Abstract
RsbR is a regulator of σB, the RNA polymerase σ factor subunit responsible for transcribing the general stress response genes when environmental stress is imposed on Bacillus subtilis. The C-terminal domain of RsbR and its paralogues is a substrate for the kinase function of another σB regulator, RsbT, but the amino acid sequence of the N-terminal domain of RsbR does not reveal any obvious biochemical function. RsbR, its paralogues, and other regulators of σB, including RsbS and RsbT, form large signaling complexes, called stressosomes. We have determined and present here the crystal structure of the N-terminal domain of RsbR. Unexpectedly, this structure belongs to the globin fold superfamily, but there is no bound cofactor. The globin domain from globin-coupled sensory systems replaces the N-terminal domain of RsbR in some bacteria, indicating a common genetic ancestry for RsbR and the globin family. We suggest that the globin fold has been “recycled” in RsbR and that one more activity can be included in the repertoire of globin functions, namely the ability to bind signaling macromolecules such as RsbT.
Keywords: signal transduction, Bacillus, crystallography, stress, globin
Complex control systems allow microbes to respond and adapt to their environment. Commonly, this adaptation is achieved in prokaryotes by variations in patterns of gene expression caused by the exchange of one σ subunit of RNA polymerase for another. Each σ factor confers a different specificity for promoter recognition on core RNA polymerase (1) and thus initiates transcription of a specific subset of genes. For instance, σB regulates the general response of Bacillus subtilis and its close phylogenetic relatives to stress (2–4) by activating the transcription of >125 genes (5, 6). The proteins encoded by these genes are responsible for the adaptation of the bacterium to a variety of stressful conditions, including variations in temperature, ionic strength, and pH, and the depletion of energy sources. The control of σB activity is extraordinarily complex and involves at least 12 proteins, only 4 of which are represented in Fig. 5, which is published as supporting information on the PNAS web site. But, in summary, the general stress response is modulated by reversible protein phosphorylation reactions and by the formation of alternative protein–protein complexes, called “partner-switching.” Partner-switching was first characterized for the sporulation sigma factor σF and its regulators SpoIIAB and SpoIIAA (7–9), and σB is regulated in a similar way by RsbW and RsbV. RsbW, a protein kinase, is the protein that switches between two mutually exclusive binding partners; one is the σ factor, and the other is the substrate of the kinase, RsbV. During normal conditions of growth, RsbV is phosphorylated by RsbW and consequently RsbW binds to σB, preventing σB from binding core RNA polymerase. During stress, phosphorylated RsbV is dephosphorylated by stress-activated phosphatases, and RsbW binds preferentially to RsbV, inducing the release of σB.
The partner switching that regulates the PP2C-type phosphatase RsbU differs from this paradigm. RsbU is activated, rather than inhibited, by the binding of the switch protein, RsbT, a kinase. The substrate for RsbT is not a single, small protein, but instead a large, ≈1-MDa structure that has a diameter of 27 nm (10), which we have named the stressosome. Stressosomes are comprised of several proteins, including but not necessarily exclusively, RsbR, RsbS, and RsbT (the RsbRST module), as well as YkoB and YojH, two paralogues of RsbR (O.D., C.-C. Chen, M. D. Yudkin & R.J.L., unpublished work; and refs. 10–13). The stoichiometry of the proteins present in stressosomes is unknown. During normal conditions, RsbT is believed to be sequestered by this large complex, but at the onset of stress it is released to activate RsbU by phosphorylating the sulfate transporter and anti-σ factor antagonist (STAS) domains of RsbR and RsbS. In B. subtilis, four paralogues of RsbR (YkoB, YqhA, YojH, and YtvA) have considerable sequence identity in their C-terminal domains to RsbR [and to the entire coding sequence of RsbS and other STAS domains (e.g., RsbV)], but have more divergent, albeit recognizably homologous, N termini. At least three of these paralogues can also form large complexes and can associate with RsbR and RsbS (O.D., C.-C. Chen, M. D. Yudkin & R.J.L., unpublished work; and refs. 10–13). The genes for the RsbR, RsbS, and RsbT proteins that form the core of the stressosome are part of an operon within the genome of B. subtilis. This RsbRST module is present in a large number of phylogenetically diverse bacterial taxa, but only in some of these bacteria is the RsbRST module associated with a σB activation pathway (14). Thus, in addition to its involvement in the stress response of B. subtilis, the RsbRST triplet appears to act as a signaling module of wider relevance. To better understand the assembly and function of the stressosome, we have obtained the crystal structure of the N-terminal domain of RsbR (N-RsbR). We now describe the structure and suggest a role for this domain in signaling.
Materials and Methods
Cloning and Site-Directed Mutagenesis. N-RsbR is defined as the initial 143 aa, based on sequence homology of the C-terminal domain of RsbR to single-domain anti-anti-σ factors such as RsbS. The coding sequences of N-rsbR, the bicistronic rsbR–rsbS complex, and rsbT were amplified by PCR from genomic DNA of B. subtilis SG38. N-rsbR and wild-type rsbR–rsbS were cloned into the NdeI and BamHI sites of pET11a (Novagen), whereas rsbT was cloned into pGEX-6P2 (General Electric Healthcare) by using the BamHI and NotI restriction sites to generate an N-terminal fusion with glutathione S-transferase (GST). The rsbR–rsbS bicistronic construct also served as the template for site-directed mutagenesis of rsbR by the QuikChange method (Stratagene), performed according to the manufacturer's instructions. Specific complementary oligonucleotide primers were used in the mutagenizing PCR to introduce single mutations in rsbR, which resulted in the conversion into alanine of the following amino acids: E60, K82, T86, E126, and N129. After amplification by PCR, the plasmid DNA template was degraded by DpnI, and the mutated PCR product was used to transform Escherichia coli DH5α. The presence of the mutations was confirmed by DNA sequencing.
Protein Expression and Purification. For crystallization purposes, selenomethionine-labeled N-RsbR was expressed in E. coli strain B834 (DE3) according to established procedures. Briefly, an E. coli B834 (DE3) strain transformed with the plasmid directing overexpression of N-RsbR was grown in 1 liter of selenomethionine medium until the culture achieved an optical density of 0.6 at 600 nm. Expression of N-RsbR was induced by the addition of isopropyl β-d-thiogalactopyranoside in the culture to a final concentration of 1 mM. Cells were harvested by centrifugation 3 h after induction before resuspending the cell pellet in 20 ml of cell lysis buffer containing 20 mM Tris·HCl (pH 8.0), 1 mM 4-(2-aminoethyl)bezenesulfonyl fluoride (AEBSF), and 1 mM EDTA, supplemented with 10 mM DTT, and lysed by sonication. Soluble proteins were separated from cell debris by centrifugation (16,000 × g for 60 min). The filtered supernatant was loaded on a 30-ml Q-Sepharose column (General Electric Healthcare) equilibrated with buffer A (20 mM Tris·HCl, pH 8.0/10 mM DTT). The bound proteins were eluted with a 100-ml linear gradient of buffer A plus 1 M NaCl. The fractions containing N-RsbR were identified by SDS/PAGE and concentrated for loading onto a Superdex 75 high-load gel filtration column. Again, fractions containing N-RsbR, which eluted from the column at a volume corresponding to a dimer (results not shown), were identified by SDS/PAGE and judged to be sufficiently pure for crystallization.
Wild-type and mutant RsbR–RsbS complexes and the GST–RsbT fusion were overexpressed in E. coli BL21 (DE3) after inducing with isopropyl β-d-thiogalactopyranoside as above before harvesting by centrifugation. Cells expressing RsbR–RsbS were disrupted by sonication in cell lysis buffer supplemented with 1 mM DTT and clarified by centrifugation. The RsbR–RsbS complex was purified first by Q-Sepharose anion-exchange chromatography and second by Superdex 200 gel-filtration chromatography. The RsbR–RsbS complex was eluted immediately after the void volume of the column and was subsequently dialyzed into a buffer of 10 mM Tris·HCl, pH 8.0/1.2 M (NH4)2SO4 overnight. The sample was loaded onto a phenyl-Sepharose column equilibrated in the same buffer and was eluted with a linear gradient decreasing in the concentration of (NH4)2SO4. Finally, those fractions that contained the RsbR–RsbS complex were further purified by gel filtration.
Cells directing the expression of the GST–RsbT fusion were disrupted by sonication into buffer B [50 mM Tris·HCl, pH 7.5/300 mM NaCl/1 mM DTT/1 mM 4-(2-aminoethyl)bezenesulfonyl fluoride/0.1 mM MgADP] and clarified by centrifugation before the supernatant was loaded onto a glutathione-Sepharose 4 Fast Flow column (General Electric Healthcare). The GST–RsbT fusion was eluted with buffer B supplemented with 50 mM reduced glutathione. To separate GST from RsbT, the GST–RsbT fusion was incubated with 3C protease at a mass ratio of 30:1 at 4°C overnight before purifying GST from RsbT by gel-filtration chromatography.
Crystallization of N-RsbR and Data Collection. Initial crystallization screening of N-RsbR was performed against a 96 solution sparse matrix screen comprising 67 conditions from a high-throughput structure determination project (15), with 29 further conditions derived from personal experience. From this screen, crystals suitable for x-ray diffraction were grown by hanging-drop vapor diffusion from the mixing of an equal volume of 10 mg/ml protein solution with 1.8–2.0 M sodium malonate, pH 8.0. Crystals of size ≈500 μm appeared in a few days, and single crystals were prepared for diffraction analysis by soaking in a cryoprotectant solution of 66% saturated sodium malonate for a few seconds before stream-freezing the crystal on an open-flow cryostat. Subsequent diffraction analysis revealed that these crystals belonged to space group P3212 with unit cell dimensions a = b = 136.06 Å, c = 113.30 Å, α = β = 90°, and γ = 120°.
Diffraction data were collected on Beamline BM14 at the European Synchrotron Radiation Facility (Grenoble, France). An energy scan over the selenium K-edge processed with chooch (16) revealed an f″ for selenium of 4.77 e–, confirming the presence of Se in the sample. A total of 360° of diffraction data were recorded at λ = 0.97889 Å, the peak selenium K absorption edge. Oscillation images were integrated and scaled to a maximum resolution of 2 Å with mosflm (17) and scala (18). Unless otherwise stated, programs from the Collaborative Computational Project 4 suite (19) were used for further data analysis.
Structure Determination and Refinement. Unmerged but scaled data were input into shelxd (20), and 12 selenium sites were readily found, corresponding to the 2 internal methionines in the 6 independent copies of N-RsbR in the asymmetric unit. Phase estimates were calculated in shelxe (21) within the hkl2map interface (22). The electron density map at this stage showed a clear boundary between protein and solvent and clearly recognizable secondary structural features. These shelxe phases were input into arp/warp (23) to build an atomic model for each of the six molecules in the asymmetric unit. The electron density maps were of sufficient quality for 764 of the expected 858 residues in the asymmetric unit to be automatically traced without recourse to real space averaging. Cycles of rebuilding in o (24) were alternated with maximum likelihood refinement in refmac (25) to produce the final structure. Further details of the refinement are given in the legend to Fig. 6, which is published as supporting information on the PNAS web site. Data collection and refinement statistics are summarized in Table 1, which is published as supporting information on the PNAS web site. The final Rwork and Rfree were 15.7% and 19.7%, respectively.
Results
Overall Structure. The structure of N-RsbR was solved to 2.0 Å by the single-wavelength anomalous dispersion technique from selenomethionine-labeled protein crystals. Characteristic electron density showing the hydrophobic core of the protein is shown in Fig. 6. Of the 143-aa N-RsbR construct, residues 3–136 were visible in the electron density. The N and C termini, residues 1–2 and 137–143, are disordered in all six molecules, and the loop between residues 102 and 107 is ordered in the electron density maps for only one of the six subunits (chain C). Overall, the structure of N-RsbR is a bundle of six α-helices: residues 4–14, 14–31, 38–56, 63–77, 80–102, and 108–136. The N-terminal two helices are in effect contiguous but are arranged with the helical axis of one almost perpendicular to the other. The first four helices are packed in an antiparallel fashion, ≈45° to the fifth and sixth helices, which also pack against each other in an antiparallel sense.
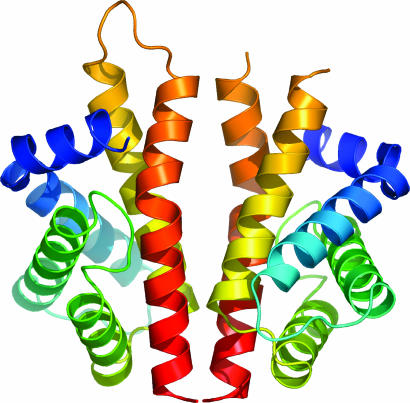
The six monomers in the asymmetric unit correspond to three essentially identical dimers. Comparisons between the six subunits by error-scaled difference distance matrices in escet (26) indicated that there were no significant differences between them, except a flexible loop region between residues 33 and 38. This loop is involved in the same crystal contacts in all six subunits but overall has crystallographic B factors 40–50% greater than the average for residues flanking this loop. Therefore, although the conclusions herein are drawn from the analysis of one dimer, they are directly applicable to the other two dimers. The biologically relevant N-RsbR dimer interface is formed by the packing of the fifth and sixth helices from each of the two protomers (Fig. 1). The buried surface area at the dimer interface is 1,040 Å2 and is composed of ≈75% nonpolar atoms, values that are consistent with those reported for the interfaces in other homodimeric proteins (27, 28).
Fig. 1.
N-RsbR dimer. A protein cartoon of the structure of the dimer of N-RsbR, with each protomer in the dimer colored in a continuous color rainbow, from blue at the N terminus to red at the C terminus. The loop between residues 102 and 107 can be traced in only one of the six independent molecules in the crystallographic asymmetric unit, drawn here in orange toward the top of the left molecule only. This figure and subsequent molecular representations were prepared with pymol (http://pymol.sourceforge.net).
Comparison with Other Structures. Structural neighbors of N-RsbR were identified by a dali (29) search and selected results are shown in Table 2, which is published as supporting information on the PNAS web site. The top match (Z score, 11.1) was to Vitreoscilla stercoraria hemoglobin (30), and there were many other matches to structures of globins in the dali database. Therefore, the N-terminal domain of RsbR has a classic globin fold, and even the characteristic perpendicularly kinked N-terminal helices of globins are maintained in N-RsbR. We have applied the standard globin helical nomenclature to N-RsbR, and in the absence of residues equivalent to the CD region of globins, the helices are thus labeled A, B, and E–H.
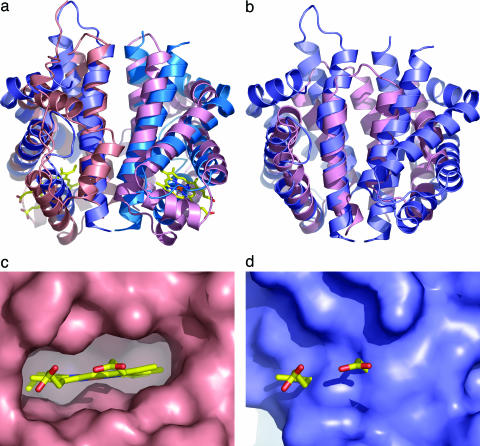
An intriguing structural match was found by dali between N-RsbR and HemAT from B. subtilis (31), with a Z score of 9.5. After structural alignment, the sequence identity between N-RsbR and HemAT is 13.5%. HemAT is a globin-coupled sensor, a recently discovered signaling system regulating, for instance, the aerotactic responses of B. subtilis and members of the Archaea (32). Globin-coupled sensors appear to have evolved from a common, ancestral globin, the protoglobin, which has been suggested to be present in organisms as far back as the last universal common ancestor, from which all life on earth is thought to be descended (33). The rms deviation (rmsd) between monomers of N-RsbR and HemAT is 2.95 Å over 117 equivalent Cα atoms. Using this means of comparison, N-RsbR is as similar to HemAT as HemAT is to horse heart myoglobin (rmsd, 2.4 Å). If the comparison of N-RsbR to HemAT is extended to the dimeric forms of these two proteins, the rmsd over 226 equivalent Cα atoms increases only slightly, to 3.08 Å. Therefore, even in the absence of meaningful sequence similarity between N-RsbR and the globin-coupled sensor HemAT, the overall folds of the monomers and their dimerization mechanisms are identical (Fig. 2a). Indeed, the use of the G and H helices to mediate dimerization in N-RsbR is reminiscent of, but dissimilar to, the interactions between β- and α-globins in the hemoglobin tetramer and those recently reported between α-globin and the α-hemoglobin stabilizing protein (34).
Fig. 2.
Comparison of N-RsbR with structural neighbors. (a) Protein cartoons of N-RsbR (shades of blue) and HemAT (shades of pink, PDB ID code 1OR4) after superimposition and in the same orientation as Fig. 1. The heme in HemAT is drawn as a “stick” figure. Each protein dimerizes through a four-helical bundle formed in each case by the C-terminal pairs of helices. Although there is little strict sequence homology between N-RsbR and HemAT residues in the dimer interface, hydrophobic amino acids in N-RsbR generally have hydrophobic equivalents in HemAT. The same trend can be observed for the polar and acidic amino acids. The Z-helix of HemAT can be seen running approximately between 1 o'clock and 7 o'clock in the figure in a groove, the floor of which comprises helices G and H, with walls of the loops between helices E and F to the left and the C–D corner to the right. (b) As a, but the structures of N-RsbR (blue) and the smaller N-RsbU (pink) are superimposed, and there is no equivalent in N-RsbU to the A–B helices of N-RsbR. (c) Close-up view of the heme pocket of HemAT, with the molecular surface depicted in pink and the bound heme drawn as a stick model, colored according to atom type. Note the complementarity of protein surface and heme and that the protrusion of surface immediately below the iron (brown) is the imidazole side chain of the proximal H123. (d) The same view as c of the equivalent region of N-RsbR (blue), with the heme from HemAT displayed for reference. Here, the surface pocket that accommodates heme in HemAT is filled in N-RsbR by the side chains of residues L37, V41, I45, E48, L70, R73, A74, I77, L79, F83, L84, and L88.
The highest dali Z score other than globins (Table 2) was the C-terminal domain of KaiA (C-KaiA, Protein DataBank ID code 1Q6A), a protein involved in the regulation of circadian rhythm in the thermophilic cyanobacterium Thermosynechococcus elongatus (35). As in partner-switching, Kai proteins are also regulated by reversible protein phosphorylation and the formation of alternative protein–protein interactions (35). We have already reported (36) that C-KaiA is structurally similar to the N-terminal domain of RsbU (N-RsbU), which was also found in our dali search, albeit with a low Z score of 3.9. This observation is of particular interest because both N-RsbU and the stressosome bind RsbT as part of the partner-switching mechanisms governing environmental stress signaling. The fact that the alternative-binding partners of RsbT are structurally similar (Fig. 2b) is in marked contrast to SpoIIAB, which binds to completely unrelated partner proteins, σF and SpoIIAA (37, 38).
Globin Heme Pocket. Despite the globin fold of N-RsbR, there is no evidence, through spectroscopy or careful inspection of the electron density maps of N-RsbR, for heme binding by RsbR. The α-helices that sandwich the heme group in globins, E and F, are brought closer together in N-RsbR than in globins, and they fill the cavity in N-RsbR (Fig. 2 c and d) in which heme would be found in globins. Furthermore, R73 and E48 (from helices E and F, respectively) form a barrier in front of the heme pocket. Finally, of the 19 residues conserved in bacterial hemoglobins identified by Tarricone et al. (30), none are present in N-RsbR. In particular, the alanine at position 74 in N-RsbR replaces the unique chemistry supplied by histidine at the heme-proximal position in globins. The absolutely invariant heme-proximal histidine is required as a ligand for the heme-bound iron, and A74 in N-RsbR has hydrophobic equivalents in the RsbR paralogues except for YqhA, where it is serine. The heme group is an intimate constituent of the globin molecule, and mutations of amino acids that line the heme-binding site in myoglobin affect protein folding and stability (39). To our knowledge, no structure has yet been published that describes the apo form of any of the subgroups of the globin family, presumably because of the conformational flexibility of residues surrounding the heme pocket.
The N-terminal domain of RsbR is highly variable in sequence in comparison with the equivalent domain in its paralogues and orthologues (Fig. 7, which is published as supporting information on the PNAS web site). The RsbRST module has been found in many bacterial species, with as yet uncharacterized functions in many of them (see ref. 14 for a full discussion; and the legend to Fig. 7). Pairwise comparisons of the paralogues against RsbR reveal sequence identities of 17–22% for the N-terminal domain and 45–50% for the C-terminal domain. The sequence similarity between N-RsbR and its paralogues is sufficient to suggest that the globin fold will be maintained and that the paralogues cannot bind heme because they lack a proximal histidine. Of the residues that are similar in the sequence alignment, the majority are hydrophobic and either form part of the core of N-RsbR or occupy the space around where the heme would be found in globins. Among the polar and charged amino acids that display some degree of conservation, D117, S121, and E126 line a solvent-filled cavity in the dimer interface.
Identification of an Interaction Surface. Although we have not been able to demonstrate binding of RsbT to N-RsbR (our unpublished observations), RsbR (10), or RsbS (10) alone, it would appear that RsbT partner switching requires the formation of RsbR and RsbS into a large, complex structure, the stressosome. Nevertheless, the distinct surface groove at the N-RsbR dimer interface may have a role in RsbT recognition; support for this hypothesis can be found in three examples of other proteins that use a corresponding surface for binding reactions. First, residues in the equivalent region of C-KaiA are involved in KaiC binding (40), and mutations at these positions have clock period-associated phenotypes (41). Second, HemAT has an N-terminal extension of ≈40 aa in comparison to other globins, part of which forms a α-helix, called the Z-helix (31). The Z-helix in HemAT folds into the same groove that is used by a partially helical peptide derived from KaiC to bind C-KaiA (40). Finally, our unpublished data reveal that mutation of I78 and I74 in RsbU prevents RsbT binding. These amino acids are at the N-RsbU dimer interface and are in equivalent positions to key residues in the C-KiaA–KaiC interface and the intramolecular packing of the Z-helix in HemAT.
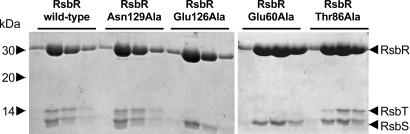
Therefore several mutations (to alanine) were made of amino acids that are surface exposed in the structure of N-RsbR and have spatial equivalents in HemAT that are involved in the intramolecular packing of the Z-helix. These variant RsbR proteins were coexpressed with wild-type RsbS from a bicistronic operon. The RsbR–RsbS complex is stable and can be purified to homogeneity from the E. coli expression host. The binding of RsbT to mutated RsbR–wild-type RsbS complexes was assessed by mixing the purified proteins before separating unbound RsbT from RsbR–RsbS–RsbT complexes by gel filtration chromatography. The wild-type RsbR–RsbS complex binds RsbT (Fig. 3) such that they coelute on gel filtration close to the void volume of the column and excess RsbT elutes as a monomeric protein (10). Similarly, both the RsbRN129A–RsbS and RsbRT86A–RsbS complexes retain the ability to bind RsbT (Fig. 3), indicating that N129 and T86 in RsbR are not crucial requirements for RsbT binding to the RsbR–RsbS complex. However, the RsbRE126A–RsbS, RsbRK82A–RsbS, and RsbRE60A–RsbS complexes are all incapable of recruiting RsbT (Fig. 3), indicating that these residues form a significant surface-exposed patch required for the binding of RsbT (Fig. 4). The Cα atoms of E60 and K82 are each within 7 Å of the Cα of E126. E60 is one of only four residues conserved in the N termini of RsbR and its paralogues in B. subtilis. Two of the others (W22 and F91) are found in the hydrophobic core of the N-RsbR monomer, indicating that E60 is a particularly critical residue for RsbT recruitment by stressosome complexes.
Fig. 3.
Gel filtration assay of mutant RsbR–RsbS complexes binding to RsbT. About 300 μg of RsbR–RsbS, RsbRN129A–RsbS, RsbRE126A–RsbS, RsbRE60A–RsbS, RsbRT86A–RsbS, or RsbRK82A–RsbS complexes was mixed with 100 μg of RsbT. The mixtures were loaded onto a Superdex 200 HR 10/30 gel filtration column, and the first four fractions of the void volume of the column, which contains the RsbR–RsbS complex, were analyzed by SDS/15% PAGE and Coomassie blue staining. RsbT alone behaves as a monomer during gel filtration (data not shown). Whereas RsbT is recruited to wild-type RsbR–RsbS complex and coelutes in the void volume, no bands corresponding to RsbT were detected in the RsbR–RsbS complexes for E60A, K82A, and E126A mutations in RsbR, indicating that these mutations affect the binding of RsbT to the RsbR–RsbS complex. Conservative mutation to alanine of amino acids in this region does not appear to affect the overall fold of RsbR because both N129 and T86 can be mutated to alanine without affecting the binding of RsbT.
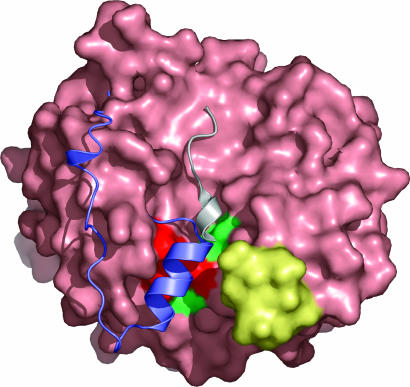
Fig. 4.
Molecular interaction surface of RsbR. The molecular surface of the N-RsbR dimer is light pink. The Z-helix and preceding ≈20 aa (both in purple) of HemAT (31) and the KaiC-derived peptide (gray) of the KaiA–KaiC complex (40) are displayed after the superimposition of HemAT and KaiA on the dimer of N-RsbR. Both of these parts of structure are found to overlap in a groove formed on the surface of N-RsbR, which is flanked on one side by the C–D corner (yellow), mutations in which, in Vitreoscilla hemoglobin, affect the binding of the flavin-binding domain of 2,4-dinitrotoluene dioxygenase (51). The positions of E60, K82, and E126, mutation of which affects RsbT binding to RsbR–RsbS, and T86 and N129, mutation of which does not affect RsbT binding to RsbR–RsbS, are highlighted on the surface of N-RsbR in red and green, respectively.
Discussion
The genes encoding RsbR, RsbS, and RsbT constitute the RsbRST module that has been identified across a wide range of bacterial taxa (14). RsbR, RsbS, and RsbT are the core components of a large signaling complex (>1 MDa), named the stressosome after its role in regulation of the response of B. subtilis to stress (10–13, 36). In other bacteria, the RsbRST cluster is found upstream of a PP2C-type phosphatase and other identifiable and conserved signaling domains, suggesting that the stressosome may control a large and disparate array of different regulators.
The structure of N-RsbR described in this paper reveals it to be closely related to the globin family. Moreover, N-RsbR dimerizes in a fashion identical to that of the globin-coupled sensor, HemAT (31). A globin-coupled sensor domain akin to HemAT replaces the N-terminal domain of RsbR in the RsbRST module in the proteobacteria Silicibacter sp. TM1040, Chromobacter violaceum ATCC 12472, and Vibrio vulnificus CMCCP6 (14). It would thus appear that RsbR has evolved by the duplication of a STAS domain and its fusion to an ancestral sensor globin. STAS domains appear to participate in regulatory protein–protein interaction networks, and some may be modulated by phosphorylation (14). As a result of this fusion, the nonheme globin RsbR has evolved novel functions, such as the binding of signaling proteins such as RsbT.
Although N-RsbR has the same fold as a globin (Fig. 2a), our structural analysis does not reveal the presence of any bound cofactor, such as heme. E. coli is clearly capable of synthesizing heme in recombinant proteins, which can be purified in their heme-bound states. The absence of heme from N-RsbR can be explained by an analysis of the structure of the “heme pocket”; the heme-proximal histidine that is invariant in all globins is absent in N-RsbR, replaced at this position, 74, by alanine, and the region occupied by a heme in hemoglobin is filled in N-RsbR with amino acid side chains (Fig. 2 c and d). This observation raises the possibility that an alternative, perhaps loosely binding cofactor, is bound by this region and is not retained during purification and subsequent structural studies. For instance, the RsbR paralogue YtvA encodes a Per–Arnt—Sim (PAS) domain that binds a flavin cofactor, which is correctly synthesized by E. coli (42). However, globins are only known to bind porphyrin–macrocycle cofactors, and furthermore, the globin-like proteins involved in light harvesting in algae also use tetrapyrroles as chromophores. The absence of a cofactor from the crystal structure of N-RsbR suggests that this domain does not utilize the canonical heme pocket to recognize prosthetic groups such as heme, or any other cofactor for that matter, because the heme pocket is closed. It would thus appear that RsbR has been customized by the recycling of an ancestral sensor globin domain to evolve new functionalities and to lose the ability to bind small-molecule ligands in the heme pocket. However, we cannot at this stage formally exclude the possibility that RsbR is also regulated by a small-molecule effector of unknown composition that binds elsewhere in RsbR.
One of the functions for RsbR is revealed by our demonstration that the dimer interface of N-RsbR provides a binding site for RsbT (Fig. 4), and that this structure is similar to the RsbT-binding surface of RsbU (Fig. 2b). Because similar regions of RsbR and RsbU are used to bind to RsbT, it would not be unreasonable to propose that the same amino acids in RsbT are used to effect both interactions. Competition between binding partners for RsbT favors the stressosome before stress, but this balance tilts toward RsbU when RsbT phosphorylates RsbR and RsbS at the onset of stress (Fig. 5). Whereas the N-RsbU–RsbT interaction can be visualized by native PAGE (36), the N-RsbR–RsbT interaction cannot, implying that other determinants exist on the surface of the stressosome for the binding of RsbT. This conclusion is not surprising because RsbT acts as a kinase toward the stressosome, where phosphorylation of residues present in the C-terminal STAS domain of RsbR stimulates the phosphorylation of RsbS (11, 43). Overall, although N-RsbR is crucial for the binding of RsbT to the stressosome (Fig. 3), the interaction of RsbT with C-RsbR (and RsbS) in the context of the complex is also important. We have thus constructed a homology model of the interaction of RsbT with RsbR (Fig. 8, which is published as supporting information on the PNAS web site), based on the crystal structure of the complex between SpoIIAB and SpoIIAA (38), functional homologues of RsbT and RsbR. This model provides a starting point for analyzing other residues that participate in stressosome assembly, such as contacts between RsbR domains, and/or structural modifications that could occur after phosphorylation. It is perhaps pertinent to note that, in this model, the T4 and T5 mutations of RsbT that block σB induction (44) are in contact with E60, K82, and E126 of N-RsbR, mutation of which destroys the binding of RsbT to the RsbR–RsbS complex.
The question that remains, however, is how is stress signaled to the stressosome, resulting in the release of RsbT to activate RsbU and hence σB? Unlike other well characterized signaling systems in B. subtilis, such as the two-component system, there are no obvious sensory domains in the proteins that compose the environmental stress signaling pathway that controls the general stress response. A potential entry point for the stress signal would be in the modification of the structure of the stressosome, resulting in the activation of the kinase function of RsbT and its dissociation from the freshly phosphorylated stressosome. The entire σB operon can be transferred to E. coli but cannot activate σB in response to environmental stress (45), indicating that there is something absent from the σB signaling system when moved to E. coli. It would appear more likely that the missing component is not a small-molecule metabolite, but something more specific to Bacillus, such as a protein.
The protein in Bacillus could be the GTPase Obg, whose role in stress signaling in B. subtilis is established (46, 47) but not understood (13, 48). We have shown here that N-RsbR binds to RsbT, and it previously has been demonstrated by a yeast two-hybrid analysis that Obg interacts with RsbT (46). It is therefore possible that a ternary complex exists between the kinase RsbT, the GTPase Obg, and the globin-like N-RsbR in the context of the stressosome. This complex would be transient and dependent on the stress status of the cell. Globins and GTPase are known to interact transiently in the stress response of humans; neuroglobin helps to protect neuronal tissue from hypoxic stress by inhibiting the exchange of GDP for GTP in the α subunit of heterotrimeric G proteins (49). Interestingly, the guanine nucleotide exchange of Gα is no longer inhibited by mutation of E53 in neuroglobin (50), and this residue is presumed to be important for the interaction between neuroglobin and Gα. E53 is found in the flexible D helix of neuroglobin; the equivalent area of N-RsbR is also flexible (amino acids 33–38) and forms one wall of the groove that we have identified as being involved in molecular interactions with RsbT. Moreover, the same region of Vitreoscilla hemoglobin is also relatively disordered in the crystal structure (30), and mutations here disrupt the binding of Vitreoscilla hemoglobin to the flavin-binding domain of 2,4-dinitrotoluene dioxygenase (51). Taking these observations together, it would appear that a disparate group of proteins that show structural homology to N-RsbR use overlapping regions of molecular surface to form binding sites for macromolecules.
The globin fold arose early in evolutionary time, and globins were likely to be present in the common universal ancestor, possibly as a sensor of toxic gaseous species (33, 52). It is a large and extremely well characterized family of proteins linked by a common requirement for porphyrin cofactors, such as heme. It is difficult to overstate the role that globins play in the history of molecular biology; the first two protein crystal structures to be determined were those of myoglobin (53) and hemoglobin (54). Nonetheless, the divergence of globins at the amino acid sequence level clearly presents difficulties in their recognition during genome annotation with commonly used protein domain recognition algorithms (55). It is thus possible that globins with functions other than binding of gaseous diatomic molecules are yet to be discovered and that N-RsbR of B. subtilis is the first member to be described of what could prove to be a large family of globins that do not bind heme.
Supplementary Material
Acknowledgments
We thank J. Pané-Farré for sharing ideas and data before publication; J. Tate for bioinformatics advice; and B. Connolly, H. Gilbert, J. Lakey, N. Robinson, and M. Yudkin for their encouragement. We thank M. Walsh and E. Lowe for support during data collection and the European Synchrotron Radiation Facility (Grenoble, France) for access to macromolecular crystallography beam lines through the Block Allocation Group. This work was supported by the Wellcome Trust and the University of Newcastle.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: N-RsbR, N-terminal domain of RsbR; N-RsbU, N-terminal domain of RsbU; STAS, sulfate transporter and anti-σ factor antagonist.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2BNL and r2BNLsf).
References
- 1.Burgess, R. R., Travers, A. A., Dunn, J. J. & Bautz, E. K. (1969) Nature 221, 43–46. [DOI] [PubMed] [Google Scholar]
- 2.Hecker, M. & Völker, U. (1998) Mol. Microbiol. 29, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 3.Hecker, M. & Völker, U. (2001) Adv. Microb. Physiol. 44, 35–91. [DOI] [PubMed] [Google Scholar]
- 4.Price, C. W. (2002) in Bacillus subtilis and Its Closest Relatives: From Genes to Cell, eds. Sonenshein, A.L., Hoch, J.A. & Losick, R. (Am. Soc. Microbiol., Washington, DC), pp. 369–384.
- 5.Petersohn, A., Brigulla, M., Haas, S., Hoheisel, J. D., Völker, U. & Hecker, M. (2001) J. Bacteriol. 183, 5617–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price, C. W., Fawcett, P., Ceremonie, H., Su, N., Murphy, C. K. & Youngman, P. (2001) Mol. Microbiol. 41, 757–774. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, L. & Losick, R. (1993) Proc. Natl. Acad. Sci. USA 90, 2325–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Min, K.-T., Hilditch, C. M., Diederich, B., Errington, J. & Yudkin, M. D. (1993) Cell 74, 735–742. [DOI] [PubMed] [Google Scholar]
- 9.Alper, S., Duncan, L. & Losick, R. (1994) Cell 77, 195–205. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C.-C., Lewis, R. J., Harris, R., Yudkin, M. D. & Delumeau, O. (2003) Mol. Microbiol. 49, 1657–1669. [DOI] [PubMed] [Google Scholar]
- 11.Kim, T. J., Gaidenko, T. A. & Price, C. W. (2004) J. Bacteriol. 186, 6124–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, T. J., Gaidenko, T. A. & Price, C. W. (2004) J. Mol. Biol. 341, 135–150. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, S., Zhang, S., Woodbury, R. L. & Haldenwang, W. G. (2004) Microbiology 150, 4125–4136. [DOI] [PubMed] [Google Scholar]
- 14.Pané-Farré, J., Engelmann, S., Hecker, M. & Stülke, J. (2005) J. Mol. Microbiol. Biotechnol., in press.
- 15.Page, R., Grzechnik, S. K., Canaves, J. M., Spraggon, G., Kreusch, A., Kuhn, P., Stevens, R. C. & Lesley, S. A. (2003) Acta Crystallogr. D 59, 1028–1037. [DOI] [PubMed] [Google Scholar]
- 16.Evans, G. & Pettifer R. F. (2001) J. Appl. Crystallogr. 34, 82–86. [Google Scholar]
- 17.Leslie, A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26.
- 18.Evans, P. (1993) CCP4 Daresbury Study Weekend: Data Collection and Processing, DL/SCI/R34 (Central Laboratory of the Research Councils, Daresbury Laboratory, Warrington, U.K.), pp. 114–122.
- 19.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 20.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 21.Sheldrick, G. M. (2002) Z. Kristallogr. 217, 644–650. [Google Scholar]
- 22.Pape, T. & Schneider, T. (2004) J. Appl. Crystallogr. 37, 843–844. [Google Scholar]
- 23.Perrakis, A., Morris, R. & Lamzin, V. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 24.Jones, T. A., Zou, J., Cowan, S. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 26.Schneider, T. R. (2002) Acta Crystallogr. D 58, 195–208. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. & Thornton, J. M. (1996) Proc. Natl. Acad. Sci. USA 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahadur, R. P., Chakrabarti, P., Rodier, F. & Janin, J. (2004) J. Mol. Biol. 336, 943–955. [DOI] [PubMed] [Google Scholar]
- 29.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 30.Tarricone, C., Galizzi, A., Coda, A., Ascenzi, P. & Bolognesi, M. (1997) Structure 5, 497–507. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, W. & Phillips, G. N. (2003) Structure 11, 1097–1110. [DOI] [PubMed] [Google Scholar]
- 32.Hou, S., Larsen, R. W., Boudko, D., Riley, C. W., Karatan, E., Zimmer, M., Ordal, G. W. & Alam, M. (2000) Nature 403, 540–544. [DOI] [PubMed] [Google Scholar]
- 33.Freitas, T. A. K., Saito, J. A., Hou, S. & Alam, M. (2005) J. Inorg. Biochem. 99, 23–33. [DOI] [PubMed] [Google Scholar]
- 34.Feng, L., Gell, D. A., Zhou, S., Gu, L., Kong, L., Li, J., Hu, M., Yan, N., Lee, C., Rich, A. M., et al. (2004) Cell 119, 629–640. [DOI] [PubMed] [Google Scholar]
- 35.Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T. & Kondo, T. (2005) Science 308, 414–415. [DOI] [PubMed] [Google Scholar]
- 36.Delumeau, O., Dutta, S., Brigulla, M., Kuhnke, G., Hardwick, S. W., Völker, U., Yudkin, M. D. & Lewis, R. J. (2004) J. Biol. Chem. 279, 40927–40937. [DOI] [PubMed] [Google Scholar]
- 37.Campbell, E. A., Masuda, S., Sun, J. L., Muzzin, O., Olson, C. A., Wang, S. & Darst, S. A. (2002) Cell 108, 795–807. [DOI] [PubMed] [Google Scholar]
- 38.Masuda, S., Murakami, K. S., Wang, S., Olson, C. A., Donigian, J., Leon, F., Darst, S. A. & Campbell, E. A. (2004) J. Mol. Biol. 340, 941–956. [DOI] [PubMed] [Google Scholar]
- 39.Hargrove, M. S., Krzywda, S., Wilkinson, A. J., Dou, Y., Ikeda-Saito, M. & Olson, J. S. (1994) Biochemistry 33, 11767–11775. [DOI] [PubMed] [Google Scholar]
- 40.Vakonakis, I. & LiWang, A. C. (2004) Proc. Natl. Acad. Sci. USA 101, 10925–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishiura, M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science 281, 1519–1523. [DOI] [PubMed] [Google Scholar]
- 42.Losi, A., Polverini, E., Quest, B. & Gartner, W. (2002) Biophys. J. 82, 2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, C.-C., Yudkin, M. D. & Delumeau, O. D. (2004) J. Bacteriol. 186, 6830–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodbury, R., Luo, T., Grant, L. & Haldenwang, W. G. (2004) J. Bacteriol. 186, 2789–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott, J. M., Smirnova, N. & Haldenwang, W. G. (1999) Biochem. Biophys. Res. Commun. 257, 106–110. [DOI] [PubMed] [Google Scholar]
- 46.Scott, J. M. & Haldenwang, W. G. (1999) J. Bacteriol. 181, 4653–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, J. M., Mitchell, T. & Haldenwang, W. G. (2000) J. Bacteriol. 182, 1452–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, S. & Haldenwang, W. G. (2004) Biochem. Biophys. Res. Commun. 322, 565–569. [DOI] [PubMed] [Google Scholar]
- 49.Wakasugi, K., Nakano, T. & Morishima, I. (2003) J. Biol. Chem. 278, 36505–36512. [DOI] [PubMed] [Google Scholar]
- 50.Wakasugi, K. & Morishima, I. (2005) Biochemistry 44, 2943–2948. [DOI] [PubMed] [Google Scholar]
- 51.Lee, S. Y., Stark, B. C. & Webster, D. A. (2004) Biochem. Biophys. Res. Commun. 316, 1101–1106. [DOI] [PubMed] [Google Scholar]
- 52.Wajcman, H. & Kiger, L. (2002) C. R. Biol. 325, 1159–1174. [DOI] [PubMed] [Google Scholar]
- 53.Kendrew, J. C., Bodo, G., Dintzis, H. M., Parrish, R. G., Wyckoff, H. & Phillips, D. C. (1958) Nature 181, 662–666. [DOI] [PubMed] [Google Scholar]
- 54.Muirhead, H. & Perutz, M. F. (1963) Nature 199, 633–638. [DOI] [PubMed] [Google Scholar]
- 55.Freitas, T. A., Hou, S. & Alam, M. (2003) FEBS Lett. 552, 99–104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.