Abstract
Various attempts to detect human pituitary growth hormone-releasing hormone receptor (pGHRH-R) in neoplastic extrapituitary tissues have thus far failed. Recently, four splice variants (SVs) of GHRH-R have been described, of which SV1 has the highest structural homology to pGHRH-R and likely plays a role in tumor growth. The aim of this study was to reinvestigate whether human tumors and normal human extrapituitary tissues express the pGHRH-R and to corroborate our previous findings on its SVs. Thus, we developed a real-time PCR method for the detection of the mRNA for the pGHRH-R, its SVs, and the GHRH peptide. Using real-time PCR, Western blotting, and radioligand-binding assays, we detected the mRNA for pGHRH-R and pGHRH-R protein in various human cancer cell lines grown in nude mice and in surgical specimens of human lung cancers. The expression of mRNA for SVs of pGHRH-R and GHRH was likewise found in xenografts of human non-Hodgkin's lymphomas, pancreatic cancer, glioblastoma, small-cell lung carcinomas, and in human nonmalignant prostate, liver, lung, kidney, and pituitary. Western blots showed that these normal and malignant human tissues contain SV1 protein and immunoreactive GHRH. Our results demonstrate that some normal human tissues and tumors express mRNA and protein for the pGHRH-R and its splice variants. These findings confirm and extend the concept that GHRH and its receptors play an important role in the pathophysiology of human cancers.
Since 1994, our laboratory has been engaged in the synthesis of GHRH antagonists for therapeutic use in the management of various cancers and for investigation of the pathophysiological role of GHRH in various malignancies (1-3). Our group demonstrated that GHRH antagonists can inhibit tumor growth through indirect and direct pathways (2-7). The indirect mechanism is based on the suppression of the pituitary GH-hepatic insulin-like growth factor (IGF)-I axis. Thus, GHRH antagonists can block the pituitary GHRH-R (pGHRH-R), inhibiting the synthesis and release of GH, which results in the reduction of hepatic IGF-I production (2, 3). However, GHRH antagonists can also exert their antiproliferative effects in vitro, where the participation of the GH-IGF-I axis is clearly excluded. Accordingly, it has been demonstrated that GHRH antagonists inhibit the autocrine production of IGF-I and IGF-II in vitro and in vivo apparently through a direct action on tumors (4-7). The synthesis of IGF-II, unlike that of IGF-I, does not depend on serum GH levels. In addition, much evidence indicates that GHRH antagonists inhibit the proliferation of various tumors by blocking the binding of autocrine GHRH to tumor cells without any involvement of IGFs (8-10). These direct effects logically imply the presence of specific receptors for GHRH and GHRH antagonists on malignant cells.
So far, various attempts to detect the pGHRH-R in human cancer cell lines have been unsuccessful (2, 11, 12-18). Moreover, GHRH-R was not detected in many normal tissues shown to produce GHRH and respond to the ligand by a rise in cAMP (11, 19). One possibility was that receptors for vasoactive intestinal peptide (VIP), pituitary adenylate cyclase-activating polypeptide, or other related receptors were targets for GHRH and its antagonists in tumors because of the structural similarities between these receptors and pGHRH-R (14, 18, 20, 21). However, it was found that the growth of a VIP and pituitary adenylate cyclase-activating polypeptide receptor-negative human pancreatic tumor (MiaPaCa-2) could be inhibited by selective antagonists for GHRH-R but not by antagonists specific for VIP receptors (14).
Subsequently our group demonstrated the presence of four truncated splice variants (SVs) of GHRH-R in nonmalignant human tissues, several human cancer cell lines, and human prostate cancer specimens (18, 22, 23). Of the four truncated receptors, SV1 of GHRH-R has the greatest structural similarity to the pGHRH-R and is probably the main SV that mediates the effects of GHRH and its antagonists in tumors (18).
The aim of the present study was to reinvestigate, through the use of sensitive real-time PCR methods, Western blotting, and binding assays, whether some human cancer specimens, human cancer cell lines xenografted into nude mice, and nonneoplastic human tissues could express the pGHRH-R and its SVs.
Materials and Methods
Cell Lines, Animals, and Cancer Xenografts. Some of our studies were performed on xenografted tumors grown in nude mice. Human cancer cell lines, including non-Hodgkin's lymphomas (HT and RL), a pancreatic cancer (MiaPaCa-2), a glioblastoma cell line (DBTRG-05), and two small-cell lung carcinoma (SCLC) lines (H-82 and H-345) were obtained from American Type Culture Collection. The culture media were purchased from GIBCO/BRL and varied depending on the requirements for each cell line as described in the instructions from American Type Culture Collection.
Male athymic (NCrnu/nu) nude mice ≈6 weeks old were purchased from the National Cancer Institute (Frederick Cancer Research and Development Center, Frederick, MD). Xenografts were initiated as described in ref. 6. All experiments were performed in accordance with institutional guidelines for the care and use of experimental animals.
Human Cancer Specimens and Human Nonmalignant Tissues. Three specimens of human non-SCLC (NSCLC) were obtained from patients 49-61 years of age at the time of therapeutic bronchoscopy for relief of airway obstruction at Tulane University Health Sciences Center. The local Institutional Review Board for use of human subjects at Tulane Medical Center and the Research and Development Committee of the Veterans Affairs Medical Center in New Orleans approved the protocol for collection and use of these specimens and authorized the study. Informed consent from patients was also obtained before specimen collection. Histopathological examination of each specimen was undertaken to confirm the presence of cancer with minimal admixed nonmalignant tissue (<20%) before Western blotting studies. Normal human pituitaries were purchased from the National Hormone and Peptide Program (A. F. Parlow, Los Angeles County Harbor-UCLA Medical Center, Torrance, CA).
RNA Isolation and Reverse Transcriptase Reaction. Total RNA isolation was performed from two representative tumors of each tested xenograft tissue and from a pool of two human pituitaries following TRI reagent protocol (Sigma-Aldrich). Messenger RNA was purified with an mRNA isolation kit (Roche, Mannheim, Germany). The yield and quality of total RNA and mRNA were determined spectrophotometrically using 260 nm and a 260/280-nm ratio, respectively. Normal human total RNA samples obtained from kidney (one person), lung (pool of three persons), liver (one person), and prostate (pool of 32 persons) were purchased from Clontech. Two micrograms of total RNA from the normal human tissues or mRNA from xenografted tumors in a final volume of 40 μl were reverse-transcribed into cDNA in the Applied Biosystems 2700 PCR system with the iScript cDNA synthesis kit from Bio-Rad.
Real-Time PCR. Gene-specific primers for all tested genes are presented in Table 1. The probes designed to evaluate the expression of pGHRH-R, GHRH and β-actin are 5′-/TexRd-XN/AGG GTG ACC CAC TCG CCA GAG CCT/BHQ_2/-3′, 5′-/HEX/TCT TGG TTG CTC TCT CCC TGC TGC CT/BHQ_1/-3′ and 5′-/6-FAM/CAG TCG GTT GGA GCG AGC ATC CCC/BHQ_1/-3′, respectively. All PCRs were analyzed by the iCycler real-time PCR instrument from Bio-Rad.
Table 1. Gene-specific primers for human GHRH, pGHRH-R, Svs, and β-actin used in this study.
| Sense primers
|
Antisense primers
|
||||||
|---|---|---|---|---|---|---|---|
| Gene | Sequence | cDNA location | Nucleotide location | Sequence | cDNA location | Nucleotide location | Product size, bp |
| SV1 | tggggagagggaaggagttgt | Intron 3 | 287-307 | gcgagaaccagccaccagaa | Exon 7 | 790-809 | 523 |
| SV2 | aggaaggccccatagtgtgtc | Intron 3 | 145-165 | ggcagccagtggagaagct | Exon 6/8 | 652-670 | 526 |
| SV3 | gccccatagggctgtgaaac | Intron 3 | 151-170 | acagctgggtgtggacgtagt | Exon 6 | 375-395 | 245 |
| SV4 | ctgaggagggctgcccgt | Exon 4/8 | 425-442 | ggccctttgatgatccaccagt | Exon 8 | 523-544 | 120 |
| pGHRH-R | atgggctgctgtgctggccaac | Exon 3 | 230-251 | taaggtggaaagggctcagacc | Exon 4 | 353-374 | 145 |
| GHRH | atgcagatgccatcttcaccaa | — | 95-116 | tgctgtctacctgacgaccaa | — | 224-244 | 150 |
| β-actin | ctggaacggtgaaggtgaca | — | 1348-1367 | aagggacttcctgtaacaatg | — | 1467-1487 | 140 |
β-actin was used as housekeeping gene. —, not applicable.
Bio-Rad iQ Supermix was used in the PCRs for pGHRH-R, GHRH, and β-actin. Thermal cycling conditions comprised an initial denaturation step at 95°C for 3 min followed by 45 cycles at 95°C for 30 sec and an annealing temperature at 60°C for GHRH and β-actin or 65°C for pGHRH-R for 1 min. As final steps, we included two cycles: one at 95°C and the other at the corresponding annealing temperature of each tested gene, both for 1 min. All samples were run in duplicate, and each well of PCR contained 25 μl as a final volume, including 2 μl of cDNA, 200 nM gene-specific primers, and 400 nM probes.
The mRNA expression of SVs was evaluated in 25-μl reactions containing 1× iQ SYBR green Supermix (Bio-Rad), 2 μl of cDNA, and 200 nM of specific primers. Duplicate samples were denatured at 95°C for 3 min followed by 45 cycles at 95°C for 30 sec and 58°C or 65°C for 1 min for SV3 and SV4, respectively. The reactions for SV1 and SV2 were set at 40 cycles at 95°C for 30 sec, 60°C for 45 sec, and 72°C for 1 min. We performed two cycles for 1 min each as final steps in all reactions. The first step was set at 95°C and the second final step was set at the corresponding annealing temperature of each tested gene.
Normal human pituitary was used as positive control, and β-actin was used as a housekeeping gene. Negative samples were run for each RT-PCR consisting of no RNA in the reverse transcriptase reaction and no cDNA in the PCR. The threshold cycle (CT), defined as the fractional PCR cycle number at which the fluorescence reaches 10 times the baseline standard deviation, was compared for the expression of GHRH, pGHRH-R, SV1, SV2, SV3, and SV4 in all tested tumors (HT, RL, MIA PaCa-2, DBTRG-05, H82, and H345) and normal human samples (pituitary, kidney, lung, liver, and prostate). The mathematical method described by Pfaffl (24) was used to evaluate the relative expression ratio for all genes compared with β-actin, with the efficiencies for each set of primers, probes, and the CT. The relative expression of SVs was based on SYBR green staining and not on molecular probes because their sequence similarities would not allow us to design specific probes for each GHRH-R SV. The efficiencies of all primers (Invitrogen) and probes (Integrated DNA Technologies, Coralville, IA) were tested before the experiments, and they were all efficient in the range of 95-105%.
Production of Antisera to the pGHRH-R. The synthesis of [Ala-28,41]pGHRH-R(23-45)-Tyr-Cys-NH2 peptide hapten (designated pGHRH-R-hapten) was described in ref. 25. This peptide is an analog of segment 23-45 of the pGHRH-R protein in which two internal cysteine residues (Cys-28 and Cys-41) were replaced by alanine residues (26, 27). Thus, the antibody raised against pGHRH-R-hapten would recognize the N terminus corresponding to the posttranslationally processed pGHRH-R protein expressed on the cell surface after signal peptide cleavage (26, 27). At the C terminus of pGHRH-R-hapten, a tyrosine residue was incorporated to allow subsequent 125I-labeling, and a cysteine residue was inserted into the molecule to make it suitable for conjugation to carriers through its free SH-functional group. The methods used for raising antisera in rabbits, their characterization by RIA, and purification were described in detail in refs. 25 and 28.
Western Blotting. Protein-matched samples (20 μg per lane) were separated by 4-20% (see Fig. 2) or 12.5% (see Fig. 1) SDS/PAGE Tris·HCl criterion precasted gels (Bio-Rad). Electroblotting was used to transfer the proteins onto nitrocellulose membranes (Bio-Rad). The membranes were incubated for 3-5 h at room temperature in 5% nonfat dry milk in TBS-Tween. Optimal dilutions of the antibodies were determined in preliminary studies. The blots were probed at 4°C overnight with polyclonal antibodies to GHRH C-16 and V-17 (1:500) (Santa Cruz Biotechnology) and to SV1-SV2-SV4 (1:2,000) and pGHRH-R (1:2,000), with antisera raised and affinity-purified in our laboratory (25). The batch numbers for SV1-SV2-SV4 and pGHRH-R antisera were 2317/7 and 2321/5, respectively. Blots were also probed with depleted antisera to prove the specificity of the signals. The depletion was done by overnight incubation at 4°C of the concentrated antisera (dilution 1:1) with the specific blocking peptides (SV1-SV2-SV4-hapten and pGHRH-R-hapten) at a concentration of 10 μg/ml, followed by centrifugation at 13,400 × g for 30 min. The signal for the immunoreactive proteins was developed with peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology) and visualized by exposure to the chemiluminescence substrate (Amersham Biosciences). The protein bands were quantified by normalizing the signals of different proteins to β-actin signal (1:2,000, Santa Cruz Biotechnology) using the Kodak EDAS 290 imaging system with Kodak 1d image analysis Software.
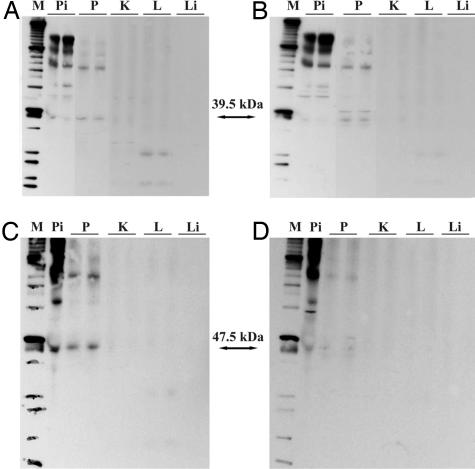
Fig. 2.
Western blot analysis of SV1-SV2-SV4 (A and B) and pGHRH-R (C and D) in samples of normal human prostate (P), kidney (K), lung (L), liver (Li), and pituitary (Pi) tissue. (A and C) Blots incubated with the purified antisera for SV1-SV2-SV4 (A) and pGHRH-R (C). (B and D) Blots with the antisera that were depleted by incubation with specific blocking peptides.
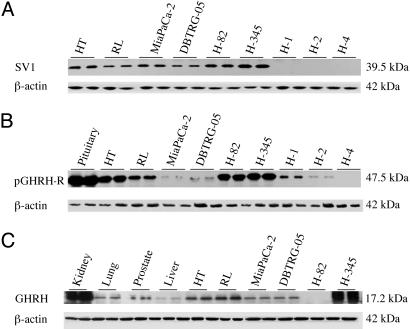
Fig. 1.
Western blot analysis of SV1 (A), pGHRH-R (B), and GHRH (C), with β-actin as control in two samples each of xenografted tumor tissues of human non-Hodgkin's lymphomas (HT and RL), pancreatic cancer (MiaPaCa-2), glioblastoma (DBTRG-05), and small cell lung cancers (H82 and H345) and in three human NSCLC specimens (H1, H2, and H4) and normal human samples of prostate, kidney, lung, liver, and pituitary. All immunoreactive signals were detected with the purified antisera for SV1-SV2-SV4 (A), pGHRH-R (B), and a commercial antiserum for GHRH (C). The molecular masses are shown.
Radioligand Binding Studies. Radioiodinated derivatives of GHRH antagonist JV-1-42 and human GHRH analog [His-1, Nle-27]hGH-RH(1-32)NH2 were prepared by the chloramine-T method as described in refs. 22 and 29. Preparation of membrane fractions from xenografted tumor samples was performed as reported in refs. 22 and 23. Binding characteristics of receptors for GHRH were determined by in vitro ligand competition assays based on the binding of radiolabeled JV-1-42 and [His-1, Nle-27]hGH-RH(1-32)NH2 to tumor membrane fractions (22, 29). Binding affinities (Kd) and capacities (Bmax) were calculated by the ligand-pc computerized curve-fitting software and by Scatchard analysis (22, 29).
Results
Real-Time PCR Analysis. The reaction efficiencies of pGHRH-R, GHRH, SV1, SV2, SV3, SV4, and β-actin were 104.6%, 99.4%, 104%, 96.7%, 102.6%, 98.6%, and 96.1%, respectively. The relative expression ratio (r) for these two genes was analyzed by a mathematical model (24), with β-actin as a housekeeping gene. For mRNA isolated from tumor samples, we calculated the ratios as compared with the expression level of glioblastoma DBTRG-05. For normal human samples, where we used total RNA, we selected the pituitary tissue as the basis of comparison. DBTRG-05 and pituitary were selected as controls because of their higher level of mRNA expression for pGHRH-R within their respective groups. PCR products were absent in the negative controls for all tested genes (Fig. 3, which is published as supporting information on the PNAS web site). HT and RL non-Hodgkin's lymphomas, glioblastoma DBTRG-05 cell line, and all normal human tissues (prostate, kidney, lung, liver, and pituitary) expressed mRNA for all SVs. H-345 SCLC expressed mRNA only for SV4, but H-82 SCLC expressed SV1, SV3, and SV4. MiaPaCa-2 pancreatic cancer expressed only SV1 and SV2 (Fig. 3). Table 2 shows the relative ratios and CT values for GHRH-R SVs of all tumor samples and normal tissues. HT non-Hodgkin's lymphoma presented the highest level of expression for SV1, SV2, and SV4 as compared with other tumor samples. The highest expression of SV3 was seen for DBTRG-05. Among the normal samples, the pituitary presented the highest level of expression for all SVs.
Table 2. Real-time PCR analysis for GHRH SV receptors in normal human tissues and human cancer lines grown in nude mice.
| CT values, mean ± SEM
|
Ratio
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Samples | SV1 | SV2 | SV3 | SV4 | β-actin | SV1 | SV2 | SV3 | SV4 |
| HT | 25.93 ± 0.41 | 28.80 ± 0.74 | 38.05 ± 0.85 | 32.98 ± 2.15 | 13.50 ± 0.35 | 2.91 | 45.27 | 0.78 | 1.30 |
| RL | 26.28 ± 0.80 | 30.30 ± 0.94 | 38.90 ± 0.30 | 34.58 ± 0.91 | 12.58 ± 0.12 | 1.22 | 8.84 | 0.23 | 0.23 |
| MiaPaCa-2 | 26.86 ± 0.60 | 32.18 ± 0.94 | — | — | 12.10 ± 0.19 | 0.58 | 1.80 | — | — |
| H82 | 25.55 ± 0.11 | — | 41.25 ± 2.35 | 35.35 ± 0.60 | 12.95 ± 0.27 | 2.64 | — | 0.06 | 0.18 |
| H345 | — | — | — | 35.05 ± 0.56 | 12.45 ± 0.37 | — | — | — | 0.15 |
| DBTRG-05 | 25.40 ± 0.46 | 32.30 ± 0.39 | 35.65 ± 0.65 | 31.25 ± 0.72 | 11.35 ± 0.20 | 1.00 | 1.00 | 1.00 | 1.00 |
| Prostate | 26.65 ± 0.15 | 30.80 ± 0.40 | 36.40 ± 0.09 | 31.60 ± 0.22 | 16.60 ± 0.01 | 0.19 | 0.063 | 0.01 | 0.0011 |
| Kidney | 30.60 ± 0.09 | 32.35 ± 0.15 | 40.70 ± 0.80 | 31.93 ± 0.35 | 16.15 ± 0.11 | 0.01 | 0.016 | 0.00041 | 0.0007 |
| Lung | 27.75 ± 0.25 | 30.55 ± 0.05 | 40.20 ± 0.50 | 31.80 ± 0.10 | 15.45 ± 0.11 | 0.04 | 0.034 | 0.00036 | 0.0005 |
| Liver | 29.20 ± 0.30 | 33.40 ± 0.09 | 41.85 ± 1.35 | 34.60 ± 0.60 | 17.65 ± 0.03 | 0.06 | 0.022 | 0.00050 | 0.0003 |
| Pituitary | 26.70 ± 1.30 | 29.25 ± 0.25 | 32.50 ± 0.30 | 24.23 ± 0.12 | 19.15 ± 0.03 | 1.00 | 1.00 | 1.00 | 1.00 |
The relative gene expression ratio values were calculated based on ref. 24. —, not detectable.
All tumors and nonneoplastic human tissues also expressed mRNA for the pGHRH-R and GHRH (Fig. 3, lanes E and F). The relative ratio and CT values are shown in Table 3. The levels of the mRNA expression for the pGHRH-R among the tumor tissues were as follows: DBTRG-05 > HT > RL ≫ H345 > H82 ≫ MiaPaCa-2. Among the normal human tissues, the mRNA expression of the pituitary was much higher than the other tissues according to the following order: pituitary ⋙ kidney ≫ lung > liver > prostate.
Table 3. Real-time PCR analysis for GHRH and pGHRH-R in normal human tissues and human cancer lines grown in nude mice.
| CT values, mean ± SEM
|
Ratio
|
||||
|---|---|---|---|---|---|
| Samples | pGHRH-R | GHRH | β-actin | pGHRH-R | GHRH |
| HT | 32.05 ± 1.75 | 36.40 ± 1.05 | 13.50 ± 0.35 | 0.478 | 0.60 |
| RL | 32.38 ± 0.14 | 31.85 ± 0.29 | 12.58 ± 0.12 | 0.204 | 7.48 |
| MiaPaCa-2 | 37.75 ± 0.88 | 35.83 ± 0.44 | 12.10 ± 0.19 | 0.003 | 0.35 |
| H82 | 34.82 ± 0.35 | 33.05 ± 0.09 | 12.95 ± 0.27 | 0.045 | 4.19 |
| H345 | 33.80 ± 0.16 | 19.33 ± 0.08 | 12.45 ± 0.37 | 0.085 | 37817.6 |
| DBTRG-05 | 29.00 ± 0.62 | 33.58 ± 0.22 | 11.35 ± 0.20 | 1 | 1 |
| Prostate | 38.47 ± 1.04 | 37.46 ± 0.49 | 16.60 ± 0.01 | 0.000003 | 0.044 |
| Kidney | 30.93 ± 0.09 | 36.80 ± 0.90 | 16.15 ± 0.11 | 0.000472 | 0.051 |
| Lung | 33.63 ± 0.04 | 37.38 ± 0.23 | 15.45 ± 0.11 | 0.000043 | 0.021 |
| Liver | 37.43 ± 1.19 | 38.40 ± 0.21 | 17.65 ± 0.03 | 0.000012 | 0.046 |
| Pituitary | 23.00 ± 0.08 | 35.40 ± 0.40 | 19.15 ± 0.03 | 1 | 1 |
The relative gene expression ratio values were calculated based on ref. 24.
The highest expression of the GHRH gene was observed in the H-345 SCLC, and it was ≈37,800 times higher than in the DBTRG-05 tumors. We noted similar results for H-69 SCLC (data not shown). For the other tumor tissues, we observed that RL showed the highest mRNA expression for GHRH, followed by H-82, DBTRG-05, HT, and, finally, MiaPaCa-2 (Table 3). Among the normal human samples, the highest mRNA expression for GHRH was found for the pituitary, followed by kidney, liver, prostate, and lung.
Production of Antisera to the pGHRH-R. Immunization of four rabbits with pGHRH-R-hapten linked to BSA or keyhole limpet hemocyanin produced 32 batches of antisera that were then evaluated for binding to the radiolabeled hapten. All batches showed a significant binding to the radiolabeled hapten at a final dilution of 1:56,000. The antisera prepared by the keyhole limpet hemocyanin method (JH-2321 and JH-2322 series) were superior to those prepared by the BSA method (JH-2319 and JH-2320 series). The best antisera were also tested at higher dilutions and still produced high or acceptable binding values at dilutions ranging up to 1:448,000 (Table 5, which is published as supporting information on the PNAS web site). Antisera JH-2321/5 and JH-2321/7 were then purified by affinity chromatography and used for the Western blotting assays.
The detectability of pGHRH-R-hapten and the crossreaction with unrelated peptides were estimated (25, 28) by using the JH-2321/5 batch at the final dilution of 1:210,000. The standard curve of the RIA was set up in the range of 2 fmol per tube to 4,000 fmol per tube and showed the following characteristics: binding of the antiserum to the tracer (percentage of bound radioactivity to total radioactivity) was 41.2%. The minimal detectable dose was 2 fmol (5.69 pg per tube) for pGHRH-R-hapten, based on the dose estimated by a 95% confidence limit for the cpm of the zero standard. Nonspecific binding was 1.2%. The coefficient of correlation was 0.9972. Interassay variation was <15%, and intraassay variation was <10%. We did not find a significant crossreaction with [Ala-23]SV1(1-25)-Tyr-Cys-NH2 peptide (SV1-SV2-SV4-hapten) (0.033%) or with the peptide [Ala-48]SV1(45-66)-Cys-NH2 equivalent to [Ala-112]pGHRH-R(109-130)-Cys-NH2 (pGHRH-R-SV1-SV2-SV4-hapten) (0.027%) (25). No detectable crossreactivity was found with luteinizing hormone-releasing hormone, somatostatin, [Tyr-4]bombesin, and GHRH(1-29)NH2. Antiserum JH-2321/5 appeared to be specific for pGHRH-R-hapten.
Western Blotting. Preliminary studies based on RIA and Western blot assays have shown that the antisera batches 2321/5 for pGHRH-R and 2317/7 for SVs SV1-SV2-SV4 were more specific (25). The purified antiserum for SV1-SV2-SV4 recognized a 39.5-kDa band in the protein fraction obtained from the whole-tissue homogenate of HT and RL lymphomas, DBTRG-05 glioblastoma, MiaPaCa-2 pancreatic tumor, and H-82 and H-345 SCLC (Fig. 1A). No bands of similar molecular size were detected in the human NSCLC specimens H-1, H-2, and H-4 (Fig. 1A).
Normal human tissues were also probed with the SV1-SV2-SV4 antiserum. A band at 39.5 kDa consistent with the size of SV1 protein was found in pituitary and prostate but not in the kidney, lung, and liver (Fig. 2A). In the lung, we detected two more bands at 10 and 24 kDa. The 10-kDa band could correspond to SV4 based on its predicted molecular mass (18, 25). The immunoreactive signals in the pituitary and lung were abolished after incubation with the blocking peptide SV1-SV2-SV4-hapten (Fig. 2B). However, the signal in the prostate tissue was not weakened and, consequently, it is not clear whether it represents the receptor protein encoded by SV1 (Fig. 2B).
The same tumor samples and normal human tissues were incubated with the purified antiserum for pGHRH-R. A band at 47.5 kDa possibly corresponding to the pGHRH-R was observed in all of the tumor samples except for the H-4 human NSCLC (Fig. 1B). Similarly, a band at 47.5 kDa was detected in the normal human pituitary and prostate tissues but not in the kidney, lung, or liver (Fig. 2C). The specific signals at 47.5 kDa were diminished in the presence of blocking peptide pGHRH-R-hapten (Fig. 2D). We also observed the presence of bands at 65 and 98 kDa in some cancer samples (data not shown) and normal tissues (Fig. 1C).
We also investigated the protein expression of GHRH peptide in the tumor models and in normal human kidney, lung, prostate, and liver. All of the proteins expressed a band at 17.2 kDa, except for H-82 SCLC (Fig. 1C). The specificity of the bands in all Western blots was confirmed by the significant decrease in the specific signals when the blots were probed with the specific blocking peptides (a representative example is shown on Fig. 2 B and D).
Radioligand Binding Studies. The results of binding studies are shown in Table 4. All samples of xenografted tumor tissues showed specific high-affinity and low-capacity binding sites for JV-1-42 ligand. The highest and lowest capacities of the binding sites for JV-1-42 were found in MiaPaCa-2 and DBTRG-05 tumors, respectively. Using [His-1, Nle-27]hGH-RH(1-32)NH2, we detected specific high-affinity and low-capacity binding sites in all tumors models tested, except for MiaPaCa-2. In accord with the levels of receptor protein detected by Western Blot, the highest receptor concentration was found in H-345 tumors and the lowest concentration was found in DBTRG-05 model.
Table 4. Binding characteristics of the radiolabeled GHRH antagonist JV-1-42 and GHRH analog [His1, Nle27]hGH-RH(1-32)NH2 for human cancer lines grown in nude mice.
| JV-1-42
|
[His1, Nle27]hGH-RH(1-32)NH2
|
|||
|---|---|---|---|---|
| Tumors | Kd | Bmax | Kd | Bmax |
| HT | 6.95 ± 0.21 | 384.7 ± 1.45 | 8.75 ± 0.16 | 290.8 ± 13.5 |
| RL | 4.11 ± 0.04 | 301.2 ± 19.5 | 4.60 ± 0.21 | 203.0 ± 5.6 |
| MiaPaCa-2 | 8.24 ± 0.40 | 523.3 ± 24.4 | ND | ND |
| H82 | 5.25 ± 0.21 | 364.2 ± 10.6 | 12.1 ± 0.95 | 320.9 ± 19.2 |
| H345 | 9.15 | 499.3 | 4.92 | 1,158.2 |
| DBTRG-05 | 1.07 ± 0.09 | 156.6 ± 8.80 | 14.3 ± 0.15 | 79.7 ± 10.3 |
Shown are values for the dissociation constant, Kd, expressed in nM (mean ± SD) and maximal binding capacity, Bmax, expressed as fmol per mg of protein (mean ± SD). ND, not detected.
Discussion
In the course of the past 4 years, our laboratory has consistently detected the expression of four SVs of GHRH-Rs in various human cancer cell lines, in human tumor tissues grown in nude mice, and in human prostate cancer specimens (3, 6, 9, 15, 17, 18, 22, 23, 25). Recently, other groups also showed the expression of these four SVs of GHRH-R in human prostate cancer cell lines and specimens of adrenocortical carcinoma (16, 30). Among these SVs, SV1 has the highest structural homology with pGHRH-R (18). The failure of many attempts to show the expression of mRNA for pGHRH-R in neoplastic tissues tended to increase the support for the view that SV1 is the main functional receptor responsible for mediating the effects of GHRH and GHRH antagonists in tumors (6, 9, 12, 15, 16, 18, 23).
To date, a nested PCR protocol has been used for the detection of the SVs of pGHRH-R. This method consisted of two consecutive PCR reactions that could accomplish the amplification of cDNA for all four SVs at the sizes of 720, 566, 390, and 335 bp corresponding to SV1, SV2, SV3, and SV4, respectively (18, 22, 23). Even though SV1 is the only SV of GHRH-R that was proven to respond physiologically to GHRH binding (31, 32), we recently established a protocol that could detect the expression of each SV separately, without using the method of nested PCR. Nonetheless, our main objective was to determine whether the mRNA and protein for the pGHRH-R could be expressed in some tumor tissues. To validate our findings, we applied real-time PCR, Western blotting, and receptor binding assays to nonmalignant human tissues, six different xenografted tumors, and three human NSCLC specimens.
Both non-Hodgkin's lymphomas (HT and RL), DBTRG-05 glioblastoma, and samples of normal human kidney, liver, lung, prostate, and pituitary tissues expressed mRNA for all SVs. However, no mRNA expression of SV1 was detected in H-345 tumors, which showed only SV4. H-82 SCLC expressed only SV1, SV3 and SV4, and Mia-Pa-Ca-2 expressed only SV1 and SV2.
Real-time protocols permitted the quantification of mRNA expression. Our study reports the semiquantitative real-time data for message levels of the receptor SVs.
Western blot experiments using an affinity-purified antibody (25) specific for SV1, SV2, and SV4 showed that all six xenografted tumors expressed a protein of 39.5 kDa, likely corresponding to SV1. However, the 12.5% SDS/PAGE gel used in our Western blots shown in Fig. 1 may not be suitable for the detection of the proteins corresponding to SV2 and SV4, with the predicted molecular masses of 15 and 7 kDa, respectively. In H-345 SCLC, the presence of Western blot signal consistent with SV1 protein conflicts with the apparent lack of its mRNA expression. Probably an unknown technical problem related to the isolation of RNA and RT-PCR amplification occured in the H-345 model. Alternatively, it is possible that the antibody crossreacts with another protein of similar size, because part of SV1-SV2-SV4-hapten (LGRGKELWLESL) shows some homology with an internal sequence (LNRGKGLWLGSL) found in the human protein KIAA0877 (GI:51475828), with a predicted size of 44 kDa (see www.ncbi.nlm.nih.gov). However, previous studies showed that the immunostaining of HEC-1A human endometrial cancer cells was abolished when the expression of SV1 was ablated by SV1-antisense RNA (32). Moreover, MDA-MB-231 human breast cancer cells that do not express SVs did not show corresponding protein bands in the Western blots, whereas RL and HT lymphomas presented a predicted protein of 40 kDa, which would also correspond to SV1 (25). Thus, the specificity of this antiserum was demonstrated in refs. 25 and 32.
By applying the Western blotting method to the normal human samples of pituitary, lung, kidney, liver, and prostate, we could detect a protein at the expected molecular size of SV1 only in the pituitary and prostate. Because the mRNA for SV1 was found by RT-PCR in the normal tissues investigated, we suggest that the level of this protein in the lung, kidney, and liver is below the detection limit of the Western blotting assay. However, further studies are necessary to confirm this result.
Confirmatory results were obtained from binding assays performed by using a radioiodinated GHRH antagonist ligand, JV-1-42, which binds with high affinity for SV1 and pGHRH-R (22, 23, 33). This ligand did not show binding to the samples that express mRNA for only SV4 or SV2, but there was a direct binding correlation when SV1 was expressed (22). High-affinity binding sites for the JV-1-42 ligand were found in all six experimental tumor models tested. The maximal binding capacity for JV-1-42 in tumors should represent the sum of binding capacities for SV1 and pGHRH-R, because this ligand is not specific for distinguishing these two receptors.
Various attempts to find the mRNA expression for pGHRH-R in neoplastic extrapituitary tissues or binding sites for the radioligand [His-1, Nle-27]hGH-RH(1-32)NH2 have previously failed (12, 15, 18, 23). This radioligand shows specific high-affinity binding mainly to tissues that express the pGHRH-R but not to those that express SV1, because it has a relatively lower binding affinity for the latter receptor (18, 23). In the present study, we established a PCR protocol using the sensitive real-time PCR equipment for the detection of mRNA of pGHRH-R. In addition, we raised a polyclonal antibody that was affinity-purified and used it for the detection of pGHRH-R protein by Western blotting. This antibody is suitable for the selective detection of pGHRH-R because it was developed against the N-terminal amino acid sequence 23-45 of pGHRH-R that is not present in SVs (18, 25).
Our results show that the expression of mRNA and protein for pGHRH-R as well as binding sites with the characteristics of this receptor could be demonstrated in five of six tumor models that were not investigated so far. In MiaPaCa-2 pancreatic tumors, which were previously found negative for the expression of pGHRH-R (18) when using the nested PCR protocol, we could detect a low level of mRNA and protein expression, which was the lowest of the six tumor models tested. In accord with previous results, specific binding sites for pGHRH-R were not found in MiaPaCa-2 by radioligand assays (14, 18). The expression of pGHRH-R protein was also observed by Western blotting in two of three human NSCLC specimens tested. Among xenografted tumors, the DBTRG-05 glioblastoma showed the highest expression of mRNA for pGHRH-R but the lowest level of GHRH-R protein and lowest maximal binding capacity. It is possible that the mRNA expression of pGHRH-R in this particular tumor is not directly correlated with the level of its product. We also investigated by real-time RT-PCR and Western blotting the expression of mRNA for pGHRH-R and its protein product in normal human pituitary, kidney, lung, prostate, and liver. We observed the expression of mRNA for pGHRH-R in all these normal human tissues, but the expression of protein was detected only in the pituitary and prostate. Interestingly, the prostate had the lowest level of mRNA for pGHRH-R among the normal human tissues tested. Thus, it appears that the protein levels for pGHRH-R are not directly correlated with the mRNA levels.
The primers used in this study for the detection of pGHRH-R are not suitable for the differentiation between pGHRH-R and its SV, which is called transcript variant 2 and is described in ref. 34. However, this SV, the expression of which was shown in the pituitary at the mRNA levels (34), lacks the 86 aa at the C terminus compared with pGHRH-R and would be expressed as a protein ≈10 kDa smaller than the pGHRH-R. Because we did not find any bands of <47.5 kDa in the Western blots with an antibody specific for pGHRH-R, we can assume that the tumors and normal tissues investigated in this study express the pGHRH-R and not transcript variant 2.
In this study, we likewise established a real-time PCR protocol for GHRH and used it for the analysis of mRNA expression of this hormone in normal and cancerous human tissues. We also tested the presence of the GHRH peptide by Western blotting and detected a band of 17.2 kDa. This band was similar in size to that previously described (35) and could represent the GHRH prohormone. We demonstrated mRNA for GHRH and the putative GHRH precursor in prostate, liver, and lung, in addition to kidney, which showed the highest expression of GHRH among these normal peripheral organs. All six tumor models expressed mRNA for GHRH, and the GHRH precursor was also detectable, except in H82 SCLC. Once again, it appears that there is no complete correlation between mRNA and the corresponding protein levels in this particular tumor model. However, H345 SCLC expressed mRNA for GHRH and immunoreactive GHRH at a much higher level than the other cancers. It is well known that various carcinoid tumors in the bronchus, gastrointestinal tract, or pancreas secrete high amounts of GHRH (3, 35). The fact that a human SCLC cancer line, such as H345, expressed high amounts of mRNA for GHRH and immunoreactive GHRH suggests that a hypersecretion of GHRH similar to that described for other tumors (36-38) can also occur in SCLC.
Our findings shed more light on the status of GHRH-Rs in neoplastic tissues. Our work demonstrates the presence of pGHRH-Rs in human lung cancer specimens and some xenografted human tumor tissues and the fact that these receptors coexist with their SVs. Further studies are necessary to evaluate the physiological and pathophysiological significance of the coexpression of isoforms of GHRH-Rs in normal tissues and tumors. pGHRH-R and its SVs should be considered as potential targets for cancer therapy based on GHRH antagonists.
Supplementary Material
Acknowledgments
We thank Mr. Ferenc Rick for participating in later phases of real-time RT-PCR studies. This work is dedicated to the late Ana Maria ComaruSchally, M.D., F.A.C.P., who died recently from thyroid cancer, for her personal contribution to this project and for the inspiration she provided for the evaluation of human tumor samples.
Abbreviations: GH, growth hormone; GHRH, GH-releasing hormone; GHRH-R, GHRH receptor; pGHRH-R, pituitary GHRH-R; SV, splice variant; SCLC, small-cell lung cancer; NSCLC, non-SCLC; CT, threshold cycle; IGF, insulin-like growth factor.
References
- 1.Schally, A. V. (2003) in Peptides and Nonpeptides of Neuroendocrine and Oncological Relevance, ed. Muller, E. E. (Springer, Milan), pp. 83-98.
- 2.Schally, A. V. & Varga, J. L. (1999) Trends Endocrinol. Metab. 10, 383-391. [DOI] [PubMed] [Google Scholar]
- 3.Schally, A. V., Comaru-Schally, A. M., Nagy, A., Kovacs, M., Szepeshazi, K., Plonowski, A., Varga, J. L. & Halmos, G. (2001) Front. Neuroendocrinol. 22, 248-291. [DOI] [PubMed] [Google Scholar]
- 4.Szepeshazi, K., Schally, A. V., Groot, K., Armatis, P., Hebert, F. & Halmos, G. (2000) Eur. J. Cancer 36, 128-136. [DOI] [PubMed] [Google Scholar]
- 5.Csernus, V. J., Schally, A.V., Kiaris, H. & Armatis, P. (1999) Proc. Natl. Acad. Sci. USA 96, 3098-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busto, R., Varga, J. L., Garcia-Fernandez, O. M., Armatis, P. & Szepeshazi, K. (2002) Proc. Natl. Acad. Sci. USA 99, 11866-11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szepeshazi, K., Schally, A. V., Groot, K., Armatis, P., Halmos, G., Hebert, F., Szende, B., Varga, J. L. & Zarandi, M. (2000) Br. J. Cancer 82, 1724-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiaris, H., Schally, A. V., Varga, J. L., Groot, K. & Armatis, P. (1999) Proc. Natl. Acad. Sci. USA 96, 14894-14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatzistamou, I., Schally, A. V., Varga, J. L., Groot, K., Busto, R., Armatis, P. & Halmos, G. (2001) Anticancer Drugs 12, 761-768. [DOI] [PubMed] [Google Scholar]
- 10.Kiaris, H., Schally, A. V. & Varga, J. L. (2000) Cancer Lett. 161, 149-155. [DOI] [PubMed] [Google Scholar]
- 11.Kineman, R. D. (2000) Proc. Natl. Acad. Sci. USA 97, 532-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahán, Z., Varga, J. L., Schally, A. V., Rekasi, Z., Armatis, P., Chatzistamou, I., Czompoly, T. & Halmos, G. (2000) Breast Cancer Res. Treat. 60, 71-79. [DOI] [PubMed] [Google Scholar]
- 13.Kahan, Z., Arencibia, J. M., Csernus, V. J., Groot, K., Kineman, R. D., Robinson, W. R. & Schally, A. V. (1999) J. Clin. Endocrinol. Metab. 84, 582-589. [DOI] [PubMed] [Google Scholar]
- 14.Rekasi, Z., Varga, J. L., Schally, A. V., Halmos, G., Armatis, P., Groot, K. & Czompoly, T. (2000) Endocrinology 141, 2120-2128. [DOI] [PubMed] [Google Scholar]
- 15.Chatzistamou, I., Schally, A. V., Varga, J. L., Groot, K., Armatis, P., Busto, R. & Halmos, G. (2001) J. Clin. Endocrinol. Metab. 86, 2144-2152. [DOI] [PubMed] [Google Scholar]
- 16.Chopin, K. L. & Herington, A. C. (2001) Prostate 49, 116-121. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Fernandez, O. M., Schally, A. V., Varga, J. L., Groot, K. & Busto, R. (2003) Breast Cancer Res. Treat. 77, 15-26. [DOI] [PubMed] [Google Scholar]
- 18.Rekasi, Z., Czompoly, T., Schally, A. V. & Halmos, G. (2000) Proc. Natl. Acad. Sci. USA 97, 10561-10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frohman, L. A. & Kineman, R. D. (1999) in Handbook of Physiology: Hormonal Control of Growth, eds. Kostyo, J. L. & Goodman, H. M. (Oxford Univ. Press, New York), pp. 189-221.
- 20.Reubi, R. C. (1995) J. Nucl. Med. 36, 1846-1853. [PubMed] [Google Scholar]
- 21.Moody, T. W. (1996) Peptides 17, 545-555. [DOI] [PubMed] [Google Scholar]
- 22.Halmos, G., Schally, A. V., Czompoly, T., Krupa, M., Varga, J. L. & Rekasi, Z. (2002) J. Clin. Endocrinol. Metab. 87, 4707-4714. [DOI] [PubMed] [Google Scholar]
- 23.Halmos, G., Schally, A. V., Varga, J. L., Plonowski, A., Rekasi, Z. & Czompoly, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaffl, M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toller, G. L., Horvath, J. E., Schally, A. V., Halmos, G., Varga, J. L., Groot, K., Chism, D. & Zarandi, M. (2004) Proc. Natl. Acad. Sci. USA 101, 15160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaylinn, B. D., Harrison, J. K., Zysk, J. R., Lyons, C. E., Jr., Lynch, K. R. & Thorner, M. D. (1993) Mol. Endocrinol. 7, 77-84. [DOI] [PubMed] [Google Scholar]
- 27.Gaylinn, B. D., DeAlmeida, V. I., Lyons, C. E., Jr., Wu, K. C., Mayo, K. E. & Thorner, M. O. (1999) Endocrinology 140, 5066-5074. [DOI] [PubMed] [Google Scholar]
- 28.Groot, K., Horvath, J., Cai, R. Z. & Schally, A. V. (1995) Int. J. Peptide. Protein Res. 45, 561-566. [DOI] [PubMed] [Google Scholar]
- 29.Halmos, G., Rekasi, Z., Szoke, B. & Schally, A.V. (1993) Receptor 3, 87-97. [PubMed] [Google Scholar]
- 30.Freddi, S., Arnaldi, G., Fazioli, F., Scarpelli, M., Appolloni, G., Mancini, T., Kola, B., Bertagna, X., Mantero, F., Collu, R. & Boscaro, M. (2005) Clin. Endocrinol. 62, 533-538. [DOI] [PubMed] [Google Scholar]
- 31.Kiaris, H., Schally, A. V., Busto, R., Halmos, G., Artavanis-Tsakonas, S. & Varga, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiaris, H., Chatzistamou, I., Schally, A. V., Halmos, G., Varga, J., Koutselini, H. & Kalofoutis, A. (2003) Proc. Nat. Acad. Sci. USA 100, 9512-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varga, J. L., Schally, A. V., Csernus, V. J., Zarandi, M., Halmos, G., Groot, K. & Rekasi, Z. (1999) Proc. Natl. Acad. Sci. USA 96, 692-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang, J., Lagace, G., Castagne, J. & Collu, R. (1995) J. Clin. Endocrinol. Metab. 80, 2381-2387. [DOI] [PubMed] [Google Scholar]
- 35.Othman, N. H., Ezzat, S., Kovacs, K., Horvath, E., Poulin, E., Smyth, H. S. & Asa, S. L. (2001) Clin. Endocrinol. 55, 135-140. [DOI] [PubMed] [Google Scholar]
- 36.Melmed, S. (1993) in Endocrine Tumors, eds. Mazzaferri, E. L. & Samaan, N. A. (Blackwell Scientific, Oxford), pp. 113-122.
- 37.Strewler, G. J. (1998) in Williams Textbook of Endocrinology, eds. Wilson, J. D., Foster, D. W., Kronenberg, H. M. & Larsen, P. R. (Saunders, Philadelphia), pp. 1693-1710.
- 38.Szabo, M. & Frohman, L. A. (1981) in Hormones in Normal and Abnormal Human Tissues, eds. Fotherby, K. & Pal, S. B. (de Gruyter, New York), pp. 415-535.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.