells. (C) TCR down-regulation by GFP+ 2C T cells in response to 3-h incubation with SIY-Kb (filled circles) or SIY peptide (open circles). MFI shown on y axis. (D) CD69 up-regulation (MFI) by GFP+ 2C T cells in response to the same treatment. (E and F) Response of indirectly exposed GFP+ 2C T cells. (E) TCR down-regulation (MFI) by GFP+ 2C T cells incubated for 3 h with GFP− 2C T cells that had been pretreated with SIY-Kb (filled circles) or SIY peptide (open circles). (F) CD69 up-regulation (MFI) of GFP+ 2C T cells in response to the same stimuli.
Discussion
The results presented here demonstrate that CD8+ cytolytic T cells and freshly isolated naïve CD8+ T cells can be activated effectively by cognate soluble peptide–MHC monomers. Rather than arising directly from monomeric engagement of the TCR, however, the activation apparently stems from a mechanism that involves transfer of peptide from monomer to T-cell MHC molecules. Support for this mechanism rests on two independent lines of evidence. First, T cells were stimulated effectively only when they expressed the same MHC as the activating monomer: 2C cells, which express Kb but not Ld, were activated by the SIY-Kb monomer but not by the more strongly binding QL9-Ld monomer; and 2C T cells that lacked Kb were not activated efficiently by the SIY-Kb monomer. Thus, T cell expression of the appropriate class I MHC molecule is required for activation by soluble peptide–MHC monomers. Second, 2C cells that were not themselves exposed to the SIY-Kb monomer were activated in trans by 2C cells that had been previously exposed to this monomer. Thus, T cell class I MHC molecules can present peptides derived from soluble peptide–MHC monomers. Both of these independent results are consistent with a peptide representation mechanism in which cognate peptides derived from soluble MHC monomers are presented on the T cells' own MHC molecules.
Although we found no evidence for an activation response triggered by monovalent engagement of the TCR, we cannot rule out the possibility that some intracellular signals result from monomeric engagement. Any such response, however, would have to be very small relative to that induced by representation of peptide and would not contribute significantly to the activation processes observed here. Because monomeric TCR engagement is ineffective in activating CD8+ T cells, it seems reasonable to conclude that CD8+ T cell triggering is due to TCR clustering, as seen for CD4+ T cells. However, we do not know how many represented peptides are needed per cell for effective representation, and there remain important differences between the activation of CD8+ and CD4+ T cells, particularly with regard to the minimal peptide–MHC density required on conventional presenting cells to induce T cell activation processes (11, 32–35).
Precisely how peptide is transferred from soluble monomers to T cell MHC molecules is unclear. The possibility that transfer results from quantitative dissociation of complexes in culture medium to yield free peptide that binds to T cell MHC is incompatible with estimates of the SIY-Kb complex's lifetime under conditions of the assay, and the observation that free peptide usually induced less response than an equivalent concentration of peptide–MHC monomer. MHC-bound peptide is protected from proteolytic degradation in culture medium, and, by being slowly released, could be more effective than free peptide in loading onto T cell MHC molecules. The efficiency of peptide loading also might be enhanced by a high local concentration of monomers at the cell surface after binding to TCR and/or CD8 molecules. Still another possibility is that TCR-bound monomers are internalized into T cell endocytic compartments where dissociating peptide is reloaded onto endogenous class I MHC molecules for recycling to the cell surface. Some evidence suggests that live cells are required for efficient representation of monomer-derived peptides (see Fig. 10, which is published as supporting information on the PNAS web site). Further studies are needed to evaluate these possibilities.
T cell adhesion to APCs is known sometimes to result in T cell acquisition of antigen from the presenting cell. This phenomenon has been observed with CD4+ and CD8+ T cells and even with B cells (36). The system described here is distinctive, however, in that APCs are absent: it consists only of purified T cells and soluble peptide–MHC monomers. It resembles most closely the situation in which addition of cognate peptide at relatively high concentration to cultured cytotoxic T lymphocyte clones can, in the absence of any other cells, result in specific T cell–T cell interactions and extensive cell lysis (“fratricide”) (21, 31). Here, however, the resulting specific T cell–T cell interaction leads not to fratricide of activated cells but to activation of naïve cells.
Much evidence indicates that, in general, activation of naive T cells requires that they recognize peptide–MHC complexes on professional APCs, usually dendritic cells (37). Our finding that naïve T cells can act as APCs able to induce maturation of naïve T cells is unexpected. In connection with a possible physiologic role of soluble peptide–MHC monomers in this unusual T cell behavior, it should be noted that soluble MHC proteins are found in human serum and synovial fluid, most likely as soluble peptide–MHC complexes (38).
In conclusion, CD8+ T cells are activated by soluble MHC monomers via a mechanism that involves T cell presentation of peptide derived from MHC–peptide complexes. The ability of CD8+ T cells to acquire and present antigenic peptide derived from soluble molecules and/or presenting cells could be important in maturation of naïve T cells in vivo.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Don C. Wiley. We thank John Altman and John Lippolis (National Institutes of Health Tetramer Facility) for Kb-bsp and Ld-bsp constructions, Carol McKinley (Massachusetts Institute of Technology) for T cell maintenance, Madelon Maurice and Hidde Ploegh (Harvard Medical School) for Kb−/−Db−/− mice, and Glenn Paradis (Massachusetts Institute of Technology) for help with flow cytometry. We also thank Hidde Ploegh, Eckart Schott, and Nicolas Bertho for helpful discussions. This work was supported by National Institutes of Health grants [NIH-AI96361 (L.J.S.), -AI44477 and CA60686 (H.N.E.), -AI44478 and -AI50631 (J.C.)], Cancer Center Core Grant CA-14051 (R. Hynes), Biotechnology Training Grant -GM08334 (J.D.S.), Anna Fuller Postdoctoral Fellowship (Q.G.), and National Institutes of Health support for the Massachusetts Institute of Technology Flow Cytometry Core Facility.
Abbreviations
- TCR
T cell receptor
- MFI
mean fluorescence intensity
- bsp
biotinylation signal peptide
- APC
antigen-presenting cell
References
- 1.Roelants G, Forni L, Pernis B. J Exp Med. 1973;137:1060–1077. doi: 10.1084/jem.137.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dintzis H M, Dintzis R Z, Vogelstein B. Proc Natl Acad Sci USA. 1976;73:3671–3675. doi: 10.1073/pnas.73.10.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger H. J Immunol. 1992;149:1477–1487. [PubMed] [Google Scholar]
- 4.Abastado J P, Lone Y C, Casrouge A, Boulot G, Kourilsky P. J Exp Med. 1995;182:439–447. doi: 10.1084/jem.182.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boniface J J, Rabinowitz J D, Wulfing C, Hampl J, Reich Z, Altman J D, Kantor R M, Beeson C, McConnell H M, Davis M M. Immunity. 1998;9:459–466. doi: 10.1016/s1074-7613(00)80629-9. [DOI] [PubMed] [Google Scholar]
- 6.Casares S, Zong C S, Radu D L, Miller A, Bona C A, Brumeanu T D. J Exp Med. 1999;190:543–554. doi: 10.1084/jem.190.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochran J R, Cameron T O, Stern L J. Immunity. 2000;12:241–250. doi: 10.1016/s1074-7613(00)80177-6. [DOI] [PubMed] [Google Scholar]
- 8.Maile R, Wang B, Schooler W, Meyer A, Collins E J, Frelinger J A. J Immunol. 2001;167:3708–3714. doi: 10.4049/jimmunol.167.7.3708. [DOI] [PubMed] [Google Scholar]
- 9.Doucey M A, Legler D F, Boucheron N, Cerottini J C, Bron C, Luescher I F. Eur J Immunol. 2001;31:1561–1570. doi: 10.1002/1521-4141(200105)31:5<1561::AID-IMMU1561>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Daniels M A, Jameson S C. J Exp Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sykulev Y, Joo M, Vturina I, Tsomides T J, Eisen H N. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 12.Delon J, Gregoire C, Malissen B, Darche S, Lemaitre F, Kourilsky P, Abastado J P, Trautmann A. Immunity. 1998;9:467–473. doi: 10.1016/s1074-7613(00)80630-5. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein J, Mostowsky H, Tung J, Hon H, Brunswick M, Kozlowski S. Eur J Immunol. 1997;27:871–878. doi: 10.1002/eji.1830270411. [DOI] [PubMed] [Google Scholar]
- 14.Sykulev Y, Brunmark A, Tsomides T J, Kageyama S, Jackson M, Peterson P A, Eisen H N. Proc Natl Acad Sci USA. 1994;91:11487–11491. doi: 10.1073/pnas.91.24.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udaka K, Wiesmuller K H, Kienle S, Jung G, Walden P. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- 16.Cho B K, Lian K C, Lee P, Brunmark A, McKinley C, Chen J, Kranz D M, Eisen H N. Proc Natl Acad Sci USA. 2001;98:1723–1727. doi: 10.1073/pnas.98.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garboczi D N, Hung D T, Wiley D C. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman-Smith A, Cronan J E., Jr Biomol Eng. 1999;16:119–125. doi: 10.1016/s1050-3862(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 19.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 20.Crawford F, Kozono H, White J, Marrack P, Kappler J. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 21.Walden P R, Eisen H N. Proc Natl Acad Sci USA. 1990;87:9015–9019. doi: 10.1073/pnas.87.22.9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sykulev Y, Vugmeyster Y, Brunmark A, Ploegh H L, Eisen H N. Immunity. 1998;9:475–483. doi: 10.1016/s1074-7613(00)80631-7. [DOI] [PubMed] [Google Scholar]
- 23.Manning T C, Rund L A, Gruber M M, Fallarino F, Gajewski T F, Kranz D M. J Immunol. 1997;159:4665–4675. [PubMed] [Google Scholar]
- 24.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 25.Cho B K, Wang C, Sugawa S, Eisen H N, Chen J. Proc Natl Acad Sci USA. 1999;96:2976–2981. doi: 10.1073/pnas.96.6.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurice M M, Gould D S, Carroll J, Vugmeyster Y, Ploegh H L. Proc Natl Acad Sci USA. 2001;98:7437–7442. doi: 10.1073/pnas.141143298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valitutti S, Muller S, Salio M, Lanzavecchia A. J Exp Med. 1997;185:1859–1864. doi: 10.1084/jem.185.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Testi R, D'Ambrosio D, De Maria R, Santoni A. Immunol Today. 1994;15:479–483. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 29.Waldmann T A. Annu Rev Biochem. 1989;58:875–911. doi: 10.1146/annurev.bi.58.070189.004303. [DOI] [PubMed] [Google Scholar]
- 30.Gakamsky D M, Boyd L F, Margulies D H, Davis D M, Strominger J L, Pecht I. Biochemistry. 1999;38:12165–12173. doi: 10.1021/bi9905821. [DOI] [PubMed] [Google Scholar]
- 31.Su M W, Walden P R, Golan D E, Eisen H N. J Immunol. 1993;151:658–667. [PubMed] [Google Scholar]
- 32.Brower R C, England R, Takeshita T, Kozlowski S, Margulies D H, Berzofsky J A, Delisi C. Mol Immunol. 1994;31:1285–1293. doi: 10.1016/0161-5890(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 33.Reay P A, Matsui K, Haase K, Wulfing C, Chien Y H, Davis M M. J Immunol. 2000;164:5626–5634. doi: 10.4049/jimmunol.164.11.5626. [DOI] [PubMed] [Google Scholar]
- 34.Sykulev Y, Cohen R J, Eisen H N. Proc Natl Acad Sci USA. 1995;92:11990–11992. doi: 10.1073/pnas.92.26.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 36.Hudrisier D, Bongrand P. FASEB J. 2002;16:477–486. doi: 10.1096/fj.01-0933rev. [DOI] [PubMed] [Google Scholar]
- 37.Mellman I, Steinman R M. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 38.Munoz-Fernandez S, Martin J, Martin-Mola E, Garcia-Rodriguez M C, Cantalejo M, Fontan G, Ferreira A. Rheumatology (Oxford) 2001;40:1365–1369. doi: 10.1093/rheumatology/40.12.1365. [DOI] [PubMed] [Google Scholar]
- 39.Ge Q, Hu H, Eisen H N, Chen J. Microbes Infect. 2002;4:555–558. doi: 10.1016/s1286-4579(02)01572-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.