Abstract
Integrase (IN) is an essential retroviral enzyme, and human transcriptional coactivator p75, which is also referred to as lens epithelium-derived growth factor (LEDGF), is the dominant cellular binding partner of HIV-1 IN. Here, we report the crystal structure of the dimeric catalytic core domain of HIV-1 IN complexed to the IN-binding domain of LEDGF. Previously identified LEDGF hotspot residues anchor the protein to both monomers at the IN dimer interface. The principal structural features of IN that are recognized by the host factor are the backbone conformation of residues 168–171 from one monomer and a hydrophobic patch that is primarily comprised of α-helices 1 and 3 of the second IN monomer. Inspection of diverse retroviral primary and secondary sequence elements helps to explain the apparent lentiviral tropism of the LEDGF-IN interaction. Because the lethal phenotypes of HIV-1 mutant viruses unable to interact with LEDGF indicate that IN function is highly sensitive to perturbations of the structure around the LEDGF-binding site, we propose that small molecule inhibitors of the protein–protein interaction might similarly disrupt HIV-1 replication.
Keywords: integration, structure, retrovirus, transcription, host factor
Like all retroviruses, HIV-1 must integrate a reverse-transcribed copy of its viral RNA genome into a host cell chromosome to establish a productive infection. Integration is mediated by the viral integrase (IN) protein acting on the DNA attachment sites at the ends of the linear reverse transcript. IN acts within the context of a higher-order preintegration complex (PIC) that is derived from the core of the infecting virion. IN catalyzes two sequential reactions, initially removing 3′ terminal GT nucleotides from both ends of HIV-1 cDNA. After nuclear entry, IN inserts the processed 3′ termini into opposing strands of chromosomal DNA. Repair of single-strand gaps by host cell enzymes completes the integration process (for a review, see ref. 1). In addition to IN, other viral and numerous cellular proteins are believed to play auxiliary roles in HIV-1 PIC assembly, transport into the nucleus, and targeting integration into transcriptionally active regions of chromatin (2–4).
HIV-1 IN is a 32-kDa protein composed of three structural domains (5, 6). The N-terminal domain (NTD; residues 1–49) is a helical bundle stabilized by the coordination of a single zinc atom (7). The catalytic core domain (CCD; residues 50–212) belongs to a superfamily of DNA/RNA strand transferases/nucleases that in addition to retroviral INs includes divergent bacterial transposases, Holiday junction resolvase RuvC, and RNase H (8, 9). Several independent retroviral IN CCD structures reveal a similar dimer interface, strongly arguing that it is physiologically relevant. The CCD contains the essential active site residues, Asp-64, Asp-116, and Glu-152 in HIV-1, collectively referred to as the DDE triad. The carboxyl-terminal domain (residues 213–288), which is the least conserved among divergent retroviruses, possesses an Src homology 3-like fold (10, 11). Each domain contributes to HIV-1 IN multimerization and is essential for 3′ processing and DNA strand transfer activities (6, 12–17). Although the functional multimeric state of IN in vivo is not known, when extracted from human cells, the HIV-1 protein is a stable tetramer (18).
Ectopically expressed HIV-1 IN accumulates in the nuclei of human cells (19, 20), associates with chromatin (21), and forms a tight complex with endogenous transcriptional coactivator p75 (2, 18), which is also referred to as lens epithelium-derived growth factor (LEDGF). Binding to LEDGF appears to account for the characteristic intracellular distribution of IN. RNA interference-mediated depletion of LEDGF or overexpression of a nuclear localization-defective mutant of LEDGF disrupted nuclear/chromosomal localization of HIV-1 IN (22–24). Conversely, the interaction-deficient IN mutant Q168A was excluded from condensed chromosomes (25). Furthermore, interaction with LEDGF protected HIV-1 IN from degradation through the ubiquitin-proteasome pathway (26). So far, LEDGF was demonstrated to bind the INs from HIV-1, HIV-2, and the divergent lentivirus feline immunodeficiency virus (FIV), but not the INs from the gammaretrovirus Moloney murine leukemia virus, alpharetrovirus Rous sarcoma virus (RSV), or deltaretrovirus human T cell leukemia virus (2, 18, 24, 27). Thus, binding to LEDGF seems to be a conserved property of lentiviral INs. Consistent with a potential role in lentiviral replication, LEDGF cofractionated with PICs isolated from HIV-1- or FIV-infected cells (24).
LEDGF binds HIV-1 IN via a small, approximate 80-residue IN-binding domain (IBD) within its C-terminal region (28, 29). The IBD is both necessary and sufficient for the interaction with HIV-1 IN (28). The three-dimensional structure of the IBD was recently determined by NMR spectroscopy, revealing an α-helical domain that is topologically similar to a pair of HEAT repeats (30). To differentiate it from HEAT repeat proteins that tend to be large and contain numerous repeated elements (31), the IBD was classified as a pseudo-HEAT repeat analogous topology (PHAT) domain, similar to the PHAT domain in Drosophila Smaug protein (30, 32). Residues Ile-365, Asp-366, and Phe-406, located in the interhelical loops within the HEAT repeats, play critical roles in HIV-1 IN recognition (30).
The CCD of IN possesses the main determinants for interacting with LEDGF, although the NTD increased the affinity of the interaction (22). Mutations within the CCD that disrupted the interaction collectively pointed to residues 165–173 as an approximate location for the interaction site on IN (2, 25, 30). The failure of these IN mutants to bind LEDGF correlated with catastrophic preintegration blocks of the corresponding mutant viruses, although the INs notably retained catalytic function. One proposed role for LEDGF in lentiviral replication is targeting PICs to transcriptionally active regions of chromatin during integration (3, 4).
To understand the structural basis for the interaction between HIV-1 IN and LEDGF, we crystallized and determined the structure of the IN CCD dimer in complex with the LEDGF IBD. The structure elucidates the mode of recognition between these proteins and reveals a potential target site on IN for the design of small molecule inhibitors of the LEDGF–IN interaction.
Materials and Methods
Recombinant Proteins. The HIV-1 IN CCD comprising residues 50–212 and containing the F185K-solubilizing mutation was produced in a His-6-tagged form and purified by affinity chromatography as described (8). The tag was removed by digestion with thrombin (Sigma-Aldrich), and the protein was further purified by gel filtration through a Superdex-200 column (Amersham Pharmacia Biosciences) in 0.5 M NaCl, 0.5 mM EDTA, and 25 mM Hepes, pH 7.2. The LEDGF IBD containing residues 347–471 was prepared as described (30). To remove 29 residues of the unstructured C terminus, LEDGF347–471 diluted to 4–6 mg/ml in 120 mM NaCl and 25 mM Tris, pH 7.4 was digested with 25 μg/ml trypsin (28). After 3-h digestion at room temperature, the protease was removed by adsorption to benzamidine Sepharose (Amersham Pharmacia Biosciences). LEDGF347–442 was further purified by chromatography on SP Sepharose (30) followed by gel filtration through Superdex-200 in 100 mM NaCl and 50 mM Na2HPO4, pH 7.2. Purified IN50–212(F185K) and LEDGF347–442 proteins were concentrated to 8–10 mg/ml, supplemented with 10 mM DTT and 10% glycerol, flash-frozen in liquid nitrogen, and stored at –70°C. For crystallization, the protein complex was prepared by mixing IN50–212(F185K) and LEDGF347–442 at a 1:2 molar ratio and final protein concentration of 6–10 mg/ml. The buffer was exchanged to 250 mM NaCl, 10 mM DTT, 0.4 mM EDTA, 20 mM Hepes, and 50 mM Na2HPO4, pH 7.0 by using BioSpin-6 columns (BioRad). Protein concentrations were determined by using the Bradford assay.
Crystallization and Structure Determination. Initial crystalline precipitates were obtained in condition 42 of Crystal Screen I (Hampton Research, Riverside, CA). The optimized well solution was 18% PEG-3350, 45.9 mM Na2HPO4, 21.6 mM NaH2PO4, and 157.5 mM KH2PO4. Crystals were grown in hanging drops by vapor diffusion at 20.5°C. Drops were made by mixing 1 μl of 8.3 mg/ml protein and 1 μl of the well solution. After growth for 5–8 days, the crystals were ≈1,000 × 90 × 50 μm. Before x-ray data collection, crystals were soaked in a cryo-protectant solution containing 22% glycerol, 16% PEG-3350, 40.8 mM Na2HPO4, 19.2 mM NaH2PO4, and 140 mM KH2PO4 for 3–5 min and flash-frozen in liquid nitrogen. X-ray diffraction data extending to 2.0-Å resolution were collected at 100 K on the X26C beam line (λ = 1.00 Å) at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) with an ADSC (Poway, CA) Quantum 4 Detector and processed with the hkl2000 package (33). The crystals belonged to the C121 space group with unit cell dimensions a = 122.42 Å, b = 60.59 Å, c = 71.13 Å, and β = 109.06°. A Matthews coefficient of 2.1 Å3·Da–1 (solvent content of ≈42%) is consistent with two IN CCDs, and probably two LEDGF molecules, in the asymmetric unit. Detailed data collection statistics and parameters are summarized in Table 1.
Table 1. Statistics for data collection and refinement.
| Data collection | |
| Space group | C121 |
| Unit cell a, b, c (Å), β (°) | 122.4, 60.6, 71.1, 109.1 |
| Resolution range | 67.27-2.02 (2.09-2.02) |
| No. of unique reflections | 32,136 (3,189) |
| Average multiplicity | 2.4 (2.3) |
| Completeness, % | 99.2 (99.2) |
| Rmerge, % | 7.2 (47) |
| I/σI | 18.9 (3.1) |
| Wilson 〈B 〉, Å2 | 35.2 |
| Solvent content, % | 41.9 |
| Refinement | |
| Resolution range, Å | 20-2.02 |
| Total no. of reflections used | 30,486 (2,223) |
| No. of reflections in working set | 28,859 |
| No. of reflections for Rfree calculation | 1,627 |
| R, % | 18.0 (21.4) |
| Rfree, % | 22.6 (27.4) |
| 〈B 〉/rms delta-B for bonded atoms, Å2 | 37.4/1.85 |
| rms deviation from ideal bond length, Å | 0.017 |
| rms deviation from ideal angle, ° | 1.602 |
Values in parentheses are for the highest-resolution shell.
The structure of the complex was determined with the molrep module of the ccp4 software suit (34) by molecular replacement methods using the structure of the IN50–212(F185K/W131E) [chain A, Protein Data Bank (PDB) code 1BIS, ref. 35] as a search model and data in the range of 15- to 3.5-Å resolution. An unambiguous solution was found with a correlation coefficient of 34.5% (18.5% for the next best hit) and an R factor of 47.9%. Visual inspection of the Fourier 2Fo – Fc and Fo – Fc maps showed a high-quality initial map for both IN chains and also revealed clear additional electron density for the two molecules of the LEDGF IBD, strongly suggesting the identification of the correct molecular replacement solution. The positions of the LEDGF subunits were found with molrep by fixing the solutions of the IN structure and using residues 348–428 of the lowest energy model from the NMR structure (PDB ID code 1ZE9, ref. 30) as a search model. Rigid body and simulated annealing model refinement were performed with the cns 1.1 package (36), followed by restrained positional, B factor, translation/libration/screw (TLS) refinement modes in refmac 5.0 (37, 38). Noncrystallographic symmetry restraints, used during the initial steps of refinement, were removed for the final steps of restrained TLS refinement. Residues 143–153 of IN chain B, two phosphate ions, four glycerol molecules, and 219 sites treated as water oxygens were modeled to fit the 2Fo – Fc and Fo – Fc maps by using programs o (39) and coot (40). In the refined structure, 95% of residues were in the most favorable regions of the Ramachandran plot, and the remaining 5% were in additionally allowed regions. Figs. 1 and 2C and Fig. 4, which is published as supporting information on the PNAS web site, were drawn with pymol (W. L. DeLano, www.pymol.org). Figs. 2 A and B and 3 were prepared with the program molmol (41).
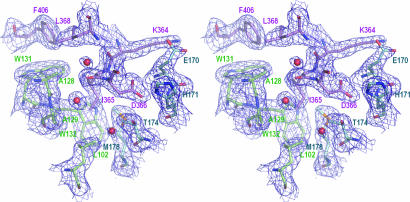
Fig. 1.
Electron density map for residues at the CCD–IBD interface. Selected interface residues are shown as sticks. The final 2Fo – Fc electron density map (dark blue) is at a 1-σ contour level. The color code for carbon atoms is: violet, LEDGF chain C; blue, IN chain A; green, IN chain B. Three water molecules buried at the protein interface are shown as red spheres.
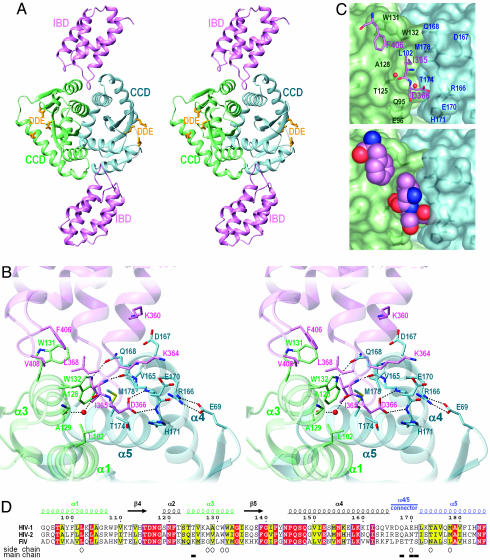
Fig. 2.
Molecular mechanism of the IN–LEDGF interaction. (A) The overall structure of the CCD–IBD complex. IN chains A and B are colored blue and green, respectively; the IBD subunits are violet. The side chains of the DDE catalytic triad are shown as yellow sticks. (B) Key intermolecular contacts at the CCD–IBD interface. Selected residues are shown as sticks. The water molecule hydrogen-bonded to main-chain carbonyl groups of LEDGF residue Ile-365 and IN residue Thr-125 is shown as a red sphere. Hydrogen bonds discussed in the text and the salt bridge between IN residues Glu-69 and Arg-166 are indicated by dotted lines. (C) The pocket at the CCD dimer interface. LEDGF hotspot residues Ile-365, Asp-366, and Phe-406 are shown as sticks (Upper) or in space-fill mode (Lower). The IN subunits are shown as semitransparent surfaces. Selected IN residues are indicated. (D) Sequence alignment of HIV-1, HIV-2, and feline immunodeficiency virus (FIV) INs. Identical residues are white on red background; residues with conserved properties are bold on yellow background. Residue numbering, secondary structure elements, and the position of the α4/5 connector in HIV-1 IN are shown above the alignment; structural elements are colorized as in A–C. Open circles and filled boxes under the alignment indicate residues that make contacts to the LEDGF IBD through side-chain and main-chain atoms, respectively. The alignment was printed by using espript-2.2 (56).
Fig. 3.
Molecular details at the dimer interfaces of HIV-1 and RSV IN. (A) Comparison of the IN CCD in complex with LEDGF to the unliganded CCD structure. The structure of the CCD–IBD complex was superimposed onto the structure of the free CCD from ref. 45 (PDB code 1BL3). The color code for IN chains A and B is the same as in Figs. 1 and 2; the unliganded structure is painted brown. Selected side chains are shown as sticks. (B) Structure of the corresponding region of RSV IN (PDB code 1C0M, ref. 48). Chain A is painted green; chain B is blue; and the loop in chain B connecting α4 and α5 is shown in pink. Side chains of exposed hydrophobic and aromatic residues at the interface are shown as sticks.
Results and Discussion
Crystallization and Structure Determination of the IN–LEDGF Complex. The CCD of HIV-1 IN (residues 50–212) is both necessary and sufficient for the interaction of IN with LEDGF (22). Furthermore, the F185K mutation, which greatly improved the solubility of recombinant HIV-1 IN protein (42), did not significantly affect the binding affinity of LEDGF (30). Consistent with these observations, the results of sedimentation equilibrium and gel filtration experiments revealed that IN50–212(F185K) readily formed a complex with the LEDGF IBD (data not shown). Crystals of the IN50–212(F185K)–LEDGF347–442 complex diffracted x-rays to ≈2-Å resolution at a synchrotron light source. The structure of the complex was solved through molecular replacement and refined to 2.02 Å with an R factor of 18.0% and an Rfree of 22.6% (Table 1). A fragment of the final electron density map showing selected residues at the CCD–IBD interface is shown in Fig. 1. The color scheme of blue and green for chains A and B of the IN dimer, respectively, and violet for the LEDGF IBD, is used throughout this work.
The asymmetric unit of the crystals contained a dimer of IN CCDs (chains A and B) and a pair of LEDGF IBD molecules (chains C and D) bound at the CCD dimer interface (Fig. 2 A). The structure has pseudo 2-fold symmetry, with the two CCD–IBD interfaces being nearly identical. The interface between the IN dimer and the upper IBD subunit as displayed in Fig. 2 A will be discussed in detail below. The CCD and IBD structures within the complex were very similar to the structures previously reported for the individual partners (8, 30). For example, the CCD formed a dimer with an extensive interface (Fig. 2 A). As previously observed in one HIV-1 structure (IN1–212, PDB code 1K6Y, ref. 43), an ordered phosphate ion is bound within each of the two active sites of the CCD dimer, coordinated by IN residues Thr-66, His-67, and Lys-159 (Fig. 4). The observed identical coordination of the phosphate ion in both structures would suggest that its position could reflect the site of coordination of a DNA backbone phosphate (43).
The IN–LEDGF Interface. The CCD–IBD interface buries ≈1,280 Å2 of protein surface. Helices α1 and α3 from the IN B chain plus α5 and the six-residue connector linking helices α4 and α5 (residues 166–171, further referred to as the α4/5 connector) from the A chain of IN form the foundation of the LEDGF-binding site (secondary structure elements of the IN CCD are numbered as in ref. 8) (Figs. 1 and 2). In agreement with the results of Ala-scanning mutagenesis and NMR studies (30), it is the interhelical loops of the IBD that are responsible for the interaction with IN.
We reported that mutation of LEDGF residue Ile-365, Asp-366, or Phe-406 ablated the interaction with HIV-1 IN and that substitution of Val-408 significantly reduced binding (30). Inspection of the CCD–IBD structure explains the molecular bases for these observations, as each of these LEDGF residues directly contacts IN in the crystal structure (Fig. 2 B and C). The side chain of LEDGF residue Ile-365 projects into a hydrophobic pocket formed by IN B-chain residues Leu-102, Ala-128, Ala-129, and Trp-132 and A-chain residues Thr-174 and Met-178. LEDGF residues Phe-406 and Val-408 contact and occlude solvent from the exposed Trp-131 in the IN B chain. Finally, LEDGF Asp-366 makes a bidentate hydrogen bond to the main-chain amides of IN residues Glu-170 and His-171 in chain A (Fig. 2B). LEDGF D366A and D336N mutants failed to bind IN (30), indicating that this double hydrogen bond is essential for recognition. These interactions presumably neutralize the negative charge of the Asp-366 carboxylate in an environment that is largely excluded from solvent. Furthermore, the backbone amide of LEDGF residue Ile-365 makes a hydrogen bond to the backbone carbonyl group of Gln-168 in chain A (Fig. 2B). Of the three water molecules that are buried within the CCD–IBD interface (Fig. 1), one is ideally positioned to make bridging hydrogen bonds to the backbone carbonyl groups of LEDGF residue Ile-365 and IN B-chain residue Thr-125 (Fig. 2 B and C). A well defined salt bridge exists between Lys-364 of LEDGF and A-chain IN residue Glu-170 (Figs. 1 and 2B). In addition, Lys-360 of LEDGF is in close proximity to A-chain residue Asp-167. However, because mutant proteins substituted with Ala at either Lys-364 or Lys-360 retained binding to IN (30), these residues would not appear to contribute significantly to overall binding affinity.
The extent of molecular surface buried at the CCD–IBD interface (1,280 Å2) is slightly smaller than the average area of protein–protein interface (1,600 Å2) compiled from a diverse group of crystallized protein complexes (44). However, the CCD–IBD interface is likely to represent a portion of the total contact area between full-length IN and LEDGF proteins. Although the CCD suffices for binding to LEDGF (22), the NTD H12N mutation or deletion of the entire Zn2+-binding domain of IN greatly reduced the affinity of the interaction (22), suggesting that the NTD contributes to interactions with LEDGF.
Structural Basis for the Recognition of Lentiviral INs by LEDGF. Extensive hydrophobic interactions and a hydrogen-bond network engaging hotspot residues of LEDGF (Fig. 2B) elucidated two key features of HIV-1 IN that are complementary to and recognized by this host factor: (i) the specific backbone conformation of α4/5 connector residues 168–171 and (ii) a hydrophobic patch accommodating the side chains of LEDGF residues Ile-365, Phe-406, and Val-408. In addition to these anchoring interactions, contacts with the CCD, and likely with the NTD, no doubt contribute to the overall affinity of the protein–protein interaction. A critical reliance on the conformation of the IN backbone in the buried protein interface explains why LEDGF can bind to diverse lentiviral INs that are expected to have similar protein folds despite their limited sequence conservation (Fig. 2D).
We and others reported that HIV-1 IN mutants V165A, R166A, L172A/K173A, Q168A, and Q168L failed to bind LEDGF (2, 25, 30). Although none of the residues altered by these mutations directly contacts the IBD in the crystal structure, we can predict that these amino acid substitutions are likely to corrupt the conformation of the α4/5 connector. For example, Arg-166 forms an intramolecular salt bridge with Glu-69, whereas the side chain of Gln-168 is hydrogen-bonded across the dimer interface to Trp-132 (Fig. 2B). Thus, both Arg-166 and Gln-168 are expected to be important for the structural integrity of the α4/5 connector. On the other hand, Val-165 and Leu-172 are buried within the IN core structure; mutating either of them to the smaller Ala residue is also expected to compromise local structure.
Typically, formation of protein complexes is not accompanied by major structural changes of the interacting proteins (44). Similarly, binding of the LEDGF IBD did not significantly alter the structure of the CCD (Fig. 3A). Rotation of the Trp-131 side chain was the only significant adjustment that occurred upon complex formation. In previous CCD structures that retained wild-type Trp at position 131, this exposed residue was involved in crystal packing interactions and its orientation varied from crystal to crystal (8, 45). On the other hand, the conformation of the α4/5 connector was preserved among all reported HIV-1 IN structures and in simian immunodeficiency virus IN (46). The LEDGF-binding site of HIV-1 IN therefore appears to adopt a stable conformation closely resembling that in the complex with LEDGF. In contrast, RSV IN, which does not bind to LEDGF (27), has an extended loop (RSV IN residues 176–182) connecting α4 and α5 that is pulled away from the potential interaction site (Fig. 3B). The conformation of this loop is preserved in the known RSV IN structures with accompanying low temperature factors of loop residues (47, 48), arguing that it reflects the native state of RSV IN. Several solvent-exposed aromatic and hydrophobic residues occupy the cleft between α4 and α5 of RSV IN (Fig. 3B). The shape and chemical features of this cleft raise the possibility of an interaction with some alternate binding partner.
Implications for IN Function and Inhibitor Design. LEDGF enhanced the in vitro enzymatic activity, solubility, and DNA-binding activity of HIV-1 IN (18, 27, 28). The interaction with LEDGF is essential for the association of ectopically expressed IN with chromatin (22, 24) and the stability of the viral protein in human cells (22, 26). Moreover, mutations within the IN α4/5 connector, which abolished interactions with LEDGF, cause catastrophic viral replication defects (25, 30, 49, 50). Intriguingly, the mutant INs retained enzymatic activity in vitro and when tested, efficiently cross-complemented the infectivity defects of IN active-site mutant viruses in cells (25, 49, 50). Although the crystal structure of the IN CCD–LEDGF IBD complex elucidates why these mutant proteins fail to bind LEDGF, we cannot unambiguously explain the replication deficiency of the corresponding mutant viruses. One obvious possibility is that the interaction with LEDGF is essential for HIV-1 replication (25). However, RNA interference-mediated depletion of endogenous LEDGF did not yield a concomitant reduction in HIV-1 replication (24), arguing against, although not ruling out, this hypothesis. Alternatively, the LEDGF-binding site on IN could mediate other essential protein–protein interactions during HIV-1 infection, such as contacts between IN protomers within higher-order complexes during PIC assembly.
Notwithstanding the final outcome of this riddle, the phenotypes of IN mutant viruses carrying changes in the α4/5 connector underscore the fact that the structural organization of the LEDGF-binding site has a profound effect on HIV-1 replication. The LEDGF-binding site pocket accommodates IBD residues Ile-365 and Asp-366 (Fig. 2C). We speculate that a small molecule inhibitor designed to fit into this pocket is likely to induce potent HIV-1 replication defects, similar to those observed for mutant viruses unable to interact with LEDGF. Persuasively, the potential inhibitor target site contains both strong nonpolar and hydrogen-bonding components (Fig. 2B). It might also be possible for an inhibitor with appropriate hydrogen-bonding groups to replace the water molecules buried at the LEDGF–IN interface. Because LEDGF binds to full-length IN even when the enzyme is in complex with its cognate DNA substrate (28, 30), we further speculate that the LEDGF-binding site may be available for drug binding during the early steps of viral replication.
Soaking IN CCD crystals in the presence of tetraphenylarsonium and its hydroxylated derivative resulted in binding of the compounds at the CCD dimer interface at a position that directly overlapped the LEDGF-binding site (51). Furthermore, acetylated analogs of l-chicoric acid inhibited HIV-1 IN activity (52) and modified Lys-173 (53), which is adjacent to the LEDGF-binding site. Because of the relatively low affinity of these compounds for IN, they did not interfere with formation of the IN-LEDGF complex (P.C. and A.E., unpublished observations). However, these known examples of small molecules that bind at or close to the LEDGF-interaction site strongly suggest that this site is suitable for interaction with drug-like compounds. It may be possible to identify molecules that bind to the LEDGF-binding site with high affinity and antagonize the protein interaction by using simple in vitro assays (22, 24, 28). Because mutations of the LEDGF-binding site did not drastically compromise the in vitro catalytic activities of HIV-1 IN (25, 49, 50), enzymatic assays are less likely to identify this class of drug. Recent successes in identifying highly specific small molecule inhibitors of protein–protein interactions (54, 55) encourage efforts to exploit virus–host protein interactions for the development of novel antiretroviral drugs.
Supplementary Material
Acknowledgments
We thank R. Craigie (National Institutes of Health, Bethesda), D. Hazuda (Merck Research Laboratories), and M. Kvaratskhelia (Ohio State University, Columbus) for sharing reagents; H. Aihara, L. Brieba, and T. Biswas (Harvard Medical School) for generous advice and helpful discussions; N. Vandegraaff, G. Maertens, and R. Lu for critical reading of the manuscript; and the staff of the X26C beam line at the National Synchrotron Light Source for assistance in x-ray data collection. This work was supported by National Institutes of Health Grants AI39394 and AI62249 (to A.E.) and GM59902 (to T.E.), and state of São Paulo Research Foundation Grant 03/00231-0 (to A.L.B.A.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CCD, catalytic core domain; IN, integrase; IBD, IN-binding domain; LEDGF, lens epithelium-derived growth factor; NTD, N-terminal domain; PDB, Protein Data Bank; PIC, preintegration complex; RSV, Rous sarcoma virus.
Data deposition: Atomic coordinates and diffraction data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2B4J).
References
- 1.Craigie, R. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol., Washington, DC), pp. 613–630.
- 2.Turlure, F., Devroe, E., Silver, P. A. & Engelman, A. (2004) Front. Biosci. 9, 3187–3208. [DOI] [PubMed] [Google Scholar]
- 3.Engelman, A. (2005) Proc. Natl. Acad. Sci. USA 102, 1275–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman, F. D. (2003) Cell 115, 135–138. [DOI] [PubMed] [Google Scholar]
- 5.Engelman, A. & Craigie, R. (1992) J. Virol. 66, 6361–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman, F. D., Engelman, A., Palmer, I., Wingfield, P. & Craigie, R. (1993) Proc. Natl. Acad. Sci. USA 90, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, M., Zheng, R., Caffrey, M., Craigie, R., Clore, G. M. & Gronenborn, A. M. (1997) Nat. Struct. Biol. 4, 567–577. [DOI] [PubMed] [Google Scholar]
- 8.Dyda, F., Hickman, A. B., Jenkins, T. M., Engelman, A., Craigie, R. & Davies, D. R. (1994) Science 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- 9.Rice, P. A. & Baker, T. A. (2001) Nat. Struct. Biol. 8, 302–307. [DOI] [PubMed] [Google Scholar]
- 10.Lodi, P. J., Ernst, J. A., Kuszewski, J., Hickman, A. B., Engelman, A., Craigie, R., Clore, G. M. & Gronenborn, A. M. (1995) Biochemistry 34, 9826–9833. [DOI] [PubMed] [Google Scholar]
- 11.Eijkelenboom, A. P., Lutzke, R. A., Boelens, R., Plasterk, R. H., Kaptein, R. & Hard, K. (1995) Nat. Struct. Biol. 2, 807–810. [DOI] [PubMed] [Google Scholar]
- 12.Vink, C., Oude Groeneger, A. M. & Plasterk, R. H. (1993) Nucleic Acids Res. 21, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman, A., Bushman, F. D. & Craigie, R. (1993) EMBO J. 12, 3269–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman, A. B., Palmer, I., Engelman, A., Craigie, R. & Wingfield, P. (1994) J. Biol. Chem. 269, 29279–29287. [PubMed] [Google Scholar]
- 15.Zheng, R., Jenkins, T. M. & Craigie, R. (1996) Proc. Natl. Acad. Sci. USA 93, 13659–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins, T. M., Engelman, A., Ghirlando, R. & Craigie, R. (1996) J. Biol. Chem. 271, 7712–7718. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. P., Xiao, J., Knutson, J. R., Lewis, M. S. & Han, M. K. (1997) Biochemistry 36, 173–180. [DOI] [PubMed] [Google Scholar]
- 18.Cherepanov, P., Maertens, G., Proost, P., Devreese, B., Van Beeumen, J., Engelborghs, Y., De Clercq, E. & Debyser, Z. (2003) J. Biol. Chem. 278, 372–381. [DOI] [PubMed] [Google Scholar]
- 19.Petit, C., Schwartz, O. & Mammano, F. (1999) J. Virol. 73, 5079–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pluymers, W., Cherepanov, P., Schols, D., De Clercq, E. & Debyser, Z. (1999) Virology 258, 327–332. [DOI] [PubMed] [Google Scholar]
- 21.Cherepanov, P., Pluymers, W., Claeys, A., Proost, P., De Clercq, E. & Debyser, Z. (2000) FASEB J. 14, 1389–1399. [DOI] [PubMed] [Google Scholar]
- 22.Maertens, G., Cherepanov, P., Pluymers, W., Busschots, K., De Clercq, E., Debyser, Z. & Engelborghs, Y. (2003) J. Biol. Chem. 278, 33528–33539. [DOI] [PubMed] [Google Scholar]
- 23.Maertens, G., Cherepanov, P., Debyser, Z., Engelborghs, Y. & Engelman, A. (2004) J. Biol. Chem. 279, 33421–33429. [DOI] [PubMed] [Google Scholar]
- 24.Llano, M., Vanegas, M., Fregoso, O., Saenz, D., Chung, S., Peretz, M. & Poeschla, E. M. (2004) J. Virol. 78, 9524–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emiliani, S., Mousnier, A., Busschots, K., Maroun, M., Van Maele, B., Tempe, D., Vandekerckhove, L., Moisant, F., Ben-Slama, L., Witvrouw, M., et al. (2005) J. Biol. Chem. 280, 25517–25523. [DOI] [PubMed] [Google Scholar]
- 26.Llano, M., Delgado, S., Vanegas, M. & Poeschla, E. M. (2004) J. Biol. Chem. 279, 55570–55577. [DOI] [PubMed] [Google Scholar]
- 27.Busschots, K., Vercammen, J., Emiliani, S., Benarous, R., Engelborghs, Y., Christ, F. & Debyser, Z. (2005) J. Biol. Chem. 280, 17841–17847. [DOI] [PubMed] [Google Scholar]
- 28.Cherepanov, P., Devroe, E., Silver, P. A. & Engelman, A. (2004) J. Biol. Chem. 279, 48883–48892. [DOI] [PubMed] [Google Scholar]
- 29.Vanegas, M., Llano, M., Delgado, S., Thompson, D., Peretz, M. & Poeschla, E. (2005) J. Cell Sci. 118, 1733–1743. [DOI] [PubMed] [Google Scholar]
- 30.Cherepanov, P., Sun, Z. Y., Rahman, S., Maertens, G., Wagner, G. & Engelman, A. (2005) Nat. Struct. Mol. Biol. 12, 526–532. [DOI] [PubMed] [Google Scholar]
- 31.Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W. & Bork, P. (2001) J. Mol. Biol. 309, 1–18. [DOI] [PubMed] [Google Scholar]
- 32.Green, J. B., Gardner, C. D., Wharton, R. P. & Aggarwal, A. K. (2003) Mol. Cell 11, 1537–1548. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 34.CCP4 (1994) Acta Crystallogr. D 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 35.Goldgur, Y., Dyda, F., Hickman, A. B., Jenkins, T. M., Craigie, R. & Davies, D. R. (1998) Proc. Natl. Acad. Sci. USA 95, 9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 38.Winn, M. D., Murshudov, G. N. & Papiz, M. Z. (2003) Methods Enzymol. 374, 300–321. [DOI] [PubMed] [Google Scholar]
- 39.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 40.Emsley, P. & Cowtan, K. (2004) Acta Crystallogr. D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 41.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins, T. M., Hickman, A. B., Dyda, F., Ghirlando, R., Davies, D. R. & Craigie, R. (1995) Proc. Natl. Acad. Sci. USA 92, 6057–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, J. Y., Ling, H., Yang, W. & Craigie, R. (2001) EMBO J. 20, 7333–7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo Conte, L., Chothia, C. & Janin, J. (1999) J. Mol. Biol. 285, 2177–2198. [DOI] [PubMed] [Google Scholar]
- 45.Maignan, S., Guilloteau, J. P., Zhou-Liu, Q., Clement-Mella, C. & Mikol, V. (1998) J. Mol. Biol. 282, 359–368. [DOI] [PubMed] [Google Scholar]
- 46.Chen, Z., Yan, Y., Munshi, S., Li, Y., Zugay-Murphy, J., Xu, B., Witmer, M., Felock, P., Wolfe, A., Sardana, V., et al. (2000) J. Mol. Biol. 296, 521–533. [DOI] [PubMed] [Google Scholar]
- 47.Lubkowski, J., Dauter, Z., Yang, F., Alexandratos, J., Merkel, G., Skalka, A. M. & Wlodawer, A. (1999) Biochemistry 38, 13512–13522. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Z. N., Mueser, T. C., Bushman, F. D. & Hyde, C. C. (2000) J. Mol. Biol. 296, 535–548. [DOI] [PubMed] [Google Scholar]
- 49.Bouyac-Bertoia, M., Dvorin, J. D., Fouchier, R. A., Jenkins, Y., Meyer, B. E., Wu, L. I., Emerman, M. & Malim, M. H. (2001) Mol. Cell 7, 1025–1035. [DOI] [PubMed] [Google Scholar]
- 50.Lu, R., Limon, A., Devroe, E., Silver, P. A., Cherepanov, P. & Engelman, A. (2004) J. Virol. 78, 12735–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molteni, V., Greenwald, J., Rhodes, D., Hwang, Y., Kwiatkowski, W., Bushman, F. D., Siegel, J. S. & Choe, S. (2001) Acta Crystallogr. D 57, 536–544. [DOI] [PubMed] [Google Scholar]
- 52.Lin, Z., Neamati, N., Zhao, H., Kiryu, Y., Turpin, J. A., Aberham, C., Strebel, K., Kohn, K., Witvrouw, M., Pannecouque, C., et al. (1999) J. Med. Chem. 42, 1401–1414. [DOI] [PubMed] [Google Scholar]
- 53.Shkriabai, N., Patil, S. S., Hess, S., Budihas, S. R., Craigie, R., Burke, T. R., Jr., Le Grice, S. F. & Kvaratskhelia, M. (2004) Proc. Natl. Acad. Sci. USA 101, 6894–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., et al. (2004) Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- 55.Lepourcelet, M., Chen, Y. N., France, D. S., Wang, H., Crews, P., Petersen, F., Bruseo, C., Wood, A. W. & Shivdasani, R. A. (2004) Cancer Cell 5, 91–102. [DOI] [PubMed] [Google Scholar]
- 56.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.